

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0093a.1
(268 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
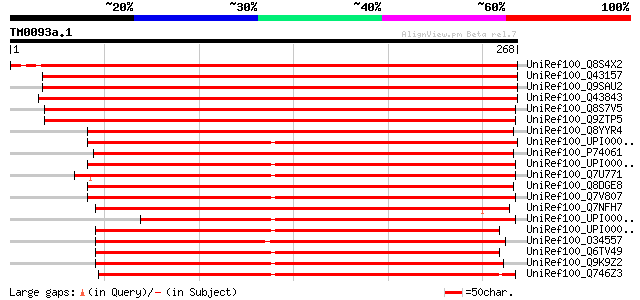
UniRef100_Q8S4X2 Ribulose-5-phosphate-3-epimerase [Pisum sativum] 474 e-132
UniRef100_Q43157 Ribulose-phosphate 3-epimerase, chloroplast pre... 472 e-132
UniRef100_Q9SAU2 Ribulose-5-phosphate-3-epimerase [Arabidopsis t... 463 e-129
UniRef100_Q43843 Ribulose-phosphate 3-epimerase, chloroplast pre... 458 e-128
UniRef100_Q8S7V5 Ribulose-5-phosphate-3-epimerase [Oryza sativa] 438 e-122
UniRef100_Q9ZTP5 Ribulose-phosphate 3-epimerase, chloroplast pre... 434 e-120
UniRef100_Q8YYR4 Ribulose-phosphate 3-epimerase [Anabaena sp.] 329 4e-89
UniRef100_UPI0000289372 UPI0000289372 UniRef100 entry 319 5e-86
UniRef100_P74061 Ribulose-phosphate 3-epimerase [Synechocystis sp.] 318 9e-86
UniRef100_UPI00002B4476 UPI00002B4476 UniRef100 entry 318 1e-85
UniRef100_Q7U771 Ribulose-5-phosphate 3-epimerase [Synechococcus... 316 3e-85
UniRef100_Q8DGE8 Pentose-5-phosphate-3-epimerase [Synechococcus ... 315 1e-84
UniRef100_Q7V807 Ribulose-phosphate 3-epimerase precursor [Proch... 311 1e-83
UniRef100_Q7NFH7 Ribulose-phosphate 3-epimerase [Gloeobacter vio... 309 6e-83
UniRef100_UPI000025C5BA UPI000025C5BA UniRef100 entry 283 3e-75
UniRef100_UPI00003CC223 UPI00003CC223 UniRef100 entry 258 1e-67
UniRef100_O34557 Ribulose-phosphate 3-epimerase [Bacillus subtilis] 258 1e-67
UniRef100_Q6TV49 Putative ribulose-5-phosphate 3-epimerase [Baci... 257 2e-67
UniRef100_Q9K9Z2 Ribulose-phosphate 3-epimerase [Bacillus halodu... 256 3e-67
UniRef100_Q746Z3 Ribulose-phosphate 3-epimerase [Geobacter sulfu... 256 3e-67
>UniRef100_Q8S4X2 Ribulose-5-phosphate-3-epimerase [Pisum sativum]
Length = 281
Score = 474 bits (1219), Expect = e-132
Identities = 243/268 (90%), Positives = 253/268 (93%), Gaps = 4/268 (1%)
Query: 1 INGPSSLHSKTAIFNNHPRSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKL 60
ING S KT++ H SLTFSRRK ST+VKASSRVDKFSKSDIIVSPSILSANFAKL
Sbjct: 17 INGLSL--RKTSLL--HSPSLTFSRRKFSTVVKASSRVDKFSKSDIIVSPSILSANFAKL 72
Query: 61 GEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVP 120
GEQVKAVELAGCDWIH+DVMDGRFVPNITIGPL+VDALRPVTDLPLDVHLMIVEP+QRVP
Sbjct: 73 GEQVKAVELAGCDWIHIDVMDGRFVPNITIGPLVVDALRPVTDLPLDVHLMIVEPDQRVP 132
Query: 121 DFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVL 180
DFIKAGADI+S+HCEQSSTIHLHR VNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVL
Sbjct: 133 DFIKAGADIISVHCEQSSTIHLHRAVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVL 192
Query: 181 IMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALV 240
IMSVNPGFGGQSFIES VKKI++LRRLC EKGVNPWIEVDGGVTPANAYKVIEAGANALV
Sbjct: 193 IMSVNPGFGGQSFIESHVKKIAELRRLCVEKGVNPWIEVDGGVTPANAYKVIEAGANALV 252
Query: 241 AGSAVFGAKDYAEAIKGIKTSKRPEPVA 268
AGSAVFGAKDYAEAIKGIK SKRPEPVA
Sbjct: 253 AGSAVFGAKDYAEAIKGIKASKRPEPVA 280
