

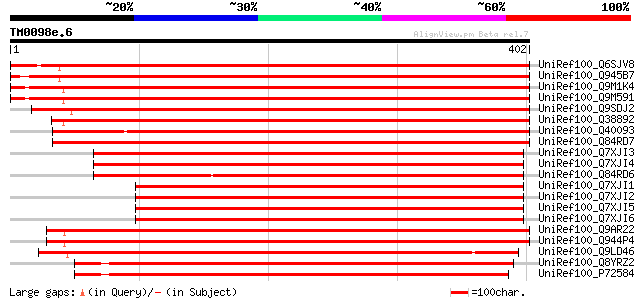
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098e.6
(402 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6SJV8 Putative leucine zipper protein [Gossypium hirs... 732 0.0
UniRef100_Q945B7 Leucine zipper-containing protein [Euphorbia es... 710 0.0
UniRef100_Q9M1K4 Leucine zipper-containing protein AT103 [Arabid... 708 0.0
UniRef100_Q9M591 Putative dicarboxylate diiron protein [Arabidop... 704 0.0
UniRef100_Q9SDJ2 ESTs D47452 [Oryza sativa] 686 0.0
UniRef100_Q38892 AT103 [Arabidopsis thaliana] 679 0.0
UniRef100_Q40093 PNIL34 [Ipomoea nil] 676 0.0
UniRef100_Q84RD7 ZIP [Nicotiana tabacum] 664 0.0
UniRef100_Q7XJI3 Putative fatty acid desaturase RDZIP [Rosa davu... 639 0.0
UniRef100_Q7XJI4 Putative fatty acid desaturase SBZIP [Salix bab... 633 e-180
UniRef100_Q84RD6 Basic leucine zipper transcription factor CAT10... 606 e-172
UniRef100_Q7XJI1 Putative fatty acid desaturase TRZIP [Trifolium... 595 e-169
UniRef100_Q7XJI2 Putative fatty acid desaturase BNZIP [Brassica ... 584 e-165
UniRef100_Q7XJI5 Putative fatty acid desaturase SOZIP [Spinacia ... 580 e-164
UniRef100_Q7XJI6 Putative fatty acid desaturase TAZIP [Triticum ... 578 e-164
UniRef100_Q9AR22 Copper target homolog 1 protein [Chlamydomonas ... 527 e-148
UniRef100_Q944P4 Copper target homolog 1 protein [Chlamydomonas ... 526 e-148
UniRef100_Q9LD46 Copper response target 1 protein [Chlamydomonas... 478 e-133
UniRef100_Q8YRZ2 Alr3300 protein [Anabaena sp.] 472 e-132
UniRef100_P72584 Sll1214 protein [Synechocystis sp.] 457 e-127
>UniRef100_Q6SJV8 Putative leucine zipper protein [Gossypium hirsutum]
Length = 424
Score = 732 bits (1890), Expect = 0.0
Identities = 362/404 (89%), Positives = 381/404 (93%), Gaps = 5/404 (1%)
Query: 1 MALVKPISKFTNAAPKFSNPRTISHSKFSTVRMSATT--TPPSSTKPSKKANKTSIKETL 58
MALVKPISKF+ ++P FSN R + KF+TVRMS+T+ T ++ K KKA KT+IKETL
Sbjct: 24 MALVKPISKFSTSSPIFSNSR---YGKFTTVRMSSTSQSTTKAAAKGGKKAAKTAIKETL 80
Query: 59 LTPRFYTTDFDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAADKLD 118
LTPRFYTTDFDEME LFNTEINKNLN+ EFE+LLQEFKTDYNQTHFVRNKEFKEAADK+D
Sbjct: 81 LTPRFYTTDFDEMEALFNTEINKNLNQSEFESLLQEFKTDYNQTHFVRNKEFKEAADKID 140
Query: 119 GPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKG 178
GPLRQIFVEF ERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKG
Sbjct: 141 GPLRQIFVEFSERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKG 200
Query: 179 LSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEYQCYPI 238
LSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLK NPEYQCYPI
Sbjct: 201 LSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKENPEYQCYPI 260
Query: 239 FKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFY 298
FKYFENWCQDENRHGDFFSAL+KAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRT FY
Sbjct: 261 FKYFENWCQDENRHGDFFSALLKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTAFY 320
Query: 299 EGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGESDDI 358
EGIGL+TKEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLD+MVEINE++LAVGE+ DI
Sbjct: 321 EGIGLDTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDKMVEINEQLLAVGETSDI 380
Query: 359 PFVKNLKRIPLIAALASELLATYLMPPIESGSVDLAEFEPQLVY 402
P VKNLKRIPLIAALASELLATYLMPPIESGSVD AEFEPQLVY
Sbjct: 381 PLVKNLKRIPLIAALASELLATYLMPPIESGSVDFAEFEPQLVY 424
>UniRef100_Q945B7 Leucine zipper-containing protein [Euphorbia esula]
Length = 405
Score = 710 bits (1832), Expect = 0.0
Identities = 355/407 (87%), Positives = 373/407 (91%), Gaps = 11/407 (2%)
Query: 1 MALVKPISKFTNAAPKFSNP-RTISHS-KFSTVRMSATT---TPPSSTKPSKKANKTSIK 55
MALVKPI+ PKF NP RT S S KFST++MSAT+ T ++TKPSKK NK I
Sbjct: 5 MALVKPIT------PKFINPMRTFSSSSKFSTIKMSATSQSNTTTTATKPSKKGNKKEIN 58
Query: 56 ETLLTPRFYTTDFDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAAD 115
ETLLTPRFYTTDFDEMETLFNTEINK LN+ EFEALLQEFKTDYNQTHFVRNKEFKEAAD
Sbjct: 59 ETLLTPRFYTTDFDEMETLFNTEINKKLNQSEFEALLQEFKTDYNQTHFVRNKEFKEAAD 118
Query: 116 KLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFL 175
K+ GPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFL
Sbjct: 119 KMQGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFL 178
