

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
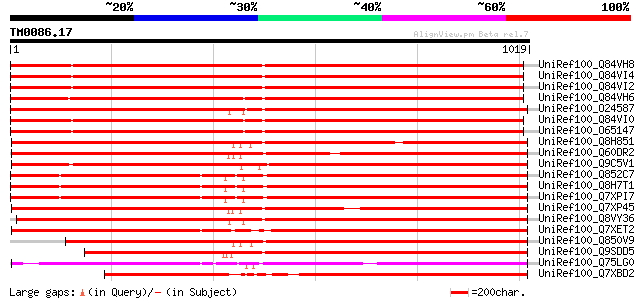
Query= TM0086.17
(1019 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84VH8 Gag-pol polyprotein [Glycine max] 974 0.0
UniRef100_Q84VI4 Gag-pol polyprotein [Glycine max] 971 0.0
UniRef100_Q84VI2 Gag-pol polyprotein [Glycine max] 967 0.0
UniRef100_Q84VH6 Gag-pol polyprotein [Glycine max] 962 0.0
UniRef100_O24587 Pol protein [Zea mays] 953 0.0
UniRef100_Q84VI0 Gag-pol polyprotein [Glycine max] 952 0.0
UniRef100_O65147 Gag-pol polyprotein [Glycine max] 952 0.0
UniRef100_Q8H851 Putative Zea mays retrotransposon Opie-2 [Oryza... 932 0.0
UniRef100_Q60DR2 Putative polyprotein [Oryza sativa] 930 0.0
UniRef100_Q9C5V1 Gag/pol polyprotein [Arabidopsis thaliana] 927 0.0
UniRef100_Q852C7 Putative gag-pol polyprotein [Oryza sativa] 917 0.0
UniRef100_Q8H7T1 Putative Zea mays retrotransposon Opie-2 [Oryza... 915 0.0
UniRef100_Q7XPI7 OSJNBb0004A17.2 protein [Oryza sativa] 915 0.0
UniRef100_Q7XP45 OSJNBa0063G07.6 protein [Oryza sativa] 914 0.0
UniRef100_Q8VY36 Opie2a pol [Zea mays] 910 0.0
UniRef100_Q7XET2 Hypothetical protein [Oryza sativa] 897 0.0
UniRef100_Q850V9 Putative polyprotein [Oryza sativa] 873 0.0
UniRef100_Q9SDD5 Similar to copia-type pol polyprotein [Oryza sa... 847 0.0
UniRef100_Q75LG0 Putative integrase [Oryza sativa] 808 0.0
UniRef100_Q7XBD2 Retrotransposon Opie-2 [Zea mays] 763 0.0
>UniRef100_Q84VH8 Gag-pol polyprotein [Glycine max]
Length = 1576
Score = 974 bits (2517), Expect = 0.0
Identities = 483/1008 (47%), Positives = 671/1008 (65%), Gaps = 6/1008 (0%)
Query: 1 QSWYLDSGCSRHMTGERRMFQELKLKPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLL 60
+ WYLDSGCSRHMTG + ++ V FG KGKI+G G + D P ++ VLL
Sbjct: 560 EDWYLDSGCSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMGKLVHDGLPSLNKVLL 619
Query: 61 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 120
V GLT NL+SISQL D+G++V F + C ++ ++ S+ K+N Y E +
Sbjct: 620 VKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWTPQETSYSS 679
Query: 121 VKCLLSVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKV 180
CL S +E +WH+R GH+ +R + ++ VRG+P LK +C CQ K K+
Sbjct: 680 T-CLSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKM 738
Query: 181 HFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHA 240
+ +TSR LELLH+DL GP++ ES+GGKRY V+VDD+SR+TWV F+ K E+
Sbjct: 739 SHQKLRHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSETFE 798
Query: 241 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVER 300
VF ++Q EK C I R+RSDHG EFEN +F S GI H+FS+ TPQ NG+VER
Sbjct: 799 VFKELSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITPQQNGIVER 858
Query: 301 KNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISY 360
KNRTLQE AR ML + WAEA+NT CYI NR+++R T YE+WK KP++ +
Sbjct: 859 KNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPSVKH 918
Query: 361 FHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 420
FH FG CY+L +++ K D KS + LGYS S+ +R +N+ +T+ ESI+V DD
Sbjct: 919 FHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDL 978
Query: 421 LDSDQSKLVEKFADLSINVSDKGKAPEEAEPEEEEPEEEAGPSDSQSQKKSRITAAHPKE 480
+ + + E L NV+D K+ E AE + +E+ + + +RI HPKE
Sbjct: 979 SPARKKDVEEDVRTLGDNVADAAKSGENAE-NSDSATDESNINQPDKRSSTRIQKMHPKE 1037
Query: 481 LILGNKDEPVRTRSAFRPSEETLLSLKGLMSLIEPKSIDEALQDKDWILAMEEELNQFSK 540
LI+G+ + V TRS E ++S +S IEPK++ EAL D+ WI AM+EEL QF +
Sbjct: 1038 LIIGDPNRGVTTRSR----EVEIVSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFKR 1093
Query: 541 NDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVA 600
N+VW LV +P+ +VIGTKW+F+NK NE+G + RNKARLVAQGY+Q EG+D+ ETFAPVA
Sbjct: 1094 NEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVA 1153
Query: 601 RLEAVKLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPGFKDEKNPDHVFKLKKS 660
RLE+++LL+ + L+QMDVKSAFLNGY++EEVYV QP GF D +PDHV++LKK+
Sbjct: 1154 RLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDHVYRLKKA 1213
Query: 661 LYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQF 720
LYGLKQAPRAWYERL+ FL + + +G +D TLF K ++++I QIYVDDI+FG +
Sbjct: 1214 LYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNE 1273
Query: 721 LCKEFSEMMQAEFEMSMMGELKYFMGIQVDQTPEGTYIHQSKYTKELLKKFNMTELVIAK 780
+ + F + MQ+EFEMS++GEL YF+G+QV Q + ++ QS+Y K ++KKF M +
Sbjct: 1274 MLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKNIVKKFGMENASHKR 1333
Query: 781 TPMHPTCILEKEDASGKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLT 840
TP L K++A V Q LYR MIGSLLYLTASRPDI ++V +CAR+Q++P+ +HLT
Sbjct: 1334 TPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCARYQANPKISHLT 1393
Query: 841 AVKRILRYLKGSTNLGLMYKKTSEYKLSGYCDADYVGDRTERKSTSGNC*FLGSNLVSWA 900
VKRIL+Y+ G+++ G+MY S L GYCDAD+ G +RKSTSG C +LG+NL+SW
Sbjct: 1394 QVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDADWAGSADDRKSTSGGCFYLGNNLISWF 1453
Query: 901 SKRQSTIALSTAEAEYIPAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPI 960
SK+Q+ ++LSTAEAEYI A +Q++WMK L++Y + + + +YCDN +AI++SKNP+
Sbjct: 1454 SKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINISKNPV 1513
Query: 961 LHSRAKHIEVKYHFIRDYVQKGVLLLKFVDIDHQWADIFTKPLAEDRF 1008
HSR KHI++++H+IRD V V+ LK VD + Q ADIFTK L ++F
Sbjct: 1514 QHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTKALDANQF 1561
>UniRef100_Q84VI4 Gag-pol polyprotein [Glycine max]
Length = 1574
Score = 971 bits (2510), Expect = 0.0
Identities = 482/1008 (47%), Positives = 670/1008 (65%), Gaps = 6/1008 (0%)
Query: 1 QSWYLDSGCSRHMTGERRMFQELKLKPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLL 60
+ WYLDSGCSRHMTG + ++ V FG KGKI+G G + D P ++ VLL
Sbjct: 558 EDWYLDSGCSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMGKLVHDGLPSLNKVLL 617
Query: 61 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 120
V GLT NL+SISQL D+G++V F + C ++ ++ S+ K+N Y E +
Sbjct: 618 VKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWTPQETSYSS 677
Query: 121 VKCLLSVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKV 180
CL S +E +WH+R GH+ +R + ++ VRG+P LK +C CQ K K+
Sbjct: 678 T-CLSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKM 736
Query: 181 HFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHA 240
+ +TSR LELLH+DL GP++ ES+GGKRY V+VDD+SR+TWVKF+ K E+
Sbjct: 737 SHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVKFIREKSETFE 796
Query: 241 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVER 300
VF ++Q EK C I R+RSDHG EFEN + S GI H+FS+ TPQ NG+VER
Sbjct: 797 VFKELSLRLQREKDCVIKRIRSDHGREFENSRLTEFCTSEGITHEFSAAITPQQNGIVER 856
Query: 301 KNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISY 360
KNRTLQE AR ML + WAEA+NT CYI NR+++R T YE+WK KP++ +
Sbjct: 857 KNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPSVKH 916
Query: 361 FHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 420
FH FG CY+L +++ K D KS + LGYS S+ +R +N+ +T+ ESI+V DD
Sbjct: 917 FHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDL 976
Query: 421 LDSDQSKLVEKFADLSINVSDKGKAPEEAEPEEEEPEEEAGPSDSQSQKKSRITAAHPKE 480
+ + + E NV+D K+ E AE + +E+ + + +RI HPKE
Sbjct: 977 SPARKKDVEEDVRTSGDNVADAAKSGENAE-NSDSATDESNINQPDKRSSTRIQKMHPKE 1035
Query: 481 LILGNKDEPVRTRSAFRPSEETLLSLKGLMSLIEPKSIDEALQDKDWILAMEEELNQFSK 540
LI+G+ + V TRS E ++S +S IEPK++ EAL D+ WI AM+EEL QF +
Sbjct: 1036 LIIGDPNRGVTTRSR----EVEIVSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFKR 1091
Query: 541 NDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVA 600
N+VW LV +P+ +VIGTKW+F+NK NE+G + RNKARLVAQGY+Q EG+D+ ETFAPVA
Sbjct: 1092 NEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVA 1151
Query: 601 RLEAVKLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPGFKDEKNPDHVFKLKKS 660
RLE+++LL+ + L+QMDVKSAFLNGY++EEVYV QP GF D +PDHV++LKK+
Sbjct: 1152 RLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDHVYRLKKA 1211
Query: 661 LYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQF 720
LYGLKQAPRAWYERL+ FL + + +G +D TLF K ++++I QIYVDDI+FG +
Sbjct: 1212 LYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNE 1271
Query: 721 LCKEFSEMMQAEFEMSMMGELKYFMGIQVDQTPEGTYIHQSKYTKELLKKFNMTELVIAK 780
+ + F + MQ+EFEMS++GEL YF+G+QV Q + ++ QS+Y K ++KKF M +
Sbjct: 1272 MLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKNIVKKFGMENASHKR 1331
Query: 781 TPMHPTCILEKEDASGKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLT 840
TP L K++A V Q LYR MIGSLLYLTASRPDI ++V +CAR+Q++P+ +HLT
Sbjct: 1332 TPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCARYQANPKISHLT 1391
Query: 841 AVKRILRYLKGSTNLGLMYKKTSEYKLSGYCDADYVGDRTERKSTSGNC*FLGSNLVSWA 900
VKRIL+Y+ G+++ G+MY S L GYCDAD+ G +RKSTSG C +LG+NL+SW
Sbjct: 1392 QVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDADWAGSADDRKSTSGGCFYLGNNLISWF 1451
Query: 901 SKRQSTIALSTAEAEYIPAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPI 960
SK+Q+ ++LSTAEAEYI A +Q++WMK L++Y + + + +YCDN +AI++SKNP+
Sbjct: 1452 SKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINISKNPV 1511
Query: 961 LHSRAKHIEVKYHFIRDYVQKGVLLLKFVDIDHQWADIFTKPLAEDRF 1008
HSR KHI++++H+IRD V V+ LK VD + Q ADIFTK L ++F
Sbjct: 1512 QHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTKALDANQF 1559
>UniRef100_Q84VI2 Gag-pol polyprotein [Glycine max]
Length = 1576
Score = 967 bits (2499), Expect = 0.0
Identities = 480/1008 (47%), Positives = 669/1008 (65%), Gaps = 6/1008 (0%)
Query: 1 QSWYLDSGCSRHMTGERRMFQELKLKPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLL 60
+ WYLDSGCSRHMTG + ++ V FG KGKI+G G + D P ++ VLL
Sbjct: 560 EDWYLDSGCSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMGKLVHDGLPSLNKVLL 619
Query: 61 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 120
V GLT NL+SISQL D+G++V F + C ++ ++ S+ K+N Y E +
Sbjct: 620 VKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWTPQETSYSS 679
Query: 121 VKCLLSVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKV 180