>UniRef100_Q43157 Ribulose-phosphate 3-epimerase, chloroplast precursor [Spinacia
oleracea]
Length = 285
Score = 472 bits (1214), Expect = e-132
Identities = 235/251 (93%), Positives = 245/251 (96%)
Query: 18 PRSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHV 77
P SLTF+RRK+ T+VKA+SRVDKFSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHV
Sbjct: 34 PNSLTFTRRKVQTLVKATSRVDKFSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHV 93
Query: 78 DVMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQS 137
DVMDGRFVPNITIGPL+VDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVS+HCE +
Sbjct: 94 DVMDGRFVPNITIGPLVVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSVHCELA 153
Query: 138 STIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQ 197
STIHLHRTVNQ+KSLGAKAGVVLNPGTPLS IEYVLDVVDLVLIMSVNPGFGGQSFIESQ
Sbjct: 154 STIHLHRTVNQIKSLGAKAGVVLNPGTPLSTIEYVLDVVDLVLIMSVNPGFGGQSFIESQ 213
Query: 198 VKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKG 257
VKKISDLR++C EKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKG
Sbjct: 214 VKKISDLRKMCVEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKG 273
Query: 258 IKTSKRPEPVA 268
IK SKRPEPVA
Sbjct: 274 IKASKRPEPVA 284
>UniRef100_Q9SAU2 Ribulose-5-phosphate-3-epimerase [Arabidopsis thaliana]
Length = 281
Score = 463 bits (1192), Expect = e-129
Identities = 233/251 (92%), Positives = 242/251 (95%)
Query: 18 PRSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHV 77
P S +FSRR+ IVKASSRVD+FSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHV
Sbjct: 30 PTSFSFSRRRTHGIVKASSRVDRFSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHV 89
Query: 78 DVMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQS 137
DVMDGRFVPNITIGPL+VDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVS+HCEQ
Sbjct: 90 DVMDGRFVPNITIGPLVVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSVHCEQQ 149
Query: 138 STIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQ 197
STIHLHRTVNQ+KSLGAKAGVVLNPGTPLSAIEYVLD+VDLVLIMSVNPGFGGQSFIESQ
Sbjct: 150 STIHLHRTVNQIKSLGAKAGVVLNPGTPLSAIEYVLDMVDLVLIMSVNPGFGGQSFIESQ 209
Query: 198 VKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKG 257
VKKISDLR++CAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKG
Sbjct: 210 VKKISDLRKMCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKG 269
Query: 258 IKTSKRPEPVA 268
IK SKRP VA
Sbjct: 270 IKASKRPAAVA 280
>UniRef100_Q43843 Ribulose-phosphate 3-epimerase, chloroplast precursor [Solanum
tuberosum]
Length = 280
Score = 458 bits (1179), Expect = e-128
Identities = 225/253 (88%), Positives = 244/253 (95%)
Query: 16 NHPRSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWI 75
++P SLTF+RR+I T+V ASSRVDKFSKSDIIVSPSILSANF+KLGEQVKA+E AGCDWI
Sbjct: 27 SNPNSLTFTRRRIQTVVNASSRVDKFSKSDIIVSPSILSANFSKLGEQVKAIEQAGCDWI 86
Query: 76 HVDVMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCE 135
HVDVMDGRFVPNITIGPL+VD+LRP+TDLPLDVHLMIVEP+QRVPDFIKAGADIVS+HCE