Query: 176 NKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEYQC 235
NKGLSDFN ALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLK NPEYQC
Sbjct: 179 NKGLSDFNYALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKENPEYQC 238
Query: 236 YPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRT 295
YPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLW+RFFCLSVYVTMYLNDCQRT
Sbjct: 239 YPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWARFFCLSVYVTMYLNDCQRT 298
Query: 296 DFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGES 355
FYEGIGL+TKEFDMHVIIETNRTTARIFPAVLDVENPEFKR+LDRMV IN+K+ AVGE+
Sbjct: 299 AFYEGIGLDTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRKLDRMVVINQKLQAVGET 358
Query: 356 DDIPFVKNLKRIPLIAALASELLATYLMPPIESGSVDLAEFEPQLVY 402
+D VKNLKR+PLIAAL SE+LA YLMPPIESGSVD AEFEP+LVY
Sbjct: 359 EDNSVVKNLKRVPLIAALVSEILAAYLMPPIESGSVDFAEFEPKLVY 405
>UniRef100_Q9M1K4 Leucine zipper-containing protein AT103 [Arabidopsis thaliana]
Length = 409
Score = 708 bits (1828), Expect = 0.0
Identities = 351/407 (86%), Positives = 376/407 (92%), Gaps = 7/407 (1%)
Query: 1 MALVKPISKFTNAAPKFSNP-RTISHSKFSTV-RMSATTTPP---SSTKPSKKANKTSIK 55
MALVKPISKF++ PK SNP + +S +FSTV RMSA+++PP ++T SKK K I+
Sbjct: 5 MALVKPISKFSS--PKLSNPSKFLSGRRFSTVIRMSASSSPPPPTTATSKSKKGTKKEIQ 62
Query: 56 ETLLTPRFYTTDFDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAAD 115
E+LLTPRFYTTDF+EME LFNTEINKNLNE EFEALLQEFKTDYNQTHFVRNKEFKEAAD
Sbjct: 63 ESLLTPRFYTTDFEEMEQLFNTEINKNLNEAEFEALLQEFKTDYNQTHFVRNKEFKEAAD 122
Query: 116 KLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFL 175
KL GPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFL
Sbjct: 123 KLQGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFL 182
Query: 176 NKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEYQC 235
NKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLK NPE+QC
Sbjct: 183 NKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKENPEFQC 242
Query: 236 YPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRT 295
YPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDW+AKLWSRFFCLSVYVTMYLNDCQRT
Sbjct: 243 YPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWQAKLWSRFFCLSVYVTMYLNDCQRT 302
Query: 296 DFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGES 355
+FYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR+LDRMV EK+LA+GE+
Sbjct: 303 NFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRKLDRMVVSYEKLLAIGET 362
Query: 356 DDIPFVKNLKRIPLIAALASELLATYLMPPIESGSVDLAEFEPQLVY 402
DD F+K LKRIPL+ +LASE+LA YLMPP+ESGSVD AEFEP LVY
Sbjct: 363 DDASFIKTLKRIPLVTSLASEILAAYLMPPVESGSVDFAEFEPNLVY 409
>UniRef100_Q9M591 Putative dicarboxylate diiron protein [Arabidopsis thaliana]
Length = 409
Score = 704 bits (1817), Expect = 0.0
Identities = 350/407 (85%), Positives = 375/407 (91%), Gaps = 7/407 (1%)
Query: 1 MALVKPISKFTNAAPKFSNP-RTISHSKFSTV-RMSATTTPP---SSTKPSKKANKTSIK 55
MALVKPISKF++ PK SNP + +S +FSTV RMSA+++PP ++T SKK K I+
Sbjct: 5 MALVKPISKFSS--PKLSNPSKFLSGRRFSTVIRMSASSSPPPPTTATSKSKKGTKKEIQ 62
Query: 56 ETLLTPRFYTTDFDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAAD 115
E+LLTPRFYTTDF+EME LFNTEINKNLNE EFEALLQEFKTDYNQTHFVRNKEFKEAAD
Sbjct: 63 ESLLTPRFYTTDFEEMEQLFNTEINKNLNEAEFEALLQEFKTDYNQTHFVRNKEFKEAAD 122
Query: 116 KLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFL 175
KL GPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFL
Sbjct: 123 KLQGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFL 182
Query: 176 NKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEYQC 235
NKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLK NPE+QC
Sbjct: 183 NKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKENPEFQC 242
Query: 236 YPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRT 295
YPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDW+AKLWSRFFCLSVYVTMYLND QRT
Sbjct: 243 YPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWQAKLWSRFFCLSVYVTMYLNDWQRT 302
Query: 296 DFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGES 355
+FYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR+LDRMV EK+LA+GE+
Sbjct: 303 NFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRKLDRMVVSYEKLLAIGET 362
Query: 356 DDIPFVKNLKRIPLIAALASELLATYLMPPIESGSVDLAEFEPQLVY 402
DD F+K LKRIPL+ +LASE+LA YLMPP+ESGSVD AEFEP LVY
Sbjct: 363 DDASFIKTLKRIPLVTSLASEILAAYLMPPVESGSVDFAEFEPNLVY 409
>UniRef100_Q9SDJ2 ESTs D47452 [Oryza sativa]
Length = 408
Score = 686 bits (1770), Expect = 0.0
Identities = 332/387 (85%), Positives = 355/387 (90%), Gaps = 2/387 (0%)
Query: 18 SNPRTISHSKFSTVRMSATTTPPSSTKPS--KKANKTSIKETLLTPRFYTTDFDEMETLF 75
+ PR +S + R++++ P + KP KK KT I+ETLLTPRFYTTDFDEME LF
Sbjct: 22 AKPRVVSSRRIVRFRVASSAAAPPAAKPGTPKKRGKTEIQETLLTPRFYTTDFDEMERLF 81
Query: 76 NTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTA 135
N EINK LN+ EF+ALLQEFKTDYNQTHFVRN EFK AADK++GPLRQIFVEFLERSCTA
Sbjct: 82 NAEINKQLNQEEFDALLQEFKTDYNQTHFVRNPEFKAAADKMEGPLRQIFVEFLERSCTA 141
Query: 136 EFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKAR 195
EFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKAR
Sbjct: 142 EFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKAR 201
Query: 196 KYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDF 255
KYTFFKPKFIFYATYLSEKIGYWRYITI+RHLKANPEYQ YPIFKYFENWCQDENRHGDF
Sbjct: 202 KYTFFKPKFIFYATYLSEKIGYWRYITIFRHLKANPEYQVYPIFKYFENWCQDENRHGDF 261
Query: 256 FSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIE 315
FSAL+KAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRT FYEGIGL+TKEFDMHVIIE
Sbjct: 262 FSALLKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTTFYEGIGLDTKEFDMHVIIE 321
Query: 316 TNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGESDDIPFVKNLKRIPLIAALAS 375
TNRTTARIFPAVLDVENPEFKR+LDRMVEIN+KI+A+GESDDIP VKNLKRIP +AAL S
Sbjct: 322 TNRTTARIFPAVLDVENPEFKRKLDRMVEINKKIIAIGESDDIPLVKNLKRIPHVAALVS 381
Query: 376 ELLATYLMPPIESGSVDLAEFEPQLVY 402
E++A YLMPPIESGSVD AEFEPQLVY
Sbjct: 382 EIIAAYLMPPIESGSVDFAEFEPQLVY 408
>UniRef100_Q38892 AT103 [Arabidopsis thaliana]
Length = 373
Score = 679 bits (1751), Expect = 0.0
Identities = 329/373 (88%), Positives = 349/373 (93%), Gaps = 3/373 (0%)
Query: 33 MSATTTPP---SSTKPSKKANKTSIKETLLTPRFYTTDFDEMETLFNTEINKNLNEHEFE 89
MSA+++PP ++T SKK K I+E+LLTPRFYTTDF+EME LFNTEINKNLNE EFE
Sbjct: 1 MSASSSPPPPTTATSKSKKGTKKEIQESLLTPRFYTTDFEEMEQLFNTEINKNLNEAEFE 60
Query: 90 ALLQEFKTDYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRR 149
ALLQEFKTDYNQTHFVRNKEFKEAADKL GPLRQIFVEFLERSCTAEFSGFLLYKELGRR
Sbjct: 61 ALLQEFKTDYNQTHFVRNKEFKEAADKLQGPLRQIFVEFLERSCTAEFSGFLLYKELGRR 120
Query: 150 LKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYAT 209
KKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYAT
Sbjct: 121 SKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYAT 180
Query: 210 YLSEKIGYWRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLND 269
YLSEKIGYWRYITIYRHLK NPE+QCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLND
Sbjct: 181 YLSEKIGYWRYITIYRHLKENPEFQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLND 240
Query: 270 WKAKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLD 329
W+AKLWSRFFCLSVYVTMYLNDCQRT+FYEGIGLNTKEFDMHVIIETNRTTARIFPAVLD
Sbjct: 241 WQAKLWSRFFCLSVYVTMYLNDCQRTNFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLD 300
Query: 330 VENPEFKRRLDRMVEINEKILAVGESDDIPFVKNLKRIPLIAALASELLATYLMPPIESG 389
VENPEFKR+LDRMV EK+LA+GE+DD F+K LKRIPL+ +LASE+LA YLMPP+ESG
Sbjct: 301 VENPEFKRKLDRMVVSYEKLLAIGETDDASFIKTLKRIPLVTSLASEILAAYLMPPVESG 360
Query: 390 SVDLAEFEPQLVY 402
SVD AEFEP LVY
Sbjct: 361 SVDFAEFEPNLVY 373
>UniRef100_Q40093 PNIL34 [Ipomoea nil]
Length = 370
Score = 676 bits (1744), Expect = 0.0
Identities = 330/369 (89%), Positives = 346/369 (93%), Gaps = 1/369 (0%)
Query: 34 SATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDFDEMETLFNTEINKNLNEHEFEALLQ 93
SATT+ ++ K SK + K IKETLLTPRFYTTDFDEME LFNTEINKNLN E +LQ
Sbjct: 3 SATTSQAAAEKGSKASTKKGIKETLLTPRFYTTDFDEMEMLFNTEINKNLNNSELR-VLQ 61
Query: 94 EFKTDYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKT 153
EFK DYNQTHFVRNKEFKE ADKL GPLR+IFVEFLERSCTAEFSGFLLYKELGRRLKKT
Sbjct: 62 EFKKDYNQTHFVRNKEFKEVADKLQGPLREIFVEFLERSCTAEFSGFLLYKELGRRLKKT 121
Query: 154 NPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSE 213
NPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSE
Sbjct: 122 NPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSE 181
