

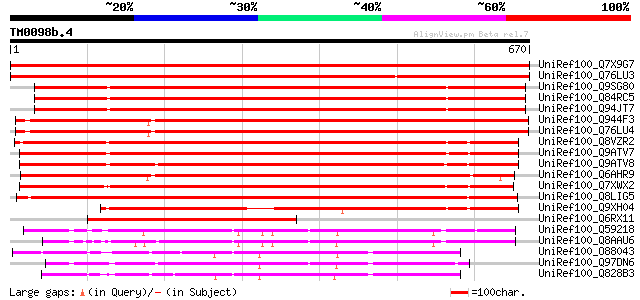
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0098b.4
(670 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7X9G7 Alpha-L-arabinofuranosidase [Malus domestica] 1010 0.0
UniRef100_Q76LU3 Alpha-L-arabinofuranosidase [Pyrus pyrifolia] 989 0.0
UniRef100_Q9SG80 Putative alpha-L-arabinofuranosidase [Arabidops... 961 0.0
UniRef100_Q84RC5 Alpha-L-arabinofuranosidase [Arabidopsis thaliana] 959 0.0
UniRef100_Q94JT7 AT3g10740/T7M13_18 [Arabidopsis thaliana] 958 0.0
UniRef100_Q944F3 Arabinosidase ARA-1 [Lycopersicon esculentum] 929 0.0
UniRef100_Q76LU4 Alpha-L-arabinofuranosidase [Lycopersicon escul... 925 0.0
UniRef100_Q8VZR2 Putative arabinosidase [Arabidopsis thaliana] 899 0.0
UniRef100_Q9ATV7 Arabinoxylan arabinofuranohydrolase isoenzyme A... 880 0.0
UniRef100_Q9ATV8 Arabinoxylan arabinofuranohydrolase isoenzyme A... 879 0.0
UniRef100_Q6AHR9 Arabinofuranosidase [Daucus carota] 852 0.0
UniRef100_Q7XWX2 OSJNBb0058J09.4 protein [Oryza sativa] 841 0.0
UniRef100_Q8LIG5 Putative arabinoxylan narabinofuranohydrolase i... 833 0.0
UniRef100_Q9XH04 T1N24.13 protein [Arabidopsis thaliana] 681 0.0
UniRef100_Q6RX11 A-arabinofuranosidase 1 [Ficus carica] 453 e-126
UniRef100_Q59218 Arabinosidase [Bacteroides ovatus] 335 3e-90
UniRef100_Q8AAU6 Alpha-L-arabinofuranosidase A [Bacteroides thet... 334 4e-90
UniRef100_O88043 Putative secreted arabinosidase [Streptomyces c... 308 3e-82
UniRef100_Q97DN6 Probable alpha-arabinofuranosidase [Clostridium... 298 5e-79
UniRef100_Q828B3 Putative secreted arabinosidase [Streptomyces a... 296 2e-78
>UniRef100_Q7X9G7 Alpha-L-arabinofuranosidase [Malus domestica]
Length = 675
Score = 1010 bits (2611), Expect = 0.0
Identities = 485/675 (71%), Positives = 577/675 (84%), Gaps = 5/675 (0%)
Query: 1 MGLYKASFIVAALQ--LCIAVSLCAIKVNADLNATLTVDAHETSGKPISETLFGLFFEEI 58
MG K+ +V L LC AI V+A+ A L +DA + S +PIS+TLFG+FFEEI
Sbjct: 1 MGSCKSPHVVLLLHVLLCSVYRCFAIGVDANQTANLLIDASQASARPISDTLFGIFFEEI 60
Query: 59 NHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIALRL 118
NHAGAGG+WAELVSNRGFEAGGPN PSNIDPW+IIGNES + V TDR+SCFDRNK+ALR+
Sbjct: 61 NHAGAGGVWAELVSNRGFEAGGPNTPSNIDPWAIIGNESSLIVSTDRSSCFDRNKVALRM 120
Query: 119 EVLCDSKGDNICPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNGV- 177
EVLCD++G N CPA GVG++NPGFWGMNIE+GK Y V YVRS+GS++++VSLTGS+G+
Sbjct: 121 EVLCDTQGANSCPAGGVGIYNPGFWGMNIEKGKTYNAVLYVRSSGSINVSVSLTGSDGLQ 180
Query: 178 -LASNVITGSASDFSNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYKG 236
LA+ I S S+ SNW K E L+A+ TN NSRLQLTTT+KGVIW DQVSA+PLDTYKG
Sbjct: 181 KLAAANIIASGSEVSNWTKFEVQLKAQGTNPNSRLQLTTTSKGVIWFDQVSAIPLDTYKG 240
Query: 237 RGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWT 296
GFR +LV MLADLKPRF RFPGGCFVEGEWLRNAFRWKE+IGPWEERPGHFGDVW YWT
Sbjct: 241 HGFRKDLVQMLADLKPRFFRFPGGCFVEGEWLRNAFRWKETIGPWEERPGHFGDVWMYWT 300
Query: 297 DDGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSK 356
DDGLGYFEFLQL+EDLG+LPIWVFNNG+SHND+VDTS+VLPFVQEALDGIEFARG P S
Sbjct: 301 DDGLGYFEFLQLSEDLGSLPIWVFNNGISHNDQVDTSSVLPFVQEALDGIEFARGSPNST 360
Query: 357 WGSIRASMGHPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSSR 416
WGS+RA+MGHPEPF+L+YVA+GNEDCGKKNYRGNYLKFYSAIR+AYPDI++ISNCDGSSR
Sbjct: 361 WGSLRAAMGHPEPFDLRYVAIGNEDCGKKNYRGNYLKFYSAIRNAYPDIKMISNCDGSSR 420
Query: 417 PLDHPADIYDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAE 476
LDHPAD+YD+HIYT+A+++FS ++ F+ R+GPKAFVSEYAVTG DAG GSLLAA+AE
Sbjct: 421 QLDHPADMYDFHIYTSASNLFSMANHFDHTSRNGPKAFVSEYAVTGKDAGRGSLLAALAE 480
Query: 477 AGFLLGLEKNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSG 536
AGFL+GLEKNSDIV+M SYAPLFVN +DRRWNPDAIVF+S Q YGTPSYWVQ F+ESSG
Sbjct: 481 AGFLIGLEKNSDIVEMASYAPLFVNTHDRRWNPDAIVFDSSQLYGTPSYWVQCLFNESSG 540
Query: 537 ATLLNSSLQTSSSSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGLDPNSLQ 596
ATL NS+LQ +SS+SL+ASAI+WQ+S ++ Y+RIKVVN G TVNLK+ ++GL+PNS+
Sbjct: 541 ATLYNSTLQMNSSTSLLASAISWQNSENENTYLRIKVVNLGTYTVNLKVFVDGLEPNSVS 600
Query: 597 PSGSTRTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDLLKESSN 656
SGST+TVLTS N MDENSF++PKKV+P +SLL++ G+EM VV+ P SF+S DLL ESS+
Sbjct: 601 LSGSTKTVLTSNNQMDENSFNEPKKVIPKRSLLESAGEEMEVVISPRSFTSIDLLMESSD 660
Query: 657 -LKMVGGGSSTRSSI 670
++ G S + SSI
Sbjct: 661 VIRTTGADSVSVSSI 675
>UniRef100_Q76LU3 Alpha-L-arabinofuranosidase [Pyrus pyrifolia]
Length = 674
Score = 989 bits (2558), Expect = 0.0
Identities = 479/675 (70%), Positives = 570/675 (83%), Gaps = 6/675 (0%)
Query: 1 MGLYKASFIVAALQ--LCIAVSLCAIKVNADLNATLTVDAHETSGKPISETLFGLFFEEI 58
MG K+ IV L LC AI V+A+ A L +DA + S +PIS+TLFG+FFEEI
Sbjct: 1 MGSCKSPHIVLLLYVLLCSVYRCFAIGVDANQTANLLIDASQASARPISDTLFGIFFEEI 60
Query: 59 NHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIALRL 118
NHAGAGG+WAELVSNRGFEAGGPN PSNIDPW+IIGNESF+ V TDR+SCFDRNK+ALR+
Sbjct: 61 NHAGAGGVWAELVSNRGFEAGGPNTPSNIDPWAIIGNESFLIVSTDRSSCFDRNKVALRM 120
Query: 119 EVLCDSKGDNICPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNGV- 177
EVLCD++G N CPA GVG++NPGFWGMNIE+GK Y V YVRS+GS++++VSLTGS+G+
Sbjct: 121 EVLCDTQGANSCPAGGVGIYNPGFWGMNIEKGKTYNAVLYVRSSGSINVSVSLTGSDGLQ 180
Query: 178 -LASNVITGSASDFSNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYKG 236
LA+ I S S+ SNW K E LEA+ TN NSRLQLTTT+KGVIW DQVSA+PLDTYKG
Sbjct: 181 KLAAANIIASGSEVSNWTKFEVQLEAQGTNPNSRLQLTTTSKGVIWFDQVSAIPLDTYKG 240
Query: 237 RGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWT 296
GFR +LV MLADLKPRF RFPGGCFVEGEWLRNAFRWKE+IGPWEERPGHFGDVWKYWT
Sbjct: 241 HGFRKDLVQMLADLKPRFFRFPGGCFVEGEWLRNAFRWKETIGPWEERPGHFGDVWKYWT 300
Query: 297 DDGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSK 356
DDGLGYFEFLQL+EDLG+LPIWVFNNG+SHND+VDTS+VLPFVQEALDGIEFARG P S
Sbjct: 301 DDGLGYFEFLQLSEDLGSLPIWVFNNGISHNDQVDTSSVLPFVQEALDGIEFARGSPNST 360
Query: 357 WGSIRASMGHPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSSR 416
WGS+RA+MGHPEPF+L+YVA+GNEDCGKKNYRGNYLKF+SAIR+AYPDI++ISNCDGSSR
Sbjct: 361 WGSLRAAMGHPEPFDLRYVAIGNEDCGKKNYRGNYLKFFSAIRNAYPDIKMISNCDGSSR 420
Query: 417 PLDHPADIYDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAE 476
LDHPAD+YD+HIYT+A+++FS ++ F+ R+GPKAFVSEYAVTG DAG GSLLAA+AE
Sbjct: 421 QLDHPADMYDFHIYTSASNLFSMANHFDHTSRNGPKAFVSEYAVTGKDAGRGSLLAALAE 480
Query: 477 AGFLLGLEKNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSG 536
AGFL+GLEKNSDIV+ P + +DRRWNPDAIVF+S Q YGTPSYWVQ F+ESSG
Sbjct: 481 AGFLIGLEKNSDIVESGHLRPP-LRQHDRRWNPDAIVFDSSQLYGTPSYWVQCLFNESSG 539
Query: 537 ATLLNSSLQTSSSSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGLDPNSLQ 596
ATL NS+LQ +SS+ L+ASAI+WQ+S ++ Y+RIKVVN G +TVNLK+ ++GLDPNS+
Sbjct: 540 ATLYNSTLQMNSSTPLLASAISWQNSENENTYLRIKVVNLGTDTVNLKVFVDGLDPNSIS 599
Query: 597 PSGSTRTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDLLKESSN 656
SGST+TVLTS N MDENSF++P KV+P QSLL++ +EM VV+ P SF+S DLL ESS+
Sbjct: 600 LSGSTKTVLTSNNQMDENSFNEPTKVIPKQSLLESASEEMEVVISPRSFTSIDLLMESSD 659
Query: 657 -LKMVGGGSSTRSSI 670
++ G S + SSI
Sbjct: 660 IIRTTGADSVSVSSI 674
>UniRef100_Q9SG80 Putative alpha-L-arabinofuranosidase [Arabidopsis thaliana]
Length = 678
Score = 961 bits (2484), Expect = 0.0
Identities = 462/636 (72%), Positives = 540/636 (84%), Gaps = 8/636 (1%)
Query: 33 TLTVDAHETSGKPISETLFGLFFEEINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSI 92
TL VDA G+PI ETLFG+FFEEINHAGAGGLWAELVSNRGFEAGG N PSNI PWSI
Sbjct: 42 TLQVDASNGGGRPIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGQNTPSNIWPWSI 101
Query: 93 IGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDNICPADGVGVFNPGFWGMNIEQGKK 152
+G+ S I V TDR+SCF+RNKIALR++VLCDSKG CP+ GVGV+NPG+WGMNIE+GKK
Sbjct: 102 VGDHSSIYVATDRSSCFERNKIALRMDVLCDSKG---CPSGGVGVYNPGYWGMNIEEGKK 158
Query: 153 YKVVFYVRSTGSLDLTVSLTGSNG--VLASNVITGSASDFSNWKKVETLLEAKATNHNSR 210
YKV YVRSTG +DL+VSLT SNG LAS I SASD S W K E LLEAKAT+ ++R
Sbjct: 159 YKVALYVRSTGDIDLSVSLTSSNGSRTLASEKIIASASDVSKWIKKEVLLEAKATDPSAR 218
Query: 211 LQLTTTAKGVIWLDQVSAMPLDTYKGRGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRN 270
LQLTTT KG IW+DQVSAMP+DT+KG GFR++L M+AD+KPRFIRFPGGCFVEGEWL N
Sbjct: 219 LQLTTTKKGSIWIDQVSAMPVDTHKGHGFRNDLFQMMADIKPRFIRFPGGCFVEGEWLSN 278
Query: 271 AFRWKESIGPWEERPGHFGDVWKYWTDDGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEV 330
AFRWKE++GPWEERPGHFGDVWKYWTDDGLG+FEF Q+AED+GA PIWVFNNG+SHNDEV
Sbjct: 279 AFRWKETVGPWEERPGHFGDVWKYWTDDGLGHFEFFQMAEDIGAAPIWVFNNGISHNDEV 338
Query: 331 DTSAVLPFVQEALDGIEFARGDPTSKWGSIRASMGHPEPFNLKYVAVGNEDCGKKNYRGN 390
+T++++PFVQEALDGIEFARGD S WGS+RA MG EPF LKYVA+GNEDCGK YRGN
Sbjct: 339 ETASIMPFVQEALDGIEFARGDANSTWGSVRAKMGRQEPFELKYVAIGNEDCGKTYYRGN 398
Query: 391 YLKFYSAIRSAYPDIQIISNCDGSSRPLDHPADIYDYHIYTNANDMFSRSSTFNTALRSG 450
Y+ FY AI+ AYPDI+IISNCDGSS PLDHPAD YDYHIYT+A+++FS F+ R G
Sbjct: 399 YIVFYDAIKKAYPDIKIISNCDGSSHPLDHPADYYDYHIYTSASNLFSMYHQFDRTSRKG 458
Query: 451 PKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLEKNSDIVKMVSYAPLFVNANDRRWNPD 510
PKAFVSEYAVTG DAGTGSLLA++AEA FL+GLEKNSDIV+M SYAPLFVN NDRRWNPD
Sbjct: 459 PKAFVSEYAVTGKDAGTGSLLASLAEAAFLIGLEKNSDIVEMASYAPLFVNTNDRRWNPD 518
Query: 511 AIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSLQTSSSSSLIASAINWQSSADKKNYIR 570
AIVFNS YGTPSYWVQ FF+ESSGATLL S+L+ +S+SL+ASAI+W+++ K+YIR
Sbjct: 519 AIVFNSSHLYGTPSYWVQRFFAESSGATLLTSTLK-GNSTSLVASAISWKNNG--KDYIR 575
Query: 571 IKVVNFGANTVNLKISLNGLDPNSLQPSGSTRTVLTSANVMDENSFSQPKKVVPIQSLLQ 630
IK VNFGAN+ N+++ + GLDPN ++ SGS +TVLTS NVMDENSFSQP+KVVP +SLL+
Sbjct: 576 IKAVNFGANSENMQVLVTGLDPNVMRVSGSKKTVLTSTNVMDENSFSQPEKVVPHESLLE 635
Query: 631 NVGKEMNVVVPPHSFSSFDLLKESSNLKMVGGGSST 666
++M VV+PPHSFSSFDLLKES+ ++M SS+
Sbjct: 636 LAEEDMTVVLPPHSFSSFDLLKESAKIRMPISDSSS 671
>UniRef100_Q84RC5 Alpha-L-arabinofuranosidase [Arabidopsis thaliana]
Length = 678
Score = 959 bits (2478), Expect = 0.0
Identities = 461/636 (72%), Positives = 539/636 (84%), Gaps = 8/636 (1%)
Query: 33 TLTVDAHETSGKPISETLFGLFFEEINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSI 92
TL VDA G+PI ETLFG+FFEEINHAGAGGLWAELVSNRGFEAGG N PSNI PWSI
Sbjct: 42 TLQVDASNGGGRPIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGQNTPSNIWPWSI 101
Query: 93 IGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDNICPADGVGVFNPGFWGMNIEQGKK 152
+G+ S I V TDR+SCF+RNKIALR++VLCDSKG CP+ GVGV+NPG+WGMNIE+GKK
Sbjct: 102 VGDHSSIYVATDRSSCFERNKIALRMDVLCDSKG---CPSGGVGVYNPGYWGMNIEEGKK 158
Query: 153 YKVVFYVRSTGSLDLTVSLTGSNG--VLASNVITGSASDFSNWKKVETLLEAKATNHNSR 210
YKV YVRSTG +DL+VSLT SNG LAS I SASD S W K E LLEAKAT+ ++R
Sbjct: 159 YKVALYVRSTGDIDLSVSLTSSNGSRTLASEKIIASASDVSKWIKKEVLLEAKATDPSAR 218
Query: 211 LQLTTTAKGVIWLDQVSAMPLDTYKGRGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRN 270
LQLTTT KG IW+DQVSAMP+DT+KG GFR++L ++AD+KPRFIRFPGGCFVEGEWL N
Sbjct: 219 LQLTTTKKGSIWIDQVSAMPVDTHKGHGFRNDLFHLMADIKPRFIRFPGGCFVEGEWLSN 278
Query: 271 AFRWKESIGPWEERPGHFGDVWKYWTDDGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEV 330
AFRWKE++GPWEERPGHFGDVWKYWTDDGLG+FEF Q+AED+GA PIWVFNNG+SHNDEV
Sbjct: 279 AFRWKETVGPWEERPGHFGDVWKYWTDDGLGHFEFFQMAEDIGAAPIWVFNNGISHNDEV 338
Query: 331 DTSAVLPFVQEALDGIEFARGDPTSKWGSIRASMGHPEPFNLKYVAVGNEDCGKKNYRGN 390
+T++++PFVQEALDGIEFARGD S WGS+RA MG EPF LKYVA+GNEDCGK YRGN
Sbjct: 339 ETASIMPFVQEALDGIEFARGDANSTWGSVRAKMGRQEPFELKYVAIGNEDCGKTYYRGN 398
Query: 391 YLKFYSAIRSAYPDIQIISNCDGSSRPLDHPADIYDYHIYTNANDMFSRSSTFNTALRSG 450
Y+ FY AI+ AYPDI+IISNCDGSS PLDHPAD YDYHIYT+A+++FS F+ R G
Sbjct: 399 YIVFYDAIKKAYPDIKIISNCDGSSHPLDHPADYYDYHIYTSASNLFSMYHQFDGTSRKG 458
Query: 451 PKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLEKNSDIVKMVSYAPLFVNANDRRWNPD 510
PKAFVSEYAVTG DAGTGSLLA++AEA FL+GLEKNSDIV M SYAPLFVN NDRRWNPD
Sbjct: 459 PKAFVSEYAVTGKDAGTGSLLASLAEAAFLIGLEKNSDIVDMASYAPLFVNTNDRRWNPD 518
Query: 511 AIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSLQTSSSSSLIASAINWQSSADKKNYIR 570
AIVFNS YGTPSYWVQ FF+ESSGATLL S+L+ +S+SL+ASAI+W+++ K+YIR
Sbjct: 519 AIVFNSSHLYGTPSYWVQRFFAESSGATLLTSTLK-GNSTSLVASAISWKNNG--KDYIR 575
Query: 571 IKVVNFGANTVNLKISLNGLDPNSLQPSGSTRTVLTSANVMDENSFSQPKKVVPIQSLLQ 630
IK VNFGAN+ N+++ + GLDPN ++ SGS +TVLTS NVMDENSFSQP+KVVP +SLL+
Sbjct: 576 IKAVNFGANSENMQVLVTGLDPNVMRVSGSKKTVLTSTNVMDENSFSQPEKVVPHESLLE 635
Query: 631 NVGKEMNVVVPPHSFSSFDLLKESSNLKMVGGGSST 666
++M VV+PPHSFSSFDLLKES+ ++M SS+
Sbjct: 636 LAEEDMTVVLPPHSFSSFDLLKESAKIRMPISDSSS 671
>UniRef100_Q94JT7 AT3g10740/T7M13_18 [Arabidopsis thaliana]
Length = 678
Score = 958 bits (2477), Expect = 0.0
Identities = 461/636 (72%), Positives = 539/636 (84%), Gaps = 8/636 (1%)
Query: 33 TLTVDAHETSGKPISETLFGLFFEEINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSI 92
TL VDA G+PI ETLFG+FFEEINHAGAGGLWAELVSNRGFEAGG N PSNI PWSI
Sbjct: 42 TLQVDASNGGGRPIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGQNTPSNIWPWSI 101
Query: 93 IGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDNICPADGVGVFNPGFWGMNIEQGKK 152
+G+ S I V TDR+SCF+RNKIALR++VLCDSKG CP+ GVGV+NPG+WGMNIE+GKK