Sbjct: 87 HVDVMDGRFVPNITIGPLVVDSLRPITDLPLDVHLMIVEPDQRVPDFIKAGADIVSVHCE 146
Query: 136 QSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIE 195
QSSTIHLHRT+NQ+KSLGAKAGVVLNPGTPL+AIEYVLD VDLVLIMSVNPGFGGQSFIE
Sbjct: 147 QSSTIHLHRTINQIKSLGAKAGVVLNPGTPLTAIEYVLDAVDLVLIMSVNPGFGGQSFIE 206
Query: 196 SQVKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAI 255
SQVKKISDLR++CAE+G+NPWIEVDGGV P NAYKVIEAGANALVAGSAVFGA DYAEAI
Sbjct: 207 SQVKKISDLRKICAERGLNPWIEVDGGVGPKNAYKVIEAGANALVAGSAVFGAPDYAEAI 266
Query: 256 KGIKTSKRPEPVA 268
KGIKTSKRPE VA
Sbjct: 267 KGIKTSKRPEAVA 279
>UniRef100_Q8S7V5 Ribulose-5-phosphate-3-epimerase [Oryza sativa]
Length = 274
Score = 438 bits (1126), Expect = e-122
Identities = 220/249 (88%), Positives = 236/249 (94%)
Query: 19 RSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHVD 78
R L FS + + V+ASSRVDKFSK+DIIVSPSILSANF+KLGEQVKAVE+AGCDWIHVD
Sbjct: 24 RRLAFSSPRKAFRVRASSRVDKFSKNDIIVSPSILSANFSKLGEQVKAVEVAGCDWIHVD 83
Query: 79 VMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSS 138
VMDGRFVPNITIGPL+VDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVS+HCEQSS
Sbjct: 84 VMDGRFVPNITIGPLVVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSVHCEQSS 143
Query: 139 TIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQV 198
TIHLHRTVNQ+KSLGAKAGVVLNP TPL+AI+YVLDVVDLVLIMSVNPGFGGQSFIESQV
Sbjct: 144 TIHLHRTVNQIKSLGAKAGVVLNPATPLTAIDYVLDVVDLVLIMSVNPGFGGQSFIESQV 203
Query: 199 KKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKGI 258
KKI++LRRLCAEKGVNPWIEVDGGV P NAYKVIEAGANA+VAGSAVFGA DYAEAIKGI
Sbjct: 204 KKIAELRRLCAEKGVNPWIEVDGGVGPKNAYKVIEAGANAIVAGSAVFGAPDYAEAIKGI 263
Query: 259 KTSKRPEPV 267
KTS++P V
Sbjct: 264 KTSQKPVAV 272
>UniRef100_Q9ZTP5 Ribulose-phosphate 3-epimerase, chloroplast precursor [Oryza
sativa]
Length = 274
Score = 434 bits (1115), Expect = e-120
Identities = 219/249 (87%), Positives = 234/249 (93%)
Query: 19 RSLTFSRRKISTIVKASSRVDKFSKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHVD 78
R L FS + + V+ASSRVDKFSK+DIIVSPSILSANF+KLGEQVKAVE+AGCDWIHVD
Sbjct: 24 RRLAFSSPRKAFRVRASSRVDKFSKNDIIVSPSILSANFSKLGEQVKAVEVAGCDWIHVD 83
Query: 79 VMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSS 138
VMDGRFVPNITIGPL+VDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVS+HCEQSS
Sbjct: 84 VMDGRFVPNITIGPLVVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSVHCEQSS 143
Query: 139 TIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQV 198
TIHLHRTVNQ+KSLGAKAGVVLNP TPL+AI+YVLDVVDLVLIMSVNPGFGGQSFIESQV
Sbjct: 144 TIHLHRTVNQIKSLGAKAGVVLNPATPLTAIDYVLDVVDLVLIMSVNPGFGGQSFIESQV 203
Query: 199 KKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKGI 258
KKI++LR+ AEKGVNPWIEVDGGV P NAYKVIEAGANA+VAGSAVFGA DYAEAIKGI
Sbjct: 204 KKIAELRKYVAEKGVNPWIEVDGGVGPKNAYKVIEAGANAIVAGSAVFGAPDYAEAIKGI 263
Query: 259 KTSKRPEPV 267
KTSKRP V
Sbjct: 264 KTSKRPVAV 272
>UniRef100_Q8YYR4 Ribulose-phosphate 3-epimerase [Anabaena sp.]
