

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0082b.8
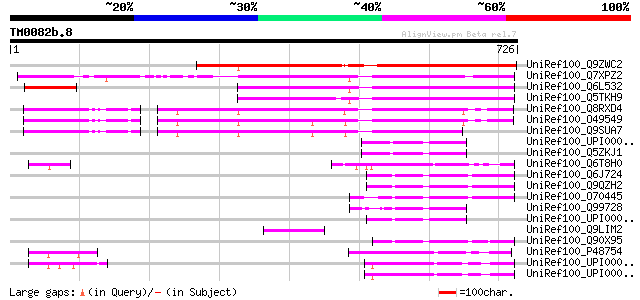
(726 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ZWC2 F21M11.4 protein [Arabidopsis thaliana] 442 e-122
UniRef100_Q7XPZ2 OSJNBa0004N05.16 protein [Oryza sativa] 425 e-117
UniRef100_Q6L532 Hypothetical protein OJ1005_B11.13 [Oryza sativa] 330 1e-88
UniRef100_Q5TKH9 Putative BRCT domain-containing protein [Oryza ... 326 1e-87
UniRef100_Q8RXD4 Hypothetical protein At4g21070 [Arabidopsis tha... 317 1e-84
UniRef100_O49549 Hypothetical protein F7J7.10 [Arabidopsis thali... 305 4e-81
UniRef100_Q9SUA7 Hypothetical protein T13K14.230 [Arabidopsis th... 281 4e-74
UniRef100_UPI00003AE4EC UPI00003AE4EC UniRef100 entry 94 2e-17
UniRef100_Q5ZKJ1 Hypothetical protein [Gallus gallus] 94 2e-17
UniRef100_Q6T8H0 Breast cancer 1 [Tetraodon nigroviridis] 93 3e-17
UniRef100_Q6J724 BRCA1-associated RING domain protein 1 beta iso... 91 2e-16
UniRef100_Q9QZH2 BRCA1-associated RING domain protein 1 [Rattus ... 91 2e-16
UniRef100_O70445 BRCA1-associated RING domain protein 1 [Mus mus... 87 2e-15
UniRef100_Q99728 BRCA1-associated RING domain protein 1 [Homo sa... 86 3e-15
UniRef100_UPI00003699A5 UPI00003699A5 UniRef100 entry 86 5e-15
UniRef100_Q9LIM2 Similarity to 26S proteasome subunit 4 [Arabido... 84 1e-14
UniRef100_Q90X95 BRCA1-associated RING domain protein [Xenopus l... 82 6e-14
UniRef100_P48754 Breast cancer type 1 susceptibility protein hom... 78 8e-13
UniRef100_UPI0000141712 UPI0000141712 UniRef100 entry 78 1e-12
UniRef100_UPI000014170B BRCA1 protein [Homo sapiens] 78 1e-12
>UniRef100_Q9ZWC2 F21M11.4 protein [Arabidopsis thaliana]
Length = 727
Score = 442 bits (1138), Expect = e-122
Identities = 227/469 (48%), Positives = 302/469 (63%), Gaps = 34/469 (7%)
Query: 268 RDLKRKKYLTKGDDHIQH--VSTHHSKLVDSHCGLDL--KSGKEPGELLPANIPIDLNPS 323
+D KRK+ +T D H V + L+ +D K + L I L +
Sbjct: 281 QDSKRKRDITASDAMENHLKVPKRENNLMQKSADIDCNGKCSANSDDQLSEKISKALEQT 340
Query: 324 TS---ICSFCQSSETSEATGPMLHYANGKSVIGDAAMQPNVIHVHRCCVDWAPQVYFVDE 380
+S IC FCQS+ SEATG MLHY+ G+ V GD + NVIHVH C++WAPQVY+ +
Sbjct: 341 SSNITICGFCQSARVSEATGEMLHYSRGRPVDGDDIFRSNVIHVHSACIEWAPQVYYEGD 400
Query: 381 TCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAMDVSTCRWDQEKYLLLC 440
T KNLKAE+ARG K+KC+ C LKGAALGC+VKSC+R+YHVPCA ++S CRWD E +LLLC
Sbjct: 401 TVKNLKAELARGMKIKCTKCSLKGAALGCFVKSCRRSYHVPCAREISRCRWDYEDFLLLC 460
Query: 441 PVHSNAKFPHEKSRPKKQATQEHPA--SAHLSSQLSNPLGAFHDDGKKVVFCGSALSDEE 498
P HS+ KFP+EKS + + P A L S P AF K++V CGSALS +
Sbjct: 461 PAHSSVKFPNEKSGHRVSRAEPLPKINPAELCSLEQTP--AF---TKELVLCGSALSKSD 515
Query: 499 KGSIENLRVMAVVVSGRGEVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKV 558