CL S +E +WH+R GH+ +R + ++ + VRG+P LK +C CQ K K+
Sbjct: 680 T-CLSSKEDEVRIWHQRFGHLHLRGMKKILDKSAVRGIPNLKIEEGRICGECQIGKQVKM 738
Query: 181 HFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHA 240
+ +TSR LELLH+DL GP++ ES+GGKRY V+VDD+SR+TWV F+ K +
Sbjct: 739 SHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSGTFE 798
Query: 241 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVER 300
VF ++Q EK C I R+RSDHG EFEN +F S GI H+FS+ TPQ NG+VER
Sbjct: 799 VFKKLSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITPQQNGIVER 858
Query: 301 KNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISY 360
KNRTLQE AR ML + WAEA+NT CYI NR+++R T YE+WK KP++ +
Sbjct: 859 KNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPSVKH 918
Query: 361 FHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 420
FH FG CY+L +++ K D KS + LGYS S+ +R +N+ +T+ ESI+V DD
Sbjct: 919 FHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDL 978
Query: 421 LDSDQSKLVEKFADLSINVSDKGKAPEEAEPEEEEPEEEAGPSDSQSQKKSRITAAHPKE 480
+ + + E NV+D K+ E AE + +E+ + + +RI HPKE
Sbjct: 979 SPARKKDVEEDVRTSGDNVADAAKSGENAE-NSDSATDESNINQPDKRSSTRIQKMHPKE 1037
Query: 481 LILGNKDEPVRTRSAFRPSEETLLSLKGLMSLIEPKSIDEALQDKDWILAMEEELNQFSK 540
LI+G+ + V TRS E ++S +S IEPK++ EAL D+ WI AM+EEL QF +
Sbjct: 1038 LIIGDPNRGVTTRSR----EVEIVSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFKR 1093
Query: 541 NDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVA 600
N+VW LV +P+ +VIGTKW+F+NK NE+G + RNKARLVAQGY+Q EG+D+ ETFAPVA
Sbjct: 1094 NEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVA 1153
Query: 601 RLEAVKLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPGFKDEKNPDHVFKLKKS 660
RLE+++LL+ + L+QMDVKSAFLNGY++EEVYV QP GF D +PDHV++LKK+
Sbjct: 1154 RLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDHVYRLKKA 1213
Query: 661 LYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQF 720
LYGLKQAPRAWYERL+ FL + + +G +D TLF K ++++I QIYVDDI+FG +
Sbjct: 1214 LYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMSNE 1273
Query: 721 LCKEFSEMMQAEFEMSMMGELKYFMGIQVDQTPEGTYIHQSKYTKELLKKFNMTELVIAK 780
+ + F + MQ+EFEMS++GEL YF+G+QV Q + ++ QS+Y K ++KKF M +
Sbjct: 1274 MLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSRYAKNIVKKFGMENASHKR 1333
Query: 781 TPMHPTCILEKEDASGKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLT 840
TP L K++A V QK YR MIGSLLYLTASRPDI ++V +CAR+Q++P+ +HL
Sbjct: 1334 TPAPTHLKLSKDEAGTSVDQKPYRSMIGSLLYLTASRPDITYAVGVCARYQANPKISHLN 1393
Query: 841 AVKRILRYLKGSTNLGLMYKKTSEYKLSGYCDADYVGDRTERKSTSGNC*FLGSNLVSWA 900
VKRIL+Y+ G+++ G+MY S L GYCDAD+ G +RKSTSG C +LG+NL+SW
Sbjct: 1394 QVKRILKYVNGTSDYGIMYCHCSSSMLVGYCDADWAGSADDRKSTSGGCFYLGNNLISWF 1453
Query: 901 SKRQSTIALSTAEAEYIPAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPI 960
SK+Q+ ++LSTAEAEYI A +Q++WMK L++Y + + + +YCDN +AI++SKNP+
Sbjct: 1454 SKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINISKNPV 1513
Query: 961 LHSRAKHIEVKYHFIRDYVQKGVLLLKFVDIDHQWADIFTKPLAEDRF 1008
HSR KHI++++H+IRD V V+ LK VD + Q ADIFTK L ++F
Sbjct: 1514 QHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQIADIFTKALDANQF 1561
>UniRef100_Q84VH6 Gag-pol polyprotein [Glycine max]
Length = 1577
Score = 962 bits (2487), Expect = 0.0
Identities = 481/1010 (47%), Positives = 668/1010 (65%), Gaps = 10/1010 (0%)
Query: 1 QSWYLDSGCSRHMTGERRMFQELKLKPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLL 60
+ WYLDSGCSRHMTG + ++ V FG KGKI G G + + P ++ VLL
Sbjct: 561 EDWYLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLVHEGLPSLNKVLL 620
Query: 61 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 120
V GLT NL+SISQL D+G++V F + C ++ ++ S+ K+N Y E + +
Sbjct: 621 VKGLTVNLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWTPQE-SSHS 679
Query: 121 VKCLLSVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKV 180
CL S +E +WH+R GH+ +R + ++ VRG+P LK +C CQ K K+
Sbjct: 680 STCLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKM 739
Query: 181 HFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHA 240
+ +TSR LELLH+DL GP++ ES+GGKRY V+VDD+SR+TWV F+ K ++
Sbjct: 740 SHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSDTFE 799
Query: 241 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVER 300
VF ++Q EK C I R+RSDHG EFEN KF S GI H+FS+ TPQ NG+VER
Sbjct: 800 VFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVER 859
Query: 301 KNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISY 360
KNRTLQE AR ML + WAEA+NT CYI NR+++R T YE+WK KP + +
Sbjct: 860 KNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPTVKH 919
Query: 361 FHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 420
FH FG CY+L +++ K D KS + LGYS S+ +R +N+ +T+ ESI+V DD
Sbjct: 920 FHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDL 979
Query: 421 LDSDQSKLVEKFADLSINVSDKGKAPEEAEPEEEEPEEEAGPSDSQSQKKS--RITAAHP 478
+ + + E NV+D K+ E AE + +E P+ +Q K+ RI HP
Sbjct: 980 TPARKKDVEEDVRTSGDNVADTAKSAENAENSDSATDE---PNINQPDKRPSIRIQKMHP 1036
Query: 479 KELILGNKDEPVRTRSAFRPSEETLLSLKGLMSLIEPKSIDEALQDKDWILAMEEELNQF 538
KELI+G+ + V TRS E ++S +S IEPK++ EAL D+ WI AM+EEL QF
Sbjct: 1037 KELIIGDPNRGVTTRSR----EIEIVSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQF 1092
Query: 539 SKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAP 598
+N+VW LV +P+ +VIGTKW+F+NK NE+G + RNKARLVAQGY+Q EG+D+ ETFAP
Sbjct: 1093 KRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAP 1152
Query: 599 VARLEAVKLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPGFKDEKNPDHVFKLK 658
VARLE+++LL+ + L+QMDVKSAFLNGY++EE YV QP GF D +PDHV++LK
Sbjct: 1153 VARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHPDHVYRLK 1212
Query: 659 KSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSAN 718
K+LYGLKQAPRAWYERL+ FL + + +G +D TLF K ++++I QIYVDDI+FG +
Sbjct: 1213 KALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMS 1272
Query: 719 QFLCKEFSEMMQAEFEMSMMGELKYFMGIQVDQTPEGTYIHQSKYTKELLKKFNMTELVI 778
+ + F + MQ+EFEMS++GEL YF+G+QV Q + ++ QSKY K ++KKF M
Sbjct: 1273 NEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYAKNIVKKFGMENASH 1332
Query: 779 AKTPMHPTCILEKEDASGKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETH 838
+TP L K++A V Q LYR MIGSLLYLTASRPDI ++V +CAR+Q++P+ +H
Sbjct: 1333 KRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGVCARYQANPKISH 1392
Query: 839 LTAVKRILRYLKGSTNLGLMYKKTSEYKLSGYCDADYVGDRTERKSTSGNC*FLGSNLVS 898
L VKRIL+Y+ G+++ G+MY S L GYCDAD+ G +RKSTSG C +LG+NL+S
Sbjct: 1393 LNQVKRILKYVNGTSDYGIMYCHCSGSMLVGYCDADWAGSADDRKSTSGGCFYLGNNLIS 1452
Query: 899 WASKRQSTIALSTAEAEYIPAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKN 958
W SK+Q+ ++LSTAEAEYI A +Q++WMK L++Y + + + +YCDN +AI++SKN
Sbjct: 1453 WFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINISKN 1512
Query: 959 PILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDIDHQWADIFTKPLAEDRF 1008
P+ HSR KHI++++H+IR+ V V+ L+ VD + Q ADIFTK L +F
Sbjct: 1513 PVQHSRTKHIDIRHHYIRELVDDKVITLEHVDTEEQIADIFTKALDAKQF 1562
>UniRef100_O24587 Pol protein [Zea mays]
Length = 1068
Score = 953 bits (2464), Expect = 0.0
Identities = 503/1058 (47%), Positives = 668/1058 (62%), Gaps = 48/1058 (4%)
Query: 2 SWYLDSGCSRHMTGERRMFQELKLKPEGK--VGFGGNEKGKIVGTGTICVDSSPCIDNVL 59
SW +DSGC+ HMTGE++MF + + + FG +GK+ G G I + + I NV
Sbjct: 10 SWIIDSGCTNHMTGEKKMFTSYVKNKDSQDSIIFGDGNQGKVKGLGKIAISNEHSISNVF 69
Query: 60 LVDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQ 119
LV+ L +NLLS+SQL + GY+ +F + DGS+ F +Y + ++ EA
Sbjct: 70 LVESLGYNLLSVSQLCNMGYNCLFTNIDVSVFRRCDGSLAFKGVLDGKLYLVDFAKEEAG 129
Query: 120 NVKCLLSVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTK 179
CL++ +WHRRL HV M+ + +L K V GL ++F D C ACQ K
Sbjct: 130 LDACLIAKTSMGWLWHRRLAHVGMKNLHKLLKGEHVIGLTNVQFEKDRPCAACQAGKQVG 189
Query: 180 VHFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESH 239
KNV++TSRPLE+LH+DLFGPV SIGG +YG+VIVDD+SR+TWV FL K E+
Sbjct: 190 GSHHTKNVMTTSRPLEMLHMDLFGPVAYLSIGGSKYGLVIVDDFSRFTWVFFLQEKSETQ 249
Query: 240 AVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVE 299
F+ + QNE ++ ++RSD+G EF+N + E + GI H+FS+P TPQ NGVVE
Sbjct: 250 GTLKRFLRRAQNEFELKVKKIRSDNGSEFKNLQVEEFLEEEGIKHEFSAPYTPQQNGVVE 309
Query: 300 RKNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNIS 359
RKNRTL +MARTML E + FW EAVNT C+ NR+ + IL T YEL KPN+S
Sbjct: 310 RKNRTLIDMARTMLGEFKTPECFWTEAVNTACHAINRVYLHRILKNTSYELLTGNKPNVS 369
Query: 360 YFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDD 419
YF FG CY+L K R KF K+ + LLGY +K +R +N + +E S V FD+
Sbjct: 370 YFRVFGSKCYILVKKGRNSKFAPKAVEGFLLGYDSNTKAYRVFNKSSGLVEVSGDVVFDE 429
Query: 420 KLDSDQSKLV-------EKFADLSINVSDKGKAPEEAEPEEEEP---------------- 456
S + ++V E +I G+ + + E E+P
Sbjct: 430 TNGSPREQVVDCDDVDEEDIPTAAIRTMAIGEVRPQEQDEREQPSPSTMVHPPTQDDEQV 489
Query: 457 -----------------EEEAGPSDSQSQKKSRITAAHPKELILGNKDEPVRTRSAFRPS 499
EEEA P+ +Q ++ I HP + ILG+ + V TRS
Sbjct: 490 HQQEVCDQGGAQDDHVLEEEAQPA-PPTQVRAMIQRDHPVDQILGDISKGVTTRSRL--- 545
Query: 500 EETLLSLKGLMSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPQSVHVIGTK 559
+S IEP ++EAL D DW+LAM+EELN F +N+VW+LV +P+ +V+GTK
Sbjct: 546 -VNFCEHNSFVSSIEPFRVEEALLDPDWVLAMQEELNNFKRNEVWTLVPRPKQ-NVVGTK 603
Query: 560 WVFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAVKLLISFSVNHNIIL 619
WVFRNK +E+G V RNKARLVA+GY+Q G+D+ ETFAPVARLE++++L++++ +H+ L
Sbjct: 604 WVFRNKQDERGVVTRNKARLVAKGYAQVAGLDFEETFAPVARLESIRILLAYAAHHSFRL 663
Query: 620 HQMDVKSAFLNGYISEEVYVHQPPGFKDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFL 679
+QMDVKSAFLNG I EEVYV QPPGF+DE+ PDHV KL K+LYGLKQAPRAWYE L FL
Sbjct: 664 YQMDVKSAFLNGPIKEEVYVEQPPGFEDERYPDHVCKLSKALYGLKQAPRAWYECLRDFL 723
Query: 680 LENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQFLCKEFSEMMQAEFEMSMMG 739
+ N F GK D TLF KT D+ + QIYVDDIIFGS NQ C+EFS +M +FEMSMMG