Sbjct: 102 VGDHSSIYVATDRSSCFERNKIALRMDVLCDSKG---CPSGGVGVYNPGYWGMNIEEGKK 158
Query: 153 YKVVFYVRSTGSLDLTVSLTGSNG--VLASNVITGSASDFSNWKKVETLLEAKATNHNSR 210
YKV YVRSTG +DL+VSLT SNG LAS I SASD S W K E LLEAKAT+ ++R
Sbjct: 159 YKVALYVRSTGDIDLSVSLTSSNGSRTLASEKIIASASDVSKWIKKEVLLEAKATDPSAR 218
Query: 211 LQLTTTAKGVIWLDQVSAMPLDTYKGRGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRN 270
LQLTTT KG IW+DQVSAMP+DT+KG GFR++ M+AD+KPRFIRFPGGCFVEGEWL N
Sbjct: 219 LQLTTTKKGSIWIDQVSAMPVDTHKGHGFRNDPFQMMADIKPRFIRFPGGCFVEGEWLSN 278
Query: 271 AFRWKESIGPWEERPGHFGDVWKYWTDDGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEV 330
AFRWKE++GPWEERPGHFGDVWKYWTDDGLG+FEF Q+AED+GA PIWVFNNG+SHNDEV
Sbjct: 279 AFRWKETVGPWEERPGHFGDVWKYWTDDGLGHFEFFQMAEDIGAAPIWVFNNGISHNDEV 338
Query: 331 DTSAVLPFVQEALDGIEFARGDPTSKWGSIRASMGHPEPFNLKYVAVGNEDCGKKNYRGN 390
+T++++PFVQEALDGIEFARGD S WGS+RA MG EPF LKYVA+GNEDCGK YRGN
Sbjct: 339 ETASIMPFVQEALDGIEFARGDANSTWGSVRAKMGRQEPFELKYVAIGNEDCGKTYYRGN 398
Query: 391 YLKFYSAIRSAYPDIQIISNCDGSSRPLDHPADIYDYHIYTNANDMFSRSSTFNTALRSG 450
Y+ FY AI+ AYPDI+IISNCDGSS PLDHPAD YDYHIYT+A+++FS F+ R G
Sbjct: 399 YIVFYDAIKKAYPDIKIISNCDGSSHPLDHPADYYDYHIYTSASNLFSMYHQFDRTSRKG 458
Query: 451 PKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLEKNSDIVKMVSYAPLFVNANDRRWNPD 510
PKAFVSEYAVTG DAGTGSLLA++AEA FL+GLEKNSDIV+M SYAPLFVN NDRRWNPD
Sbjct: 459 PKAFVSEYAVTGKDAGTGSLLASLAEAAFLIGLEKNSDIVEMASYAPLFVNTNDRRWNPD 518
Query: 511 AIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSLQTSSSSSLIASAINWQSSADKKNYIR 570
AIVFNS YGTPSYWVQ FF+ESSGATLL S+L+ +S+SL+ASAI+W+++ K+YIR
Sbjct: 519 AIVFNSSHLYGTPSYWVQRFFAESSGATLLTSTLK-GNSTSLVASAISWKNNG--KDYIR 575
Query: 571 IKVVNFGANTVNLKISLNGLDPNSLQPSGSTRTVLTSANVMDENSFSQPKKVVPIQSLLQ 630
IK VNFGAN+ N+++ + GLDPN ++ SGS +TVLTS NVMDENSFSQP+KVVP +SLL+
Sbjct: 576 IKAVNFGANSENMQVLVTGLDPNVMRVSGSKKTVLTSTNVMDENSFSQPEKVVPHESLLE 635
Query: 631 NVGKEMNVVVPPHSFSSFDLLKESSNLKMVGGGSST 666
++M VV+PPHSFSSFDLLKES+ ++M SS+
Sbjct: 636 LAEEDMTVVLPPHSFSSFDLLKESAKIRMPISDSSS 671
>UniRef100_Q944F3 Arabinosidase ARA-1 [Lycopersicon esculentum]
Length = 674
Score = 929 bits (2400), Expect = 0.0
Identities = 449/666 (67%), Positives = 538/666 (80%), Gaps = 12/666 (1%)
Query: 8 FIVAALQLCIAVSLCAIKVNADLNATLTVDAHETSGKPISETLFGLFFEEINHAGAGGLW 67
F ++AL C A V A+ A L V+A E S + I +TLFG+FFEEINHAGAGGLW
Sbjct: 16 FGLSALCQCSATG-----VEANQTAVLLVNASEASARRIPDTLFGIFFEEINHAGAGGLW 70
Query: 68 AELVSNRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGD 127
AELV+NRGFE GGPN+PSNIDPWSIIG+ES + V TDR+SCFDRNKIA++++VLCD G
Sbjct: 71 AELVNNRGFEGGGPNVPSNIDPWSIIGDESKVIVSTDRSSCFDRNKIAVQVQVLCDHTGA 130
Query: 128 NICPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNGV---LASNVIT 184
NICP GVG++NPGFWGMNIEQGK YK+V YVRS S++++V+LTGSNG+ A+N++
Sbjct: 131 NICPDGGVGIYNPGFWGMNIEQGKSYKLVLYVRSEESVNVSVALTGSNGLQKLAAANIV- 189
Query: 185 GSASDFSNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYKGRGFRSELV 244
A+D S+W KVE LLEA T+ NSRL+L ++ KGVIW DQVS MP DTYKG GFR +L
Sbjct: 190 --AADVSSWTKVEILLEATGTDPNSRLELRSSKKGVIWFDQVSLMPTDTYKGHGFRKDLF 247
Query: 245 GMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWTDDGLGYFE 304
GML DLKP FIRFPGGCFVEG+WLRNAFRWKE+IG WEERPGHFGDVW YWTDDGLG+FE
Sbjct: 248 GMLKDLKPAFIRFPGGCFVEGDWLRNAFRWKETIGQWEERPGHFGDVWNYWTDDGLGHFE 307
Query: 305 FLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSKWGSIRASM 364
FLQLAEDL +LP+WVFNNGVSH+D+VDTS++LPFVQE LDG+EFARGDPTS WGSIRA M
Sbjct: 308 FLQLAEDLDSLPVWVFNNGVSHHDQVDTSSILPFVQEILDGLEFARGDPTSTWGSIRAKM 367
Query: 365 GHPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSSRPLDHPADI 424
GHPEPF+L+YVA+GNEDCGK YRGNYLKFY+AI+ YPDI+IISNCDGS+RPLDHPAD+
Sbjct: 368 GHPEPFDLRYVAIGNEDCGKTQYRGNYLKFYTAIKDKYPDIKIISNCDGSTRPLDHPADL 427
Query: 425 YDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLE 484
YD+HIY++A+ +FS + F++A R GPKAFVSEYAVTGNDAG GSLLAA+ EAGFL+G+E
Sbjct: 428 YDFHIYSSASSVFSNARHFDSAPRRGPKAFVSEYAVTGNDAGKGSLLAALGEAGFLIGVE 487
Query: 485 KNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSL 544
KNS+ ++M SYAPLFVN NDRRWNPDAIVF S Q YGTPSYW+Q FF ES+GATLL+SSL
Sbjct: 488 KNSEAIEMASYAPLFVNDNDRRWNPDAIVFTSSQMYGTPSYWMQHFFKESNGATLLSSSL 547
Query: 545 QTSSSSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGLDPNSLQP-SGSTRT 603
Q + S+SLIASAI W++S D +Y+RIKVVNFG V KISL GL NSL+ G+ T
Sbjct: 548 QANPSNSLIASAITWRNSLDNNDYLRIKVVNFGTTAVITKISLTGLGQNSLETLFGAVMT 607
Query: 604 VLTSANVMDENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDLLKESSNLKMVGGG 663
LTS NVMDENSF +P KV+P+++ ++ V M+VV+ P S +S D L S V
Sbjct: 608 ELTSNNVMDENSFREPNKVIPVKTQVEKVSDNMDVVLAPRSLNSIDFLLRKSINNNVDTA 667
Query: 664 SSTRSS 669
S +SS
Sbjct: 668 SVLKSS 673
>UniRef100_Q76LU4 Alpha-L-arabinofuranosidase [Lycopersicon esculentum]
Length = 674
Score = 925 bits (2390), Expect = 0.0
Identities = 447/666 (67%), Positives = 537/666 (80%), Gaps = 12/666 (1%)
Query: 8 FIVAALQLCIAVSLCAIKVNADLNATLTVDAHETSGKPISETLFGLFFEEINHAGAGGLW 67
F ++AL C A V A+ A L V+A E S + I +TLFG+FFEEINHAGAGGLW
Sbjct: 16 FGLSALCQCSATG-----VEANQTAVLLVNASEASARRIPDTLFGIFFEEINHAGAGGLW 70
Query: 68 AELVSNRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGD 127
AELV+NRGFE GGPN+PSNIDPWSIIG+ES + V TDR+SCFDRNKIA++++VLCD G
Sbjct: 71 AELVNNRGFEGGGPNVPSNIDPWSIIGDESKVIVSTDRSSCFDRNKIAVQVQVLCDHTGA 130
Query: 128 NICPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNGV---LASNVIT 184
NICP GVG++NPGFWGMNIEQGK YK+V YVRS S++++V+LTGSNG+ A+N++
Sbjct: 131 NICPDGGVGIYNPGFWGMNIEQGKSYKLVLYVRSEESVNVSVALTGSNGLQKLAAANIV- 189
Query: 185 GSASDFSNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYKGRGFRSELV 244
A+D S+W KVE LLEA T+ NSRL+L ++ KGVIW DQVS MP DTYKG GFR +L
Sbjct: 190 --AADVSSWTKVEILLEATGTDPNSRLELRSSKKGVIWFDQVSLMPTDTYKGHGFRKDLF 247
Query: 245 GMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWTDDGLGYFE 304
GML DLKP FIRFPGGCFVEG+WLRNAFRWKE+IG WEERPGHFGDVW YWTDDGLG+FE
Sbjct: 248 GMLKDLKPAFIRFPGGCFVEGDWLRNAFRWKETIGQWEERPGHFGDVWNYWTDDGLGHFE 307
Query: 305 FLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSKWGSIRASM 364
FLQLAEDL +LP+WVFNNGVSH+D+VDTS++LPFVQE LDG+EFARGDPTS WGSIRA M
Sbjct: 308 FLQLAEDLDSLPVWVFNNGVSHHDQVDTSSILPFVQEILDGLEFARGDPTSTWGSIRAKM 367
Query: 365 GHPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSSRPLDHPADI 424
GHPEPF+L+YVA+GNEDCGK YRGNYLKFY+AI+ YPDI+IISNCDGS+RPLDHPAD+
Sbjct: 368 GHPEPFDLRYVAIGNEDCGKTQYRGNYLKFYTAIKDKYPDIKIISNCDGSTRPLDHPADL 427