K +E+L V + AT++++W VTHVIA+TD GAC+RTLKV
Sbjct: 516 KKLMESLAV------------------RFNATISRYWNPSVTHVIASTDEKGACTRTLKV 557
Query: 559 LMAILNGRWVLKIDWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKLFS 618
LM ILNG+W++ W+KA +K V+EEP+EI +D QGC GPK RL A N+PKLF
Sbjct: 558 LMGILNGKWIINAAWMKASLKASQPVDEEPFEIQIDTQGCQDGPKTARLRAETNKPKLFE 617
Query: 619 GLKFYFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKL--EAKRHEFEVTSNLLIVYNLDPP 676
GLKFYF GD+ YKEDL++LV+V GG +L ++D+L E+ + + S+ ++VYN+DPP
Sbjct: 618 GLKFYFFGDFYKGYKEDLQNLVKVAGGTILNTEDELGAESSNNVNDQRSSSIVVYNIDPP 677
Query: 677 QGSKLGDEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAACKLQPFI 725
G LG+EV+ +WQR N+AE LA+ TGS+++GHTW+LESIA KL P I
Sbjct: 678 HGCALGEEVTIIWQRANDAEALASQTGSRLVGHTWVLESIAGYKLHPVI 726
>UniRef100_Q7XPZ2 OSJNBa0004N05.16 protein [Oryza sativa]
Length = 629
Score = 425 bits (1093), Expect = e-117
Identities = 269/724 (37%), Positives = 374/724 (51%), Gaps = 116/724 (16%)
Query: 12 KLMNPWMLHFQKLALELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSGSECAVCKTKY 71
+ +NP +L+ QK+ LEL CP+CL L P +LPC H C C ++ G CA+CK+ Y
Sbjct: 6 RFLNPLVLNLQKMELELTCPVCLKLLNAPTMLPCYHTSCSKCATTRTMDGYSCAICKSAY 65
Query: 72 AQTDIRNVPFVENMVAIYRSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADN 131
D+R +E +V I+RSL S S V QQ + ++I A
Sbjct: 66 RSQDLRPASHLEAIVNIHRSLS-----------STVSSMVTQQ------EAQADIPVAKT 108
Query: 132 FSQSSP---NSNGFGVGENRKSMITMHVKPEELEMSSGGRAGFRNDVKPY-PMQRSRVEI 187
Q +P N NG + KS + K S+G G + VK P +R
Sbjct: 109 SFQGTPESGNRNGAEKSDQVKSYTPVASKLA-YNQSTGLAYGNVDGVKERNPALETRGAA 167
Query: 188 GDYVEMDVNQVTQAAVYSPPFCDTKGSDNDCSELDSDHPFNPEILENSSLKRASTGKGNL 247
DV + V P C ++ SD +LD D LE + S+ + L
Sbjct: 168 ------DVTAMPTILVQKGP-CRSQSSDGP-RDLDCDS----NDLEGELITSRSSPQSVL 215
Query: 248 KERKSQFRSESSASETDKPTRDLKRKKYLTKGDDHIQHVSTHHSKLVDSHCGLDLKSGKE 307
K R ++A++ + R+LKR+K T DD V+
Sbjct: 216 K------REPNTANDDN---RELKRQKS-TDQDDRQPAVAGAWK---------------- 249
Query: 308 PGELLPANIPIDLNPSTSICSFCQSSETSEATGPMLHYANGKSVIGDAAMQPNVIHVHRC 367
C FC SS+T+E+TGP+ HY +G+ + + A +PNV+HVH
Sbjct: 250 -------------------CEFCHSSKTTESTGPLSHYLHGEPLEDNQAWKPNVLHVHEK 290
Query: 368 CVDWAPQVYFVDETCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAMDVS 427
C++WAPQ +F + NL+ E+AR +K+KCS CGLKGAALGC VKSC++++HVPCA +S
Sbjct: 291 CIEWAPQAFFTGDIANNLEPELARASKIKCSVCGLKGAALGCLVKSCRKSFHVPCAHGIS 350
Query: 428 TCRWDQEKYLLLCPVHSNAKFPHEKSRPKKQATQEHPASAHLSSQLSNPLGAFHDDG--- 484
CRWD E +++LCP HS+ K P E+S+ K + T +S+ N H DG
Sbjct: 351 GCRWDDENFVMLCPSHSSKKLPCERSKSKNKKTSLQRSSSDTMLDDLNSPSTIHMDGLWT 410
Query: 485 ------KKVVFCGSALSDEEKGSIENLRVMAVVVSGRGEVLLINFASKVGATVTKFWTSD 538
+ V CGSALS +EK +L F + G TVT W S+