Sbjct: 724 IANAFKVGKADPTLFTKTCDGDLFVCQIYVDDIIFGSTNQKSCEEFSRVMTQKFEMSMMG 783
Query: 740 ELKYFMGIQVDQTPEGTYIHQSKYTKELLKKFNMTELVIAKTPMHPTCILEKEDASGKVC 799
EL YF+G QV Q +GT+I Q+KYT++LLK+F M + AKTPM + V
Sbjct: 784 ELNYFLGFQVKQLKDGTFISQTKYTQDLLKRFGMKDAKPAKTPMGTDGHTDLNKGGKSVD 843
Query: 800 QKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGSTNLGLMY 859
QK YR MIGSLLYL ASRPDI+ SV +CARFQSDP+E HL AVKRILRYL + GL Y
Sbjct: 844 QKAYRSMIGSLLYLCASRPDIMLSVCMCARFQSDPKECHLVAVKRILRYLVATPCFGLWY 903
Query: 860 KKTSEYKLSGYCDADYVGDRTERKSTSGNC*FLGSNLVSWASKRQSTIALSTAEAEYIPA 919
K S + L GY D+DY G + +RKSTSG C FLG +LVSW SK+Q+++ALSTAEAEY+ A
Sbjct: 904 PKGSTFDLVGYSDSDYAGCKVDRKSTSGTCQFLGRSLVSWNSKKQTSVALSTAEAEYVAA 963
Query: 920 AICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYV 979
C Q+LWM+ L D+ S +P+ CDN +AI +++NP+ HSR KHI++++HF+RD+
Sbjct: 964 GQCCAQLLWMRQTLRDFGYNLSKVPLLCDNESAIRMAENPVEHSRTKHIDIRHHFLRDHQ 1023
Query: 980 QKGVLLLKFVDIDHQWADIFTKPLAEDRFNFILKNLNM 1017
QKG + + V ++Q ADIFTKPL E F + LN+
Sbjct: 1024 QKGDIEVFHVSTENQLADIFTKPLDEKTFCRLRSELNV 1061
>UniRef100_Q84VI0 Gag-pol polyprotein [Glycine max]
Length = 1576
Score = 952 bits (2461), Expect = 0.0
Identities = 477/1010 (47%), Positives = 662/1010 (65%), Gaps = 10/1010 (0%)
Query: 1 QSWYLDSGCSRHMTGERRMFQELKLKPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLL 60
+ WYLDSGCSRHMTG + ++ V FG KGKI G G + D P ++ VLL
Sbjct: 560 EDWYLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLVHDGLPSLNKVLL 619
Query: 61 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 120
V GLT NL+SISQL D+G++V F + C ++ ++ S+ K+N Y E +
Sbjct: 620 VKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWTPQETSYSS 679
Query: 121 VKCLLSVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKV 180
CL S +E +WH+R GH+ +R + ++ VRG+P LK +C CQ K K+
Sbjct: 680 T-CLSSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKM 738
Query: 181 HFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHA 240
+ +TS LELLH+DL GP++ ES+GGKRY V+VDD+SR+TWV F+ K ++
Sbjct: 739 SHQKLQHQTTSMVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSDTFE 798
Query: 241 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVER 300
VF ++Q EK C I R+RSDHG EFEN KF S GI H+FS+ TPQ NG+VER
Sbjct: 799 VFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVER 858
Query: 301 KNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISY 360
KNRTLQE R ML + WAEA+NT CYI NR+++R T YE+WK KP + +
Sbjct: 859 KNRTLQEATRVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPTVKH 918
Query: 361 FHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 420
FH FG CY+L +++ K D KS + LGYS S+ +R +N+ +T+ ESI+V DD
Sbjct: 919 FHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDL 978
Query: 421 LDSDQSKLVEKFADLSINVSDKGKAPEEAEPEEEEPEEEAGPSDSQSQKKS--RITAAHP 478
+ + + E NV+D K+ E AE + +E P+ +Q K RI P
Sbjct: 979 TPARKKDVEEDVRTSEDNVADTAKSAENAEKSDSTTDE---PNINQPDKSPFIRIQKMQP 1035
Query: 479 KELILGNKDEPVRTRSAFRPSEETLLSLKGLMSLIEPKSIDEALQDKDWILAMEEELNQF 538
KELI+G+ + V TRS E ++S +S IEPK++ EAL D+ WI AM+EEL QF
Sbjct: 1036 KELIIGDPNRGVTTRSR----EIEIVSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQF 1091
Query: 539 SKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAP 598
+N+VW LV +P+ +VIGTKW+F+NK NE+G + RNKARLVAQGY+Q EG+D+ ETFAP
Sbjct: 1092 KRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAP 1151
Query: 599 VARLEAVKLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPGFKDEKNPDHVFKLK 658
VARLE+++LL+ + L+QMDVKSAFLNGY++EE YV QP GF D + DHV++LK
Sbjct: 1152 VARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHLDHVYRLK 1211
Query: 659 KSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSAN 718
K+LYGLKQAPRAWYERL+ FL + + +G +D TLF K ++++I QIYVDDI+FG +
Sbjct: 1212 KALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMS 1271
Query: 719 QFLCKEFSEMMQAEFEMSMMGELKYFMGIQVDQTPEGTYIHQSKYTKELLKKFNMTELVI 778
+ + F MQ+EFEMS++GEL YF+G+QV Q + ++ QSKY K ++KKF M
Sbjct: 1272 NEMLRHFVPQMQSEFEMSLVGELHYFLGLQVKQMEDSIFLSQSKYAKNIVKKFGMENASH 1331
Query: 779 AKTPMHPTCILEKEDASGKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETH 838
+TP L K++A V Q LYR MIGSLLYLTASRPDI F+V +CAR+Q++P+ +H
Sbjct: 1332 KRTPAPTHLKLSKDEAGTSVDQNLYRSMIGSLLYLTASRPDITFAVGVCARYQANPKISH 1391
Query: 839 LTAVKRILRYLKGSTNLGLMYKKTSEYKLSGYCDADYVGDRTERKSTSGNC*FLGSNLVS 898
L VKRIL+Y+ G+++ G+MY S+ L GYCDAD+ G +RK TSG C +LG+NL+S
Sbjct: 1392 LNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGSADDRKCTSGGCFYLGTNLIS 1451
Query: 899 WASKRQSTIALSTAEAEYIPAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKN 958
W SK+Q+ ++LSTAEAEYI A +Q++WMK L++Y + + + +YCDN +AI++SKN
Sbjct: 1452 WFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNMSAINISKN 1511
Query: 959 PILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDIDHQWADIFTKPLAEDRF 1008
P+ H+R KHI++++H+IRD V ++ L+ VD + Q ADIFTK L ++F
Sbjct: 1512 PVQHNRTKHIDIRHHYIRDLVDDKIITLEHVDTEEQVADIFTKALDANQF 1561
>UniRef100_O65147 Gag-pol polyprotein [Glycine max]
Length = 1550
Score = 952 bits (2460), Expect = 0.0
Identities = 478/1010 (47%), Positives = 663/1010 (65%), Gaps = 10/1010 (0%)
Query: 1 QSWYLDSGCSRHMTGERRMFQELKLKPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLL 60
+ WYLDSGCSRHMTG + ++ V FG KGKI G G + D P ++ VLL
Sbjct: 534 EDWYLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLVHDGLPSLNKVLL 593
Query: 61 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 120
V GLT NL+SISQL D+G++V F + C ++ ++ S+ K+N Y E +
Sbjct: 594 VKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWTPQETSYSS 653
Query: 121 VKCLLSVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKV 180
CL S +E +WH+R GH+ +R + ++ VRG+P LK +C CQ K K+
Sbjct: 654 T-CLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKM 712
Query: 181 HFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHA 240
+ +TSR LELLH+DL GP++ ES+G KRY V+VDD+SR+TWV F+ K ++
Sbjct: 713 SNQKLQHQTTSRVLELLHMDLMGPMQVESLGRKRYAYVVVDDFSRFTWVNFIREKSDTFE 772
Query: 241 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVER 300
VF ++Q EK C I R+RSDHG EFEN KF S GI H+FS+ TPQ NG+VER
Sbjct: 773 VFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVER 832
Query: 301 KNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISY 360
KNRTLQE AR ML + WAEA+NT CYI NR+++R T YE+WK KP + +
Sbjct: 833 KNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPTVKH 892
Query: 361 FHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 420
FH G CY+L +++ K D KS + LGYS S+ +R +N+ +T+ ESI+V DD
Sbjct: 893 FHICGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDL 952
Query: 421 LDSDQSKLVEKFADLSINVSDKGKAPEEAEPEEEEPEEEAGPSDSQSQKKS--RITAAHP 478
+ + + E NV+D K+ E AE + +E P+ +Q K+ RI HP
Sbjct: 953 TPARKKDVEEDVRTSGDNVADTAKSAENAENSDSATDE---PNINQPDKRPSIRIQKMHP 1009
Query: 479 KELILGNKDEPVRTRSAFRPSEETLLSLKGLMSLIEPKSIDEALQDKDWILAMEEELNQF 538
KELI+G+ + V TRS E ++S +S IEPK++ EAL D+ WI AM+EEL QF
Sbjct: 1010 KELIIGDPNRGVTTRSR----EIEIISNSCFVSKIEPKNVKEALTDEFWINAMQEELEQF 1065
Query: 539 SKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAP 598
+N+VW LV +P+ +VIGTKW+F+NK NE+G + RNKARLVAQGY+Q EG+D+ ETFAP
Sbjct: 1066 KRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAP 1125
Query: 599 VARLEAVKLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPGFKDEKNPDHVFKLK 658
ARLE+++LL+ + L+QMDVKSAFLNGY++EE YV QP GF D +PDHV++LK
Sbjct: 1126 GARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHPDHVYRLK 1185
Query: 659 KSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSAN 718
K+LYGLKQAPRAWYERL+ FL + + +G +D TLF K ++++I QIYVDDI+FG +
Sbjct: 1186 KALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFGGMS 1245
Query: 719 QFLCKEFSEMMQAEFEMSMMGELKYFMGIQVDQTPEGTYIHQSKYTKELLKKFNMTELVI 778
+ + F + MQ+EFEMS++GEL YF+G+QV Q + ++ QSKY K ++KKF M
Sbjct: 1246 NEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYAKNIVKKFGMENASH 1305
Query: 779 AKTPMHPTCILEKEDASGKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETH 838
+TP L K++A V Q LYR MIGSLLYLTASRPDI ++V CAR+Q++P+ +H
Sbjct: 1306 KRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAVGGCARYQANPKISH 1365
Query: 839 LTAVKRILRYLKGSTNLGLMYKKTSEYKLSGYCDADYVGDRTERKSTSGNC*FLGSNLVS 898
L VKRIL+Y+ G+++ G+MY S+ L GYCDAD+ G +RKST G C +LG+N +S
Sbjct: 1366 LNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGSVDDRKSTFGGCFYLGTNFIS 1425
Query: 899 WASKRQSTIALSTAEAEYIPAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKN 958
W SK+Q+ ++LSTAEAEYI A +Q++WMK L++Y + + + +YCDN +AI++SKN
Sbjct: 1426 WFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMTLYCDNLSAINISKN 1485
Query: 959 PILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDIDHQWADIFTKPLAEDRF 1008
P+ HSR KHI++++H+IRD V V+ L+ VD + Q ADIFTK L ++F
Sbjct: 1486 PVQHSRTKHIDIRHHYIRDLVDDKVITLEHVDTEEQIADIFTKALDANQF 1535
>UniRef100_Q8H851 Putative Zea mays retrotransposon Opie-2 [Oryza sativa]
Length = 2011
Score = 932 bits (2408), Expect = 0.0
Identities = 494/1042 (47%), Positives = 665/1042 (63%), Gaps = 45/1042 (4%)
Query: 3 WYLDSGCSRHMTGERRMFQELKL--KPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLL 60
W LDSGC++HMTG+R MF ++ + KV FG N KGK++G G I + + IDNV L
Sbjct: 558 WVLDSGCTQHMTGDRAMFTTFEVGGNEQEKVTFGDNSKGKVIGLGKIAISNDLSIDNVSL 617
Query: 61 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 120
V L NLLS++Q+ D F + S +D S +F R N+Y + EA
Sbjct: 618 VKSLNFNLLSVAQICDLVLSCAFFPQEVIVSSLLDKSCVFKGFRYGNLYLGDFNSSEANL 677
Query: 121 VKCLLSVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKV 180
CL++ +WHRRL HV M ++S+LSK +LV GL +KF D LC ACQ K
Sbjct: 678 KTCLVAKTSLGWLWHRRLAHVGMNQLSKLSKRDLVVGLKDVKFEKDKLCSACQAGKQVAC 737
Query: 181 HFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHA 240
K+++STSRPLELLH++ FGP +SIGG + +VIVDDYSR+TW+ FL K
Sbjct: 738 SHPTKSIMSTSRPLELLHMEFFGPTTYKSIGGNSHCLVIVDDYSRYTWMFFLHDKSIVAE 797
Query: 241 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVER 300