Query: 425 YDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLE 484
YD+HIY++A+ +FS + F++A R GPKAFVSEYAVTGNDAG GSLLAA+ EAGFL+G+E
Sbjct: 428 YDFHIYSSASSVFSNARHFDSAPRRGPKAFVSEYAVTGNDAGKGSLLAALGEAGFLIGVE 487
Query: 485 KNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSL 544
KNS+ ++M SYAPLFVN NDRRWNPDAIVF S Q YGTPSYW+Q FF E +GATLL+SSL
Sbjct: 488 KNSEAIEMASYAPLFVNDNDRRWNPDAIVFTSSQMYGTPSYWMQHFFKEPNGATLLSSSL 547
Query: 545 QTSSSSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGLDPNSLQP-SGSTRT 603
Q + S+SLIASAI W++S D +Y+RIKVVNFGA KISL GL +SL+ G+ T
Sbjct: 548 QANPSNSLIASAITWRNSLDNNDYLRIKVVNFGATAAITKISLTGLGQSSLETLFGAVMT 607
Query: 604 VLTSANVMDENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDLLKESSNLKMVGGG 663
LTS NVMDENSF +P KV+P+++ ++ V M+VV+ P S +S D L S V
Sbjct: 608 ELTSNNVMDENSFREPNKVIPVKTQVEKVSDNMDVVLAPRSLNSIDFLLRKSINNNVDTA 667
Query: 664 SSTRSS 669
S +SS
Sbjct: 668 SVLKSS 673
>UniRef100_Q8VZR2 Putative arabinosidase [Arabidopsis thaliana]
Length = 674
Score = 899 bits (2324), Expect = 0.0
Identities = 448/654 (68%), Positives = 524/654 (79%), Gaps = 14/654 (2%)
Query: 7 SFIVAALQLCIAVSLCAIKVNADLNATLTVDAHETSGKPISETLFGLFFEEINHAGAGGL 66
SFI+ + + +LC + D TL VDA + +PI ETLFG+FFEEINHAGAGGL
Sbjct: 17 SFILGSFS--VYQTLCLVDAQEDAIVTLQVDASNVTRRPIPETLFGIFFEEINHAGAGGL 74
Query: 67 WAELVSNRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKG 126
WAELVSNRGFEAGG IPSNI PWSIIG+ES I V TDR+SCF+RNKIALR+EVLCDS
Sbjct: 75 WAELVSNRGFEAGGQIIPSNIWPWSIIGDESSIYVVTDRSSCFERNKIALRMEVLCDS-- 132
Query: 127 DNICPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNG--VLASNVIT 184
N CP GVGV+NPG+WGMNIE+GKKYKVV YVRSTG +D++VS T SNG LAS I
Sbjct: 133 -NSCPLGGVGVYNPGYWGMNIEEGKKYKVVLYVRSTGDIDVSVSFTSSNGSVTLASENII 191
Query: 185 GSASDFSNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYKGRGFRSELV 244
ASD NW K E LLEA T++ +RLQ TTT KG IW DQVSAMP+DTYKG GFR++L
Sbjct: 192 ALASDLLNWTKKEMLLEANGTDNGARLQFTTTKKGSIWFDQVSAMPMDTYKGHGFRNDLF 251
Query: 245 GMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWTDDGLGYFE 304
M+ DLKPRFIRFPGGCFVEG+WL NAFRWKE++ WEERPGH+GDVWKYWTDDGLG+FE
Sbjct: 252 QMMVDLKPRFIRFPGGCFVEGDWLGNAFRWKETVRAWEERPGHYGDVWKYWTDDGLGHFE 311
Query: 305 FLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSKWGSIRASM 364
F QLAEDLGA PIWVFNNG+SHND+V+T V+PFVQEA+DGIEFARGD S WGS+RA+M
Sbjct: 312 FFQLAEDLGASPIWVFNNGISHNDQVETKNVMPFVQEAIDGIEFARGDSNSTWGSVRAAM 371
Query: 365 GHPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSSRPLDHPADI 424
GHPEPF LKYVAVGNEDC K YRGNYL+FY+AI+ AYPDI+IISNCD S++PLDHPAD
Sbjct: 372 GHPEPFELKYVAVGNEDCFKSYYRGNYLEFYNAIKKAYPDIKIISNCDASAKPLDHPADY 431
Query: 425 YDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLE 484
+DYHIYT A D+FS+S F+ R+GPKAFVSEYAV DA G+LLAA+ EA FLLGLE
Sbjct: 432 FDYHIYTLARDLFSKSHDFDNTPRNGPKAFVSEYAVNKADAKNGNLLAALGEAAFLLGLE 491
Query: 485 KNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSL 544
KNSDIV+MVSYAPLFVN NDRRW PDAIVFNS YGTPSYWVQ FF+ESSGATLLNS+L
Sbjct: 492 KNSDIVEMVSYAPLFVNTNDRRWIPDAIVFNSSHLYGTPSYWVQHFFTESSGATLLNSTL 551
Query: 545 QTSSSSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGLDPNSLQPSGSTRTV 604
+ +SS+ ASAI++Q++ K+YI+IK VNFG +VNLK+++ GL + GS + V
Sbjct: 552 K-GKTSSVEASAISFQTNG--KDYIQIKAVNFGEQSVNLKVAVTGL---MAKFYGSKKKV 605
Query: 605 LTSANVMDENSFSQPKKVVPIQSLLQNVGKE-MNVVVPPHSFSSFDLLKESSNL 657
LTSA+VMDENSFS P +VP +SLL+ +E + V+PPHSFSSFDLL ES N+
Sbjct: 606 LTSASVMDENSFSNPNMIVPQESLLEMTEQEDLMFVLPPHSFSSFDLLTESENV 659
>UniRef100_Q9ATV7 Arabinoxylan arabinofuranohydrolase isoenzyme AXAH-II [Hordeum
vulgare]
Length = 656
Score = 880 bits (2274), Expect = 0.0
Identities = 423/645 (65%), Positives = 501/645 (77%), Gaps = 8/645 (1%)
Query: 13 LQLCIAVSLCAIKVNADLNATLTVDAHETSGKPISETLFGLFFEEINHAGAGGLWAELVS 72
L C A ++ A ATL VDA + I +TLFG+FFEEINHAGAGG+WAELVS
Sbjct: 16 LLFCFGCKCIASELQATQTATLKVDASSQLARKIPDTLFGMFFEEINHAGAGGIWAELVS 75
Query: 73 NRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDNICPA 132
NRGFEAGG + PSNIDPWSIIG++S I V TDRTSCF RN IALR+EVLCD CPA
Sbjct: 76 NRGFEAGGLHTPSNIDPWSIIGDDSSIFVATDRTSCFSRNIIALRMEVLCDD-----CPA 130
Query: 133 DGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNGVLASNVITGSASDFSN 192
GVG++NPGFWGMNIE GK Y +V YV+S + +LTVSL S+G+ +T + SN
Sbjct: 131 SGVGIYNPGFWGMNIEDGKTYNLVMYVKSAEAAELTVSLASSDGLQNLASVTVPVAGTSN 190
Query: 193 WKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYKGRGFRSELVGMLADLKP 252
W KVE L AK TN SRLQ+T+ KGV+W DQVS MP DT+KG GFR+EL+ ML DLKP
Sbjct: 191 WTKVEQKLIAKGTNRTSRLQITSNKKGVVWFDQVSLMPSDTFKGHGFRTELISMLLDLKP 250
Query: 253 RFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWTDDGLGYFEFLQLAEDL 312
RF+RFPGGCFVEGEWLRNAFRW+ESIGPWEERPGHFGDVW YWTDDGLGY+EFLQL+EDL
Sbjct: 251 RFLRFPGGCFVEGEWLRNAFRWRESIGPWEERPGHFGDVWHYWTDDGLGYYEFLQLSEDL 310
Query: 313 GALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSKWGSIRASMGHPEPFNL 372
GA PIWVFNNG+SHNDEV T+A+ PFV++ LD +EFARG S WGS+RA+MGHPEPF +
Sbjct: 311 GAAPIWVFNNGISHNDEVSTAAIAPFVKDVLDSLEFARGSANSTWGSVRAAMGHPEPFPV 370
Query: 373 KYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSSRPLDHPADIYDYHIYTN 432
KYVA+GNEDCGKK Y GNYLKFY+AIR +YPDIQ+ISNCDGSS+PLDHPAD+YD+H+YT+
Sbjct: 371 KYVAIGNEDCGKKYYLGNYLKFYNAIRESYPDIQMISNCDGSSKPLDHPADLYDFHVYTD 430
Query: 433 ANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLEKNSDIVKM 492
+ +F+ TF+ R+GPKAFVSEYAV DAG GSLL ++AEA FL GLEKNSDIV M
Sbjct: 431 SKTLFNMKGTFDKTSRTGPKAFVSEYAVWRTDAGRGSLLGSLAEAAFLTGLEKNSDIVHM 490
Query: 493 VSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSLQTSSSSSL 552
SYAPLFVN ND+ WNPDAIVFNS+Q YGTPSYW+Q FF ESSGA + ++ +S S SL
Sbjct: 491 ASYAPLFVNDNDQTWNPDAIVFNSWQQYGTPSYWMQKFFRESSGAMIHPITISSSYSGSL 550
Query: 553 IASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGLDPNSLQPSGSTRTVLTSANVMD 612
ASAI WQ S + N++R+K+VNFG++TV+L IS++GL S+ GS TVLTS+NV D
Sbjct: 551 AASAITWQDSGN--NFLRVKIVNFGSDTVSLTISVSGLQA-SINALGSNATVLTSSNVKD 607
Query: 613 ENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDLLKESSNL 657
ENSFS P KVVP+ S L N ++M V + HSFSSFDL S L
Sbjct: 608 ENSFSNPTKVVPVTSQLHNAAEQMQVTLAAHSFSSFDLALAQSEL 652
>UniRef100_Q9ATV8 Arabinoxylan arabinofuranohydrolase isoenzyme AXAH-I [Hordeum
vulgare]
Length = 658
Score = 879 bits (2272), Expect = 0.0
Identities = 421/647 (65%), Positives = 507/647 (78%), Gaps = 12/647 (1%)
Query: 13 LQLCIAVSLCAIKVNADLNATLTVDAHETSGKPISETLFGLFFEEINHAGAGGLWAELVS 72
L C++ A + A+L VD+ + I +TLFG+FFEEINHAGAGG+WAELVS
