

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
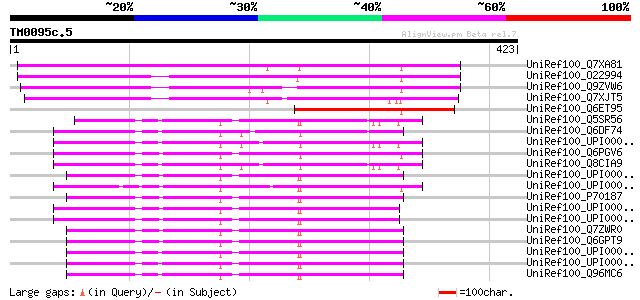
Query= TM0095c.5
(423 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XA81 At2g16980 [Arabidopsis thaliana] 204 3e-51
UniRef100_O22994 Putative tetracycline transporter protein [Arab... 194 5e-48
UniRef100_Q9ZVW6 Putative tetracycline transporter protein [Arab... 186 1e-45
UniRef100_Q7XJT5 Putative tetracycline transporter protein [Arab... 180 8e-44
UniRef100_Q6ET95 Tetracycline transporter protein-like [Oryza sa... 126 1e-27
UniRef100_Q5SR56 Hippocampus abundant transcript-like 1 [Homo sa... 94 7e-18
UniRef100_Q6DF74 LOC445835 protein [Xenopus laevis] 92 3e-17
UniRef100_UPI00001D10AB UPI00001D10AB UniRef100 entry 91 8e-17
UniRef100_Q6PGV6 Hypothetical protein zgc:63758 [Brachydanio rerio] 90 1e-16
UniRef100_Q8CIA9 RIKEN cDNA 5730414C17 [Mus musculus] 90 1e-16
UniRef100_UPI00003AE9E0 UPI00003AE9E0 UniRef100 entry 86 2e-15
UniRef100_UPI0000365D33 UPI0000365D33 UniRef100 entry 85 4e-15
UniRef100_P70187 Hippocampus abundant transcript 1 protein [Mus ... 84 6e-15
UniRef100_UPI00002F9BB3 UPI00002F9BB3 UniRef100 entry 84 8e-15
UniRef100_UPI0000362B7F UPI0000362B7F UniRef100 entry 84 1e-14
UniRef100_Q7ZWR0 Hiat1-prov protein [Xenopus laevis] 83 1e-14
UniRef100_Q6GPT9 MGC82622 protein [Xenopus laevis] 83 2e-14
UniRef100_UPI00001CEF36 UPI00001CEF36 UniRef100 entry 82 2e-14
UniRef100_UPI00003681F2 UPI00003681F2 UniRef100 entry 82 2e-14
UniRef100_Q96MC6 Hippocampus abundant transcript 1 protein [Homo... 82 2e-14
>UniRef100_Q7XA81 At2g16980 [Arabidopsis thaliana]
Length = 461
Score = 204 bits (520), Expect = 3e-51
Identities = 131/429 (30%), Positives = 213/429 (49%), Gaps = 59/429 (13%)
Query: 7 LIELRPLFHLLFPLSIHWIAEEMTVSVLVDVTTSALCPG-GSTCPKVIYINGLQQTIVGI 65
+ L L HLL + + +AE + V+ DVT +A+C G +C +Y+ G+QQ VG+
Sbjct: 3 VFRLGELRHLLVTVFLSGLAEYLIRPVMTDVTVAAVCSGLDDSCSLAVYLTGVQQVTVGM 62
Query: 66 FKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQ 125
MV++P++G LSD +G K +L + M ++ A+L + + F YA+YV++T ++ Q
Sbjct: 63 GTMVMMPVIGNLSDRYGIKAMLTLPMCLSVLPPAILGYRRDTNFFYAFYVIKTLFDMVCQ 122
Query: 126 GSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSIALLTF 185
G+I C++ AYVA V+ +KR ++F + G+ S S V ++ ARFL F V+ L
Sbjct: 123 GTIDCLANAYVAKNVHGTKRISMFGILAGVSSISGVCASLSARFLSIASTFQVAAISLFI 182
Query: 186 CPVYMQFFLVETVTPAPKKNQV-SGFCTK-----------------------VVDVLHKR 221
VYM+ FL E + A ++ SG C V + +
Sbjct: 183 GLVYMRVFLKERLQDADDDDEADSGGCRSHQEVHNGGDLKMLTEPILRDAPTKTHVFNSK 242
Query: 222 YKSMRNAAEILDNLIIAVQ--------------------YYLKAVFGFNKNQFSELLMMV 261
Y ++ +++N I +Q Y ++ F +N F+EL ++V
Sbjct: 243 YSPWKDVVSLINNSTILIQALVVTFFATFSESGRGSALMYLSESSFWVQQNDFAELFLLV 302
Query: 262 GIGSIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVL 321
I SQ+ +LP L+ +GE+ +LS LL A +AW+PWVPY +
Sbjct: 303 TIIGSISQLFILPTLSSTIGERKVLSTGLLMEFFNATCLSVAWSPWVPYAMTMLVPGAMF 362
Query: 322 VKPS--------------GKAQTFIAGAQSISDLLSPIAMSPLTSWFLSSNAPFDCKGFS 367
V PS GK Q I+G ++ + +++P SPLT+ FLS NAPF GFS
Sbjct: 363 VMPSVCGIASRQVGSSEQGKVQGCISGVRAFAQVVAPFVYSPLTALFLSENAPFYFPGFS 422
Query: 368 IICASVSMV 376
I+C ++S++
Sbjct: 423 ILCIAISLM 431
>UniRef100_O22994 Putative tetracycline transporter protein [Arabidopsis thaliana]
Length = 415
Score = 194 bits (492), Expect = 5e-48
Identities = 129/397 (32%), Positives = 201/397 (50%), Gaps = 41/397 (10%)
Query: 7 LIELRPLFHLLFPLSIHWIAEEMTVSVLVDVTTSALCPG-GSTCPKVIYINGLQQTIVGI 65
+ L L HLL + + +AE + V+ DVT +A+C G +C +Y+ G+QQ VG+
Sbjct: 3 VFRLGELRHLLVTVFLSGLAEYLIRPVMTDVTVAAVCSGLDDSCSLAVYLTGVQQVTVGM 62