+F F + QNE C +V++RSD+G +F+N E D GI H+ S+ +PQ NGVVE
Sbjct: 798 LFKKFAKRGQNEFNCTLVKIRSDNGSKFKNTNIEDYCDDLGIKHELSATYSPQQNGVVEM 857
Query: 301 KNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISY 360
KNRTL EMARTML E G++ FWAEA+NT C+ NR+ + +L KT YEL KPN++Y
Sbjct: 858 KNRTLIEMARTMLDEYGVSDSFWAEAINTACHATNRLYLHRLLKKTSYELIVGRKPNVAY 917
Query: 361 FHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 420
F FGC CY+ RL KF+++ + LLGY+ SK +R YN + +EE+ V+FD+
Sbjct: 918 FRVFGCKCYIYRKGVRLTKFESRCDEGFLLGYASNSKAYRVYNKNKGIVEETADVQFDET 977
Query: 421 LDSDQS-KLVEKFADLSI-----NVSDKGKAPEEAEPE-----EEEPEEEAGPSDSQSQK 469
S + + ++ D + N+S P E E + ++EP A PS +Q +
Sbjct: 978 NGSQEGHENLDDVGDEGLMRAMKNMSIGDVKPIEVEDKPSTSTQDEPSTSATPSQAQVEV 1037
Query: 470 K--------------SRITAAHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLMSLIEP 515
+ + ++ HP + +LG+ + V+TRS ++ +S +EP
Sbjct: 1038 EEEKAQDLPMPPRIHTALSKDHPIDQVLGDISKGVQTRSRV----ASICEHYSFVSCLEP 1093
Query: 516 KSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRN 575
K +DEAL D DWI AM +ELN F++N VW+LV++ + +VIGTKWVFRNK +E G VVRN
Sbjct: 1094 KHVDEALCDPDWINAMHKELNNFARNKVWTLVERLRDHNVIGTKWVFRNKQDENGLVVRN 1153
Query: 576 KARLVAQGYSQQEGIDYTETFAPVARLEAVKLLISFSVNHNIILHQMDVKSAFLNGYISE 635
KAR VAQG++Q EG+D+ ETFAPV RLEA+ +L++F+ NI L QMDVKSAFLNG I+E
Sbjct: 1154 KARFVAQGFTQVEGLDFGETFAPVTRLEAICILLAFASCFNIKLFQMDVKSAFLNGEIAE 1213
Query: 636 EVYVHQPPGFKDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFC 695
V+V QPPGF+D K P+HV+KL K+LYGLKQAPRAWYERL FLL +F GKVDTTLF
Sbjct: 1214 LVFVEQPPGFEDPKYPNHVYKLSKALYGLKQAPRAWYERLRDFLLSKDFKIGKVDTTLFT 1273
Query: 696 KTYKDDILIVQIYVDDIIFGSANQFLCKEFSEMMQAEFEMSMMGELKYFMGIQVDQTPEG 755
K DD + QIYVDDIIFG N+ CKEF +MM EFEMSM+GEL +F G+Q+ Q +G
Sbjct: 1274 KIIGDDFFVCQIYVDDIIFGCTNEVFCKEFGDMMSREFEMSMIGELSFFHGLQIKQLKDG 1333
Query: 756 TYIHQSKYTKELLKKFNMTELVIAKTPMHPTCILEKEDASGKVCQKLYRGMIGSLLYLTA 815
T F + + KTPM L+ ++ V KLYR MIGSLLYLTA
Sbjct: 1334 T--------------FGLEDAKPIKTPMATNGHLDLDEGGKPVDLKLYRSMIGSLLYLTA 1379
Query: 816 SRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGSTNLGLMYKKTSEYKLSGYCDADY 875
SRPDI+FSV +CARFQ+ P+E HL AVKRILRYLK S+ +GL Y K +++KL GY D+DY
Sbjct: 1380 SRPDIMFSVCMCARFQAAPKECHLVAVKRILRYLKHSSTIGLWYPKGAKFKLVGYSDSDY 1439
Query: 876 VGDRTERKSTSGNC*FLGSNLVSWASKRQSTIALSTAEAEYIPAAICSTQMLWMKHQLED 935
G + +RKSTSG+C LG +LVSW+SK+Q+ +AL AEAEY+ A C Q+LWMK L D
Sbjct: 1440 AGCKVDRKSTSGSCQMLGRSLVSWSSKKQNFVALFIAEAEYVSAGSCCAQLLWMKQILLD 1499
Query: 936 YQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDIDHQW 995
Y I + P+ C+N +AI ++ NP+ HSR KHI++++HF+RD+V K +++ + + Q
Sbjct: 1500 YGISFTKTPLLCENDSAIKIANNPVQHSRTKHIDIRHHFLRDHVAKCDIVISHIRTEDQL 1559
Query: 996 ADIFTKPLAEDRFNFILKNLNM 1017
ADIFTKPL E RF + LN+
Sbjct: 1560 ADIFTKPLDETRFCKLRNELNL 1581
>UniRef100_Q60DR2 Putative polyprotein [Oryza sativa]
Length = 1577
Score = 930 bits (2404), Expect = 0.0
Identities = 491/1042 (47%), Positives = 660/1042 (63%), Gaps = 47/1042 (4%)
Query: 3 WYLDSGCSRHMTGERRMFQ--ELKLKPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLL 60
W LDSGC++HMTG+R MF E+ + KV FG N KGK++G G I + + IDNV L
Sbjct: 549 WVLDSGCTQHMTGDRAMFTTFEVGRNEQEKVTFGDNSKGKVIGLGKIAISNDLSIDNVSL 608
Query: 61 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 120
V L NLLS++Q+ D F + S +D S +F R N+Y + + EA
Sbjct: 609 VKSLNFNLLSVAQICDLSLSCAFFPQEVIVSSLLDKSCVFKGFRYGNLYLVDFNSSEANL 668
Query: 121 VKCLLSVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKV 180
CL++ +WHRRL HV M ++S+ SK +LV GL +KF D LC ACQ K
Sbjct: 669 KTCLVAKTSLGWLWHRRLAHVGMNQLSKFSKRDLVMGLKDVKFEKDKLCSACQAGKQVAC 728
Query: 181 HFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHA 240
K+++STS+PLELLH+DLF P +SIGG + +VIVDDYSR+TWV FL K
Sbjct: 729 SHPTKSIMSTSKPLELLHMDLFDPTTYKSIGGNSHCLVIVDDYSRYTWVFFLHDKSIVAD 788
Query: 241 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVER 300
+F F + QNE +C +V++RS+ G EF+N E D GI H+ + +PQ NGVVER
Sbjct: 789 LFKKFAKRAQNEFSCTLVKIRSNIGSEFKNTNIEDYCDDLGIKHELFATYSPQQNGVVER 848
Query: 301 KNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISY 360
KNRTL EMARTML E G++ FWAEA+NT C+ NR+ + +L KT YE+ KPNI+Y
Sbjct: 849 KNRTLIEMARTMLDEYGVSDSFWAEAINTACHATNRLYLHRVLKKTSYEVIVGRKPNIAY 908
Query: 361 FHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 420
F FGC CY+ RL KF+++ + LLGY+ +SK +R YN + +EE+ V+FD+
Sbjct: 909 FRVFGCKCYIHRKGVRLTKFESRCDEGFLLGYASKSKAYRVYNKNKGIVEETADVQFDET 968
Query: 421 LDSDQSK----------LVEKFADLSIN------VSDKGKAPEEAEP---------EEEE 455
S + L+ ++SI V DK + EP + E
Sbjct: 969 NGSQEGHENLDDVGDEGLMRVMKNMSIGDVKPIEVEDKPSTSTQDEPSTSAMPSQAQVEV 1028
Query: 456 PEEEAGPSDSQSQKKSRITAAHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLMSLIEP 515
EE+A + + ++ HP + +LG+ + V+TRS ++ +S +E
Sbjct: 1029 EEEKAQEPPMPPRIHTALSKDHPIDQVLGDISKGVQTRSRVT----SICEHYSFVSCLER 1084
Query: 516 KSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRN 575
K +DEAL D DW+ AM EEL F++N VW+LV++P+ +VIGTKWVFRNK +E G VVRN
Sbjct: 1085 KHVDEALCDPDWMNAMHEELKNFARNKVWTLVERPRDHNVIGTKWVFRNKQDENGLVVRN 1144
Query: 576 KARLVAQGYSQQEGIDYTETFAPVARLEAVKLLISFSVNHNIILHQMDVKSAFLNGYISE 635
KARLVAQG++Q EG+D+ ETFAPVARLEA+ +L++F+ +I L QMDVKSAFLN
Sbjct: 1145 KARLVAQGFTQVEGLDFGETFAPVARLEAICILLAFASCFDIKLFQMDVKSAFLN----- 1199
Query: 636 EVYVHQPPGFKDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFC 695
D K P+HV+KL K+LYGL+QAPRAWYERL FLL +F GKVD TLF
Sbjct: 1200 -----------DTKYPNHVYKLSKALYGLRQAPRAWYERLRDFLLSKDFKIGKVDITLFT 1248
Query: 696 KTYKDDILIVQIYVDDIIFGSANQFLCKEFSEMMQAEFEMSMMGELKYFMGIQVDQTPEG 755
K DD + QIYVDDIIFGS N+ CKEF +MM EFEMSM+GEL +F+G+Q+ Q G
Sbjct: 1249 KIIGDDFFVYQIYVDDIIFGSTNEVFCKEFGDMMSREFEMSMIGELSFFLGLQIKQLKNG 1308
Query: 756 TYIHQSKYTKELLKKFNMTELVIAKTPMHPTCILEKEDASGKVCQKLYRGMIGSLLYLTA 815
T++ Q+KY K+LLK+F + + KTPM L+ ++ V KLYR MIGSLLYLT
Sbjct: 1309 TFVSQTKYIKDLLKRFGLEDAKPIKTPMATNGHLDLDEGGKPVDLKLYRSMIGSLLYLTV 1368
Query: 816 SRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGSTNLGLMYKKTSEYKLSGYCDADY 875
SRPDI+FSV +CARFQ+ P+E HL AVKRILRYLK S+ +GL Y K +++KL GY D DY
Sbjct: 1369 SRPDIMFSVCMCARFQAAPKECHLVAVKRILRYLKHSSTIGLWYPKGAKFKLVGYSDPDY 1428
Query: 876 VGDRTERKSTSGNC*FLGSNLVSWASKRQSTIALSTAEAEYIPAAICSTQMLWMKHQLED 935
G + +RKSTS +C LG +LVSW+SK+Q+++ALSTAE EY+ A C Q+LWMK L D
Sbjct: 1429 AGCKVDRKSTSSSCQMLGRSLVSWSSKKQNSVALSTAETEYVSAGSCCAQLLWMKQTLLD 1488
Query: 936 YQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDIDHQW 995
Y I + P+ CDN AI ++ NP+ HSR KHI++++HF+RD+V K +++ + + Q
Sbjct: 1489 YGISFTKTPLLCDNDGAIKIANNPVQHSRTKHIDIRHHFLRDHVAKCDIVISHIRTEDQL 1548
Query: 996 ADIFTKPLAEDRFNFILKNLNM 1017
ADIFTKPL E RF + LN+
Sbjct: 1549 ADIFTKPLDETRFCKLRNELNI 1570
>UniRef100_Q9C5V1 Gag/pol polyprotein [Arabidopsis thaliana]
Length = 1643
Score = 927 bits (2395), Expect = 0.0
Identities = 479/1035 (46%), Positives = 668/1035 (64%), Gaps = 28/1035 (2%)
Query: 3 WYLDSGCSRHMTGERRMFQELKLKPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLLVD 62
WY DSG SRHMTG + E V FGG KG+I G G + P + NV V+
Sbjct: 614 WYFDSGASRHMTGSQANLNNYSSVKESNVMFGGGAKGRIKGKGDLTETEKPHLTNVYFVE 673
Query: 63 GLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQNVK 122
GLT NL+S+SQL D+G V FN+ C A ++ + + L + NN Y ++
Sbjct: 674 GLTANLISVSQLCDEGLTVSFNKVKCWATNERNQNTLTGVRTGNNCYMWEEPKI------ 727
Query: 123 CLLSVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKVHF 182
CL + E+ +WH+RLGH++ R +S+L +VRG+P LK +C AC + K +V
Sbjct: 728 CLRAEKEDPVLWHQRLGHMNARSMSKLVNKEMVRGVPELKHIEKIVCGACNQGKQIRVQH 787
Query: 183 KAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHAVF 242
K + T++ L+L+H+DL GP++TESI GKRY V+VDD+SR+ WV+F+ K E+ F
Sbjct: 788 KRVEGIQTTQVLDLIHMDLMGPMQTESIAGKRYVFVLVDDFSRYAWVRFIREKSETANSF 847
Query: 243 STFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVERKN 302
Q++NEK I ++RSD GGEF N+ F S +S GI H +S+PRTPQ NGVVERKN
Sbjct: 848 KILALQLKNEKKMGIKQIRSDRGGEFMNEAFNSFCESQGIFHQYSAPRTPQSNGVVERKN 907
Query: 303 RTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISYFH 362
RTLQEMAR M+ G+ + FWAEA++T CY+ NR+ VR +KT YE+WK KPN+SYF
Sbjct: 908 RTLQEMARAMIHGNGVPEKFWAEAISTACYVINRVYVRLGSDKTPYEIWKGKKPNLSYFR 967
Query: 363 PFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLD 422
FGCVCY++N KD+L KFD++S + LGY+ S +R +N IEES++V FDD
Sbjct: 968 VFGCVCYIMNDKDQLGKFDSRSEEGFFLGYATNSLAYRVWNKQRGKIEESMNVVFDDGSM 1027
Query: 423 SDQSKLVEKFADLSINVSDK-GKAPEEAEPEE-------EEPEEEAGPSDSQSQKKSRIT 474
+ +V + ++S+ G+ + + + EE +EE P+ S+
Sbjct: 1028 PELQIIVRNRNEPQTSISNNHGEERNDNQFDNGDINKSGEESDEEVPPAQVHRDHASKDI 1087
Query: 475 AAHP--KELILGNKDE---------PVRTRSAFRPSEETLLSLKGLMSLIEPKSIDEALQ 523
P + + G K + R ++F EE + S +S++EPK++ EAL+
Sbjct: 1088 IGDPSGERVTRGVKQDYRQLAGIKQKHRVMASFACFEEIMFSC--FVSIVEPKNVKEALE 1145
Query: 524 DKDWILAMEEELNQFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAQG 583
D WILAMEEEL +FS++ VW LV +P V+VIGTKW+F+NK +E G++ RNKARLVAQG
Sbjct: 1146 DHFWILAMEEELEEFSRHQVWDLVPRPPQVNVIGTKWIFKNKFDEVGNITRNKARLVAQG 1205
Query: 584 YSQQEGIDYTETFAPVARLEAVKLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPP 643
Y+Q EG+D+ ETFAPVARLE ++ L+ + LHQMDVK AFLNG I EEVYV QP
Sbjct: 1206 YTQVEGLDFDETFAPVARLECIRFLLGTACGMGFKLHQMDVKCAFLNGIIEEEVYVEQPK 1265
Query: 644 GFKDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDIL 703
GF++ + P++V+KLKK+LYGLKQAPRAWYERL++FL+ + RG VD TLF K I+
Sbjct: 1266 GFENLEFPEYVYKLKKALYGLKQAPRAWYERLTTFLIVQGYTRGSVDKTLFVKNDVHGII 1325
Query: 704 IVQIYVDDIIFGSANQFLCKEFSEMMQAEFEMSMMGELKYFMGIQVDQTPEGTYIHQSKY 763
I+QIYVDDI+FG + L K F + M EF MSM+GELKYF+G+Q++QT EG I QS Y
Sbjct: 1326 IIQIYVDDIVFGGTSDKLVKTFVKTMTTEFRMSMVGELKYFLGLQINQTDEGITISQSTY 1385
Query: 764 TKELLKKFNMTELVIAKTPMHPTCILEKEDASGKVCQKLYRGMIGSLLYLTASRPDILFS 823
+ L+K+F M A TPM T L K++ KV +KLYRGMIGSLLYLTA+RPD+ S
Sbjct: 1386 AQNLVKRFGMCSSKPAPTPMSTTTKLFKDEKGVKVDEKLYRGMIGSLLYLTATRPDLCLS 1445