Sbjct: 18 LLFCVSCKCLASEFEITQVASLGVDSSPHLARKIPDTLFGIFFEEINHAGAGGIWAELVS 77
Query: 73 NRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDNICPA 132
NRGFEAGGP+ PSNI+PWSIIG++S I V TDRTSCF RNK+ALR+EVLCD+ CP
Sbjct: 78 NRGFEAGGPHTPSNINPWSIIGDDSSIFVGTDRTSCFSRNKVALRMEVLCDN-----CPV 132
Query: 133 DGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNG--VLASNVITGSASDF 190
GVG++NPGFWGMNIE GK Y +V +V+S ++++TVSLT S+G VLAS I
Sbjct: 133 GGVGIYNPGFWGMNIEDGKTYNLVMHVKSPETIEMTVSLTSSDGLQVLASAAIRVPGG-- 190
Query: 191 SNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYKGRGFRSELVGMLADL 250
SNW K++ L AK T+ SRLQ+T KGV+WLDQVS MP DTYKG GFR+EL+ ML DL
Sbjct: 191 SNWIKLDQKLVAKGTDRTSRLQITARTKGVVWLDQVSLMPSDTYKGHGFRTELISMLLDL 250
Query: 251 KPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWTDDGLGYFEFLQLAE 310
KPRF+RFPGGCFVEG WLRNAFRW++SIGPWEERPGH+GDVW YWTDDGLGY+EFLQLAE
Sbjct: 251 KPRFLRFPGGCFVEGSWLRNAFRWRDSIGPWEERPGHYGDVWNYWTDDGLGYYEFLQLAE 310
Query: 311 DLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSKWGSIRASMGHPEPF 370
DLGA P+WVFNNG+SH+DEVDT+A+ PFV++ LD +EFARG S WGS+RA+MGHPEPF
Sbjct: 311 DLGAAPVWVFNNGISHHDEVDTTAIAPFVKDILDSLEFARGSAESTWGSVRAAMGHPEPF 370
Query: 371 NLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSSRPLDHPADIYDYHIY 430
+KYVA+GNEDCGK+NYRGNYLKFY+AIR AYPDIQ+ISNCDGSS PLDHPAD+YD+H+Y
Sbjct: 371 PVKYVAIGNEDCGKENYRGNYLKFYNAIREAYPDIQMISNCDGSSTPLDHPADLYDFHVY 430
Query: 431 TNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLEKNSDIV 490
T ++ +FS +TF+ RSGPKAFVSEYAVTGNDAG GSLLA++AEA FL GLEKNSD+V
Sbjct: 431 TGSSALFSMKNTFDNTSRSGPKAFVSEYAVTGNDAGRGSLLASLAEAAFLTGLEKNSDVV 490
Query: 491 KMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSLQTSSSS 550
+M SYAPLF+N NDR WNPDAIVFNS+Q YGTPSYW+Q F ESSGAT+ SL +S S
Sbjct: 491 QMASYAPLFINDNDRTWNPDAIVFNSWQQYGTPSYWMQTLFRESSGATVHPISLSSSCSG 550
Query: 551 SLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGLDPNSLQPSGSTRTVLTSANV 610
L ASAI W D+ +++R+K+VNFG + V L IS GL S+ GST TVLTS V
Sbjct: 551 LLAASAITWHD--DENSFLRVKIVNFGPDAVGLTISATGLQ-GSINAFGSTATVLTSGGV 607
Query: 611 MDENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDLLKESSNL 657
MDENSF+ P KVVP+ L++ +EM V +PPHS ++FDL S L
Sbjct: 608 MDENSFANPNKVVPVTVELRDAAEEMQVTLPPHSLTAFDLALAQSRL 654
>UniRef100_Q6AHR9 Arabinofuranosidase [Daucus carota]
Length = 654
Score = 852 bits (2202), Expect = 0.0
Identities = 419/647 (64%), Positives = 511/647 (78%), Gaps = 16/647 (2%)
Query: 15 LCIAVSLCAIKV--NADLNATLTVDAHETSGKPISETLFGLFFEEINHAGAGGLWAELVS 72
+CI V LC+ D+ A L V+A + S + I +TLFG+FFEEINHAGAGGLWAELV
Sbjct: 12 VCILVGLCSFYPCSGTDITAQLFVNASKQSARTIPDTLFGIFFEEINHAGAGGLWAELVD 71
Query: 73 NRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDNICPA 132
NRGFEAGG PS+I PW IIG+ES + + TDR+SCFDRNKIALR++VLCDSKG ICPA
Sbjct: 72 NRGFEAGGQATPSDIAPWFIIGDESLVVLSTDRSSCFDRNKIALRMDVLCDSKGPLICPA 131
Query: 133 DGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNG---VLASNVITGSASD 189
GVGV+NPGFWGMNIEQGK YK++ Y+RS ++++VSLT S G + SN+I AS
Sbjct: 132 GGVGVYNPGFWGMNIEQGKSYKLILYLRSYNPINVSVSLTDSTGSQPLATSNII---ASS 188
Query: 190 FSNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYKGRGFRSELVGMLAD 249
SNW ++E L AKATN NSRLQ TTT GVIW DQVS MP+DTYKG GFR +L +LAD
Sbjct: 189 VSNWTRMEFLFVAKATNPNSRLQFTTTQAGVIWFDQVSLMPVDTYKGLGFREDLFQLLAD 248
Query: 250 LKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWTDDGLGYFEFLQLA 309
LKP F+RFPGGCFVEG L NA RWK++IGPWEERPGH+GDVW YWTDDGLG+FEF QLA
Sbjct: 249 LKPAFLRFPGGCFVEGGRLPNAVRWKDTIGPWEERPGHYGDVWDYWTDDGLGHFEFFQLA 308
Query: 310 EDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSKWGSIRASMGHPEP 369
EDLGALPIWVFNNG+S + EV+ S +LPF+ E LDG+EFA+GD ++ WGS+RA++GHPEP
Sbjct: 309 EDLGALPIWVFNNGISLHGEVNMSVILPFIHEVLDGLEFAKGDASTTWGSVRAALGHPEP 368
Query: 370 FNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSSRPLDHPADIYDYHI 429
F+L+YVA+GNE+C + YRGNYLKFY AI+ YP+I+IISNCDGSS+ LDHPAD+YDYHI
Sbjct: 369 FDLRYVAIGNEECPIQGYRGNYLKFYQAIKHYYPEIKIISNCDGSSQKLDHPADLYDYHI 428
Query: 430 YTNANDMFSRSSTFNTALRSGPKAFVSEYAVT-GNDAGTGSLLAAIAEAGFLLGLEKNSD 488
YT+AN+MFS+ + F+ R GPKAFVSEYAVT DAGTGSLLAA+AEAGFLLGLE+NSD
Sbjct: 429 YTSANNMFSKYNAFDYVARDGPKAFVSEYAVTPKEDAGTGSLLAALAEAGFLLGLERNSD 488
Query: 489 IVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSLQTSS 548
IV+M SYAPLFVN NDR WNPDAIV NS ++YGTPSYW+Q FF ES+GATLLNS+LQ SS
Sbjct: 489 IVEMASYAPLFVNTNDRHWNPDAIVLNSAKAYGTPSYWMQQFFIESNGATLLNSTLQASS 548
Query: 549 SSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGLDPNSLQPSGSTRTVLTSA 608
++SL ASAI W+++ D Y+R+K VN G+ V++KIS++G+ N+ + TVLTS
Sbjct: 549 TNSLHASAIIWKNTDDATYYLRLKFVNIGSVPVSMKISIDGVKINT---KSALSTVLTSD 605
Query: 609 NVMDENSFSQPKKVVPIQS-LLQNV---GKEMNVVVPPHSFSSFDLL 651
NVMDENSF +P KV P+++ L NV G EM + HS +S D+L
Sbjct: 606 NVMDENSFDKPAKVTPVRAPLTANVELDGTEMTFTLAQHSLTSIDVL 652
>UniRef100_Q7XWX2 OSJNBb0058J09.4 protein [Oryza sativa]
Length = 654
Score = 841 bits (2172), Expect = 0.0
Identities = 404/638 (63%), Positives = 492/638 (76%), Gaps = 8/638 (1%)
Query: 13 LQLCIAVSLCAIKVNADLNATLTVDAHETSGKPISETLFGLFFEEINHAGAGGLWAELVS 72
L LC++ +VN A L VDA + I ETLFG+FFEEINHAGAGG+WAELVS
Sbjct: 16 LLLCVSCKCLTSEVNTTQLAVLKVDASPQHARKIPETLFGIFFEEINHAGAGGIWAELVS 75
Query: 73 NRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDNICPA 132
NRGFEAGGPN PS+IDPWSIIGNES I+V TDR+SCF RN IALR+EVLC GD C A
Sbjct: 76 NRGFEAGGPNTPSSIDPWSIIGNESVISVATDRSSCFSRNIIALRMEVLC---GD--CQA 130
Query: 133 DGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNGVLASNVITGSASDFSN 192
GVG++NPGFWGMNIE GK Y +V Y +S + +LTVSLT S+G+ + T + SN
Sbjct: 131 GGVGIYNPGFWGMNIEDGKNYSLVMYAKSLENTELTVSLTSSDGLQNLSSATIQVAGTSN 190
Query: 193 WKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYKGRGFRSELVGMLADLKP 252
W K+E L AK TN SRLQ+TT KGVIWLDQ+S MP DTYKG GFR ELV ML DLKP
Sbjct: 191 WTKLEQKLVAKGTNRTSRLQITTNKKGVIWLDQISLMPSDTYKGHGFRKELVSMLLDLKP 250
Query: 253 RFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWTDDGLGYFEFLQLAEDL 312
+F+RFPGGCFVEG+WLRNAFRW+ES+GPWEERPGHFGDVW YWTDDGLGY+EFLQL+EDL
Sbjct: 251 QFMRFPGGCFVEGQWLRNAFRWRESVGPWEERPGHFGDVWGYWTDDGLGYYEFLQLSEDL 310
Query: 313 GALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSKWGSIRASMGHPEPFNL 372
GA PIWVFNNG SHN+E++T+A+ PFV++ LD +EFARG S WGS+R +MGHPEPF +
Sbjct: 311 GAAPIWVFNNGFSHNEEINTTAIAPFVKDILDSLEFARGSTNSTWGSLRVAMGHPEPFPV 370
Query: 373 KYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSSRPLDHPADIYDYHIYTN 432
KYV +GNEDC KK Y GNYLKF+ AIR AYPDIQIISNCDGSS+PLDHPADIYD+H+Y +
Sbjct: 371 KYVTIGNEDCTKKFYHGNYLKFHRAIREAYPDIQIISNCDGSSKPLDHPADIYDFHVYGD 430
Query: 433 ANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLEKNSDIVKM 492
+N +FS + F++ R+G KAFVSEYAV+ N G G+LLA++AEA FL GLEKNSD+V+M
Sbjct: 431 SNTLFSMRNKFDSTPRNGTKAFVSEYAVSSNGVGRGTLLASLAEAAFLTGLEKNSDVVQM 490
Query: 493 VSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSLQTSSSSSL 552
SYAPLF+N ND WNP A+VFNS++ YG+PSYW+Q F ESSGA L ++ + S+SL
Sbjct: 491 ASYAPLFMNDNDLSWNPAAVVFNSWKQYGSPSYWMQTIFRESSGAVLHPVTINSMYSNSL 550
Query: 553 IASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGLDPNSLQPSGSTRTVLTSANVMD 612
ASAI W++S +++R+K+VN G+N VNL +S GL+ + ST T+LTS N+ D
Sbjct: 551 AASAITWKAS--NSSFLRVKIVNIGSNPVNLIVSTTGLEA-LVNMRKSTITILTSKNLSD 607
Query: 613 ENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDL 650
ENSFS+P VVP+ L N G+EM + P+SF+SFDL
Sbjct: 608 ENSFSKPTNVVPVTRELPNAGEEMFAFLGPYSFTSFDL 645
>UniRef100_Q8LIG5 Putative arabinoxylan narabinofuranohydrolase isoenzyme AXAH-I
[Oryza sativa]
Length = 664
Score = 833 bits (2152), Expect = 0.0
Identities = 395/649 (60%), Positives = 502/649 (76%), Gaps = 6/649 (0%)
Query: 9 IVAALQLCIAVSLCAIKVNADLNATLTVDAHETSGKPISETLFGLFFEEINHAGAGGLWA 68
+V +C VSL + A L VDA + + I + +FG+FFEEINHAGAGGLWA
Sbjct: 15 LVLVYAMCWTVSLGFV---AGQTGQLNVDASPQNARKIPDKMFGIFFEEINHAGAGGLWA 71
Query: 69 ELVSNRGFEAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDN 128
ELVSNRGFEAGGPN PSNIDPW IIGNES I V TDRTSCF++N +ALR+EVLCDSKG N
Sbjct: 72 ELVSNRGFEAGGPNTPSNIDPWLIIGNESSIIVGTDRTSCFEKNPVALRMEVLCDSKGTN 131
Query: 129 ICPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNGV--LASNVITGS 186
CP+ GVGV+NPG+WGMNIE+ + YKV ++RS+ ++ LTVSLT S+G+ LAS+ IT S
Sbjct: 132 NCPSGGVGVYNPGYWGMNIERRRVYKVGLHIRSSDAVSLTVSLTSSDGLQKLASHTITAS 191
Query: 187 ASDFSNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYKGRGFRSELVGM 246
F+ W K+E L++ TN NSRLQLTT+ GVIWLDQVS MP DTY G GFR +L M
Sbjct: 192 KKQFAKWTKIEFHLKSSQTNTNSRLQLTTSKSGVIWLDQVSVMPSDTYMGHGFRKDLASM 251
Query: 247 LADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYWTDDGLGYFEFL 306
LA+LKP+F++FPGG + G +LRNAFRW E++GPWEERPGHF D W YWTDDGLG+FEFL
Sbjct: 252 LANLKPQFLKFPGGNYAMGNYLRNAFRWSETVGPWEERPGHFNDAWGYWTDDGLGFFEFL 311
Query: 307 QLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTSKWGSIRASMGH 366
QLAEDLGA P+WV N+G S N+EV T+ + V++ +DGIEFARG P + WGS+RA+MGH
Sbjct: 312 QLAEDLGASPVWVVNDGASQNEEVSTATIASLVKDVVDGIEFARGGPKTTWGSVRAAMGH 371
Query: 367 PEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSSRPLDHPADIYD 426
P+PFNL YV++GN++C YRGNY KFYSAI++AYPDI ++S+CD S+ +PAD+YD
Sbjct: 372 PQPFNLDYVSIGNQECWMLYYRGNYQKFYSAIKAAYPDINVVSSCDKSTISPSNPADLYD 431
Query: 427 YHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLEKN 486
H+YT+++DMFSR+S F+ RSGPKA VSEYAVTG DAG G+L+ A+AEA FL+GLE+N
Sbjct: 432 VHVYTSSSDMFSRTSMFDNTTRSGPKAIVSEYAVTGKDAGKGTLVEALAEAAFLVGLERN 491
Query: 487 SDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSLQT 546
SD+V+M S APLFVN NDRRW+PDAIVFNS+Q+YG P+YW+ FF +S GAT S++Q
Sbjct: 492 SDVVEMASCAPLFVNDNDRRWSPDAIVFNSWQNYGCPNYWMLHFFKDSCGATFHPSNIQI 551
Query: 547 SSSSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGLDPNSLQPSGSTRTVLT 606
SS + L+ASAI WQ+S DK Y++IK+VNFG VNL IS++GLD ++ SGS +TVLT
Sbjct: 552 SSYNQLVASAITWQNSKDKSTYLKIKLVNFGNQAVNLSISVSGLD-EGIKSSGSKKTVLT 610
Query: 607 SANVMDENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDLLKESS 655
S+ +DENSF QP+KV P+ S + N ++M V+V P+S +SFDLL + S
Sbjct: 611 SSGPLDENSFQQPQKVAPVSSPVDNANEQMGVLVDPYSLTSFDLLLQPS 659
>UniRef100_Q9XH04 T1N24.13 protein [Arabidopsis thaliana]
Length = 521
Score = 681 bits (1757), Expect = 0.0
Identities = 349/548 (63%), Positives = 410/548 (74%), Gaps = 50/548 (9%)
Query: 118 LEVLCDSKGDNICPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNG- 176
+EVLCDS N CP GVGV+NPG+WGMNIE+GKKYKVV YVRSTG +D++VS T SNG
Sbjct: 1 MEVLCDS---NSCPLGGVGVYNPGYWGMNIEEGKKYKVVLYVRSTGDIDVSVSFTSSNGS 57
Query: 177 -VLASNVITGSASDFSNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYK 235
LAS I ASD NW K E LLEA T++ +RLQ TTT KG IW DQVSAMP+DTYK
Sbjct: 58 VTLASENIIALASDLLNWTKKEMLLEANGTDNGARLQFTTTKKGSIWFDQVSAMPMDTYK 117
Query: 236 GRGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKYW 295
G GFR++L M+ DLKPRFIRFPGGCFVEG+WL NAFRWKE++ WEERPGH+GDVWKYW
Sbjct: 118 GHGFRNDLFQMMVDLKPRFIRFPGGCFVEGDWLGNAFRWKETVRAWEERPGHYGDVWKYW 177
Query: 296 TDDGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGIEFARGDPTS 355
TDDGLG+FEF Q EA+DGIEFARGD S
Sbjct: 178 TDDGLGHFEFFQ---------------------------------EAIDGIEFARGDSNS 204
Query: 356 KWGSIRASMGHPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSS 415
WGS+RA+MGHPEPF LKYVAVGNEDC K YRGNYL+FY+AI+ AYPDI+IISNCD S+
Sbjct: 205 TWGSVRAAMGHPEPFELKYVAVGNEDCFKSYYRGNYLEFYNAIKKAYPDIKIISNCDASA 264
Query: 416 RPLDHPADIYDYH-----IYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSL 470
+PLDHPAD +DYH IYT A D+FS+S F+ R+GPKAFVSEYAV DA G+L
Sbjct: 265 KPLDHPADYFDYHNLVIQIYTLARDLFSKSHDFDNTPRNGPKAFVSEYAVNKADAKNGNL 324
Query: 471 LAAIAEAGFLLGLEKNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLF 530
LAA+ EA FLLGLEKNSDIV+MVSYAPLFVN NDRRW PDAIVFNS YGTPSYWVQ F
Sbjct: 325 LAALGEAAFLLGLEKNSDIVEMVSYAPLFVNTNDRRWIPDAIVFNSSHLYGTPSYWVQHF 384
Query: 531 FSESSGATLLNSSLQTSSSSSLIASAINWQSSADKKNYIRIKVVNFGANTVNLKISLNGL 590
F+ESSGATLLNS+L+ +SS+ ASAI++Q++ K+YI+IK VNFG +VNLK+++ GL
Sbjct: 385 FTESSGATLLNSTLK-GKTSSVEASAISFQTNG--KDYIQIKAVNFGEQSVNLKVAVTGL 441
Query: 591 DPNSLQPSGSTRTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKE-MNVVVPPHSFSSFD 649
+ GS + VLTSA+VMDENSFS P +VP +SLL+ +E + V+PPHSFSSFD
Sbjct: 442 ---MAKFYGSKKKVLTSASVMDENSFSNPNMIVPQESLLEMTEQEDLMFVLPPHSFSSFD 498
Query: 650 LLKESSNL 657
LL ES N+
Sbjct: 499 LLTESENV 506
>UniRef100_Q6RX11 A-arabinofuranosidase 1 [Ficus carica]
Length = 272
Score = 453 bits (1165), Expect = e-126
Identities = 208/272 (76%), Positives = 236/272 (86%), Gaps = 2/272 (0%)
Query: 101 VETDRTSCFDRNKIALRLEVLCDSKGDNICPADGVGVFNPGFWGMNIEQGKKYKVVFYVR 160
V TDR+SCF+RNKIAL++EVLCDSKG NICP +GVG++NPG+WGMNIEQGK YKV+ YVR
Sbjct: 1 VSTDRSSCFERNKIALKMEVLCDSKGPNICPPEGVGIYNPGYWGMNIEQGKSYKVILYVR 60
Query: 161 STGSLDLTVSLTGSNGV--LASNVITGSASDFSNWKKVETLLEAKATNHNSRLQLTTTAK 218
S +++++V+LTGSNG LAS I ++ S+W K E LLEAK TN NSRLQLTTT K
Sbjct: 61 SDDAINVSVALTGSNGSQKLASTNIIALVNEISDWTKKEFLLEAKGTNSNSRLQLTTTRK 120