Query: 66 FKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQ 125
MV++P++G LSD +G K +L + M ++ A+L + + F YA+YV++T
Sbjct: 63 GTMVMMPVIGNLSDRYGIKAMLTLPMCLSVLPPAILGYRRDTNFFYAFYVIKT------- 115
Query: 126 GSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSIALLTF 185
+ +A V+ +KR ++F + G+ S S V ++ ARFL F V+ L
Sbjct: 116 -------LFDMAKNVHGTKRISMFGILAGVSSISGVCASLSARFLSIASTFQVAAISLFI 168
Query: 186 CPVYMQFFLVETVTPAPKKNQV-SGFCTKVVDVLHKRYKSMRNAAEILDNLIIA------ 238
VYM+ FL E + A ++ SG C +V + M + D
Sbjct: 169 GLVYMRVFLKERLQDADDDDEADSGGCRSHQEVHNGGDLKMLTEPILRDAPTKTHVFNSK 228
Query: 239 -----VQYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAALLAS 293
Y+LKA FGFNKN F+EL ++V I SQ+ +LP L+ +GE+ +LS LL
Sbjct: 229 YSSWKDMYFLKARFGFNKNDFAELFLLVTIIGSISQLFILPTLSSTIGERKVLSTGLLME 288
Query: 294 IAYAWLYGLAWAPWVPYFSASFGIIYVLVKPS--------------GKAQTFIAGAQSIS 339
A +AW+PWVPY + V PS GK Q I+G ++ +
Sbjct: 289 FFNATCLSVAWSPWVPYAMTMLVPGAMFVMPSVCGIASRQVGSSEQGKVQGCISGVRAFA 348
Query: 340 DLLSPIAMSPLTSWFLSSNAPFDCKGFSIICASVSMV 376
+++P SPLT+ FLS NAPF GFSI+C ++S++
Sbjct: 349 QVVAPFVYSPLTALFLSENAPFYFPGFSILCIAISLM 385
>UniRef100_Q9ZVW6 Putative tetracycline transporter protein [Arabidopsis thaliana]
Length = 414
Score = 186 bits (471), Expect = 1e-45
Identities = 129/406 (31%), Positives = 198/406 (47%), Gaps = 65/406 (16%)
Query: 10 LRPLFHLLFPLSIHWIAEEMTVSVLVDVTTSALCPG-GSTCPKVIYINGLQQTIVGIFKM 68
L L HLL + + +E + V+ DVT +A+C G TC +Y+ G++Q VG+ M
Sbjct: 6 LGELRHLLTTVFLSGFSEFLVKPVMTDVTVAAVCSGLNETCSLAVYLTGVEQVTVGLGTM 65
Query: 69 VVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSI 128
V++P++G LSD +G K LL + M +I A+LA+ + F YA+Y+ +
Sbjct: 66 VMMPVIGNLSDRYGIKTLLTLPMCLSILPPAILAYRRDTNFFYAFYITKI---------- 115
Query: 129 FCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSIALLTFCPV 188
+ +A V KR ++F + G+ S S V AR LP IF V+ F V
Sbjct: 116 ----LFDMAKNVCGRKRISMFGVLAGVRSISGVCATFSARLLPIASIFQVAAISFFFGLV 171
Query: 189 YMQFFLVETV-----TPAPKKNQVSG-------------------FCTKVVDVLHKRYKS 224
YM+ FL E + + + SG TK+ VL+ +Y S
Sbjct: 172 YMRVFLKERLHDDDEDDCDEDDNTSGRNHHDGGDLTMLAEPILRDAPTKIHIVLNTKYSS 231
Query: 225 MRNAAEILDNLIIAVQYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKV 284
+++ Y+LKA FGFNKN F+EL+++V I SQ+ +LP L +GE+
Sbjct: 232 LKD------------MYFLKARFGFNKNDFAELILLVTIIGSISQLFILPKLVSAIGERR 279
Query: 285 ILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKPS--------------GKAQT 330
+LS LL A ++W+ WVPY + + + V PS GK Q
Sbjct: 280 VLSTGLLMDSVNAACLSVSWSAWVPYATTVLVPVTMFVMPSVCGIASRQVGPGEQGKVQG 339
Query: 331 FIAGAQSISDLLSPIAMSPLTSWFLSSNAPFDCKGFSIICASVSMV 376
I+G +S S +++P SPLT+ FLS APF GFS++C + S++
Sbjct: 340 CISGVKSFSGVVAPFIYSPLTALFLSEKAPFYFPGFSLLCVTFSLM 385
>UniRef100_Q7XJT5 Putative tetracycline transporter protein [Arabidopsis thaliana]
Length = 418
Score = 180 bits (456), Expect = 8e-44
Identities = 119/398 (29%), Positives = 196/398 (48%), Gaps = 54/398 (13%)
Query: 13 LFHLLFPLSIHWIAEEMTVSVLVDVTTSALCPG-GSTCPKVIYINGLQQTIVGIFKMVVL 71
L H+L + + A M V V+ DVT +A+C G +C +Y+ G QQ +G+ M+++
Sbjct: 8 LRHMLATVFLSAFAGFMVVPVITDVTVAAVCSGPDDSCSLAVYLTGFQQVAIGMGTMIMM 67
Query: 72 PLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCI 131
P++G LSD +G K +L + M +I +L + + +F Y +Y+ +
Sbjct: 68 PVIGNLSDRYGIKTILTLPMCLSIVPPVILGYRRDIKFFYVFYISKI------------- 114
Query: 132 SVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEYIFVVSIALLTFCPVYMQ 191
+ +A ++ S R + F + G+ + + + G ++ARFLP F VS VYM+
Sbjct: 115 -LTSMAVNIHGSTRISAFGILAGIKTIAGLFGTLVARFLPIALTFQVSAISFFVGLVYMR 173