Sbjct: 411 ASPFLTSEWVICGSALSSQEK------------------EILDQFEHQTGITVTNGWRSN 452
Query: 539 VTHVIAATDANGACSRTLKVLMAILNGRWVLKIDWIKACMKVMSLVEEEPYEISLDNQGC 598
VTHVIA TD GAC+RTLKVLMAIL G+WVL I+W+KACM+ V EEPYEIS D G
Sbjct: 453 VTHVIANTDECGACARTLKVLMAILAGKWVLNINWLKACMEAKEPVPEEPYEISSDVHGS 512
Query: 599 NGGPKAGRLSALANEPKLFSGLKFYFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKR 658
GP+ GRL A+ N P LF+GL FYFSG ++ +YK LEDL+ GG++L D
Sbjct: 513 FDGPRMGRLRAMQNAPHLFAGLTFYFSGHFMPNYKVHLEDLITAAGGSILDKAD------ 566
Query: 659 HEFEVTSNLLIVYNLDPPQGSKLGDEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAA 718
++S LI+Y+++PPQGS + +R EAE+LAA GS+ + HT +L+SIA+
Sbjct: 567 ----LSSTSLIIYSMEPPQGSDPDTLNEVIRKRKAEAEELAATIGSRAVPHTCVLDSIAS 622
Query: 719 CKLQ 722
C +Q
Sbjct: 623 CTVQ 626
>UniRef100_Q6L532 Hypothetical protein OJ1005_B11.13 [Oryza sativa]
Length = 995
Score = 330 bits (845), Expect = 1e-88
Identities = 161/407 (39%), Positives = 237/407 (57%), Gaps = 28/407 (6%)
Query: 327 CSFCQSSETSEATGPMLHYANGKSVIGDAAMQPNVIHVHRCCVDWAPQVYFVDETCKNLK 386
C+FCQ+ +E +G M+HY NGK V + NV+H H+ C++WAP VYF D++ NL
Sbjct: 606 CAFCQTDVITEESGEMVHYQNGKQVPAEFNGGANVVHSHKNCLEWAPDVYFEDDSAFNLT 665
Query: 387 AEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAMDVSTCRWDQEKYLLLCPVHSNA 446
E+AR ++KC+ CG+KGAALGC+ SC+R++H CA + CRWD E +++LCP+H +
Sbjct: 666 TELARSRRIKCACCGIKGAALGCFEMSCRRSFHFTCAKLIPECRWDNENFVMLCPLHRST 725
Query: 447 KFPHEKSRPKKQATQEHPASAHLSSQLSNPLGAFHDDG----------KKVVFCGSALSD 496
K P+E S +KQ ++ S+ +G+ D G +K V C S+LS
Sbjct: 726 KLPNENSEQQKQPKRKTTLKGITGFSRSSQIGSNQDCGNNWKWPSGSPQKWVLCCSSLSS 785
Query: 497 EEKGSIENLRVMAVVVSGRGEVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTL 556
EK L+ FA G ++ W+ +VTHVIA+TD +GAC RTL
Sbjct: 786 SEKE------------------LVSEFAKLAGVPISATWSPNVTHVIASTDLSGACKRTL 827
Query: 557 KVLMAILNGRWVLKIDWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKL 616
K LMAILNGRW++ IDW+K CM+ M ++E +E++ D G GP+ GR + +PKL
Sbjct: 828 KFLMAILNGRWIVSIDWVKTCMECMEPIDEHKFEVATDVHGITDGPRLGRCRVIDRQPKL 887
Query: 617 FSGLKFYFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNLLIVYNLDPP 676
F ++FY GDY SY+ L+DLV GG VL K ++ + +S+LLIVY+ +
Sbjct: 888 FDSMRFYLHGDYTKSYRGYLQDLVVAAGGIVLQRKPVSRDQQKLLDDSSDLLIVYSFENQ 947
Query: 677 QGSKLGDEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAACKLQP 723
+K E +R +A+ LA +G +V+ W+++SIAAC LQP
Sbjct: 948 DRAKSKAETKAADRRQADAQALACASGGRVVSSAWVIDSIAACNLQP 994
Score = 62.4 bits (150), Expect = 5e-08
Identities = 28/76 (36%), Positives = 49/76 (63%), Gaps = 2/76 (2%)
Query: 22 QKLALELKCPLC--LSLFEKPVLLPCDHLFCDSCLVDSSLSGSECAVCKTKYAQTDIRNV 79
+K+ ELK + LSL V + C+H+FC+ CL +S S S C VCK + + ++R
Sbjct: 8 EKMGRELKNLVLGSLSLLSSAVSISCNHVFCNDCLTESMKSTSSCPVCKVPFRRREMRPA 67