Length = 235
Score = 329 bits (844), Expect = 4e-89
Identities = 155/225 (68%), Positives = 192/225 (84%)
Query: 42 SKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPV 101
S+ I++SPSILSA+F++LG++++AV+ AG DWIHVDVMDGRFVPNITIGPL+V+A+RPV
Sbjct: 6 SEKPIVISPSILSADFSRLGDEIRAVDAAGADWIHVDVMDGRFVPNITIGPLVVEAIRPV 65
Query: 102 TDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLN 161
T PLDVHLMIVEPE+ V DF KAGADI+S+HCE +++ HLHRT+ Q+K LG +AGVVLN
Sbjct: 66 TKKPLDVHLMIVEPEKYVEDFAKAGADIISVHCEHNASPHLHRTLGQIKELGKQAGVVLN 125
Query: 162 PGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDG 221
PGTPL IEYVL++ DL+LIMSVNPGFGGQSFI V KI LR++C E+G++PWIEVDG
Sbjct: 126 PGTPLELIEYVLELCDLILIMSVNPGFGGQSFIPGVVPKIRQLRQMCDERGLDPWIEVDG 185
Query: 222 GVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTSKRPEP 266
G+ N ++V+EAGANA+VAGSAVF AKDYAEAI I+ SKRP P
Sbjct: 186 GLKANNTWQVLEAGANAIVAGSAVFNAKDYAEAITNIRNSKRPTP 230
>UniRef100_UPI0000289372 UPI0000289372 UniRef100 entry
Length = 227
Score = 319 bits (817), Expect = 5e-86
Identities = 155/227 (68%), Positives = 189/227 (82%), Gaps = 2/227 (0%)
Query: 42 SKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPV 101
S+ +++SPSILSA+F+KLGE V+AV+ AG DWIHVDVMDGRFVPNITIGP+IV ALRPV
Sbjct: 2 SQKSLVISPSILSADFSKLGEDVRAVDQAGADWIHVDVMDGRFVPNITIGPMIVKALRPV 61
Query: 102 TDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLN 161
T PLDVHLMIVEPE+ V DF KAGADI+S+ E + HLHR + Q+K LG KAG VLN
Sbjct: 62 TSKPLDVHLMIVEPERYVEDFAKAGADIISVQVEACT--HLHRNLAQIKDLGKKAGAVLN 119
Query: 162 PGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDG 221
PGTP+ +EY L++ DLVL+MSVNPGFGGQSFIE+QV KI +LRR+C E+G++PWIEVDG
Sbjct: 120 PGTPIDTLEYCLELCDLVLVMSVNPGFGGQSFIENQVDKIRNLRRMCDERGLDPWIEVDG 179
Query: 222 GVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTSKRPEPVA 268
G+ ANA+KVIEAGANA+V+GS VF +YA+AI+GI+ SKRPE A
Sbjct: 180 GIKAANAWKVIEAGANAIVSGSGVFNQPNYADAIQGIRDSKRPELAA 226
>UniRef100_P74061 Ribulose-phosphate 3-epimerase [Synechocystis sp.]
Length = 229
Score = 318 bits (815), Expect = 9e-86
Identities = 152/222 (68%), Positives = 188/222 (84%)
Query: 45 DIIVSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDL 104
+I+V+PSILSA+F++LGE++KAV+ AG DWIHVDVMDGRFVPNITIGPLIVDA+RP+T
Sbjct: 3 NIVVAPSILSADFSRLGEEIKAVDEAGADWIHVDVMDGRFVPNITIGPLIVDAIRPLTKK 62
Query: 105 PLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGT 164
LDVHLMIVEPE+ V DF KAGADI+S+H E +++ HLHRT+ Q++ LG KAG VLNP T
Sbjct: 63 TLDVHLMIVEPEKYVEDFAKAGADIISVHVEHNASPHLHRTLCQIRELGKKAGAVLNPST 122
Query: 165 PLSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVT 224
PL +EYVL V DL+LIMSVNPGFGGQSFI + KI LR++C E+G++PWIEVDGG+
Sbjct: 123 PLDFLEYVLPVCDLILIMSVNPGFGGQSFIPEVLPKIRALRQMCDERGLDPWIEVDGGLK 182
Query: 225 PANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTSKRPEP 266
P N ++V+EAGANA+VAGSAVF A +YAEAI G++ SKRPEP
Sbjct: 183 PNNTWQVLEAGANAIVAGSAVFNAPNYAEAIAGVRNSKRPEP 224
>UniRef100_UPI00002B4476 UPI00002B4476 UniRef100 entry
Length = 258
Score = 318 bits (814), Expect = 1e-85
Identities = 157/226 (69%), Positives = 185/226 (81%), Gaps = 2/226 (0%)
Query: 42 SKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPV 101
S +++SPSILSA+F++LG++VKAV+ AG DWIHVDVMDGRFVPNITIGPLIV+ALRPV
Sbjct: 33 STKPLVISPSILSADFSRLGDEVKAVDEAGADWIHVDVMDGRFVPNITIGPLIVEALRPV 92
Query: 102 TDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLN 161
T PLDVHLMIVEPE+ VPDF KAGADI+S+ E HLHR + Q+K LG AG VLN
Sbjct: 93 TQKPLDVHLMIVEPEKYVPDFAKAGADIISVQVEACP--HLHRNLAQIKDLGKMAGAVLN 150
Query: 162 PGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDG 221
P TPL +EY L++ DLVLIMSVNPGFGGQSFIE+QV+KI DLRR+C E+G++PWIEVDG
Sbjct: 151 PSTPLDTLEYCLELCDLVLIMSVNPGFGGQSFIENQVQKIRDLRRMCDERGLDPWIEVDG 210
Query: 222 GVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTSKRPEPV 267
GV NA+KVIEAGANA+V+GS VF DYAEAIKGI+ S E V
Sbjct: 211 GVKAGNAWKVIEAGANAIVSGSGVFNQPDYAEAIKGIRNSSSKEAV 256
>UniRef100_Q7U771 Ribulose-5-phosphate 3-epimerase [Synechococcus sp.]