Query: 824 VHLCARFQSDPRETHLTAVKRILRYLKGSTNLGLMYKKTSEYKLSGYCDADYVGDRTERK 883
V LCAR+QS+P+ +HL AVKRI++Y+ G+ N GL Y + + L GYCDAD+ G+ +R+
Sbjct: 1446 VGLCARYQSNPKASHLLAVKRIIKYVSGTINYGLNYTRDTSLVLVGYCDADWGGNLDDRR 1505
Query: 884 STSGNC*FLGSNLVSWASKRQSTIALSTAEAEYIPAAICSTQMLWMKHQLEDY-QILESN 942
ST+G FLGSNL+SW SK+Q+ ++LS+ ++EYI C TQ+LWM+ DY
Sbjct: 1506 STTGGVFFLGSNLISWHSKKQNCVSLSSTQSEYIALGSCCTQLLWMRQMGLDYGMTFPDP 1565
Query: 943 IPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDIDHQWADIFTKP 1002
+ + CDN +AI++SKNP+ HS KHI +++HF+R+ V++ + ++ V + Q DIFTKP
Sbjct: 1566 LLVKCDNESAIAISKNPVQHSVTKHIAIRHHFVRELVEEKQITVEHVPTEIQLVDIFTKP 1625
Query: 1003 LAEDRFNFILKNLNM 1017
L + F + K+L +
Sbjct: 1626 LDLNTFVNLQKSLGI 1640
>UniRef100_Q852C7 Putative gag-pol polyprotein [Oryza sativa]
Length = 1969
Score = 917 bits (2369), Expect = 0.0
Identities = 481/1045 (46%), Positives = 657/1045 (62%), Gaps = 38/1045 (3%)
Query: 2 SWYLDSGCSRHMTGERRMFQEL-KLKPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLL 60
SW +DSGCSRHMTGE + F L + + + FG G+++ GTI V+ + +V L
Sbjct: 753 SWLVDSGCSRHMTGEAKWFTSLTRASGDETITFGDASSGRVMAKGTIKVNDKFMLKDVAL 812
Query: 61 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 120
V L +NLLS+SQL D+ +V F + R + + V F+ R ++
Sbjct: 813 VSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVFFANFDSSAPGP 871
Query: 121 VKCLL-SVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTK 179
+CL+ S N + WHRRLGH+ +S++S ++L+RGLP LK D +C C+ K T
Sbjct: 872 SRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVQKDLVCAPCRHGKMTS 931
Query: 180 VHFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESH 239
K +V T P +LLH+D GP + +S+GGK Y +V+VDD+SR++WV FL K+E+
Sbjct: 932 SSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWYVLVVVDDFSRYSWVYFLESKEETF 991
Query: 240 AVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVE 299
F + + E + +RSD+G EF+N FES DS G+ H FSSP PQ NGVVE
Sbjct: 992 GFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQFSSPYVPQQNGVVE 1051
Query: 300 RKNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNIS 359
RKNRTL EMARTML E + FW EA++ C+I NR+ +R IL+KT YEL +P +S
Sbjct: 1052 RKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPYELRFGRRPKVS 1111
Query: 360 YFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDD 419
+ FGC C+VL + + L KF+++S + LGY+ S+ +R Y I E+ V FD+
Sbjct: 1112 HLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIVETCEVTFDE 1170
Query: 420 KL---------------------DSDQSKLVEKFADLSINVSDKGKAPEEAEPEEEEPE- 457
D D + D + V + G +P P + P
Sbjct: 1171 ASPGARPEISGVPDESIFVDEDSDDDDDDSIPPPLDSTPPVQETG-SPSTTSPSGDAPTT 1229
Query: 458 -----EEAGPSDSQSQKKSRITAAHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLMSL 512
EE S I HP + ++G E V ++ L ++
Sbjct: 1230 SSSAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYE------LVNSAFVAS 1283
Query: 513 IEPKSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDV 572
EPK++ AL D++W+ AM EEL F +N VWSLV+ P +VIGTKWVF+NKL E G +
Sbjct: 1284 FEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTKWVFKNKLGEDGSI 1343
Query: 573 VRNKARLVAQGYSQQEGIDYTETFAPVARLEAVKLLISFSVNHNIILHQMDVKSAFLNGY 632
VRNKARLVAQG++Q EG+D+ ETFAPVARLEA+++L++F+ + L QMDVKSAFLNG
Sbjct: 1344 VRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQMDVKSAFLNGV 1403
Query: 633 ISEEVYVHQPPGFKDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTT 692
I EEVYV QPPGF++ K P+HVFKL+K+LYGLKQAPRAWYERL +FLL+N F G VD T
Sbjct: 1404 IEEEVYVKQPPGFENPKFPNHVFKLEKALYGLKQAPRAWYERLKTFLLQNGFEMGAVDKT 1463
Query: 693 LFCKTYKDDILIVQIYVDDIIFGSANQFLCKEFSEMMQAEFEMSMMGELKYFMGIQVDQT 752
LF D L+VQIYVDDIIFG ++ L +FS++M EFEMSMMGEL +F+G+Q+ QT
Sbjct: 1464 LFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMGELTFFLGLQIKQT 1523
Query: 753 PEGTYIHQSKYTKELLKKFNMTELVIAKTPMHPTCILEKEDASGKVCQKLYRGMIGSLLY 812
EG ++HQ+KY+KELLKKF+M + TPM T L ++ +V Q+ YR MIGSLLY
Sbjct: 1524 KEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQREYRSMIGSLLY 1583
Query: 813 LTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGSTNLGLMYKKTSEYKLSGYCD 872
LTASRPDI FSV LCARFQ+ PR +H AVKRI RY+K + G+ Y +S + + D
Sbjct: 1584 LTASRPDIHFSVCLCARFQASPRTSHRQAVKRIFRYIKSTLEYGIWYSCSSALSVRAFSD 1643
Query: 873 ADYVGDRTERKSTSGNC*FLGSNLVSWASKRQSTIALSTAEAEYIPAAICSTQMLWMKHQ 932
AD+ G + +RKSTSG C FLG++LVSW+S++QS++A STAEAEY+ AA +Q+LWM
Sbjct: 1644 ADFAGCKIDRKSTSGTCHFLGTSLVSWSSRKQSSVAQSTAEAEYVAAASACSQVLWMIST 1703
Query: 933 LEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDID 992
L+DY + S +P+ CDNT+AI+++KNP+ HSR KHIE++YHF+RD V+KG ++L+FV+ +
Sbjct: 1704 LKDYGLSFSGVPLLCDNTSAINIAKNPVQHSRTKHIEIRYHFLRDNVEKGTIVLEFVESE 1763
Query: 993 HQWADIFTKPLAEDRFNFILKNLNM 1017
Q ADIFTKPL RF F+ L +
Sbjct: 1764 KQLADIFTKPLDRSRFEFLRSELGV 1788
>UniRef100_Q8H7T1 Putative Zea mays retrotransposon Opie-2 [Oryza sativa]
Length = 2145
Score = 915 bits (2366), Expect = 0.0
Identities = 480/1045 (45%), Positives = 657/1045 (61%), Gaps = 38/1045 (3%)
Query: 2 SWYLDSGCSRHMTGERRMFQEL-KLKPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLL 60
SW +DSGCSRHMTGE + F L + + + FG G+++ GTI V+ + +V L
Sbjct: 753 SWLVDSGCSRHMTGEAKWFTSLTRASSDETITFGDASSGRVMAKGTIKVNDKFMLKDVAL 812
Query: 61 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 120
V L +NLLS+SQL D+ +V F + R + + V F+ R ++
Sbjct: 813 VSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVFFANFDSSAPGP 871
Query: 121 VKCLL-SVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTK 179
+CL+ S N + WHRRLGH+ +S++S ++L+RGLP LK D +C C+ K T
Sbjct: 872 SRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVPKDLVCAPCRHGKMTS 931
Query: 180 VHFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESH 239
K +V T P +LLH+D GP + +S+GGK Y +V+VDD+SR++WV FL K+E+
Sbjct: 932 SSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWYVLVVVDDFSRYSWVYFLESKEETF 991
Query: 240 AVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVE 299
F + + E + +RSD+G EF+N FES DS G+ H FSSP PQ NGVVE
Sbjct: 992 GFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQFSSPYVPQQNGVVE 1051
Query: 300 RKNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNIS 359
RKNRTL EMARTML E + FW EA++ C+I NR+ +R IL+KT YEL +P +S
Sbjct: 1052 RKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPYELRFGRRPKVS 1111
Query: 360 YFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDD 419
+ FGC C+VL + + L KF+++S + LGY+ S+ +R Y I E+ V FD+
Sbjct: 1112 HLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIVETCEVTFDE 1170
Query: 420 KL---------------------DSDQSKLVEKFADLSINVSDKGKAPEEAEPEEEEPE- 457
D D + D + V + G +P P + P
Sbjct: 1171 ASPGARPEISGVPDESIFVDEDSDDDDDDSIPPPLDSTPPVQETG-SPSTTSPSGDAPTT 1229
Query: 458 -----EEAGPSDSQSQKKSRITAAHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLMSL 512
EE S I HP + ++G E V ++ L ++
Sbjct: 1230 SSSAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYE------LVNSAFVAS 1283
Query: 513 IEPKSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDV 572
EPK++ AL D++W+ AM EEL F +N VWSLV+ P +VIGTKWVF+NKL E G +
Sbjct: 1284 FEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTKWVFKNKLGEDGSI 1343
Query: 573 VRNKARLVAQGYSQQEGIDYTETFAPVARLEAVKLLISFSVNHNIILHQMDVKSAFLNGY 632
VRNKARLVAQG++Q EG+D+ ETFAPVARLEA+++L++F+ + L QMDVKSAFLNG
Sbjct: 1344 VRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQMDVKSAFLNGV 1403
Query: 633 ISEEVYVHQPPGFKDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTT 692
I EEVYV QPPGF++ K P+HVFKL+K+LYGLKQAPRAWYERL +FLL+N F G VD T
Sbjct: 1404 IEEEVYVKQPPGFENPKFPNHVFKLEKALYGLKQAPRAWYERLKTFLLQNGFEMGAVDKT 1463
Query: 693 LFCKTYKDDILIVQIYVDDIIFGSANQFLCKEFSEMMQAEFEMSMMGELKYFMGIQVDQT 752
LF D L+VQIYVDDIIFG ++ L +FS++M EFEMSMMGEL +F+G+Q+ QT
Sbjct: 1464 LFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMGELTFFLGLQIKQT 1523
Query: 753 PEGTYIHQSKYTKELLKKFNMTELVIAKTPMHPTCILEKEDASGKVCQKLYRGMIGSLLY 812
EG ++HQ+KY+KELLKKF+M + TPM T L ++ +V Q+ YR MIGSLLY
Sbjct: 1524 KEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQREYRSMIGSLLY 1583
Query: 813 LTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGSTNLGLMYKKTSEYKLSGYCD 872
LTASRPDI FSV LCARFQ+ PR +H AVKR+ RY+K + G+ Y +S + + D
Sbjct: 1584 LTASRPDIHFSVCLCARFQASPRTSHRQAVKRMFRYIKSTLEYGIWYSCSSALSVRAFSD 1643
Query: 873 ADYVGDRTERKSTSGNC*FLGSNLVSWASKRQSTIALSTAEAEYIPAAICSTQMLWMKHQ 932
AD+ G + +RKSTSG C FLG++LVSW+S++QS++A STAEAEY+ AA +Q+LWM
Sbjct: 1644 ADFAGCKIDRKSTSGTCHFLGTSLVSWSSRKQSSVAQSTAEAEYVAAASACSQVLWMIST 1703
Query: 933 LEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDID 992
L+DY + S +P+ CDNT+AI+++KNP+ HSR KHIE++YHF+RD V+KG ++L+FV+ +
Sbjct: 1704 LKDYGLSFSGVPLLCDNTSAINIAKNPVQHSRTKHIEIRYHFLRDNVEKGTIVLEFVESE 1763
Query: 993 HQWADIFTKPLAEDRFNFILKNLNM 1017
Q ADIFTKPL RF F+ L +
Sbjct: 1764 KQLADIFTKPLDRSRFEFLRSELGV 1788
>UniRef100_Q7XPI7 OSJNBb0004A17.2 protein [Oryza sativa]
Length = 1877
Score = 915 bits (2366), Expect = 0.0
Identities = 480/1045 (45%), Positives = 657/1045 (61%), Gaps = 38/1045 (3%)
Query: 2 SWYLDSGCSRHMTGERRMFQEL-KLKPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLL 60
SW +DSGCSRHMTGE + F L + + + FG G+++ GTI V+ + +V L
Sbjct: 835 SWLVDSGCSRHMTGEAKWFTSLTRASSDETITFGDASSGRVMAKGTIKVNDKFMLKDVAL 894
Query: 61 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 120
V L +NLLS+SQL D+ +V F + R + + V F+ R ++
Sbjct: 895 VSKLKYNLLSVSQLCDENLEVRFKKDRSRVLDASESPV-FDISRVGRVFFANFDSSAPGP 953
Query: 121 VKCLL-SVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTK 179
+CL+ S N + WHRRLGH+ +S++S ++L+RGLP LK D +C C+ K T
Sbjct: 954 SRCLIASENRDLFFWHRRLGHIGFDHLSRISGMDLIRGLPKLKVPKDLVCAPCRHGKMTS 1013
Query: 180 VHFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESH 239
K +V T P +LLH+D GP + +S+GGK Y +V+VDD+SR++WV FL K+E+
Sbjct: 1014 SSHKPVTMVMTDGPGQLLHMDTVGPARVQSVGGKWYVLVVVDDFSRYSWVYFLESKEETF 1073
Query: 240 AVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVE 299