Query: 192 FFLVETVTPAPKKNQVSGFCTK---------------------VVDVLHKRYKSMRNAAE 230
FL E + + G + V HK+Y S+++
Sbjct: 174 VFLKEKLNDDEDDDLHHGTYHQEDHDSINTTMLAEPILNDRPIKTQVFHKKYSSLKD--- 230
Query: 231 ILDNLIIAVQYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAAL 290
+ +L+ Y+LKA FGF+K QF++LL+++ I SQ+ +LP +GE +LS L
Sbjct: 231 -MISLMKTRYYFLKARFGFDKKQFADLLLLITIVGSISQLFVLPRFASAIGECKLLSTGL 289
Query: 291 LASIAYAWLYGLAWAPWVPYFSASF--GIIYVL----------VKP--SGKAQTFIAGAQ 336
+ ++WAPWVPY + F G ++V+ V P GK Q I+G +
Sbjct: 290 FMEFINMAIVSISWAPWVPYLTTVFVPGALFVMPSVCGIASRQVGPGEQGKVQGCISGVR 349
Query: 337 SISDLLSPIAMSPLTSWFLSSNAPFDCKGFSIICASVS 374
S +++P SPLT+ FLS NAPF GFS++C S+S
Sbjct: 350 SFGKVVAPFVFSPLTALFLSKNAPFYFPGFSLLCISLS 387
>UniRef100_Q6ET95 Tetracycline transporter protein-like [Oryza sativa]
Length = 211
Score = 126 bits (316), Expect = 1e-27
Identities = 63/148 (42%), Positives = 91/148 (60%), Gaps = 14/148 (9%)
Query: 238 AVQYYLKAVFGFNKNQFSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAALLASIAYA 297
A+ YYLKA FG++K++F+ LL++ G + SQ+ ++P+L VGE ++L LL +
Sbjct: 31 ALLYYLKAQFGYSKDEFANLLLIAGAAGMLSQLTVMPVLARFVGEDILLIIGLLGGCTHV 90
Query: 298 WLYGLAWAPWVPYFSASFGIIYVLVKPS--------------GKAQTFIAGAQSISDLLS 343
+LYG+AW+ WVPY SA F I+ V PS G AQ I+G S + +L+
Sbjct: 91 FLYGIAWSYWVPYLSAVFIILSAFVHPSIRTNVSKSVGSNEQGIAQGCISGISSFASILA 150
Query: 344 PIAMSPLTSWFLSSNAPFDCKGFSIICA 371
P+ +PLT+W LS APF KGFSI+CA
Sbjct: 151 PLIFTPLTAWVLSETAPFKFKGFSIMCA 178
>UniRef100_Q5SR56 Hippocampus abundant transcript-like 1 [Homo sapiens]
Length = 506
Score = 94.0 bits (232), Expect = 7e-18
Identities = 90/327 (27%), Positives = 137/327 (41%), Gaps = 50/327 (15%)
Query: 55 INGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQSEEFVYAYY 114
+NGL Q + G+ + PL+G LSD GRKP LL T+ T F ++ + Y+
Sbjct: 81 MNGLIQGVKGLLSFLSAPLIGALSDVWGRKPFLLGTVFFTCFPIPLMRISP-----WWYF 135
Query: 115 VLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVLARFLPEEY 174
+ + S + S F + AYVADV E +R+ + W++ F+AS V + +L Y
Sbjct: 136 AMISVSGVFS--VTFSVIFAYVADVTQEHERSTAYGWVSATFAASLVSSPAIGAYLSASY 193
Query: 175 ------IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHKRYKSMRNA 228
+ +ALL C F LV P+K + + ++ + S++
Sbjct: 194 GDSLVVLVATVVALLDIC-----FILVAVPESLPEKMRPVSWGAQISWKQADPFASLKKV 248
Query: 229 AEILDNLIIAV-----------QY-----YLKAVFGFNKNQFSELLMMVGIGSIFSQMVL 272
+ L+I + QY YL+ V GF + + + MVGI SI +Q
Sbjct: 249 GKDSTVLLICITVFLSYLPEAGQYSSFFLYLRQVIGFGSVKIAAFIAMVGILSIVAQTAF 308
Query: 273 LPILNPKVGEK-VILSAALLASIAYAWLYGL---AWAPW-----VPYFSASFGIIYVLVK 323
L IL +G K +L + AW YG AW W S +F I LV
Sbjct: 309 LSILMRSLGNKNTVLLGLGFQMLQLAW-YGFGSQAWMMWAAGTVAAMSSITFPAISALVS 367
Query: 324 ------PSGKAQTFIAGAQSISDLLSP 344
G AQ I G + + + L P
Sbjct: 368 RNAESDQQGVAQGIITGIRGLCNGLGP 394
>UniRef100_Q6DF74 LOC445835 protein [Xenopus laevis]
Length = 626
Score = 92.0 bits (227), Expect = 3e-17
Identities = 81/314 (25%), Positives = 136/314 (42%), Gaps = 33/314 (10%)
Query: 37 VTTSALCPGGSTCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTI 95
+TT L T P+ ++ NGL Q + G + PL+G LSD +GRK LL+T+ T
Sbjct: 186 LTTPMLVVLHKTFPQHTFLMNGLIQGVKGFLSFMCAPLIGALSDVYGRKSFLLLTVFFTC 245
Query: 96 FSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGL 155
F +++ + Y+ + + S S F + AYVAD+ E +R+ + ++
Sbjct: 246 FPIPLMSISP-----WWYFAMISVSGAFS--VTFSVIFAYVADITQEHERSTAYGLVSAT 298
Query: 156 FSASHVVGNVLARFLPEEY------IFVVSIALLTFCPVYMQF--FLVETVTPAPKKNQV 207
F+AS V + F+ E Y + +ALL C + + L E + P
Sbjct: 299 FAASLVTSPAIGAFISEYYGDNLVVLLATVVALLDICFILLAVPESLHEKIKPT---TWG 355