Length = 258
Score = 316 bits (810), Expect = 3e-85
Identities = 159/236 (67%), Positives = 190/236 (80%), Gaps = 5/236 (2%)
Query: 35 SSRVDKF---SKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIG 91
S R+ ++ S +++SPSILSA+F++LG++VKAV+ AG DWIHVDVMDGRFVPNITIG
Sbjct: 23 SPRIPRYFYMSTKPLVISPSILSADFSRLGDEVKAVDEAGADWIHVDVMDGRFVPNITIG 82
Query: 92 PLIVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKS 151
PLIV+ALRPVT PLDVHLMIVEPE+ VPDF KAGADI+S+ E HLHR + Q+K
Sbjct: 83 PLIVEALRPVTQKPLDVHLMIVEPEKYVPDFAKAGADIISVQVEACP--HLHRNLAQIKD 140
Query: 152 LGAKAGVVLNPGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEK 211
LG AG VLNP T L +EY L++ DLVLIMSVNPGFGGQSFIE+QV+KISDLRR+C E+
Sbjct: 141 LGKLAGAVLNPSTRLDTLEYCLELCDLVLIMSVNPGFGGQSFIENQVQKISDLRRMCDER 200
Query: 212 GVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTSKRPEPV 267
G++PWIEVDGGV NA+KVIEAGANA+V+GS VF DYAEAIKGI+ S E V
Sbjct: 201 GLDPWIEVDGGVKAGNAWKVIEAGANAIVSGSGVFNQPDYAEAIKGIRNSSSKEAV 256
>UniRef100_Q8DGE8 Pentose-5-phosphate-3-epimerase [Synechococcus elongatus]
Length = 231
Score = 315 bits (806), Expect = 1e-84
Identities = 150/225 (66%), Positives = 188/225 (82%)
Query: 42 SKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPV 101
S ++++PSILSA+F++LGE+++AV+ AG DWIHVDVMDGRFVPNITIGPLIV+A+RP+
Sbjct: 2 SAKPVVIAPSILSADFSRLGEEIQAVDRAGADWIHVDVMDGRFVPNITIGPLIVEAIRPL 61
Query: 102 TDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLN 161
T PLDVHLMIVEPE+ V DF KAGADI+S+H E +++ HLHRT+ Q++ LG +AGVVLN
Sbjct: 62 TQKPLDVHLMIVEPEKYVADFAKAGADIISVHAEHNASPHLHRTLCQIRELGKQAGVVLN 121
Query: 162 PGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDG 221
P +PL IEYVL+V DLVLIMSVNPGFGGQ FI + + KI LR+LC E+G++PWIEVDG
Sbjct: 122 PSSPLELIEYVLEVCDLVLIMSVNPGFGGQKFIPAVLPKIRRLRQLCEERGLDPWIEVDG 181
Query: 222 GVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTSKRPEP 266
G+ N ++V+EAGANA+VAGSAVF A DYA AI GI+ SKRP P
Sbjct: 182 GLKVDNTWQVLEAGANAIVAGSAVFNAPDYAAAIAGIRNSKRPSP 226
>UniRef100_Q7V807 Ribulose-phosphate 3-epimerase precursor [Prochlorococcus marinus]
Length = 227
Score = 311 bits (797), Expect = 1e-83
Identities = 153/226 (67%), Positives = 185/226 (81%), Gaps = 2/226 (0%)
Query: 42 SKSDIIVSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPV 101
S ++++PSILSA+F++LGE V+AV+ AG DWIHVDVMDGRFVPNITIGPLIV+ALRPV
Sbjct: 2 STKPLVIAPSILSADFSRLGEDVQAVDQAGADWIHVDVMDGRFVPNITIGPLIVEALRPV 61
Query: 102 TDLPLDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLN 161
T PLDVHLMIVEPE+ V DF KAGAD + + E HLHR + Q+K LG KAG VLN
Sbjct: 62 TSKPLDVHLMIVEPERYVGDFAKAGADHIYVQVEACP--HLHRNLAQIKDLGKKAGAVLN 119
Query: 162 PGTPLSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDG 221