F + + E + +RSD+G EF+N FES DS G+ H FSSP PQ NGVVE
Sbjct: 1074 GFFQSLARSLALEFPGALRAIRSDNGSEFKNSAFESFCDSSGVEHQFSSPYVPQQNGVVE 1133
Query: 300 RKNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNIS 359
RKNRTL EMARTML E + FW EA++ C+I NR+ +R IL+KT YEL +P +S
Sbjct: 1134 RKNRTLVEMARTMLDEFTTPRKFWTEAISAACFISNRVFLRTILHKTPYELRFGRRPKVS 1193
Query: 360 YFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDD 419
+ FGC C+VL + + L KF+++S + LGY+ S+ +R Y I E+ V FD+
Sbjct: 1194 HLRVFGCKCFVLKSGN-LDKFESRSLDGIFLGYATHSRAYRVYVLSTNKIVETCEVTFDE 1252
Query: 420 KL---------------------DSDQSKLVEKFADLSINVSDKGKAPEEAEPEEEEPE- 457
D D + D + V + G +P P + P
Sbjct: 1253 ASPGARPEISGVPDESIFVDEDSDDDDDDSIPPPLDSTPPVQETG-SPSTTSPSGDAPTT 1311
Query: 458 -----EEAGPSDSQSQKKSRITAAHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLMSL 512
EE S I HP + ++G E V ++ L ++
Sbjct: 1312 SSSAAEEIDGGTSGPTAPRHIQNRHPPDSMIGGLGERVTRNRSYE------LVNSAFVAS 1365
Query: 513 IEPKSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDV 572
EPK++ AL D++W+ AM EEL F +N VWSLV+ P +VIGTKWVF+NKL E G +
Sbjct: 1366 FEPKNVCHALSDENWVNAMHEELENFERNKVWSLVEPPLGFNVIGTKWVFKNKLGEDGSI 1425
Query: 573 VRNKARLVAQGYSQQEGIDYTETFAPVARLEAVKLLISFSVNHNIILHQMDVKSAFLNGY 632
VRNKARLVAQG++Q EG+D+ ETFAPVARLEA+++L++F+ + L QMDVKSAFLNG
Sbjct: 1426 VRNKARLVAQGFTQVEGLDFEETFAPVARLEAIRILLAFAASKGFKLFQMDVKSAFLNGV 1485
Query: 633 ISEEVYVHQPPGFKDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTT 692
I EEVYV QPPGF++ K P+HVFKL+K+LYGLKQAPRAWYERL +FLL+N F G VD T
Sbjct: 1486 IEEEVYVKQPPGFENPKFPNHVFKLEKALYGLKQAPRAWYERLKTFLLQNGFEMGAVDKT 1545
Query: 693 LFCKTYKDDILIVQIYVDDIIFGSANQFLCKEFSEMMQAEFEMSMMGELKYFMGIQVDQT 752
LF D L+VQIYVDDIIFG ++ L +FS++M EFEMSMMGEL +F+G+Q+ QT
Sbjct: 1546 LFTLHSGIDFLLVQIYVDDIIFGGSSHALVAQFSDVMSREFEMSMMGELTFFLGLQIKQT 1605
Query: 753 PEGTYIHQSKYTKELLKKFNMTELVIAKTPMHPTCILEKEDASGKVCQKLYRGMIGSLLY 812
EG ++HQ+KY+KELLKKF+M + TPM T L ++ +V Q+ YR MIGSLLY
Sbjct: 1606 KEGIFVHQTKYSKELLKKFDMADCKPIATPMATTSSLGPDEDGEEVDQREYRSMIGSLLY 1665
Query: 813 LTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGSTNLGLMYKKTSEYKLSGYCD 872
LTASRPDI FSV LCARFQ+ PR +H AVKR+ RY+K + G+ Y +S + + D
Sbjct: 1666 LTASRPDIHFSVCLCARFQASPRTSHRQAVKRMFRYIKSTLEYGIWYSCSSALSVRAFSD 1725
Query: 873 ADYVGDRTERKSTSGNC*FLGSNLVSWASKRQSTIALSTAEAEYIPAAICSTQMLWMKHQ 932
AD+ G + +RKSTSG C FLG++LVSW+S++QS++A STAEAEY+ AA +Q+LWM
Sbjct: 1726 ADFAGCKIDRKSTSGTCHFLGTSLVSWSSRKQSSVAQSTAEAEYVAAASACSQVLWMIST 1785
Query: 933 LEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDID 992
L+DY + S +P+ CDNT+AI+++KNP+ HSR KHIE++YHF+RD V+KG ++L+FV+ +
Sbjct: 1786 LKDYGLSFSGVPLLCDNTSAINIAKNPVQHSRTKHIEIRYHFLRDNVEKGTIVLEFVESE 1845
Query: 993 HQWADIFTKPLAEDRFNFILKNLNM 1017
Q ADIFTKPL RF F+ L +
Sbjct: 1846 KQLADIFTKPLDRSRFEFLRSELGV 1870
>UniRef100_Q7XP45 OSJNBa0063G07.6 protein [Oryza sativa]
Length = 1539
Score = 914 bits (2362), Expect = 0.0
Identities = 485/1042 (46%), Positives = 655/1042 (62%), Gaps = 60/1042 (5%)
Query: 3 WYLDSGCSRHMTGERRMFQELKL--KPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLL 60
W LDSGC++HMTG+R MF ++ + KV F N K K++G G I + + IDNV
Sbjct: 524 WVLDSGCTQHMTGDRAMFTTFEVGGNEQEKVTFVDNSKRKVIGLGKIAISNDLSIDNVSF 583
Query: 61 VDGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQN 120
V L NLLS++Q+ D G F + S +D S +F R N+Y + + EA
Sbjct: 584 VKSLNFNLLSVAQICDLGLSCAFFPQEVIVSSLLDKSCVFKGFRYGNLYFVDFNSSEANL 643
Query: 121 VKCLLSVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKV 180
CL++ +WHRRL HV M ++S+LSK +LV GL +KF D LC ACQ K
Sbjct: 644 KTCLVAKTSLGWLWHRRLAHVGMNQLSKLSKRDLVVGLKDVKFEKDKLCSACQAGKQVAC 703
Query: 181 HFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHA 240
K+++STSRPLELLH+DLFGP +SIGG + +VIVDDYSR+TWV FL K
Sbjct: 704 SHPTKSIMSTSRPLELLHMDLFGPTTYKSIGGNSHCLVIVDDYSRYTWVFFLHDKSIVAE 763
Query: 241 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVER 300
+F + QNE +C +V++RSD+G EF+N E D GI H+ S+ +PQ NGVVER
Sbjct: 764 LFKKIAKRAQNEFSCTLVKIRSDNGSEFKNTNIEDYCDDLGIKHELSATYSPQQNGVVER 823
Query: 301 KNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISY 360
KNRTL EMARTML E G++ FWAEA+NT C+ NR + +L T YEL KPN++Y
Sbjct: 824 KNRTLIEMARTMLDEYGVSDSFWAEAINTACHATNRFYLHRLLKNTSYELIVGRKPNVAY 883
Query: 361 FHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 420
F FGC CY+ RL KF+++ + LLGY+ SK +R YN + T+EE+ V+FD+
Sbjct: 884 FRVFGCKCYIYRKGVRLTKFESRCDEGFLLGYASNSKAYRVYNKNKGTVEETADVQFDET 943
Query: 421 LDSDQSK----------LVEKFADLS------INVSDKGKAPEEAEP---------EEEE 455
S + L+ ++S I V DK + EP + E
Sbjct: 944 NGSQEGHENLDDVGDEGLIRAMKNMSFGDVKPIEVEDKPSTSTQDEPSTFATPSQAQVEV 1003
Query: 456 PEEEAGPSDSQSQKKSRITAAHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLMSLIEP 515
EE+A + + ++ HP + +LG+ + V+T S ++ +S +EP
Sbjct: 1004 EEEKAQDPPIPPRIHTTLSKDHPIDQVLGDISKGVQTLSRV----ASICEHYSFVSCLEP 1059
Query: 516 KSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRN 575
K +DEAL D DW+ AM EELN F++N VW+LV++P+ +VIGTKWVFRNK +E G VVRN
Sbjct: 1060 KHVDEALCDPDWMNAMHEELNNFARNKVWTLVERPRDHNVIGTKWVFRNKQDENGLVVRN 1119
Query: 576 KARLVAQGYSQQEGIDYTETFAPVARLEAVKLLISFSVNHNIILHQMDVKSAFLNGYISE 635
KARLVAQG++Q EG+D+ ETFAPVARLEA+ +L++F+ +I L QMDVKSAFLNG I+E
Sbjct: 1120 KARLVAQGFTQVEGLDFGETFAPVARLEAICILLAFASWFDIKLFQMDVKSAFLNGEIAE 1179
Query: 636 EVYVHQPPGFKDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFC 695
V+V QPPGF+D K P+H FK+ GKVDTTLF
Sbjct: 1180 LVFVEQPPGFEDPKYPNHDFKI-----------------------------GKVDTTLFT 1210
Query: 696 KTYKDDILIVQIYVDDIIFGSANQFLCKEFSEMMQAEFEMSMMGELKYFMGIQVDQTPEG 755
K DD + QIYVDDIIFGS N+ CKEF +MM EFEMSM+GEL +F+G+Q+ Q +G
Sbjct: 1211 KIIGDDFFVCQIYVDDIIFGSTNEVFCKEFGDMMSREFEMSMIGELSFFLGLQIKQLKDG 1270
Query: 756 TYIHQSKYTKELLKKFNMTELVIAKTPMHPTCILEKEDASGKVCQKLYRGMIGSLLYLTA 815
T++ Q+KY K+LLK+F + + KTPM L+ ++ V KLYR MIGSLLYLTA
Sbjct: 1271 TFVSQTKYIKDLLKRFGLEDAKPIKTPMATNGHLDLDEGGKPVDLKLYRSMIGSLLYLTA 1330
Query: 816 SRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGSTNLGLMYKKTSEYKLSGYCDADY 875
SRPDI+FSV +CA FQ+ P+E HL AVKRILRYLK S+ +GL Y K +++KL GY D+DY
Sbjct: 1331 SRPDIMFSVCMCAWFQAAPKECHLVAVKRILRYLKYSSTIGLWYPKGAKFKLVGYSDSDY 1390
Query: 876 VGDRTERKSTSGNC*FLGSNLVSWASKRQSTIALSTAEAEYIPAAICSTQMLWMKHQLED 935
G + +R STSG+C LG +LVSW+SK+Q+++ALSTAEAEY+ A C Q+LWMK L D
Sbjct: 1391 AGCKVDRNSTSGSCQMLGRSLVSWSSKKQNSVALSTAEAEYVSAGSCCAQLLWMKQTLLD 1450
Query: 936 YQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDIDHQW 995
Y I + P+ CDN +AI ++ NP+ HSR KHI++++HF+RD+V K +++ + + Q
Sbjct: 1451 YGISFTKTPLLCDNDSAIKIANNPVQHSRTKHIDIRHHFLRDHVAKCDIVISHIRTEDQL 1510
Query: 996 ADIFTKPLAEDRFNFILKNLNM 1017
ADIFTKPL E RF + LN+
Sbjct: 1511 ADIFTKPLDETRFCKLRNELNV 1532
>UniRef100_Q8VY36 Opie2a pol [Zea mays]
Length = 1048
Score = 910 bits (2353), Expect = 0.0
Identities = 483/1046 (46%), Positives = 648/1046 (61%), Gaps = 46/1046 (4%)
Query: 13 MTGERRMFQELKLKPEGK--VGFGGNEKGKIVGTGTICVDSSPCIDNVLLVDGLTHNLLS 70
MTGE++MF + + + FG +GK+ G G I + S I NV LV+ L +N LS
Sbjct: 1 MTGEKKMFTSYVKNKDSQDSIIFGDGNQGKVKGLGKIAISSEHSISNVFLVESLGYNFLS 60
Query: 71 ISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQNVKCLLSVNEE 130
+SQL + GY+ +F + DGS+ F +Y + ++ EA CL++
Sbjct: 61 VSQLCNMGYNCLFTNVDVSVFRRSDGSLAFKGVLDGKLYLVDFAKEEAGLDACLIAKTSM 120
Query: 131 Q*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKVHFKAKNVVST 190
+WHRRL HV M+ + +L K V GL ++F D C ACQ K + KNV++T
Sbjct: 121 GWLWHRRLAHVGMKNLHKLLKGEHVIGLTNVQFKKDRPCVACQAGKQVRGSHHTKNVMTT 180
Query: 191 SRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHAVFSTFIAQVQ 250
SRPLE+LH+DLFGPV SIGG +YG+VIVDD+SR+TWV FL K E+ ++ + Q
Sbjct: 181 SRPLEMLHMDLFGPVAYLSIGGSKYGLVIVDDFSRFTWVFFLQDKSETQGTLKRYLRRAQ 240
Query: 251 NEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVERKNRTLQEMAR 310
NE ++ ++RSD+G EF+N + E GI H+FS+P TPQ NGVVERKNRTL +MAR
Sbjct: 241 NEFELKVKKIRSDNGSEFKNLQVEEFLVEEGIKHEFSAPYTPQQNGVVERKNRTLMDMAR 300
Query: 311 TMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISYFHPFGCVCYV 370
TML E + FW+EAVNT C+ NR+ + +L T YEL KPN+SYF FG CY+
Sbjct: 301 TMLGEFKTPERFWSEAVNTACHSINRVYLHRLLKNTSYELLTGNKPNVSYFRVFGSKCYI 360
Query: 371 LNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQSKLV- 429
L K R KF K+ + LLGY +K +R +N + +E S V FD+ S + ++V
Sbjct: 361 LVKKGRTSKFAPKAVEGFLLGYDSNTKAYRVFNKSSGLVEVSSDVVFDETNGSLREQVVN 420
Query: 430 ------EKFADLSINVSDKGKAPEEAEPEEEEP--------------------------- 456
E ++ G + E+++P
Sbjct: 421 LDDVDEEDVPTAAMRTMAIGDVRPQEHLEQDQPSSTTMVHPPTQDDEQAPQVGAHDQGGA 480
Query: 457 -----EEEAGPSDSQSQKKSRITAAHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLMS 511
EEE P +Q ++ I HP ILG+ + V TRS +S
Sbjct: 481 QDVQVEEEEAPQAPPTQVRATIQRNHPVNQILGDISKGVTTRSRL----VNFCEHYSFVS 536
Query: 512 LIEPKSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGD 571
IEP ++E L D DW+LAM+EELN F +N+VW+LV +P+ +V+GTKWVFRNK +E G
Sbjct: 537 SIEPFRVEEVLLDPDWVLAMQEELNNFKRNEVWTLVPRPKQ-NVVGTKWVFRNKQDEHGV 595
Query: 572 VVRNKARLVAQGYSQQEGIDYTETFAPVARLEAVKLLISFSVNHNIILHQMDVKSAFLNG 631
V RNKARLVA+GY+Q G+D+ ETFAPVARL+++++ ++++ +H+ L+QMDVKSAFLNG
Sbjct: 596 VTRNKARLVAKGYAQVAGLDFEETFAPVARLKSIRIWLAYAAHHSFRLYQMDVKSAFLNG 655
Query: 632 YISEEVYVHQPPGFKDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDT 691
I EEVYV QPPGF+DE+ PDHV KL K+LYGLKQAPRAWYE L FL+ N F GK D
Sbjct: 656 PIKEEVYVEQPPGFEDERFPDHVCKLSKALYGLKQAPRAWYECLRDFLIANAFKVGKADP 715
Query: 692 TLFCKTYKDDILIVQIYVDDIIFGSANQFLCKEFSEMMQAEFEMSMMGELKYFMGIQVDQ 751
TLF KT D+ + QIYVDDIIFGS NQ C+EFS +M +FEMSMMGEL YF+G QV Q
Sbjct: 716 TLFTKTCDGDLFVCQIYVDDIIFGSTNQNSCEEFSRVMTQKFEMSMMGELSYFLGFQVRQ 775
Query: 752 TPEGTYIHQSKYTKELLKKFNMTELVIAKTPMHPTCILEKEDASGKVCQKLYRGMIGSLL 811
+GT+I Q+KYT++L+K+F M + AKTPM + V QK YR IGSLL
Sbjct: 776 LKDGTFISQTKYTQDLIKRFGMKDAKPAKTPMGTDGHTDLNKGGKSVDQKAYRSTIGSLL 835
Query: 812 YLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGSTNLGLMYKKTSEYKLSGYC 871
YL ASRPDI+ SV +CARFQSDPRE HL AVKRILRYL + G+ Y K S + L GY