Query: 208 SGFCTKVVDVLHKRYKSMRNAAEILDNLIIAVQY------------YLKAVFGFNKNQFS 255
+ F + D K ++ +L + + + Y YL+ + GFN +
Sbjct: 356 APFSWEQADPFASLKKIGKDTTVLLICITVFLSYLPEAGQYSSFFLYLRQIIGFNPGSIA 415
Query: 256 ELLMMVGIGSIFSQMVLLPILNPKVGEK-VILSAALLASIAYAWLYGLAWAPWVPYFSAS 314
+ +VGI SI +Q VLL IL +G K +L AW YG PW+ + + +
Sbjct: 416 AFIAVVGILSIVAQTVLLSILMRSIGNKNTVLLGLGFQMFQLAW-YGFGSQPWMMWAAGA 474
Query: 315 FGIIYVLVKPSGKA 328
+ + P+ A
Sbjct: 475 VAAMSSITFPAVSA 488
>UniRef100_UPI00001D10AB UPI00001D10AB UniRef100 entry
Length = 484
Score = 90.5 bits (223), Expect = 8e-17
Identities = 92/343 (26%), Positives = 142/343 (40%), Gaps = 45/343 (13%)
Query: 37 VTTSALCPGGSTCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTI 95
+TT L T P+ ++ NGL Q + G+ + PL+G LSD GRKP LL T+ T
Sbjct: 41 LTTPMLTVLHETFPQHTFLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKPFLLGTVFFTC 100
Query: 96 FSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGL 155
F ++ + Y+ + + S + S F + AYVAD E +R+ + W++
Sbjct: 101 FPIPLMRISP-----WWYFGMISVSGVFS--VTFSVIFAYVADFTQEHERSTAYGWVSAT 153
Query: 156 FSASHVVGNVLARFLPEEY------IFVVSIALLTFCPVYMQF--FLVETVTPAPKKNQV 207
F+AS V + +L Y + +ALL C + + L E + PA Q+
Sbjct: 154 FAASLVSSPAIGTYLSSNYGDSLVVLVATVVALLDICFILVAVPESLPEKIRPASWGAQI 213
Query: 208 SGFCTKVVDVLHKRYKSMRNAAEILDNLIIAVQY------------YLKAVFGFNKNQFS 255
S K D K +++ +L + + + Y YL+ V GF +
Sbjct: 214 S---WKQADPFASLKKVGKDSTVLLICITVFLSYLPEAGQYSSFFLYLRQVIGFGSVKIV 270
Query: 256 ELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGL---AWAPW----- 307
+ MVGI SI +Q V L L +G K + L I YG AW W
Sbjct: 271 AFIAMVGILSILAQTVFLSKLMRSLGNKNTVLLGLGFQILQLAWYGFGAQAWMMWAAGTV 330
Query: 308 VPYFSASFGIIYVLVK------PSGKAQTFIAGAQSISDLLSP 344
S +F + L+ G AQ I G + + + L P
Sbjct: 331 AAMSSITFPAVSALISRNAESDQQGVAQGIITGIRGLCNGLGP 373
>UniRef100_Q6PGV6 Hypothetical protein zgc:63758 [Brachydanio rerio]
Length = 493
Score = 90.1 bits (222), Expect = 1e-16
Identities = 84/345 (24%), Positives = 141/345 (40%), Gaps = 49/345 (14%)
Query: 37 VTTSALCPGGSTCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTI 95
+TT L T P+ ++ NGL + G+ + PL+G LSD GRK LL+T+ T
Sbjct: 54 LTTPMLAVLRQTFPQHTFLMNGLIHGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTC 113
Query: 96 FSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGL 155
++ + Y+ + + S + + F + AYVAD+ E +R+ + ++
Sbjct: 114 APIPLMKISP-----WWYFAVISMSGVFAV--TFSVIFAYVADITQEHERSTAYGLVSAT 166
Query: 156 FSASHVVGNVLARFLPEEY------IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSG 209
F+AS V + +L E Y I +IALL C F LV P+K + +
Sbjct: 167 FAASLVTSPAIGAYLSEVYGDTLVVILATAIALLDIC-----FILVAVPESLPEKMRPAS 221
Query: 210 FCTKVVDVLHKRYKSMRNAAEILDNLIIAVQY----------------YLKAVFGFNKNQ 253
+ + + S+R + L+I + YL+ V GF
Sbjct: 222 WGAPISWEQADPFASLRKVGQDSTVLLICITVFLSYLPEAGPYSSFFLYLRQVIGFTSET 281
Query: 254 FSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSA 313
+ + +VGI SI +Q V+L IL +G K + L I YG PW+ + +
Sbjct: 282 VAAFIAVVGILSILAQTVVLGILMRSIGNKNTILLGLGFQILQLAWYGFGSQPWMMWAAG 341
Query: 314 SFGIIYVLVKPS--------------GKAQTFIAGAQSISDLLSP 344
+ + + P+ G Q I G + + + L P
Sbjct: 342 AVAAMSSITFPAISAIVSRNADPDQQGVVQGMITGIRGLCNGLGP 386
>UniRef100_Q8CIA9 RIKEN cDNA 5730414C17 [Mus musculus]
Length = 484
Score = 90.1 bits (222), Expect = 1e-16
Identities = 92/344 (26%), Positives = 145/344 (41%), Gaps = 47/344 (13%)
Query: 37 VTTSALCPGGSTCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTI 95
+TT L T P+ ++ NGL Q + G+ + PL+G LSD GRKP LL T+ T