P TPL +EY L++ DLVLIMSVNPGFGGQSFI++QV+KI +LRR+C E+G++PWIEVDG
Sbjct: 120 PSTPLDTLEYCLELCDLVLIMSVNPGFGGQSFIDNQVQKIRELRRICDEQGLDPWIEVDG 179
Query: 222 GVTPANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTSKRPEPV 267
G+ NA+KVIEAGANA+V+GS VF DYAEAI+GI+ SKRPE V
Sbjct: 180 GIKADNAWKVIEAGANAIVSGSGVFNQPDYAEAIRGIRNSKRPEGV 225
>UniRef100_Q7NFH7 Ribulose-phosphate 3-epimerase [Gloeobacter violaceus]
Length = 233
Score = 309 bits (791), Expect = 6e-83
Identities = 145/221 (65%), Positives = 185/221 (83%), Gaps = 2/221 (0%)
Query: 46 IIVSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLP 105
++V+PSILSA+FA+LGE++ A++ AG DWIHVDVMDGRFVPNITIGPLIV A+RPVT
Sbjct: 5 VVVAPSILSADFARLGEEIHAIDEAGADWIHVDVMDGRFVPNITIGPLIVKAIRPVTQKV 64
Query: 106 LDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTP 165
LDVHLMIVEPE+ + DF KAGADI+++HCE +ST+HLHR ++Q+K LG +AGV LNP TP
Sbjct: 65 LDVHLMIVEPEKYIADFAKAGADIITVHCEHNSTVHLHRALSQIKELGKQAGVSLNPSTP 124
Query: 166 LSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTP 225
LS IEYVLD+ DLVL+MSVNPGFGGQSFI + KI+ L+ +C E+G++PWIEVDGG+ P
Sbjct: 125 LSLIEYVLDLCDLVLVMSVNPGFGGQSFIPQVLPKIARLKSMCEERGLDPWIEVDGGLKP 184
Query: 226 ANAYKVIEAGANALVAGSAVFGA--KDYAEAIKGIKTSKRP 264
+N ++V+E GANA+V+GS VFG YAEAI+G++ S RP
Sbjct: 185 SNTWQVLEVGANAIVSGSGVFGVGPARYAEAIEGVRASHRP 225
>UniRef100_UPI000025C5BA UPI000025C5BA UniRef100 entry
Length = 198
Score = 283 bits (724), Expect = 3e-75
Identities = 140/198 (70%), Positives = 160/198 (80%), Gaps = 2/198 (1%)
Query: 70 AGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLDVHLMIVEPEQRVPDFIKAGADI 129
+G DWIHVDVMDGRFVPNITIGPLIV+ALRPVT PLDVHLMIVEPE+ VPDF KAGADI
Sbjct: 1 SGADWIHVDVMDGRFVPNITIGPLIVEALRPVTQKPLDVHLMIVEPEKYVPDFAKAGADI 60
Query: 130 VSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLSAIEYVLDVVDLVLIMSVNPGFG 189
+S+ E HLHR + Q+K LG AG VLNP TPL +EY L++ DLVLIMSVNPGFG
Sbjct: 61 ISVQVEACP--HLHRNLAQIKDLGKMAGAVLNPSTPLDTLEYCLELCDLVLIMSVNPGFG 118
Query: 190 GQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTPANAYKVIEAGANALVAGSAVFGAK 249
GQSFIE+QV+KI DLRR+C E+G++PWIEVDGGV NA+KVIEAGANA+V+GS VF
Sbjct: 119 GQSFIENQVQKIRDLRRMCDERGLDPWIEVDGGVKAGNAWKVIEAGANAIVSGSGVFNQP 178
Query: 250 DYAEAIKGIKTSKRPEPV 267
DYAEAIK I+ S E V
Sbjct: 179 DYAEAIKAIRNSSSKEAV 196
>UniRef100_UPI00003CC223 UPI00003CC223 UniRef100 entry
Length = 214
Score = 258 bits (659), Expect = 1e-67
Identities = 129/214 (60%), Positives = 162/214 (75%), Gaps = 2/214 (0%)
Query: 46 IIVSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLP 105
I ++PSILSA+F+KLGE++K VE G D+IHVDVMDG FVPNITIGPLIV+A+RP+T LP
Sbjct: 2 IKIAPSILSADFSKLGEEIKDVEKGGADYIHVDVMDGHFVPNITIGPLIVEAIRPITSLP 61