Sbjct: 836 YLCASRPDIMLSVCMCARFQSDPRECHLVAVKRILRYLVATPCFGIWYPKGSTFDLIGYS 895
Query: 872 DADYVGDRTERKSTSGNC*FLGSNLVSWASKRQSTIALSTAEAEYIPAAICSTQMLWMKH 931
D+DY + +RKSTS C FLG +LVSW SK+Q+++ALSTAEAEY+ C Q+LWM+
Sbjct: 896 DSDYARCKVDRKSTSRMCQFLGRSLVSWNSKKQTSVALSTAEAEYVAVGQCCAQLLWMRQ 955
Query: 932 QLEDYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDI 991
L D+ S +P+ CDN +AI +++NP+ HSR KHI++++HF+RD+ QKGV+ + V
Sbjct: 956 TLRDFGYNLSEVPLLCDNESAIRIAENPVEHSRTKHIDIRHHFLRDHQQKGVIEVFHVSS 1015
Query: 992 DHQWADIFTKPLAEDRFNFILKNLNM 1017
++ ADIFTKPL E F + LN+
Sbjct: 1016 ENHLADIFTKPLDEQTFCKLHSELNV 1041
>UniRef100_Q7XET2 Hypothetical protein [Oryza sativa]
Length = 1419
Score = 897 bits (2317), Expect = 0.0
Identities = 469/1017 (46%), Positives = 650/1017 (63%), Gaps = 35/1017 (3%)
Query: 3 WYLDSGCSRHMTGERRMFQELKLKPEGK-VGFGGNEKGKIVGTGTICVDSSPCIDNVLLV 61
W +DSG SRHMTG++ F LK + + + F +V TG + V+ + NV LV
Sbjct: 429 WIVDSGDSRHMTGDKNWFSSLKKASKTESIVFADASTSAVVATGLVKVNEKFELKNVALV 488
Query: 62 DGLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQNV 121
L +NLLS+SQ+ D+ ++V F + + SVL N R ++K + +
Sbjct: 489 KDLKYNLLSVSQIVDENFEVHFKKTGSKVFDSCGDSVL-NISRYERVFKADFENSVSLVI 547
Query: 122 KCLLS-VNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKV 180
CL++ +++ WHRRLGHV +++LS L+LVRGL LK D +C C+ K
Sbjct: 548 TCLVAKFDKDVMFWHRRLGHVGFDHLTRLSGLDLVRGLSKLKKDLDLICTPCRHAKMVST 607
Query: 181 HFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHA 240
V T P +LLH+D GP + +S+GGK Y +VIVDD+SR++WV F++ KDE+
Sbjct: 608 SHAPIVSVMTDAPGQLLHMDTVGPARVQSVGGKWYVLVIVDDFSRYSWVFFMTTKDEAFQ 667
Query: 241 VFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVER 300
F +++ E + R+RSD+GGEF+N FE + G+ H+FSSPR PQ N VVER
Sbjct: 668 HFRGLFLRLELEFPGSLKRIRSDNGGEFKNASFEQFCNERGLEHEFSSPRVPQQNSVVER 727
Query: 301 KNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISY 360
KNR L EMARTML E + FWAEA+NT CYI NR+ +R L KT YEL +P +S+
Sbjct: 728 KNRVLVEMARTMLDEYKTTRKFWAEAINTACYISNRVFLRSKLGKTSYELRFGHQPKVSH 787
Query: 361 FHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDK 420
FGC C+VL + + L KF+A+S+ L LGY ++G+R I E+ V FD+
Sbjct: 788 LRVFGCKCFVLKSGN-LDKFEARSTDGLFLGYPAHTRGYRVLILGTNKIVETCEVSFDEA 846
Query: 421 LDSDQSKLVEKFADLSINVSDKGKAPEEAEPEEEEPEEEAGPSDSQSQKKSRITAAHPKE 480
+ + + + +G+ E E + +++ S + +S++T
Sbjct: 847 SPGTRPDIAGTLSQV------QGEDGRIFEDESDYDDDDEVGSAGERTTRSKVTT----- 895
Query: 481 LILGNKDEPVRTRSAFRPSEETLLSLKGLMSLIEPKSIDEALQDKDWILAMEEELNQFSK 540
V SAF S EPK + AL D+ WI AM EEL F +
Sbjct: 896 -------HDVCANSAFVAS-------------FEPKDVSHALTDESWINAMHEELENFER 935
Query: 541 NDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVA 600
N VW+LV+ P +VIGTKWVF+NK NE G +VRNKARLVAQG++Q EG+D+ ETFAPVA
Sbjct: 936 NKVWTLVEPPSGHNVIGTKWVFKNKQNEDGLIVRNKARLVAQGFTQVEGLDFDETFAPVA 995
Query: 601 RLEAVKLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPGFKDEKNPDHVFKLKKS 660
R+EA++LL++F+ + L+QMDVKSAFLNG+I EEVYV QPPGF++ P+HVFKL K+
Sbjct: 996 RIEAIRLLLAFAASKGFKLYQMDVKSAFLNGFIQEEVYVKQPPGFENPDFPNHVFKLSKA 1055
Query: 661 LYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQF 720
LYGLKQAPRAWY+RL +FLL F GKVD TLF + D+ L VQIYVDDIIFG +
Sbjct: 1056 LYGLKQAPRAWYDRLKNFLLAKGFTMGKVDKTLFVLKHGDNQLFVQIYVDDIIFGCSTHA 1115
Query: 721 LCKEFSEMMQAEFEMSMMGELKYFMGIQVDQTPEGTYIHQSKYTKELLKKFNMTELVIAK 780
+ +F+E M+ EFEMSMMGEL YF+G+Q+ QTP+GT++HQ+KYTK+LL++F M
Sbjct: 1116 VVVDFAETMRREFEMSMMGELSYFLGLQIKQTPQGTFVHQTKYTKDLLRRFKMENCKPIS 1175
Query: 781 TPMHPTCILEKEDASGKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLT 840
TP+ T +L+ ++ V QK YR MIGSLLYLTASRPDI F+V LCARFQ+ PR +H
Sbjct: 1176 TPIGSTAVLDPDEDGEAVDQKEYRSMIGSLLYLTASRPDIQFAVCLCARFQASPRASHRQ 1235
Query: 841 AVKRILRYLKGSTNLGLMYKKTSEYKLSGYCDADYVGDRTERKSTSGNC*FLGSNLVSWA 900
AVKRI+RYL + G+ Y +S LSGY DAD+ G R +RKSTSG C FLG++L++W+
Sbjct: 1236 AVKRIMRYLNHTLEFGIWYSTSSSICLSGYSDADFGGCRIDRKSTSGTCHFLGTSLIAWS 1295
Query: 901 SKRQSTIALSTAEAEYIPAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPI 960
S++QS++A STAE+EY+ AA C +Q+LW+ L+DY I +P++CDNT+AI+++KNP+
Sbjct: 1296 SRKQSSVAQSTAESEYVAAASCCSQILWLLSTLKDYGITFEKVPLFCDNTSAINIAKNPV 1355
Query: 961 LHSRAKHIEVKYHFIRDYVQKGVLLLKFVDIDHQWADIFTKPLAEDRFNFILKNLNM 1017
HSR KHI++++HF+RD+V+KG + L+F+D Q ADIFTKPL +RF F+ L +
Sbjct: 1356 QHSRTKHIDIRFHFLRDHVEKGDVELQFLDTKLQIADIFTKPLDSNRFAFLRGELGV 1412
>UniRef100_Q850V9 Putative polyprotein [Oryza sativa]
Length = 1128
Score = 873 bits (2255), Expect = 0.0
Identities = 455/935 (48%), Positives = 617/935 (65%), Gaps = 31/935 (3%)
Query: 110 KIRLSELEAQNVKCLLSVNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALC 169
K+ + +NV L++ +WHRRL HV M ++S+LSK +LV GL +KF D LC
Sbjct: 191 KVTFGDNSKRNVIGLVAKTSFGWLWHRRLAHVGMNQLSKLSKRDLVVGLKDVKFEKDKLC 250
Query: 170 EACQKDKFTKVHFKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWV 229
ACQ K K+++STSRPLELLH+DLFGP +SIGG + +VIVDDYS +TWV
Sbjct: 251 SACQASKQVACSHPTKSIMSTSRPLELLHMDLFGPTTYKSIGGNSHCLVIVDDYSCYTWV 310
Query: 230 KFLSRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSP 289
FL K +F F + QNE +C +V++RSD+G +F+N E D I H+ S+
Sbjct: 311 FFLHDKCIVAELFKKFAKRAQNEFSCTLVKIRSDNGSKFKNTNIEDYCDDLSIKHELSAT 370
Query: 290 RTPQ*NGVVERKNRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYE 349
+PQ NGVVERKNRTL EMARTML E G++ FWAEA+NT C+ NR+ + +L KT YE
Sbjct: 371 YSPQQNGVVERKNRTLIEMARTMLDEYGVSDSFWAEAINTACHATNRLYLHRLLKKTSYE 430
Query: 350 LWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTI 409
L KPN++YF FGC CY+ RL KF+++ + LLGY+ SK +R YN + +
Sbjct: 431 LIVGRKPNVAYFRVFGCKCYIYRKGVRLTKFESRCDEGFLLGYASNSKAYRVYNKNKGIV 490
Query: 410 EESIHVRFDDKLDSDQS-KLVEKFADLSI-----NVSDKGKAPEEAEPE-----EEEPEE 458
EE+ V+FD+ S + + ++ D + N+S P E E + ++EP
Sbjct: 491 EETADVQFDETNGSQEGHENLDDVGDEGLMRAMKNMSIGDVKPIEVEDKPSTSTQDEPST 550
Query: 459 EAGPSDSQSQKKSR--------------ITAAHPKELILGNKDEPVRTRSAFRPSEETLL 504
A PS +Q + + ++ HP + +LG+ + V+TRS ++
Sbjct: 551 SASPSQAQVEVEKEKAQDPPMPPRIYTALSKDHPIDQVLGDISKGVQTRSPVA----SIC 606
Query: 505 SLKGLMSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPQSVHVIGTKWVFRN 564
+S +EPK +DEAL D DW+ A+ EELN F++N VW+LV++P+ +VIGTKWVFRN
Sbjct: 607 EHYSFVSCLEPKHVDEALYDPDWMNAIHEELNNFARNKVWTLVERPRDHNVIGTKWVFRN 666
Query: 565 KLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAVKLLISFSVNHNIILHQMDV 624
K +E VVRNKARLVAQG++Q E +D+ ETF PVARLEA+++L++F+ +I L QMDV
Sbjct: 667 KQDENRLVVRNKARLVAQGFTQVEDLDFGETFGPVARLEAIRILLAFASCFDIKLFQMDV 726
Query: 625 KSAFLNGYISEEVYVHQPPGFKDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEF 684
KSAFLNG I+E V+V QPPGF D K P+HV+KL K+LYGLKQAPRAWYERL FLL +F
Sbjct: 727 KSAFLNGEIAELVFVEQPPGFDDPKYPNHVYKLSKALYGLKQAPRAWYERLRDFLLSKDF 786
Query: 685 VRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQFLCKEFSEMMQAEFEMSMMGELKYF 744
GKVDTTLF K DD + QIYVDDIIFGS N+ CKEF +MM EFEMSM+ EL +F
Sbjct: 787 KIGKVDTTLFTKIIGDDFFVCQIYVDDIIFGSTNEVFCKEFGDMMSREFEMSMIEELSFF 846
Query: 745 MGIQVDQTPEGTYIHQSKYTKELLKKFNMTELVIAKTPMHPTCILEKEDASGKVCQKLYR 804
+G+Q+ Q +GT++ Q+KY K+LLK+F + + KTPM L+ ++ V KLYR
Sbjct: 847 LGLQIKQLKDGTFVSQTKYIKDLLKRFGLEDAKPIKTPMATNWHLDLDEGGKPVDLKLYR 906
Query: 805 GMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGSTNLGLMYKKTSE 864
MIGSLLYLTASRPDI+FSV + ARFQ+ P+E HL AVKRILRYLK S+ + L Y K ++
Sbjct: 907 SMIGSLLYLTASRPDIMFSVCMYARFQAAPKECHLVAVKRILRYLKHSSTISLWYPKGAK 966
Query: 865 YKLSGYCDADYVGDRTERKSTSGNC*FLGSNLVSWASKRQSTIALSTAEAEYIPAAICST 924
+KL GY D+DY G + +RKSTSG+C LG +LVSW+SK+Q+++ALSTAEAEYI A C
Sbjct: 967 FKLVGYSDSDYAGYKVDRKSTSGSCQMLGRSLVSWSSKKQNSVALSTAEAEYISAGSCCA 1026
Query: 925 QMLWMKHQLEDYQI--LESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKG 982
Q+LWMK L DY I E+ P+ C+N + I ++ NP+ H R KHI++++HF+ D+V K
Sbjct: 1027 QLLWMKQILLDYGISFTETQTPLLCNNDSTIKIANNPVQHFRTKHIDIRHHFLTDHVAKC 1086
Query: 983 VLLLKFVDIDHQWADIFTKPLAEDRFNFILKNLNM 1017
+++ + + Q ADIFTKPL E RF + LN+
Sbjct: 1087 DIVISHIRTEDQLADIFTKPLDETRFCKLRNELNV 1121
Score = 42.0 bits (97), Expect = 0.095
Identities = 20/43 (46%), Positives = 28/43 (64%), Gaps = 2/43 (4%)
Query: 3 WYLDSGCSRHMTGERRMFQ--ELKLKPEGKVGFGGNEKGKIVG 43
W LDS C++ MTG+R MF E++ K + KV FG N K ++G
Sbjct: 162 WVLDSVCTQRMTGDRAMFTTFEVEGKEQEKVTFGDNSKRNVIG 204
>UniRef100_Q9SDD5 Similar to copia-type pol polyprotein [Oryza sativa]
Length = 940
Score = 847 bits (2189), Expect = 0.0
Identities = 447/905 (49%), Positives = 592/905 (65%), Gaps = 39/905 (4%)
Query: 147 SQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKVHFKAKNVVSTSRPLELLHIDLFGPVK 206
SQ SK + GL ++F D +C ACQ K H KNV++T+RPLELLH+DLFGP+
Sbjct: 34 SQTSKARHILGLTNIQFEKDRVCSACQAGKQIGAHHPVKNVMTTTRPLELLHMDLFGPIA 93
Query: 207 TESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHAVFSTFIAQVQNEKACRIVRVRSDHGG 266
SIGG +YG+VIVDD+S +TWV FL K E+ A+F F + QNE +I +RSD+
Sbjct: 94 YLSIGGNKYGLVIVDDFSCFTWVFFLHDKSETQAIFKKFARRAQNEFDLKIKNIRSDNVK 153
Query: 267 EFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVERKNRTLQEMARTMLQETGMAKYFWAEA 326
EF+N ES D GI H+FS+P +PQ NGV ERKNRTL E+ARTML E + FWAEA
Sbjct: 154 EFKNTCIESFLDEEGIKHEFSAPYSPQQNGVAERKNRTLIEIARTMLDEYKTSDRFWAEA 213
Query: 327 VNTVCYIQNRISVRPILNKTRYELWKNIKPNISYFHPFGCVCYVLNTKDRLHKFDAKSSK 386
VNTVC+ NR+ + +L KT YEL KPN+SYF FG CY+LN K R KF K
Sbjct: 214 VNTVCHDINRLYLHRLLKKTPYELLTGNKPNVSYFRVFGSKCYILNKKARSSKFAPKVDG 273
Query: 387 CLLLGYSDRSKGFRFYNTDAKTIEESIHVRFD-------DKLDS-------DQSKLVEKF 432
LLGY +R +N + +E + V FD +++DS D + +++
Sbjct: 274 GFLLGYGSNECAYRVFNKTSGIVEIAPDVTFDETNGSQVEQVDSHVLGEEEDPREAIKRL 333
Query: 433 A-----------------DLSINVSDKGKAPEEAEPEEEEPEEEAGPSD---SQSQKKSR 472
A + S + P + ++ E E+ PS + +
Sbjct: 334 ALGDVRPREPQQGASSSTQVEPPTSTQANDPSTSSLDQGEEGEQVPPSSINLAHPRIHQS 393
Query: 473 ITAAHPKELILGNKDEPVRTRSAFRPSEETLLSLKGLMSLIEPKSIDEALQDKDWILAME 532
I HP + ILG+ ++ V TRS +S +EP ++EAL D DW++AM+
Sbjct: 394 IQRDHPTDNILGDINKGVSTRSHI----ANFCEHYSFVSSLEPLRVEEALNDPDWVMAMQ 449