Sbjct: 41 LTTPMLTVLHETFPQHTFLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKPFLLGTVFFTC 100
Query: 96 FSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGL 155
F ++ + + Y+ + + S + S F + AYVAD E +R+ + W++
Sbjct: 101 FPIPLMRINP-----WWYFGMISVSGVFS--VTFSVIFAYVADFTQEHERSTAYGWVSAT 153
Query: 156 FSASHVVGNVLARFLPEEY------IFVVSIALLTFCPVYMQF--FLVETVTPAPKKNQV 207
F+AS V + +L Y + +ALL C + + L E + PA Q+
Sbjct: 154 FAASLVSSPAIGTYLSANYGDSLVVLVATLVALLDICFILIAVPESLSEKIRPASWGAQI 213
Query: 208 SGFCTKVVDVLHKRYKSMRNAAEILDNLIIAVQY------------YLKAVFGFNKNQFS 255
S K D K +++ +L + + + Y YL+ V GF +
Sbjct: 214 S---WKQADPFASLKKVGKDSTVLLICITVFLSYLPEAGQYSSFFLYLRQVIGFGSVKIV 270
Query: 256 ELLMMVGIGSIFSQMVLLPILNPKVGEK-VILSAALLASIAYAWLYGL---AWAPW---- 307
+ MVGI SI +Q V L L +G K +L + AW YG AW W
Sbjct: 271 AFIAMVGILSIVAQTVFLSKLMRSLGNKNTVLLGLGFQMLQLAW-YGFGSQAWMMWAAGT 329
Query: 308 -VPYFSASFGIIYVLVK------PSGKAQTFIAGAQSISDLLSP 344
S +F + L+ G AQ + G + + + L P
Sbjct: 330 VAAMSSITFPAVSALISRNAESDQQGVAQGIVTGIRGLCNGLGP 373
>UniRef100_UPI00003AE9E0 UPI00003AE9E0 UniRef100 entry
Length = 491
Score = 85.9 bits (211), Expect = 2e-15
Identities = 73/304 (24%), Positives = 132/304 (43%), Gaps = 35/304 (11%)
Query: 48 TCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQS 106
T PK ++ NGL Q + G+ + PL+G LSD GRK LL+T+ T ++
Sbjct: 66 TFPKHTFLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTCAPIPLMKISP- 124
Query: 107 EEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVL 166
+ Y+ + + S + + F + AYVAD+ E +R+ + ++ F+AS V +
Sbjct: 125 ----WWYFAVISVSGVFAV--TFSVVFAYVADITQEHERSMAYGLVSATFAASLVTSPAI 178
Query: 167 ARFLPEEY------IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHK 220
+L Y + +IALL C F LV P+K + + + +
Sbjct: 179 GAYLGRVYGDSLVVVLATAIALLDIC-----FILVAVPESLPEKMRPASWGAPISWEQAD 233
Query: 221 RYKSMRNAAEILDNLIIAV-----------QY-----YLKAVFGFNKNQFSELLMMVGIG 264
+ S++ + L+I + QY YL+ + GF+ + + ++GI
Sbjct: 234 PFASLKKVGQDSIVLLICITVFLSYLPEAGQYSSFFLYLRQIMGFSSESVAAFIAVLGIL 293
Query: 265 SIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKP 324
SI +Q ++L +L +G K + L I YG PW+ + + + + + P
Sbjct: 294 SIIAQTIVLSLLMRSIGNKNTILLGLGFQILQLAWYGFGSEPWMMWAAGAVAAMSSITFP 353
Query: 325 SGKA 328
+ A
Sbjct: 354 AVSA 357
>UniRef100_UPI0000365D33 UPI0000365D33 UniRef100 entry
Length = 462
Score = 84.7 bits (208), Expect = 4e-15
Identities = 84/341 (24%), Positives = 143/341 (41%), Gaps = 42/341 (12%)
Query: 37 VTTSALCPGGSTCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTI 95
+TT L T P+ ++ NGL Q + G + PL+G LSD GRK LL+T+
Sbjct: 27 LTTPMLTVLHETFPRHTFLMNGLVQGVKGFLSFLSAPLIGALSDIWGRKSFLLLTV---F 83
Query: 96 FSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGL 155
F+ A + + + + Y+ L + S I + F + AYVAD+ E +R+ + ++
Sbjct: 84 FTCAPIPFMRISP--WCYFALISLSGIFAV--TFSVIFAYVADITEEQERSTAYGLVSAT 139
Query: 156 FSASHVVGNVLARFLPEEY---IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCT 212
F+AS V + +L +Y + + +++ + FF+V P + GF
Sbjct: 140 FAASLVTSPAIGAYLSAQYGDSLVALVATVISVIDIAFVFFVVPESLPDKMRLTSWGFPI 199
Query: 213 KVVDVLHKRYKSMRNAAEILDNLIIAV-----------QY-----YLKAVFGFNKNQFSE 256
+ S+R + L+I V QY YL+ V F+ +
Sbjct: 200 SWEQA--DPFASLRRVGKDTTVLLICVTVFLSYLPEAGQYSSFFLYLRQVIEFSPAAIAA 257
Query: 257 LLMMVGIGSIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFG 316
+ MVGI SI +Q + L IL +G K + L + YG PW+ + + +
Sbjct: 258 FIAMVGILSIVAQTLFLSILMRTIGNKNTVLLGLGFQLLQLTWYGFGSEPWMMWAAGTIA 317
Query: 317 IIYVLVKPS-------------GKAQTFIAGAQSISDLLSP 344
+ + P+ G AQ I G + + + L P
Sbjct: 318 AMSSITFPAVSALVSHCASPDQGVAQGMITGIRGLCNGLGP 358