Query: 214 KIGYWRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAK 273
KIGYWRYITIYRHLKANPE+QCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAK
Sbjct: 182 KIGYWRYITIYRHLKANPEFQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAK 241
Query: 274 LWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENP 333
LW+RFFCLSVYVTMYLNDCQRT FYEGIGL+TKEFDMHVIIETNRTTARIFPAVLDVENP
Sbjct: 242 LWARFFCLSVYVTMYLNDCQRTAFYEGIGLDTKEFDMHVIIETNRTTARIFPAVLDVENP 301
Query: 334 EFKRRLDRMVEINEKILAVGESDDIPFVKNLKRIPLIAALASELLATYLMPPIESGSVDL 393
EFKR+LDRMVEIN+K+LAVGE+DDIP VKNLKRIPL+AAL SE+LA YLM P+ESGSVDL
Sbjct: 302 EFKRKLDRMVEINQKLLAVGETDDIPLVKNLKRIPLVAALVSEILAAYLMKPVESGSVDL 361
Query: 394 AEFEPQLVY 402
AEFEPQL Y
Sbjct: 362 AEFEPQLGY 370
>UniRef100_Q84RD7 ZIP [Nicotiana tabacum]
Length = 371
Score = 664 bits (1713), Expect = 0.0
Identities = 328/369 (88%), Positives = 341/369 (91%)
Query: 34 SATTTPPSSTKPSKKANKTSIKETLLTPRFYTTDFDEMETLFNTEINKNLNEHEFEALLQ 93
SATT+ ++ K SK + K IKETLLTPRFYTTDFDEM E N ++ EFEALLQ
Sbjct: 3 SATTSQAAAEKGSKASTKKGIKETLLTPRFYTTDFDEMTDFEEMEPLFNTDQAEFEALLQ 62
Query: 94 EFKTDYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKT 153
EFKTDYNQTHFVRNKEFKEAADK+ G LR+IFVEFLERSCTAEFSGFLLYKELGRRLKKT
Sbjct: 63 EFKTDYNQTHFVRNKEFKEAADKMQGALREIFVEFLERSCTAEFSGFLLYKELGRRLKKT 122
Query: 154 NPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSE 213
NPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSE
Sbjct: 123 NPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSE 182
Query: 214 KIGYWRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAK 273
KIGYWRYITIYRHLKANPE+QCYPIFKYFENWCQDENRHGDFFSAL+KAQPQFLNDWKAK
Sbjct: 183 KIGYWRYITIYRHLKANPEFQCYPIFKYFENWCQDENRHGDFFSALLKAQPQFLNDWKAK 242
Query: 274 LWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENP 333
LWSRFFCLSVYVTMYLND QRT FYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENP
Sbjct: 243 LWSRFFCLSVYVTMYLNDWQRTAFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENP 302
Query: 334 EFKRRLDRMVEINEKILAVGESDDIPFVKNLKRIPLIAALASELLATYLMPPIESGSVDL 393
EFKR+LDRMVEIN+KILAVGE+DDIP VKN KRIPLIAALASELLA YLM PIESGSVD
Sbjct: 303 EFKRKLDRMVEINQKILAVGETDDIPLVKNFKRIPLIAALASELLAAYLMKPIESGSVDF 362
Query: 394 AEFEPQLVY 402
AEFEPQLVY
Sbjct: 363 AEFEPQLVY 371
>UniRef100_Q7XJI3 Putative fatty acid desaturase RDZIP [Rosa davurica]
Length = 333
Score = 639 bits (1649), Expect = 0.0
Identities = 308/333 (92%), Positives = 320/333 (95%)
Query: 66 TDFDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAADKLDGPLRQIF 125
TDF+EME LFNTEINKNLNE EF ALLQEFKTDYNQTHFVRNKEFKEAADKL GPLRQIF
Sbjct: 1 TDFEEMEQLFNTEINKNLNEEEFIALLQEFKTDYNQTHFVRNKEFKEAADKLQGPLRQIF 60
Query: 126 VEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLA 185
VEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLA
Sbjct: 61 VEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLA 120
Query: 186 LDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEYQCYPIFKYFENW 245
LDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLK NPE+QCYPIFKYFENW
Sbjct: 121 LDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKQNPEFQCYPIFKYFENW 180
Query: 246 CQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNT 305
CQDENRHGDFFSALMKAQPQFLNDW+AKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNT
Sbjct: 181 CQDENRHGDFFSALMKAQPQFLNDWQAKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNT 240
Query: 306 KEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGESDDIPFVKNLK 365
KEFDMHVIIETNRTTARIFPAVLDVENPEFKR+LDRMV I EK++AVG++DD FVKNLK
Sbjct: 241 KEFDMHVIIETNRTTARIFPAVLDVENPEFKRKLDRMVVIYEKLMAVGKTDDPSFVKNLK 300
Query: 366 RIPLIAALASELLATYLMPPIESGSVDLAEFEP 398
++PLIA L SE+LA YLMPP+ESGSVD AEFEP
Sbjct: 301 KVPLIAGLVSEILAAYLMPPVESGSVDFAEFEP 333
>UniRef100_Q7XJI4 Putative fatty acid desaturase SBZIP [Salix babylonica]
Length = 333
Score = 633 bits (1632), Expect = e-180
Identities = 305/333 (91%), Positives = 318/333 (94%)
Query: 66 TDFDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAADKLDGPLRQIF 125
TDF+EME LFNTEINKN NE EF ALLQEFKTDYNQTHFVRNKEFKEAAD+L GPLRQIF
Sbjct: 1 TDFEEMEQLFNTEINKNPNEEEFIALLQEFKTDYNQTHFVRNKEFKEAADRLQGPLRQIF 60
Query: 126 VEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLA 185