Query: 106 LDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTP 165
LDVHLMI P+ +P F KAGADI+++H E HLHRT+ +KS G KAGVVLNP TP
Sbjct: 62 LDVHLMIENPDNYIPTFAKAGADIITVHVEACP--HLHRTIQLIKSHGIKAGVVLNPHTP 119
Query: 166 LSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTP 225
+S IE+VL+ +D+VL+M+VNPGFGGQ FI S + KI + + E+ + IEVDGGV P
Sbjct: 120 VSMIEHVLEDIDMVLLMTVNPGFGGQKFIHSVLPKIKQVAEMVKERNLEVEIEVDGGVNP 179
Query: 226 ANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIK 259
A +EAGAN LVAGSAV+ KD EAI+ I+
Sbjct: 180 ETARLCVEAGANVLVAGSAVYNQKDRGEAIRVIR 213
>UniRef100_O34557 Ribulose-phosphate 3-epimerase [Bacillus subtilis]
Length = 217
Score = 258 bits (659), Expect = 1e-67
Identities = 136/218 (62%), Positives = 164/218 (74%), Gaps = 3/218 (1%)
Query: 46 IIVSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLP 105
I V+PSILSA+FA LG ++K VE G D IH+DVMDG FVPNITIGPLIV+A+RPVTDLP
Sbjct: 2 IKVAPSILSADFAALGNEIKDVEKGGADCIHIDVMDGHFVPNITIGPLIVEAVRPVTDLP 61
Query: 106 LDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTP 165
LDVHLMI EP++ +P F KAGADI+S+H E + HLHRT+ +K G KAGVVLNP TP
Sbjct: 62 LDVHLMIEEPDRYIPAFAKAGADILSVHAE--ACPHLHRTIQLIKEQGVKAGVVLNPHTP 119
Query: 166 LSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNP-WIEVDGGVT 224
+ IE+V D +DLVL+M+VNPGFGGQ FI S + KI +++R+ EKG IEVDGGV
Sbjct: 120 VQVIEHVFDDLDLVLLMTVNPGFGGQKFIHSVLPKIKEVKRMADEKGKKDLLIEVDGGVN 179
Query: 225 PANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTSK 262
A VIEAGAN LVAGSAV+G D +AI I+ SK
Sbjct: 180 KETAPLVIEAGANLLVAGSAVYGQSDRKKAISEIRGSK 217
>UniRef100_Q6TV49 Putative ribulose-5-phosphate 3-epimerase [Bacillus methanolicus]
Length = 214
Score = 257 bits (657), Expect = 2e-67
Identities = 131/214 (61%), Positives = 164/214 (76%), Gaps = 2/214 (0%)
Query: 46 IIVSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLP 105
I ++PSILSANFA+L E++K VE G D+IHVDVMDG FVPNITIGPLIV+A+RPVT+LP
Sbjct: 2 IKIAPSILSANFARLEEEIKDVERGGADYIHVDVMDGHFVPNITIGPLIVEAIRPVTNLP 61
Query: 106 LDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTP 165
LDVHLMI P+Q + F KAGADI+S+H E + HLHRT+ +KS G KAGVVLNP TP
Sbjct: 62 LDVHLMIENPDQYIGTFAKAGADILSVHVEACT--HLHRTIQYIKSEGIKAGVVLNPHTP 119
Query: 166 LSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTP 225
+S IE+V++ VDLVL+M+VNPGFGGQSFI S + KI + + EK + IEVDGGV P
Sbjct: 120 VSMIEHVIEDVDLVLLMTVNPGFGGQSFIHSVLPKIKQVANIVKEKNLQVEIEVDGGVNP 179
Query: 226 ANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIK 259
A +EAGAN LVAGSA++ +D ++AI I+
Sbjct: 180 ETAKLCVEAGANVLVAGSAIYNQEDRSQAIAKIR 213
>UniRef100_Q9K9Z2 Ribulose-phosphate 3-epimerase [Bacillus halodurans]
Length = 216