Query: 533 EELNQFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGIDY 592
EELN F++N+VW+LV++ + +VIGTKW+FRNK +E G V+RNKARLVAQG++Q EG+D+
Sbjct: 450 EELNNFTRNEVWTLVERSRQ-NVIGTKWIFRNKQDEAGVVIRNKARLVAQGFTQIEGLDF 508
Query: 593 TETFAPVARLEAVKLLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPGFKDEKNPD 652
ETFAPVARLE++++L++F+ N N L+QMDVKSAFLNG I+E VYV QPPGFKD K P+
Sbjct: 509 GETFAPVARLESIRILLTFATNLNFKLYQMDVKSAFLNGLINELVYVEQPPGFKDPKYPN 568
Query: 653 HVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDI 712
HV+KL K+LY LKQAPRAWYE L +FL++N F GK D+TLF K + +DI + QIYVDDI
Sbjct: 569 HVYKLHKALYELKQAPRAWYECLRNFLVKNGFEIGKADSTLFTKRHDNDIFVCQIYVDDI 628
Query: 713 IFGSANQFLCKEFSEMMQAEFEMSMMGELKYFMGIQVDQTPEGTYIHQSKYTKELLKKFN 772
IFGS N+ +EFS MM FEMSMMGELK+F+G+Q+ Q EGT+I Q+KY K++LKKF
Sbjct: 629 IFGSTNKSFSEEFSRMMTKRFEMSMMGELKFFLGLQIKQLKEGTFICQTKYLKDMLKKFG 688
Query: 773 MTELVIAKTPMHPTCILEKEDASGKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQS 832
M TPM L+ + V QK+YR +IGSLLYL ASRPDI+ SV +CARFQ+
Sbjct: 689 MENAKPIHTPMPSNGHLDLNEQGKDVDQKVYRSIIGSLLYLCASRPDIMLSVCMCARFQA 748
Query: 833 DPRETHLTAVKRILRYLKGSTNLGLMYKKTSEYKLSGYCDADYVGDRTERKSTSGNC*FL 892
P+E HL AVKRILRYL + NLGL Y K + + L GY DADY G + +RKSTSG C FL
Sbjct: 749 APKECHLVAVKRILRYLVHTPNLGLWYPKGARFDLIGYADADYAGCKVDRKSTSGTCQFL 808
Query: 893 GSNLVSWASKRQSTIALSTAEAEYIPAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAA 952
G +LVSW+SK+Q+++ALSTAEAEYI C Q++WMK L DY + S IP+ CDN +A
Sbjct: 809 GRSLVSWSSKKQNSVALSTAEAEYISTGSCCAQLIWMKQTLRDYGLNVSKIPLLCDNESA 868
Query: 953 ISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDIDHQWADIFTKPLAEDRFNFIL 1012
I ++ NP+ HSR KHI++++HF+RD+ +G + ++ V ID Q ADIFTKPL E RF +
Sbjct: 869 IKIANNPVQHSRTKHIDIRHHFLRDHSTRGDIDIQHVRIDKQLADIFTKPLDEARFCELR 928
Query: 1013 KNLNM 1017
LN+
Sbjct: 929 SELNI 933
>UniRef100_Q75LG0 Putative integrase [Oryza sativa]
Length = 1507
Score = 808 bits (2087), Expect = 0.0
Identities = 442/1043 (42%), Positives = 625/1043 (59%), Gaps = 93/1043 (8%)
Query: 3 WYLDSGCSRHMTGERRMFQELKLKPEGKVGFGGNEKGKIVGTGTICVDSSPCIDNVLLVD 62
W +DSGC RHMTG ++ ++ +LK NV LV+
Sbjct: 523 WIVDSGCLRHMTGMVKVNEKFELK------------------------------NVALVE 552
Query: 63 GLTHNLLSISQLADKGYDVIFNQKSCRAVSQIDGSVLFNSKRKNNIYKIRLSELEAQNVK 122
L +NLLS+ Q+ + ++V F + + SVL N R ++K + +
Sbjct: 553 DLNYNLLSVLQIVYENFEVHFKKTRSKVFDSCGDSVL-NISRYGRVFKADFENPVSPVIT 611
Query: 123 CLLS-VNEEQ*VWHRRLGHVSMRKISQLSKLNLVRGLPTLKFSSDALCEACQKDKFTKVH 181
CL++ +++ WHRRLGHV +++LS L+LVRGLP LK D C C K
Sbjct: 612 CLVAKFDKDVMFWHRRLGHVGFDHLTRLSGLDLVRGLPKLKKDLDLDCAPCHHAKMVASS 671
Query: 182 FKAKNVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHAV 241
V T P +LLH+D GP + +S+GGK Y +VI+DD+SR++WV F++ KDE+
Sbjct: 672 HAPIVSVMTDAPRQLLHMDTVGPARVQSVGGKWYVLVIIDDFSRYSWVFFMATKDEAFQH 731
Query: 242 FSTFIAQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVERK 301
F +++ E + R+RSD+G E++N FE + G+ H+FSSPR PQ NGVVERK
Sbjct: 732 FRGLFLRLELEFPGSLKRIRSDNGSEYKNASFEQFCNERGLEHEFSSPRVPQQNGVVERK 791
Query: 302 NRTLQEMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISYF 361
N L EM RTML E + FWAEA+NT YI NR+ +R L K+ YEL +P +S+
Sbjct: 792 NHVLVEMVRTMLDEYKTPRKFWAEAINTAYYISNRVFLRSKLGKSSYELRFGHQPKVSHL 851
Query: 362 HPFGCVCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKL 421
FGC C+VL + + L KF+A+S+ L LGY ++G+R T ++ E + F+D+
Sbjct: 852 RVFGCKCFVLKSGN-LDKFEARSTDGLFLGYPAHTRGYR---TLSQVQGEDGRI-FEDES 906
Query: 422 DSDQSKLVEKFADLSINVSDKGKAPEEAEPEEEEPEEEAGPS------------------ 463
D + V P P EE + G S
Sbjct: 907 DDNDDDEVGSAGQTGRQAGQTAGTPP-VRPAHEERSDRPGLSAEGSVDAVRDGPLEITTS 965
Query: 464 ---DSQSQKKSRITAA------HPKELILGNKDEPVRTRSAFRPSEETLLSLKGLMSLIE 514
D++ S + A HP E I+GN E TRS + + + + ++ E
Sbjct: 966 TSTDTEHGSTSEVVAPLHIQQRHPPEQIIGNIGERT-TRS--KVTTHDVCANYAFVASFE 1022
Query: 515 PKSIDEALQDKDWILAMEEELNQFSKNDVWSLVKKPQSVHVIGTKWVFRNKLNEKGDVVR 574
PK + AL D+ WI AM EEL F +N VW+LV+ P ++IGTKWVF+NK NE +VR
Sbjct: 1023 PKDVSHALTDESWINAMHEELENFERNKVWTLVEPPSGHNIIGTKWVFKNKQNEDDLIVR 1082
Query: 575 NKARLVAQGYSQQEGIDYTETFAPVARLEAVKLLISFSVNHNIILHQMDVKSAFLNGYIS 634
NKARLVAQG++Q EG+D+ ETFAPVAR+EA++L ++F+ + L+QMDVKSAFLNG+I
Sbjct: 1083 NKARLVAQGFTQVEGLDFDETFAPVARIEAIRLFLAFASSKGFKLYQMDVKSAFLNGFIQ 1142
Query: 635 EEVYVHQPPGFKDEKNPDHVFKLKKSLYGLKQAPRAWYERLSSFLLENEFVRGKVDTTLF 694
EEVYV QPPGF++ P++VFKL K+LYGLKQAPRAWY+RL +FLL F GKVD TLF
Sbjct: 1143 EEVYVKQPPGFENPDFPNYVFKLSKALYGLKQAPRAWYDRLKNFLLAKGFTMGKVDKTLF 1202
Query: 695 CKTYKDDILIVQIYVDDIIFGSANQFLCKEFSEMMQAEFEMSMMGELKYFMGIQVDQTPE 754
L +F+E M+ EFEMSMMGEL YF+G+Q+ QTP+
Sbjct: 1203 V-------------------------LKHDFAETMRREFEMSMMGELSYFLGLQIKQTPQ 1237
Query: 755 GTYIHQSKYTKELLKKFNMTELVIAKTPMHPTCILEKEDASGKVCQKLYRGMIGSLLYLT 814
GT++HQ+KYTK+LL++F M TP+ T +L+ ++ V QK YR MIGSLLYLT
Sbjct: 1238 GTFVHQTKYTKDLLRRFKMENCKPISTPIDSTAVLDPDEDGEAVDQKEYRSMIGSLLYLT 1297
Query: 815 ASRPDILFSVHLCARFQSDPRETHLTAVKRILRYLKGSTNLGLMYKKTSEYKLSGYCDAD 874
ASRP+I F+V LCARFQ+ PR +H AVKRI+RYL + G+ Y +S LSGY DA+
Sbjct: 1298 ASRPEIQFAVCLCARFQASPRASHRQAVKRIMRYLNHTLEFGIWYSTSSSLCLSGYSDAN 1357
Query: 875 YVGDRTERKSTSGNC*FLGSNLVSWASKRQSTIALSTAEAEYIPAAICSTQMLWMKHQLE 934
+ G R +RKSTSG C FLG++L++W+S++QS++A STAE+EY+ AA C +Q+LW+ L+
Sbjct: 1358 FGGCRIDRKSTSGTCHFLGTSLIAWSSRKQSSVAQSTAESEYVAAASCCSQILWLLSTLK 1417
Query: 935 DYQILESNIPIYCDNTAAISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDIDHQ 994
+Y + +P++CDNT+AI+++KN + HSR KH+++++HF+RD+V+KG + L+F+D Q
Sbjct: 1418 NYGLTFEKVPLFCDNTSAINIAKNLVQHSRTKHVDIRFHFLRDHVEKGDVELQFLDTKLQ 1477
Query: 995 WADIFTKPLAEDRFNFILKNLNM 1017
ADIFTKPL + F F+ L +
Sbjct: 1478 IADIFTKPLDSNCFTFLCGELGI 1500
>UniRef100_Q7XBD2 Retrotransposon Opie-2 [Zea mays]
Length = 1512
Score = 763 bits (1970), Expect = 0.0
Identities = 402/831 (48%), Positives = 536/831 (64%), Gaps = 56/831 (6%)
Query: 187 VVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFLSRKDESHAVFSTFI 246
+V PLELLH+DLFGPV SIGG +YG+VIVDD+SR+TWV FL K E+ F+
Sbjct: 731 IVFLQIPLELLHMDLFGPVAYLSIGGSKYGLVIVDDFSRFTWVFFLQDKSETQGTLKRFL 790
Query: 247 AQVQNEKACRIVRVRSDHGGEFENDKFESLFDSYGIAHDFSSPRTPQ*NGVVERKNRTLQ 306
+ QNE ++ ++RSD+G EF+N + E + GI H+FS+P TPQ NGVVERKNRTL
Sbjct: 791 RRAQNEFELKVKKIRSDNGSEFKNLQVEEFLEEEGIKHEFSAPYTPQQNGVVERKNRTLI 850
Query: 307 EMARTMLQETGMAKYFWAEAVNTVCYIQNRISVRPILNKTRYELWKNIKPNISYFHPFGC 366
+MARTML E + FW+EAVNT C+ NR+ + +L KT YEL KPN+SYF FG
Sbjct: 851 DMARTMLGEFKTPECFWSEAVNTACHAINRVYLHRLLKKTSYELLTGNKPNVSYFRVFGS 910
Query: 367 VCYVLNTKDRLHKFDAKSSKCLLLGYSDRSKGFRFYNTDAKTIEESIHVRFDDKLDSDQS 426
CY+L K R KF K+ + LLGY +K +R +N + +E S V FD+ S +
Sbjct: 911 KCYILVKKGRNSKFAPKAVEGFLLGYDSNTKAYRVFNKSSGLVEVSSDVVFDETNGSPRE 970
Query: 427 KLVEKFADLSINVSDKGKAPEEAEPEEEEPEEEAGPSDSQSQKKSRITAAHPKELILGNK 486
++V+ ++ +EE P+ + + I P+E +
Sbjct: 971 QVVDC----------------------DDVDEEDVPT--AAIRTMAIGEVRPQEQ--DER 1004
Query: 487 DEPVRTRSAFRPSEETLLSLKGLMSLIEPKSIDEALQDKDWILAMEEELNQFSKNDVWSL 546
D+P + + P+++ +P ++EAL D DW+LAM+EELN F +N+VWSL
Sbjct: 1005 DQPSSSTTVHPPTQDDE----------QPFRVEEALLDLDWVLAMQEELNNFKRNEVWSL 1054
Query: 547 VKKPQSVHVIGTKWVFRNKLNEKGDVVRNKARLVAQGYSQQEGIDYTETFAPVARLEAVK 606
+E G V RNKARLVA+GY+Q G+D+ ETFAPVARLE+++
Sbjct: 1055 --------------------DEHGVVTRNKARLVAKGYAQVAGLDFEETFAPVARLESIR 1094
Query: 607 LLISFSVNHNIILHQMDVKSAFLNGYISEEVYVHQPPGFKDEKNPDHVFKLKKSLYGLKQ 666
+L++++ +H+ L+QMDVKSAFLNG I EEVYV QPPGF+DE+ PDHV KL K+LYGLKQ
Sbjct: 1095 ILLAYAAHHSFRLYQMDVKSAFLNGPIKEEVYVEQPPGFEDERYPDHVCKLAKALYGLKQ 1154
Query: 667 APRAWYERLSSFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANQFLCKEFS 726
APRAWYE L FL+ N F GK D TLF KT D+ + QI+VDDIIFGS NQ C+EFS
Sbjct: 1155 APRAWYECLRDFLIANAFKVGKADPTLFTKTCNGDLFVCQIFVDDIIFGSTNQKSCEEFS 1214
Query: 727 EMMQAEFEMSMMGELKYFMGIQVDQTPEGTYIHQSKYTKELLKKFNMTELVIAKTPMHPT 786
+M +FEMSMMG+L YF+G QV Q +GT+I Q KYT++LLK+F M + AKTPM
Sbjct: 1215 RVMTQKFEMSMMGKLNYFLGFQVKQLKDGTFISQMKYTQDLLKRFGMKDAKPAKTPMGTD 1274
Query: 787 CILEKEDASGKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRIL 846
+ V QK YR MIGSLLYL ASRPDI+ SV +CARFQS+P+E HL AVKRIL
Sbjct: 1275 GHTDLNKGGKSVDQKAYRSMIGSLLYLCASRPDIMLSVCMCARFQSEPKECHLVAVKRIL 1334
Query: 847 RYLKGSTNLGLMYKKTSEYKLSGYCDADYVGDRTERKSTSGNC*FLGSNLVSWASKRQST 906
RYL + GL Y K S + L GY D+D G + +RKSTSG C FLG +LVSW SK+Q++
Sbjct: 1335 RYLVATPCFGLWYPKGSTFDLVGYSDSDNAGCKVDRKSTSGTCQFLGRSLVSWNSKKQTS 1394
Query: 907 IALSTAEAEYIPAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHSRAK 966
+ALSTAEAEY+ A C Q+LWM+ L D+ S +P+ CDN +AI +++NP+ H+R K
Sbjct: 1395 VALSTAEAEYVAAGQCCAQLLWMRQTLRDFGYNLSKVPLLCDNESAIRMAENPVEHNRTK 1454
Query: 967 HIEVKYHFIRDYVQKGVLLLKFVDIDHQWADIFTKPLAEDRFNFILKNLNM 1017
HI++++HF+RD+ QKG + + V ++Q ADIFTKPL E F + LN+
Sbjct: 1455 HIDIRHHFLRDHQQKGDIEVFHVSTENQLADIFTKPLDEKTFCRLRSELNV 1505
Score = 44.7 bits (104), Expect = 0.015
Identities = 22/49 (44%), Positives = 31/49 (62%), Gaps = 2/49 (4%)
Query: 2 SWYLDSGCSRHMTGERRMFQE-LKLK-PEGKVGFGGNEKGKIVGTGTIC 48
SW +DSGC+ HMTGE++MF +K K + + FG +GK+ TIC
Sbjct: 671 SWIIDSGCTNHMTGEKKMFTSYVKNKDSQDSIIFGDGNQGKLSLLATIC 719
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.136 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,697,982,563
Number of Sequences: 2790947
Number of extensions: 72810582
Number of successful extensions: 247448
Number of sequences better than 10.0: 3291
Number of HSP's better than 10.0 without gapping: 2285
Number of HSP's successfully gapped in prelim test: 1012
Number of HSP's that attempted gapping in prelim test: 238366
Number of HSP's gapped (non-prelim): 5534
length of query: 1019
length of database: 848,049,833
effective HSP length: 138
effective length of query: 881
effective length of database: 462,899,147
effective search space: 407814148507
effective search space used: 407814148507
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0086.17