VEFLERSCTAEFSGFLLYKELGRR KKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLA
Sbjct: 61 VEFLERSCTAEFSGFLLYKELGRRPKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLA 120
Query: 186 LDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEYQCYPIFKYFENW 245
LDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLK NPE+QCYPIFKYFENW
Sbjct: 121 LDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKQNPEFQCYPIFKYFENW 180
Query: 246 CQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNT 305
CQDENRHGDFFSALMKAQPQFLNDW+AKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNT
Sbjct: 181 CQDENRHGDFFSALMKAQPQFLNDWQAKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNT 240
Query: 306 KEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGESDDIPFVKNLK 365
KEFDMHVIIETNRTTARIFPAVLDVENPEFKR+LDRMV I EK++AVG++DD FVKNLK
Sbjct: 241 KEFDMHVIIETNRTTARIFPAVLDVENPEFKRKLDRMVVIYEKLMAVGKTDDPSFVKNLK 300
Query: 366 RIPLIAALASELLATYLMPPIESGSVDLAEFEP 398
++PLIA L SE+LA YLMPP+ESGSVD AEFEP
Sbjct: 301 KVPLIAGLVSEILAAYLMPPVESGSVDFAEFEP 333
>UniRef100_Q84RD6 Basic leucine zipper transcription factor CAT103 [Cucumis sativus]
Length = 332
Score = 606 bits (1562), Expect = e-172
Identities = 295/333 (88%), Positives = 311/333 (92%), Gaps = 1/333 (0%)
Query: 66 TDFDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAADKLDGPLRQIF 125
TDF+EME LFNTEINKNLN+ EFEALLQEFKTDYNQTHFVRNKEFKEAADK+ G LR+IF
Sbjct: 1 TDFEEMEPLFNTEINKNLNQAEFEALLQEFKTDYNQTHFVRNKEFKEAADKMQGALREIF 60
Query: 126 VEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLA 185
VEFLERSCTAEFSGFLLYKELGRRL+ + V AEIFSLMSRDEARHAGFLNKG
Sbjct: 61 VEFLERSCTAEFSGFLLYKELGRRLENQSSV-AEIFSLMSRDEARHAGFLNKGYRISTWP 119
Query: 186 LDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEYQCYPIFKYFENW 245
LDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPE+QCYPIFKYFENW
Sbjct: 120 LDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKANPEFQCYPIFKYFENW 179
Query: 246 CQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNT 305
CQDENRHGDFFSAL+KAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRT FYEGIGLNT
Sbjct: 180 CQDENRHGDFFSALLKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTAFYEGIGLNT 239
Query: 306 KEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKILAVGESDDIPFVKNLK 365
KEFDMHVIIETNRTTARIFPAV DVENPEFKR+LDRMVEINEK++AVGE++D+P VK+LK
Sbjct: 240 KEFDMHVIIETNRTTARIFPAVPDVENPEFKRKLDRMVEINEKLIAVGETNDLPLVKSLK 299
Query: 366 RIPLIAALASELLATYLMPPIESGSVDLAEFEP 398
RIPL AAL SE+LA YLMPPIESGSVD AEFEP
Sbjct: 300 RIPLAAALVSEILAAYLMPPIESGSVDFAEFEP 332
>UniRef100_Q7XJI1 Putative fatty acid desaturase TRZIP [Trifolium repens]
Length = 301
Score = 595 bits (1534), Expect = e-169
Identities = 289/301 (96%), Positives = 293/301 (97%)
Query: 98 DYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVV 157
DYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVV
Sbjct: 1 DYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVV 60
Query: 158 AEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGY 217
AEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGY
Sbjct: 61 AEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGY 120
Query: 218 WRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSR 277
WRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSR
Sbjct: 121 WRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSR 180
Query: 278 FFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR 337
FFCLSVYVTMYLND QRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR
Sbjct: 181 FFCLSVYVTMYLNDWQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR 240
Query: 338 RLDRMVEINEKILAVGESDDIPFVKNLKRIPLIAALASELLATYLMPPIESGSVDLAEFE 397
+LDRMVEIN+KILA+GESDD VKNLK IPL+AAL SELLA YLMPPIESGSVD AEFE
Sbjct: 241 KLDRMVEINQKILAIGESDDNSVVKNLKGIPLVAALVSELLAAYLMPPIESGSVDFAEFE 300
Query: 398 P 398
P
Sbjct: 301 P 301
>UniRef100_Q7XJI2 Putative fatty acid desaturase BNZIP [Brassica napus]
Length = 308
Score = 584 bits (1506), Expect = e-165
Identities = 280/301 (93%), Positives = 289/301 (95%)
Query: 98 DYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVV 157
DYNQTHFVRNKEFKEAADKL GPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVV
Sbjct: 1 DYNQTHFVRNKEFKEAADKLQGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVV 60
Query: 158 AEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGY 217
AEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGY
Sbjct: 61 AEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGY 120
Query: 218 WRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSR 277
WRYITIYRHLK NPE+QCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDW+AKLWSR
Sbjct: 121 WRYITIYRHLKQNPEFQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWQAKLWSR 180
Query: 278 FFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR 337
FFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR
Sbjct: 181 FFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR 240
Query: 338 RLDRMVEINEKILAVGESDDIPFVKNLKRIPLIAALASELLATYLMPPIESGSVDLAEFE 397
+LDRMV INEK++AVG++DD FVKNLKRIPLIA L SE+LA YLMPP+ESGS D E E
Sbjct: 241 KLDRMVVINEKLMAVGQTDDPSFVKNLKRIPLIAGLVSEILAAYLMPPVESGSTDFEEME 300
Query: 398 P 398
P
Sbjct: 301 P 301
>UniRef100_Q7XJI5 Putative fatty acid desaturase SOZIP [Spinacia oleracea]
Length = 301
Score = 580 bits (1494), Expect = e-164
Identities = 278/301 (92%), Positives = 289/301 (95%)
Query: 98 DYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVV 157
DYNQTHFVRNKEFKEAADKL GPLRQIFVEFLERSCTAEFSGFLLYKELG+RLKKTNPVV
Sbjct: 1 DYNQTHFVRNKEFKEAADKLQGPLRQIFVEFLERSCTAEFSGFLLYKELGKRLKKTNPVV 60
Query: 158 AEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGY 217
AEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFI YATYLSEKIGY
Sbjct: 61 AEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIXYATYLSEKIGY 120
Query: 218 WRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSR 277
WRYITIYRHLK NPE+QCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDW+AKLWSR
Sbjct: 121 WRYITIYRHLKQNPEFQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWQAKLWSR 180
Query: 278 FFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR 337
FFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR
Sbjct: 181 FFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR 240
Query: 338 RLDRMVEINEKILAVGESDDIPFVKNLKRIPLIAALASELLATYLMPPIESGSVDLAEFE 397
+LDRMV I EK++AVG++DD FVKNLK++PLIA L SE+LA YLMPP ESGSVD AEFE
Sbjct: 241 KLDRMVVIYEKLMAVGKTDDPSFVKNLKKVPLIAGLVSEILAAYLMPPAESGSVDFAEFE 300
Query: 398 P 398
P
Sbjct: 301 P 301
>UniRef100_Q7XJI6 Putative fatty acid desaturase TAZIP [Triticum aestivum]
Length = 301
Score = 578 bits (1491), Expect = e-164
Identities = 276/301 (91%), Positives = 288/301 (94%)
Query: 98 DYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVV 157
DYNQTHFVRNKEFKEAADKL GPLRQIFVEFLERSCTAEFSGFLLY+ELGRRLKKTNPVV
Sbjct: 1 DYNQTHFVRNKEFKEAADKLQGPLRQIFVEFLERSCTAEFSGFLLYEELGRRLKKTNPVV 60
Query: 158 AEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGY 217
AEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGY
Sbjct: 61 AEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGY 120
Query: 218 WRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSR 277
WRYITIYRHLK NPE+QCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDW+AKLWSR
Sbjct: 121 WRYITIYRHLKENPEFQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWQAKLWSR 180
Query: 278 FFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR 337
FFCLSVYVTMYLNDCQRT FYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR
Sbjct: 181 FFCLSVYVTMYLNDCQRTSFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKR 240
Query: 338 RLDRMVEINEKILAVGESDDIPFVKNLKRIPLIAALASELLATYLMPPIESGSVDLAEFE 397
+LDRMV EK+LA+GE+DD F+K LKRIPL+ +LA E+LA YLMPP+ESGSVD AEFE
Sbjct: 241 KLDRMVVSYEKLLAIGETDDASFIKTLKRIPLVTSLAPEILAAYLMPPVESGSVDFAEFE 300
Query: 398 P 398
P
Sbjct: 301 P 301
>UniRef100_Q9AR22 Copper target homolog 1 protein [Chlamydomonas reinhardtii]
Length = 407
Score = 527 bits (1358), Expect = e-148
Identities = 252/376 (67%), Positives = 305/376 (81%), Gaps = 2/376 (0%)
Query: 29 STVRMSATTTPPS--STKPSKKANKTSIKETLLTPRFYTTDFDEMETLFNTEINKNLNEH 86
S+VR++AT P K + K + ETLLTPRFYTTDFDEME LF+ E+NKN++
Sbjct: 32 SSVRVAATAAPQEVEGFKVMRDGIKVASDETLLTPRFYTTDFDEMERLFSLELNKNMDME 91
Query: 87 EFEALLQEFKTDYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKEL 146
EFEA+L EFK DYNQ HFVRN+ FKEAA+K+ GP R+IF+EFLERSCTAEFSGFLLYKEL