Score = 256 bits (655), Expect = 3e-67
Identities = 132/216 (61%), Positives = 164/216 (75%), Gaps = 2/216 (0%)
Query: 46 IIVSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLP 105
I ++PSILSA+FA LG +++ VE G D+IHVDVMDG FVPNITIGPLIVDA+RPVT LP
Sbjct: 2 IKIAPSILSADFANLGNEIQDVERGGADYIHVDVMDGHFVPNITIGPLIVDAIRPVTTLP 61
Query: 106 LDVHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTP 165
LDVHLMI +P+ +P F KAGADI+++H E HLHRT++ +K G KAGVVLNP TP
Sbjct: 62 LDVHLMIEQPDGYIPAFAKAGADIITVHVEACP--HLHRTLHLIKESGVKAGVVLNPATP 119
Query: 166 LSAIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTP 225
+S+I++VL VD+VL M+VNPGFGGQ FI S + K+ +L L E+G+ IEVDGGV
Sbjct: 120 VSSIQHVLSDVDMVLFMTVNPGFGGQRFIPSVLPKLKELASLKKEQGLTFEIEVDGGVNE 179
Query: 226 ANAYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTS 261
A + +EAGAN LVAGSAVF +D A AIKGI+ S
Sbjct: 180 ETAKQCVEAGANVLVAGSAVFNEEDRAAAIKGIRGS 215
>UniRef100_Q746Z3 Ribulose-phosphate 3-epimerase [Geobacter sulfurreducens]
Length = 221
Score = 256 bits (655), Expect = 3e-67
Identities = 126/220 (57%), Positives = 168/220 (76%), Gaps = 3/220 (1%)
Query: 48 VSPSILSANFAKLGEQVKAVELAGCDWIHVDVMDGRFVPNITIGPLIVDALRPVTDLPLD 107
++PSILSA+F++LG++V+AV AG D+IHVDVMDG FVPNITIGPL+V+A+R VT+LPLD
Sbjct: 4 IAPSILSADFSRLGDEVRAVAAAGADYIHVDVMDGHFVPNITIGPLVVEAVRRVTELPLD 63
Query: 108 VHLMIVEPEQRVPDFIKAGADIVSIHCEQSSTIHLHRTVNQVKSLGAKAGVVLNPGTPLS 167
VHLMI P++ +PDF AG+DI+ +H E S+ HLHRT+ +KSLG KAGV LNP TPL+
Sbjct: 64 VHLMIEHPDRYIPDFAAAGSDIIVVHAEAST--HLHRTIQLIKSLGKKAGVSLNPATPLN 121
Query: 168 AIEYVLDVVDLVLIMSVNPGFGGQSFIESQVKKISDLRRLCAEKGVNPWIEVDGGVTPAN 227
++YVL+ +DLVL+M+VNPGFGGQSFIE+ + KI LR + +G +EVDGGV N
Sbjct: 122 CLDYVLEDLDLVLLMTVNPGFGGQSFIEACIPKIQSLRAMLDRRGCEAELEVDGGVKIDN 181
Query: 228 AYKVIEAGANALVAGSAVFGAKDYAEAIKGIKTSKRPEPV 267
++ AGA+ VAGSA+FG+ DY IK +K + EP+
Sbjct: 182 IARIAHAGADVFVAGSAIFGSPDYEATIKELK-RRAKEPI 220
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.136 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 427,357,639
Number of Sequences: 2790947
Number of extensions: 17151292
Number of successful extensions: 46892
Number of sequences better than 10.0: 911
Number of HSP's better than 10.0 without gapping: 860
Number of HSP's successfully gapped in prelim test: 51
Number of HSP's that attempted gapping in prelim test: 44957
Number of HSP's gapped (non-prelim): 933
length of query: 268
length of database: 848,049,833
effective HSP length: 125
effective length of query: 143
effective length of database: 499,181,458
effective search space: 71382948494
effective search space used: 71382948494
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0093a.1