Query: 219 GVIWLDQVSAMPLDTYKGRGFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESI 278
GVIW DQVS MPLDTYKG GFR++LV MLA+LKPRF RFPGGCFVEGEWLRNAFRWKE++
Sbjct: 121 GVIWFDQVSVMPLDTYKGHGFRTDLVQMLAELKPRFFRFPGGCFVEGEWLRNAFRWKETV 180
Query: 279 GPWEERPGHFGDVWKYWTDDGLGYFEFLQLAEDLGALPIWVFNNGVSHNDEVDTSAVLPF 338
GPWEERPGHFGDVW YWTDDG+GYFEFLQLAEDLGALP+WVFNNG H DEV TS VLPF
Sbjct: 181 GPWEERPGHFGDVWFYWTDDGIGYFEFLQLAEDLGALPVWVFNNGNGHRDEVATSTVLPF 240
Query: 339 VQEALDGIEFARGDPTSKWGSIRASMGHPEPF 370
VQEALDGIEFARG P SKWGS+RA+MGHP+PF
Sbjct: 241 VQEALDGIEFARGSPNSKWGSLRANMGHPQPF 272
>UniRef100_Q59218 Arabinosidase [Bacteroides ovatus]
Length = 660
Score = 335 bits (859), Expect = 3e-90
Identities = 223/678 (32%), Positives = 346/678 (50%), Gaps = 68/678 (10%)
Query: 19 VSLCAIKVNADLNATLTVDAHETS--GKPISETLFGLFFEEINHAGAGGLWAELVSNRGF 76
+++ A+ N L+A +T G I T++GLFFE+IN+A GGL+AELV NR F
Sbjct: 8 LAVLALSTNLALHAQTNELVIQTKKLGAEIQPTMYGLFFEDINYAADGGLYAELVKNRSF 67
Query: 77 EAGGPNIPSNIDPWSIIGNESFINVETDRTSCFDRNKIALRLEVLCDSKGDNICPADGVG 136
E P ++ W G S +N F+RN +RL D G
Sbjct: 68 E-----FPQHLMGWKTYGKVSLMN-----DGPFERNPHYVRLS-------DPGHAHKHTG 110
Query: 137 VFNPGFWGMNIEQGKKYKVVFYVR-STGSLDLTVSL----TGSNGVLASNVITGSASDFS 191
+ N GF+G+ +++G++Y+ + R GS T+ + T S G + V D
Sbjct: 111 LDNEGFFGIGVKKGEEYRFSVWARLPQGSTKETLRIELVDTQSMGERQALVAGNLTIDSK 170
Query: 192 NWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYKGR--GFRSELVGMLAD 249
+WKK + +L+ +T+ S L++ T+KG + L+ VS P+DT+KG G R +L LAD
Sbjct: 171 DWKKYQMILKPGSTHPKSVLRIFLTSKGTVDLEHVSLFPVDTWKGHENGLRKDLAQALAD 230
Query: 250 LKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWEERPGHFGDVWKY----------WTDDG 299
+ P RFPGGC VEG L + WK+S+GP E RP + + W+Y + G
Sbjct: 231 IHPGVFRFPGGCIVEGTDLETRYDWKKSVGPVENRPLN-ENRWQYTFTHRFFPDYYQSYG 289
Query: 300 LGYFEFLQLAEDLGALPIWVFNNGVS----HNDEVDTSAVLP---FVQEALDGIEFARGD 352
LG++E+ L+E++GA P+ + N G+S +ND AV ++Q+ALD IEFA G+
Sbjct: 290 LGFYEYFLLSEEMGAAPLPILNCGLSCQYQNNDPKAHVAVCDLDNYIQDALDLIEFANGN 349
Query: 353 PTSKWGSIRASMGHPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCD 412
+ WG +RA MGHP PFNLK++ +GNE GK+ Y F AIR A+P+I+I+ +
Sbjct: 350 VNTTWGKVRADMGHPAPFNLKFIGIGNEQWGKE-YPERLEPFIKAIRKAHPEIKIVGSSG 408
Query: 413 GSSRPLDHP----------ADIYDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTG 462
+S D D+ D H Y + ++ + ++ R GPK F EYA G
Sbjct: 409 PNSEGKDFDYLWPEMKRLKVDLVDEHFYRPESWFLAQGARYDNYDRKGPKVFAGEYACHG 468
Query: 463 NDAGTGSLLAAIAEAGFLLGLEKNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGT 522
AA+ EA F+ GLE+N+DIV M +YAPLF + +W PD I F++ S T
Sbjct: 469 KGKKWNHYHAALLEAAFMTGLERNADIVHMATYAPLFAHVEGWQWRPDMIWFDNLNSVRT 528
Query: 523 PSYWVQLFFSESSGATLLNSSLQ------TSSSSSLIASAINWQSSADK-KNYIRIKVVN 575
SY+VQ ++++ G +L ++ + L ASA+ DK KN + +KV N
Sbjct: 529 TSYYVQQLYAQNKGTNVLPLTMNKKNVTGAEGQNGLFASAV-----YDKGKNELIVKVAN 583
Query: 576 FGANTVNLKISLNGLDPNSLQPSGSTRTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKE 635
A + ++ GL + +G L S ++ +N+ QP +VP ++ + G
Sbjct: 584 TSATIQPISLNFEGLKKQDVLSNGRC-IKLRSLDLDKDNTLEQPFGIVPQETPVSIEGNV 642
Query: 636 MNVVVPPHSFSSFDLLKE 653
+ P +F+ + K+
Sbjct: 643 FTTELEPTTFAVYKFTKK 660
>UniRef100_Q8AAU6 Alpha-L-arabinofuranosidase A [Bacteroides thetaiotaomicron]
Length = 660
Score = 334 bits (857), Expect = 4e-90
Identities = 220/656 (33%), Positives = 341/656 (51%), Gaps = 74/656 (11%)
Query: 43 GKPISETLFGLFFEEINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNESFINVE 102
G I T++GLFFE+IN+A GGL+AELV NR FE P + W G + ++
Sbjct: 34 GAEIQPTMYGLFFEDINYAADGGLYAELVKNRSFE-----FPQRLMGWKTYGK---VTLQ 85
Query: 103 TDRTSCFDRNKIALRLEVLCDSKGDNICPADGVGVFNPGFWGMNIEQGKKYKVVFYVR-- 160
D F+RN +RL D+ G G+ N GF+G+ ++QG++Y+ + R
Sbjct: 86 DDGP--FERNPHYVRL----DNPGH---AHKHTGLDNEGFFGIGVKQGEEYRFSVWARLP 136
Query: 161 --STGSLDLTVSLT-----GSNGVLASNVITGSASDFSNWKKVETLLEAKATNHNSRLQL 213
TG + V L G + AS +T + D WKK + +L+A TN + L++
Sbjct: 137 HGGTGE-KIRVELVDTKSMGEHQAFASQTLTIDSKD---WKKYQVILKAGVTNPKATLRI 192
Query: 214 TTTAKGVIWLDQVSAMPLDTYKGR--GFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNA 271
++G + L+ +S P+DT+KG G R +L LAD+ P RFPGGC VEG L
Sbjct: 193 FLASQGTVDLEHISLFPVDTWKGHENGLRKDLAQALADIHPGVFRFPGGCIVEGTDLETR 252
Query: 272 FRWKESIGPWEERPGHFGDVWKY----------WTDDGLGYFEFLQLAEDLGALPIWVFN 321
+ WK+S+GP E RP + + W+Y + GLG++E+ L+E++GA P+ + N
Sbjct: 253 YDWKKSVGPVENRPLN-ENRWQYTFTHRFYPDYYQSYGLGFYEYFLLSEEMGAAPLPILN 311
Query: 322 NGVS----HNDEVDTSAVLP---FVQEALDGIEFARGDPTSKWGSIRASMGHPEPFNLKY 374
G++ +N+E AV ++Q+ALD IEFA GD + WG +RA MGHP PFNLKY
Sbjct: 312 CGLACQYQNNEEKAHVAVCDLDSYIQDALDLIEFANGDVNTTWGKVRADMGHPAPFNLKY 371
Query: 375 VAVGNEDCGKKNYRGNYLKFYSAIRSAYPDIQIISNCDGSS--RPLDH--------PADI 424
+ +GNE GK+ Y F A+R A+P+I I+ + +S + D+ AD+
Sbjct: 372 LGIGNEQWGKE-YPERLEPFIKALRKAHPEIMIVGSSGPNSEGKEFDYLWPEMKRLKADL 430
Query: 425 YDYHIYTNANDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLE 484
D H Y + S+ + ++ R GPK F EYA G AA+ EA F+ GLE
Sbjct: 431 VDEHFYRPESWFLSQGARYDNYDRKGPKVFAGEYACHGKGKKWNHFHAALLEAAFMTGLE 490
Query: 485 KNSDIVKMVSYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSL 544
+N+DIV M +YAPLF + +W PD I F++ S T SY+VQ ++++ G +L ++
Sbjct: 491 RNADIVHMATYAPLFAHVEGWQWRPDMIWFDNLNSVRTVSYYVQQLYAQNKGTNVLPLTM 550
Query: 545 Q------TSSSSSLIASAINWQSSADK-KNYIRIKVVNFGANTVNLKISLNGLDPNSLQP 597
+ L ASA+ DK KN + +KV N T + ++ GL +
Sbjct: 551 NKKPVTGAEGQNGLFASAV-----YDKDKNELIVKVANTSDKTQPVSLTFEGLKKQDVLS 605
Query: 598 SGSTRTVLTSANVMDENSFSQPKKVVPIQSLLQNVGKEMNVVVPPHSFSSFDLLKE 653
G T L+S + +N+ QP + P ++ + G + + P++F+ + K+
Sbjct: 606 EGRCIT-LSSLDQDKDNTLEQPFAITPQETPVTINGHALTTELGPNTFAVYKFTKK 660
>UniRef100_O88043 Putative secreted arabinosidase [Streptomyces coelicolor]
Length = 824
Score = 308 bits (789), Expect = 3e-82
Identities = 209/609 (34%), Positives = 311/609 (50%), Gaps = 59/609 (9%)
Query: 4 YKASFIVAALQLCIAVSLCAIKVNADLNATLTVDAHETSGKPISETLFGLFFEEINHAGA 63
++ V AL A+ A + T+TVD + G I +T++G+F+E+IN A
Sbjct: 6 WRLGLSVTALLTAAALLPSPAHAEAVADYTITVDPSDR-GPAIDDTMYGVFYEDINRAAD 64
Query: 64 GGLWAELVSNRGFEAGGPNIPSN--IDPWSIIGNESFINVETDRTSCFDRNKIALRLEVL 121
GGL+AELV NR FE + S + W+ G +N D +RN+ L L
Sbjct: 65 GGLYAELVQNRSFEYSTADNASYTPLTAWAADGTAEVVN---DGGRLNERNRNYLSL--- 118
Query: 122 CDSKGDNICPADGVGVFNPGF-WGMNIEQGKKYKVVFYVRSTGSLDLTVSLTGSNGVLAS 180
A G V N G+ G+ +E+G++Y + R+ LTV+LT + G LA
Sbjct: 119 ----------AAGSTVTNAGYNTGIRVEKGERYDFSVWSRAGHGTTLTVTLTDAAGTLAK 168
Query: 181 NVITGSASDFSNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLDQVSAMPLDTYKGR--G 238
+ W+K A T++ RL +TTTA LD VS P DTY+ + G
Sbjct: 169 ---ARQVAAKGQWRKYTATFTATRTSNRGRLAVTTTAPAA--LDMVSLFPRDTYRHQPGG 223
Query: 239 FRSELVGMLADLKPRFIRFPGGCFV----------EGEWLRN-AFRWKESIGPWEERPGH 287
R +L + L P F+RFPGGC V + W R +++WK++IGP EER +
Sbjct: 224 LRKDLAEKIEALHPGFLRFPGGCLVNTGSMEDYSADSGWQRKRSYQWKDTIGPVEERATN 283
Query: 288 FGDVWKYWTDDGLGYFEFLQLAEDLGALPIWVFN---NGVSHNDEVDTSAVLP-FVQEAL 343
+ W Y GLGY+E+ + AED+GA+P+ V G N VD A+L +Q+ L
Sbjct: 284 -ANFWGYNQSYGLGYYEYFRFAEDIGAMPLPVVPALVTGCGQNKAVDDDALLKRHIQDTL 342
Query: 344 DGIEFARGDPTSKWGSIRASMGHPEPFNLKYVAVGNEDCGKKNYRGNYLKFYSAIRSAYP 403
D IEFA G TS+WG RA MGHP+PF+L ++ VGNE+ K + + +F +AI + YP
Sbjct: 343 DLIEFANGPRTSEWGGKRAEMGHPKPFHLTHLEVGNEENLPKEFFARFEQFRTAIEAKYP 402
Query: 404 DIQIISNC--DGSSRPLDHP--------ADIYDYHIYTNANDMFSRSSTFNTALRSGPKA 453
D+ ++SN D S D D+ D H Y + + +++ R+GPK
Sbjct: 403 DVTVVSNSGPDDSGATFDTAWQLNRDAGVDMVDEHYYNSPQWFLQNNDRYDSYDRNGPKV 462
Query: 454 FVSEYAVTGNDAGTGSLLAAIAEAGFLLGLEKNSDIVKMVSYAPLFVNANDRRWNPDAIV 513
F+ EYA GN + A+AEA F+ GLE+N+D+VK+ SYAPL N + +W+PD I
Sbjct: 463 FLGEYASQGN-----AWKNALAEAAFMTGLERNADVVKLASYAPLLSNEDYVQWSPDLIW 517
Query: 514 FNSFQSYGTPSYWVQLFFSESSGATLLNSSLQ-TSSSSSLIASAINWQSSADKKNYIRIK 572
FN+ S+G+ +Y VQ F ++G ++ S+ T S S I + + A Y +K
Sbjct: 518 FNNHASWGSANYEVQKLFMNNTGDRVVPSTATGTPSVSGPITGGVGLSTWATSAAYDDVK 577
Query: 573 VVNFGANTV 581
V + T+
Sbjct: 578 VTSADGETL 586
>UniRef100_Q97DN6 Probable alpha-arabinofuranosidase [Clostridium acetobutylicum]
Length = 835
Score = 298 bits (762), Expect = 5e-79
Identities = 195/574 (33%), Positives = 292/574 (49%), Gaps = 50/574 (8%)
Query: 47 SETLFGLFFEEINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNESFINVETDRT 106
S+ L GLFFE+INH GGL++EL+ N+ FE ++ W++ S + T
Sbjct: 51 SDMLTGLFFEDINHGADGGLYSELLQNQSFE-----FKDSLSSWTVDKTGSTSSTAEVCT 105
Query: 107 SCFDRNKIALRLEVLCDSKGDNICPADGVGVFNPGFWGMNIEQGKKYKVVFYVRSTGSLD 166
S + LE+ C ++ + N G+ G+ + KY F+ R+ G +
Sbjct: 106 SKPLNSNNTHYLELNCPDNNSSL------KLVNSGYKGITVNNNAKYDFYFWARNVGKGN 159
Query: 167 -LTVSLTGSNG-VLASNVITGSASDFSNWKKVETLLEAKATNHNSRLQLTTTAKGVIWLD 224
+T+ L NG ++ + G + W+K E L A + N++L ++ T + + LD
Sbjct: 160 KVTIQLEDENGNAISEDKTIGKIN--GQWRKYEGHLRATKSTSNAKLAVSITGEAKMNLD 217
Query: 225 QVSAMPLDTYKGR--GFRSELVGMLADLKPRFIRFPGGCFVEGEWLRNAFRWKESIGPWE 282
VS P DT+K R G R +LV L DLKPRF+RFPGGC VEG + WK++IG E
Sbjct: 218 MVSLFPQDTWKNRKYGLRKDLVERLKDLKPRFLRFPGGCIVEGNSKEEIYNWKDTIGNVE 277
Query: 283 ERPGHFGDVWKYWTDDGLGYFEFLQLAEDLGALPIWVFN----------NGVSHNDEVDT 332
ER + ++W Y GLG++E+ QL ED+GA P+ V N NGV +
Sbjct: 278 ERKENT-NLWGYNQSYGLGFYEYFQLCEDIGATPVPVLNCGMTCQARGVNGVPNYMAPVG 336
Query: 333 SAVLPFVQEALDGIEFARGDPTSKWGSIRASMGHPEPFNLKYVAVGNEDCGKKNYRGNYL 392
+ P++Q A+D +E+A GD ++ WG R GH +PFNLKY+A+GNE G + Y +
Sbjct: 337 PDLDPYIQNAVDLVEYANGDASTYWGRKRIESGHKKPFNLKYMAIGNEQWGPE-YHKRFE 395
Query: 393 KFYSAIRSAYPDIQIISNC--DGSSRPLDH---------PADIYDYHIYTNANDMFSRSS 441
F + P I +ISN S D P + D H Y + + S ++
Sbjct: 396 AFQKVLNQKCPGITLISNAGTSPSGSTFDDNWNWIKEKAPNTVVDEHYYMSPDWFLSNTN 455
Query: 442 TFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLEKNSDIVKMVSYAPLFVN 501
++ R+ K FV EYA N +L +A+AE +L GLE+NSDIVKM SYAPLF
Sbjct: 456 RYDKYDRNSNKVFVGEYASQSN-----TLKSALAEGAYLTGLERNSDIVKMASYAPLFAK 510
Query: 502 ANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSL--QTSSSSSLIASAINW 559
A+D +W+PD I FN +Y TP+Y VQ FS + G ++ L T+ + I +
Sbjct: 511 ADDYQWSPDMIWFNGNTNYVTPNYNVQKLFSTNLGTQIVKGDLIKPTAHVKNDIKGGVLL 570
Query: 560 QSSADKKNYIRIKVVNFGANTVNLKISLNGLDPN 593
+ + + Y +KV + NT +I + D N
Sbjct: 571 GAWSTQVQYDNVKVTD---NTTGKEIFSDSFDNN 601
>UniRef100_Q828B3 Putative secreted arabinosidase [Streptomyces avermitilis]
Length = 710
Score = 296 bits (757), Expect = 2e-78
Identities = 197/569 (34%), Positives = 292/569 (50%), Gaps = 56/569 (9%)
Query: 42 SGKPISETLFGLFFEEINHAGAGGLWAELVSNRGFEAGGPNIPSN--IDPWSIIGNESFI 99
SG I +T++G+FFE+IN A GGL+AELV NR FE + S + W+ G +
Sbjct: 45 SGAKIDDTMYGVFFEDINRAADGGLYAELVQNRSFEYATADNTSYTPLTSWNTSGTADVV 104
Query: 100 NVETDRTSCFDRNKIALRLEVLCDSKGDNICPADGVGVFNPGF-WGMNIEQGKKYKVVFY 158
+ + R + +R+ +AL GD+ V N G+ G+ +E GK Y +
Sbjct: 105 S-DDGRLNARNRSYLALG--------GDS-------SVTNSGYNTGIAVESGKVYDFSVW 148
Query: 159 VRSTGSLDLTVSLTGSNGVLASNVITGSASDFSNWKKVETLLEAKATNHNSRLQLTTTAK 218
R+ + L+V+L ++G LA + W K A T+ RL T A
Sbjct: 149 ARADQADPLSVTLHDTDGDLAR---ARRVTVRGGWAKYTARFTAGRTSTTGRL--TVAAA 203
Query: 219 GVIWLDQVSAMPLDTYKGRGFRSELVGMLADLKPRFIRFPGGCFVE-----------GEW 267
G + LD VS +P DTY G G R +L +A L P F+RFPGGC V G
Sbjct: 204 GAVALDMVSLIPHDTYMGHGLRKDLAEKIAALHPGFVRFPGGCLVNTGSMRGYDEASGYE 263
Query: 268 LRNAFRWKESIGPWEERPGHFGDVWKYWTDDGLGYFEFLQLAEDLGALPIWVFN---NGV 324
+ +++WK++IGP E+R + + W Y GLGY+E+ Q AED GA+P+ V G
Sbjct: 264 RKRSYQWKDTIGPVEQRATN-ANFWGYNQSYGLGYYEYFQFAEDTGAMPLPVVPALVTGC 322
Query: 325 SHNDEVDTSAVLP-FVQEALDGIEFARGDPTSKWGSIRASMGHPEPFNLKYVAVGNEDCG 383
N VD A+L +Q+ LD IEFA G TS+WG RA MGHPEPF+L ++ VGNE+
Sbjct: 323 GQNKAVDDDALLERHIQDTLDLIEFANGPVTSEWGRKRARMGHPEPFHLTHLEVGNEENL 382
Query: 384 KKNYRGNYLKFYSAIRSAYPDIQIISNC-DGSSRPL---------DHPADIYDYHIYTNA 433
+ + +F +AI + YPD+ +ISN S P D D+ D H Y +
Sbjct: 383 PDEFFARFTRFRAAIEAKYPDVTVISNAGPDDSGPTFDTAWKLNRDADVDMVDEHYYNSP 442
Query: 434 NDMFSRSSTFNTALRSGPKAFVSEYAVTGNDAGTGSLLAAIAEAGFLLGLEKNSDIVKMV 493
+ ++ R+GPK F+ EYA GN + A+AEA ++ GLE+N+D+VK+
Sbjct: 443 QWFLQNNDRYDAYDRNGPKVFLGEYASGGN-----TFKNALAEAAYMTGLERNADVVKLA 497
Query: 494 SYAPLFVNANDRRWNPDAIVFNSFQSYGTPSYWVQLFFSESSGATLLNSSLQ-TSSSSSL 552
SYAPL N + +W PD I F++ S+G+ Y VQ F ++G ++ S+ T + S
Sbjct: 498 SYAPLLANEDYVQWRPDMIWFDNHASWGSADYEVQKLFMTNTGDRVVPSTASGTPALSGP 557
Query: 553 IASAINWQSSADKKNYIRIKVVNFGANTV 581
I+ A+ + A Y ++V T+
Sbjct: 558 ISGAVGLSTWATTAAYDDVRVTGEDGTTL 586
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.134 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,160,015,223
Number of Sequences: 2790947
Number of extensions: 51277711
Number of successful extensions: 122093
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 61
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 121620
Number of HSP's gapped (non-prelim): 176
length of query: 670
length of database: 848,049,833
effective HSP length: 134
effective length of query: 536
effective length of database: 474,062,935
effective search space: 254097733160
effective search space used: 254097733160
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0098b.4