Sbjct: 92 EFEAMLNEFKLDYNQRHFVRNETFKEAAEKIQGPTRKIFIEFLERSCTAEFSGFLLYKEL 151
Query: 147 GRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIF 206
GRRLK TNPVVAEIF+LMSRDEARHAGFLNK +SDFNLALDLGFLTK RKYTFFKPKFIF
Sbjct: 152 GRRLKATNPVVAEIFTLMSRDEARHAGFLNKAMSDFNLALDLGFLTKNRKYTFFKPKFIF 211
Query: 207 YATYLSEKIGYWRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQF 266
YATYLSEKIGYWRYI+IYRHL+ NP+ Q YP+F+YFENWCQDENRHGDFF+A++KA+P+
Sbjct: 212 YATYLSEKIGYWRYISIYRHLQRNPDNQLYPLFEYFENWCQDENRHGDFFTAVLKARPEM 271
Query: 267 LNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPA 326
+NDW AKLWSRFFCLSVY+TMYLND QR FY +GLNT +F+ HVIIETN++T RIFPA
Sbjct: 272 VNDWAAKLWSRFFCLSVYITMYLNDHQRDAFYSSLGLNTTQFNQHVIIETNKSTERIFPA 331
Query: 327 VLDVENPEFKRRLDRMVEINEKILAVGESDDIPFVKNLKRIPLIAALASELLATYLMPPI 386
V DVENPEF RR+D +V+ N +++ +G + +K + + P++ + +E+ ++M P
Sbjct: 332 VPDVENPEFFRRMDLLVKYNAQLVNIGSMNLPSPIKAIMKAPILERMVAEVFQVFIMTPK 391
Query: 387 ESGSVDLAEFEPQLVY 402
ESGS DL + LVY
Sbjct: 392 ESGSYDLDANKTALVY 407
>UniRef100_Q944P4 Copper target homolog 1 protein [Chlamydomonas reinhardtii]
Length = 407
Score = 526 bits (1354), Expect = e-148
Identities = 251/376 (66%), Positives = 304/376 (80%), Gaps = 2/376 (0%)
Query: 29 STVRMSATTTPPS--STKPSKKANKTSIKETLLTPRFYTTDFDEMETLFNTEINKNLNEH 86
S+VR++ T P K + K + ETLLTPRFYTTDFDEME LF+ E+NKN++
Sbjct: 32 SSVRVAVTAAPQEVEGFKVMRDGIKVASDETLLTPRFYTTDFDEMERLFSLELNKNMDME 91
Query: 87 EFEALLQEFKTDYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKEL 146
EFEA+L EFK DYNQ HFVRN+ FKEAA+K+ GP R+IF+EFLERSCTAEFSGFLLYKEL
Sbjct: 92 EFEAMLNEFKLDYNQRHFVRNETFKEAAEKIQGPTRKIFIEFLERSCTAEFSGFLLYKEL 151
Query: 147 GRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIF 206
GRRLK TNPVVAEIF+LMSRDEARHAGFLNK +SDFNLALDLGFLTK RKYTFFKPKFIF
Sbjct: 152 GRRLKATNPVVAEIFTLMSRDEARHAGFLNKAMSDFNLALDLGFLTKNRKYTFFKPKFIF 211
Query: 207 YATYLSEKIGYWRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQF 266
YATYLSEKIGYWRYI+IYRHL+ NP+ Q YP+F+YFENWCQDENRHGDFF+A++KA+P+
Sbjct: 212 YATYLSEKIGYWRYISIYRHLQRNPDNQLYPLFEYFENWCQDENRHGDFFTAVLKARPEM 271
Query: 267 LNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPA 326
+NDW AKLWSRFFCLSVY+TMYLND QR FY +GLNT +F+ HVIIETN++T RIFPA
Sbjct: 272 VNDWAAKLWSRFFCLSVYITMYLNDHQRDAFYSSLGLNTTQFNQHVIIETNKSTERIFPA 331
Query: 327 VLDVENPEFKRRLDRMVEINEKILAVGESDDIPFVKNLKRIPLIAALASELLATYLMPPI 386
V DVENPEF RR+D +V+ N +++ +G + +K + + P++ + +E+ ++M P
Sbjct: 332 VPDVENPEFFRRMDLLVKYNAQLVDIGSMNLPSPIKAIMKAPILERMVAEVFQVFIMTPK 391
Query: 387 ESGSVDLAEFEPQLVY 402
ESGS DL + LVY
Sbjct: 392 ESGSYDLDANKTALVY 407
>UniRef100_Q9LD46 Copper response target 1 protein [Chlamydomonas reinhardtii]
Length = 407
Score = 478 bits (1230), Expect = e-133
Identities = 233/375 (62%), Positives = 289/375 (76%), Gaps = 5/375 (1%)
Query: 23 ISHSKFSTVRMSATTTPPSST---KPSKKANKTSIKETLLTPRFYTTDFDEMETLFNTEI 79
++ + STVR+ A+ P + + + K + KETLLTPRFYTTDFDEME LF+ EI
Sbjct: 25 VARPRRSTVRVQASAAPLNDGLGFETMRDGIKVAAKETLLTPRFYTTDFDEMEQLFSKEI 84
Query: 80 NKNLNEHEFEALLQEFKTDYNQTHFVRNKEFKEAADKLDGPLRQIFVEFLERSCTAEFSG 139
N NL+ E A L EF+ DYN+ HFVRN+ FK AADK+ G R+IF+EFLERSCTAEFSG
Sbjct: 85 NPNLDMEELNACLNEFRNDYNKVHFVRNETFKAAADKVTGETRRIFIEFLERSCTAEFSG 144
Query: 140 FLLYKELGRRLKKTNPVVAEIFSLMSRDEARHAGFLNKGLSDFNLALDLGFLTKARKYTF 199
FLLYKEL RR+K ++P VAE+F LMSRDEARHAGFLNK LSDFNLALDLGFLTK R YT+
Sbjct: 145 FLLYKELARRMKASSPEVAEMFLLMSRDEARHAGFLNKALSDFNLALDLGFLTKNRTYTY 204
Query: 200 FKPKFIFYATYLSEKIGYWRYITIYRHLKANPEYQCYPIFKYFENWCQDENRHGDFFSAL 259
FKPKFI YAT+LSEKIGYWRYITIYRHL+ NP+ Q YP+F+YFENWCQDENRHGDF +A
Sbjct: 205 FKPKFIIYATFLSEKIGYWRYITIYRHLQRNPDNQFYPLFEYFENWCQDENRHGDFLAAC 264
Query: 260 MKAQPQFLNDWKAKLWSRFFCLSVYVTMYLNDCQRTDFYEGIGLNTKEFDMHVIIETNRT 319
+KA+P+ LN ++AKLWS+FFCLSVY+TMYLND QRT FYE +GLNT++F+ HVIIETNR
Sbjct: 265 LKAKPELLNTFEAKLWSKFFCLSVYITMYLNDHQRTKFYESLGLNTRQFNQHVIIETNRA 324
Query: 320 TARIFPAVLDVENPEFKRRLDRMVEINEKILAVGESDDIPFVKNLKRIPLIAALASELLA 379
T R+FP V DVE+P F L++MV++N K++ + S + L+++PL+ +AS L
Sbjct: 325 TERLFPVVPDVEDPRFFEILNKMVDVNAKLVELSASSSP--LAGLQKLPLLERMASYCLQ 382
Query: 380 TYLMPPIESGSVDLA 394
+ GSVD+A
Sbjct: 383 LLFFKEKDVGSVDIA 397
>UniRef100_Q8YRZ2 Alr3300 protein [Anabaena sp.]