>UniRef100_P70187 Hippocampus abundant transcript 1 protein [Mus musculus]
Length = 490
Score = 84.3 bits (207), Expect = 6e-15
Identities = 73/304 (24%), Positives = 132/304 (43%), Gaps = 35/304 (11%)
Query: 48 TCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQS 106
T PK ++ NGL Q + G+ + PL+G LSD GRK LL+T+ T ++
Sbjct: 65 TFPKHTFLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTCAPIPLMKISP- 123
Query: 107 EEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVL 166
+ Y+ + + S + + F + AYVAD+ E +R+ + ++ F+AS V +
Sbjct: 124 ----WWYFAVISVSGVFAV--TFSVVFAYVADITQEHERSMAYGLVSATFAASLVTSPAI 177
Query: 167 ARFLPEEY------IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHK 220
+L + Y + +IALL C F LV P+K + + + +
Sbjct: 178 GAYLGQMYGDSLVVVLATAIALLDIC-----FILVAVPESLPEKMRPASWGAPISWEQAD 232
Query: 221 RYKSMRNAAEILDNLIIAV-----------QY-----YLKAVFGFNKNQFSELLMMVGIG 264
+ S++ + L+I + QY YLK + F+ + + ++GI
Sbjct: 233 PFASLKKVGQDSIVLLICITVFLSYLPEAGQYSSFFLYLKQIMKFSPESVAAFIAVLGIL 292
Query: 265 SIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKP 324
SI +Q ++L +L +G K + L I YG PW+ + + + + + P
Sbjct: 293 SIIAQTIVLSLLMRSIGNKNTILLGLGFQILQLAWYGFGSEPWMMWAAGAVAAMSSITFP 352
Query: 325 SGKA 328
+ A
Sbjct: 353 AVSA 356
>UniRef100_UPI00002F9BB3 UPI00002F9BB3 UniRef100 entry
Length = 491
Score = 84.0 bits (206), Expect = 8e-15
Identities = 78/312 (25%), Positives = 133/312 (42%), Gaps = 35/312 (11%)
Query: 37 VTTSALCPGGSTCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTI 95
+TT L T P+ ++ NGL + G+ + PL+G LSD GRK LL+T+ T
Sbjct: 54 LTTPMLTVLHQTFPQHTFLMNGLIHGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTC 113
Query: 96 FSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGL 155
++ + Y+ + + S + + F + AYVAD+ E +R+ + ++
Sbjct: 114 APIPLMKISP-----WWYFAVISVSGVFAV--TFSVIFAYVADITQEHERSTAYGLVSAT 166
Query: 156 FSASHVVGNVLARFLPEEY------IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSG 209
F+AS V + +L E Y I +IALL C F LV P+K + +
Sbjct: 167 FAASLVTSPAIGAYLSEAYGDTLVVILATAIALLDIC-----FILVAVPESLPEKMRPAS 221
Query: 210 FCTKVVDVLHKRYKSMRNAAEILDNLIIAV-----------QY-----YLKAVFGFNKNQ 253
+ + + S+R + L+I + QY YLK V F+
Sbjct: 222 WGAPISWEQADPFASLRKVGQDSTVLLICITVFLSYLPEAGQYSSFFLYLKQVIRFSSET 281
Query: 254 FSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSA 313
+ + +VGI SI +Q ++L IL +G K + L I YG W+ + +
Sbjct: 282 VAAFIAVVGILSILAQTLVLGILMRSIGNKNSILLGLGFQILQLAWYGFGSQHWMMWAAG 341
Query: 314 SFGIIYVLVKPS 325
+ + + P+
Sbjct: 342 AVAAMSSITFPA 353
>UniRef100_UPI0000362B7F UPI0000362B7F UniRef100 entry
Length = 496
Score = 83.6 bits (205), Expect = 1e-14
Identities = 77/312 (24%), Positives = 133/312 (41%), Gaps = 35/312 (11%)
Query: 37 VTTSALCPGGSTCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTI 95
+TT L T P+ ++ NGL + G+ + PL+G LSD GRK LL+T+ T
Sbjct: 54 LTTPMLTVLRQTFPQHTFLMNGLIHGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTC 113
Query: 96 FSFAVLAWDQSEEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGL 155
++ + Y+ + + S + + F + AYVAD+ E +R+ + ++
Sbjct: 114 APIPLMKISP-----WWYFAVISMSGVFAV--TFSVIFAYVADITQEHERSTAYGLVSAT 166
Query: 156 FSASHVVGNVLARFLPEEY------IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSG 209
F+AS V + +L + Y I +IALL C F LV P+K + +
Sbjct: 167 FAASLVTSPAIGAYLSDAYGDTLVVILATAIALLDIC-----FILVAVPESLPEKMRPAS 221
Query: 210 FCTKVVDVLHKRYKSMRNAAEILDNLIIAV-----------QY-----YLKAVFGFNKNQ 253
+ + + S+R + L+I + QY YLK V F+
Sbjct: 222 WGAPISWEQADPFASLRKVGQDSTVLLICITVFLSYLPEAGQYSSFFLYLKQVIRFSSET 281
Query: 254 FSELLMMVGIGSIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSA 313
+ + +VGI SI +Q ++L IL +G K + L I YG W+ + +
Sbjct: 282 VAAFIAVVGILSILAQTLVLGILMRSIGNKNTILLGLGFQILQLAWYGFGSQHWMMWAAG 341
Query: 314 SFGIIYVLVKPS 325
+ + + P+
Sbjct: 342 AVAAMSSITFPA 353
>UniRef100_Q7ZWR0 Hiat1-prov protein [Xenopus laevis]
Length = 484
Score = 83.2 bits (204), Expect = 1e-14
Identities = 73/304 (24%), Positives = 131/304 (43%), Gaps = 35/304 (11%)
Query: 48 TCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQS 106
T PK ++ NGL Q + G+ + PL+G LSD GRK LL+T+ T ++
Sbjct: 64 TFPKHTFLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTCAPIPLMKISP- 122
Query: 107 EEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVL 166
+ Y+ + + S + + F + AYVAD+ E +R+ + ++ F+AS V +
Sbjct: 123 ----WWYFAVISVSGVFAV--TFSVVFAYVADITQEHERSMAYGLVSATFAASLVTSPAI 176
Query: 167 ARFLPEEY------IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHK 220
+L Y + +IALL C F LV P+K + + + +
Sbjct: 177 GAYLSRVYGDNLVVLLATAIALLDIC-----FILVAVPESLPEKMRPASWGAPISWEQAD 231
Query: 221 RYKSMRNAAEILDNLIIAV-----------QY-----YLKAVFGFNKNQFSELLMMVGIG 264
+ S++ + L+I + QY YL+ + F+ + + ++GI
Sbjct: 232 PFASLKKVGQDSIVLLICITVFLSYLPEAGQYSSFFLYLRQIMDFSPESVAAFIAVLGIL 291
Query: 265 SIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKP 324
SI +Q V+L +L +G K + L I YG PW+ + + + + + P
Sbjct: 292 SIIAQTVVLSLLMRSIGNKNTILLGLGFQILQLAWYGFGSEPWMMWAAGAVAAMSSITFP 351
Query: 325 SGKA 328
+ A
Sbjct: 352 AVSA 355
>UniRef100_Q6GPT9 MGC82622 protein [Xenopus laevis]
Length = 484
Score = 82.8 bits (203), Expect = 2e-14
Identities = 73/304 (24%), Positives = 131/304 (43%), Gaps = 35/304 (11%)
Query: 48 TCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQS 106
T PK ++ NGL Q + G+ + PL+G LSD GRK LL+T+ T ++
Sbjct: 64 TFPKHTFLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTCAPIPLMKISP- 122
Query: 107 EEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVL 166
+ Y+ + + S + + F + AYVAD+ E +R+ + ++ F+AS V +
Sbjct: 123 ----WWYFAVISVSGVFAV--TFSVVFAYVADITEEHERSMAYGLVSATFAASLVTSPAI 176
Query: 167 ARFLPEEY------IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHK 220
+L Y + +IALL C F LV P+K + + + +
Sbjct: 177 GAYLGHVYGDNLVVLLATAIALLDIC-----FILVAVPESLPEKMRPASWGAPISWEQAD 231
Query: 221 RYKSMRNAAEILDNLIIAV-----------QY-----YLKAVFGFNKNQFSELLMMVGIG 264
+ S++ + L+I + QY YL+ + F+ + + ++GI
Sbjct: 232 PFASLKKVGQDSIVLLICITVFLSYLPEAGQYSSFFLYLRQIMDFSPESVAAFIAVLGIL 291
Query: 265 SIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKP 324
SI +Q V+L +L +G K + L I YG PW+ + + + + + P
Sbjct: 292 SIIAQTVVLSLLMRSIGNKNTILLGLGFQILQLAWYGFGSEPWMMWAAGAVAAMSSITFP 351
Query: 325 SGKA 328
+ A
Sbjct: 352 AVSA 355
>UniRef100_UPI00001CEF36 UPI00001CEF36 UniRef100 entry
Length = 641
Score = 82.4 bits (202), Expect = 2e-14
Identities = 72/304 (23%), Positives = 131/304 (42%), Gaps = 35/304 (11%)
Query: 48 TCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQS 106
T PK ++ NGL Q + G+ + PL+G LSD GRK LL+T+ T ++
Sbjct: 216 TFPKHTFLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTCAPIPLMKISP- 274
Query: 107 EEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVL 166
+ Y+ + + S + + F + AYVAD+ E +R+ + ++ F+AS V +
Sbjct: 275 ----WWYFAVISVSGVFA--VTFSVVFAYVADITQEHERSMAYGLVSATFAASLVTSPAI 328
Query: 167 ARFLPEEY------IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHK 220
+L Y + +IALL C F LV P+K + + + +
Sbjct: 329 GAYLGRVYGDSLVVVLATAIALLDIC-----FILVAVPESLPEKMRPASWGAPISWEQAD 383
Query: 221 RYKSMRNAAEILDNLIIAV-----------QY-----YLKAVFGFNKNQFSELLMMVGIG 264