Length = 358
Score = 472 bits (1214), Expect = e-132
Identities = 224/340 (65%), Positives = 279/340 (81%), Gaps = 5/340 (1%)
Query: 51 KTSIKETLLTPRFYTTDFDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEF 110
K KETLLTPRFYTTDFDEM ++ ++NE E A+L+EF+TDYN+ HFVR+ EF
Sbjct: 18 KVPAKETLLTPRFYTTDFDEM-----ARMDISVNEDELLAILEEFRTDYNRHHFVRDAEF 72
Query: 111 KEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEAR 170
+++ D +DG R++FVEFLERSCTAEFSGFLLYKELGRRLK +PV+AE F+LMSRDEAR
Sbjct: 73 EKSWDHIDGDTRRLFVEFLERSCTAEFSGFLLYKELGRRLKDKSPVLAECFNLMSRDEAR 132
Query: 171 HAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKAN 230
HAGFLNK +SDFNL+LDLGFLTK+R YTFFKPKFIFYATYLSEKIGYWRYITIYRHL+A+
Sbjct: 133 HAGFLNKAMSDFNLSLDLGFLTKSRNYTFFKPKFIFYATYLSEKIGYWRYITIYRHLEAH 192
Query: 231 PEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLN 290
PE + YPIF++FENWCQDENRHGDFF A+MK+QPQ LNDWKAKLWSRFF LSV+ TMYLN
Sbjct: 193 PEDRIYPIFRFFENWCQDENRHGDFFDAIMKSQPQMLNDWKAKLWSRFFLLSVFATMYLN 252
Query: 291 DCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKIL 350
D QR DFY IGL+ +++D++VI +TN T R+FP +LDV++PEF RLD VE +EK+
Sbjct: 253 DIQRKDFYATIGLDARDYDIYVIKKTNETAGRVFPIILDVDSPEFYERLDICVENSEKLS 312
Query: 351 AVGESDDIPFVKNLKRIPLIAALASELLATYLMPPIESGS 390
A+ S+ F++ +++P++A+ +LL YLM PI++ S
Sbjct: 313 AIASSNTPKFLQFFQKLPVLASTGWQLLRLYLMKPIDAVS 352
>UniRef100_P72584 Sll1214 protein [Synechocystis sp.]
Length = 358
Score = 457 bits (1176), Expect = e-127
Identities = 218/336 (64%), Positives = 262/336 (77%), Gaps = 5/336 (1%)
Query: 51 KTSIKETLLTPRFYTTDFDEMETLFNTEINKNLNEHEFEALLQEFKTDYNQTHFVRNKEF 110
KT KET+LTPRFYTTDFDEM +++ + NE E A+L+EF+ DYN+ HFVRN+ F
Sbjct: 18 KTPAKETILTPRFYTTDFDEM-----AKMDISPNEDELRAILEEFRVDYNRHHFVRNESF 72
Query: 111 KEAADKLDGPLRQIFVEFLERSCTAEFSGFLLYKELGRRLKKTNPVVAEIFSLMSRDEAR 170
++ D +DG RQ+FVEFLERSCTAEFSGFLLYKELGRRLK NP++AE F+LMSRDEAR
Sbjct: 73 NKSWDHIDGEKRQLFVEFLERSCTAEFSGFLLYKELGRRLKNKNPLLAECFNLMSRDEAR 132
Query: 171 HAGFLNKGLSDFNLALDLGFLTKARKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLKAN 230
HAGFLNK +SDFNL+LDLGFLTK+RKYTFFKPKFIFYATYLSEKIGYWRYITIYRHL+ N
Sbjct: 133 HAGFLNKAMSDFNLSLDLGFLTKSRKYTFFKPKFIFYATYLSEKIGYWRYITIYRHLEKN 192
Query: 231 PEYQCYPIFKYFENWCQDENRHGDFFSALMKAQPQFLNDWKAKLWSRFFCLSVYVTMYLN 290
P YPIF++FENWCQDENRHGDFF A+M+AQP LNDWKAKLW RFF LSV+ TMYLN
Sbjct: 193 PNDCIYPIFEFFENWCQDENRHGDFFDAIMRAQPHTLNDWKAKLWCRFFLLSVFATMYLN 252
Query: 291 DCQRTDFYEGIGLNTKEFDMHVIIETNRTTARIFPAVLDVENPEFKRRLDRMVEINEKIL 350
D QR DFY +GL + +D VI +TN T R+FP +LDV NPEF RL+ V NE++
Sbjct: 253 DTQRADFYACLGLEARSYDKEVIEKTNETAGRVFPIILDVNNPEFYNRLETCVSNNEQLR 312
Query: 351 AVGESDDIPFVKNLKRIPLIAALASELLATYLMPPI 386
A+ S +K L+++P+ A+ + + YLM PI
Sbjct: 313 AIDASGAPGVIKALRKLPIFASNGWQFIKLYLMKPI 348
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.137 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 664,413,686
Number of Sequences: 2790947
Number of extensions: 28135706
Number of successful extensions: 81424
Number of sequences better than 10.0: 119
Number of HSP's better than 10.0 without gapping: 94
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 81185
Number of HSP's gapped (non-prelim): 137
length of query: 402
length of database: 848,049,833
effective HSP length: 130
effective length of query: 272
effective length of database: 485,226,723
effective search space: 131981668656
effective search space used: 131981668656
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0098e.6