+ S++ + L+I + QY YL+ + F+ + + ++GI
Sbjct: 384 PFASLKKVGQDSIVLLICITVFLSYLPEAGQYSSFFLYLRQIMKFSPESVAAFIAVLGIL 443
Query: 265 SIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKP 324
SI +Q ++L +L +G K + L I YG PW+ + + + + + P
Sbjct: 444 SIIAQTIVLSLLMRSIGNKNTILLGLGFQILQLAWYGFGSEPWMMWAAGAVAAMSSITFP 503
Query: 325 SGKA 328
+ A
Sbjct: 504 AVSA 507
>UniRef100_UPI00003681F2 UPI00003681F2 UniRef100 entry
Length = 489
Score = 82.4 bits (202), Expect = 2e-14
Identities = 72/304 (23%), Positives = 131/304 (42%), Gaps = 35/304 (11%)
Query: 48 TCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQS 106
T PK ++ NGL Q + G+ + PL+G LSD GRK LL+T+ T ++
Sbjct: 64 TFPKHTFLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTCAPIPLMKISP- 122
Query: 107 EEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVL 166
+ Y+ + + S + + F + AYVAD+ E +R+ + ++ F+AS V +
Sbjct: 123 ----WWYFAVISVSGVFA--VTFSVVFAYVADITQEHERSMAYGLVSATFAASLVTSPAI 176
Query: 167 ARFLPEEY------IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHK 220
+L Y + +IALL C F LV P+K + + + +
Sbjct: 177 GAYLGRVYGDSLVVVLATAIALLDIC-----FILVAVPESLPEKMRPASWGAPISWEQAD 231
Query: 221 RYKSMRNAAEILDNLIIAV-----------QY-----YLKAVFGFNKNQFSELLMMVGIG 264
+ S++ + L+I + QY YL+ + F+ + + ++GI
Sbjct: 232 PFASLKKVGQDSIVLLICITVFLSYLPEAGQYSSFFLYLRQIMKFSPESVAAFIAVLGIL 291
Query: 265 SIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKP 324
SI +Q ++L +L +G K + L I YG PW+ + + + + + P
Sbjct: 292 SIIAQTIVLSLLMRSIGNKNTILLGLGFQILQLAWYGFGSEPWMMWAAGAVAAMSSITFP 351
Query: 325 SGKA 328
+ A
Sbjct: 352 AVSA 355
>UniRef100_Q96MC6 Hippocampus abundant transcript 1 protein [Homo sapiens]
Length = 490
Score = 82.4 bits (202), Expect = 2e-14
Identities = 72/304 (23%), Positives = 131/304 (42%), Gaps = 35/304 (11%)
Query: 48 TCPKVIYI-NGLQQTIVGIFKMVVLPLLGQLSDEHGRKPLLLITMSTTIFSFAVLAWDQS 106
T PK ++ NGL Q + G+ + PL+G LSD GRK LL+T+ T ++
Sbjct: 65 TFPKHTFLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTCAPIPLMKISP- 123
Query: 107 EEFVYAYYVLRTFSYIISQGSIFCISVAYVADVVNESKRAAVFSWITGLFSASHVVGNVL 166
+ Y+ + + S + + F + AYVAD+ E +R+ + ++ F+AS V +
Sbjct: 124 ----WWYFAVISVSGVFA--VTFSVVFAYVADITQEHERSMAYGLVSATFAASLVTSPAI 177
Query: 167 ARFLPEEY------IFVVSIALLTFCPVYMQFFLVETVTPAPKKNQVSGFCTKVVDVLHK 220
+L Y + +IALL C F LV P+K + + + +
Sbjct: 178 GAYLGRVYGDSLVVVLATAIALLDIC-----FILVAVPESLPEKMRPASWGAPISWEQAD 232
Query: 221 RYKSMRNAAEILDNLIIAV-----------QY-----YLKAVFGFNKNQFSELLMMVGIG 264
+ S++ + L+I + QY YL+ + F+ + + ++GI
Sbjct: 233 PFASLKKVGQDSIVLLICITVFLSYLPEAGQYSSFFLYLRQIMKFSPESVAAFIAVLGIL 292
Query: 265 SIFSQMVLLPILNPKVGEKVILSAALLASIAYAWLYGLAWAPWVPYFSASFGIIYVLVKP 324
SI +Q ++L +L +G K + L I YG PW+ + + + + + P
Sbjct: 293 SIIAQTIVLSLLMRSIGNKNTILLGLGFQILQLAWYGFGSEPWMMWAAGAVAAMSSITFP 352
Query: 325 SGKA 328
+ A
Sbjct: 353 AVSA 356
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 662,074,403
Number of Sequences: 2790947
Number of extensions: 25918977
Number of successful extensions: 127311
Number of sequences better than 10.0: 784
Number of HSP's better than 10.0 without gapping: 170
Number of HSP's successfully gapped in prelim test: 614
Number of HSP's that attempted gapping in prelim test: 126545
Number of HSP's gapped (non-prelim): 982
length of query: 423
length of database: 848,049,833
effective HSP length: 130
effective length of query: 293
effective length of database: 485,226,723
effective search space: 142171429839
effective search space used: 142171429839
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0095c.5