Query: 80 PFVENMVAIYRSLDAS 95
P ++N+V+I++S++A+
Sbjct: 68 PHMDNLVSIFKSMEAA 83
>UniRef100_Q5TKH9 Putative BRCT domain-containing protein [Oryza sativa]
Length = 799
Score = 326 bits (836), Expect = 1e-87
Identities = 161/407 (39%), Positives = 237/407 (57%), Gaps = 34/407 (8%)
Query: 327 CSFCQSSETSEATGPMLHYANGKSVIGDAAMQPNVIHVHRCCVDWAPQVYFVDETCKNLK 386
C+FCQ+ +E +G M+HY NGK V + NV+H H+ C++WAP VYF D++ NL
Sbjct: 416 CAFCQTDVITEESGEMVHYQNGKQVPAEFNGGANVVHSHKNCLEWAPDVYFEDDSAFNLT 475
Query: 387 AEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAMDVSTCRWDQEKYLLLCPVHSNA 446
E+AR ++KC+ CG+KGAALGC+ SC+R++H CA + CRWD E +++LCP+H +
Sbjct: 476 TELARSRRIKCACCGIKGAALGCFEMSCRRSFHFTCAKLIPECRWDNENFVMLCPLHRST 535
Query: 447 KFPHEKSRPKKQATQEHPASAHLSSQLSNPLGAFHDDG----------KKVVFCGSALSD 496
K P+E S +KQ ++ S+ +G+ D G +K V C S+LS
Sbjct: 536 KLPNENSEQQKQPKRKTTLKG------SSQIGSNQDCGNNWKWPSGSPQKWVLCCSSLSS 589
Query: 497 EEKGSIENLRVMAVVVSGRGEVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTL 556
EK L+ FA G ++ W+ +VTHVIA+TD +GAC RTL
Sbjct: 590 SEKE------------------LVSEFAKLAGVPISATWSPNVTHVIASTDLSGACKRTL 631
Query: 557 KVLMAILNGRWVLKIDWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKL 616
K LMAILNGRW++ IDW+K CM+ M ++E +E++ D G GP+ GR + +PKL
Sbjct: 632 KFLMAILNGRWIVSIDWVKTCMECMEPIDEHKFEVATDVHGITDGPRLGRCRVIDRQPKL 691
Query: 617 FSGLKFYFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNLLIVYNLDPP 676
F ++FY GDY SY+ L+DLV GG VL K ++ + +S+LLIVY+ +
Sbjct: 692 FDSMRFYLHGDYTKSYRGYLQDLVVAAGGIVLQRKPVSRDQQKLLDDSSDLLIVYSFENQ 751
Query: 677 QGSKLGDEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAACKLQP 723
+K E +R +A+ LA +G +V+ W+++SIAAC LQP
Sbjct: 752 DRAKSKAETKAADRRQADAQALACASGGRVVSSAWVIDSIAACNLQP 798
>UniRef100_Q8RXD4 Hypothetical protein At4g21070 [Arabidopsis thaliana]
Length = 941
Score = 317 bits (811), Expect = 1e-84
Identities = 194/544 (35%), Positives = 270/544 (48%), Gaps = 69/544 (12%)
Query: 212 KGSDNDCSELDSDHPFNPEILENSSLK---RASTGKGNLKERKSQFRSESSASETDKPTR 268
KG + HP E SLK R S +LK+ + + ++S + +
Sbjct: 429 KGDQDQAHGPSDTHPEKRSPTEKPSLKKRGRKSNASSSLKDLSGKTQKKTSEKKLKLDSH 488
Query: 269 DLKRKKYLTKGDDHIQHVSTHHSKLVDSHCGLDLKSGKEPGELLPANIPIDLNPSTS--- 325
+ K G+ + DS GK+ + +N S+S
Sbjct: 489 MISSKATQPHGNGILTAGLNQGGDKQDSRNNRKSTVGKDDHTMQVIEKCSTINKSSSGGS 548
Query: 326 --------------ICSFCQSSETSEATGPMLHYANGKSVIGDAAMQPNVIHVHRCCVDW 371
C+FCQ SE +EA+G M HY G+ V D VIHVH+ C +W
Sbjct: 549 AHLRRCNGSLTKKFTCAFCQCSEDTEASGEMTHYYRGEPVSADFNGGSKVIHVHKNCAEW 608
Query: 372 APQVYFVDETCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAMDVSTCRW 431
AP VYF D T NL E+ R ++ CS CGLKGAALGCY KSCK ++HV CA + CRW
Sbjct: 609 APNVYFNDLTIVNLDVELTRSRRISCSCCGLKGAALGCYNKSCKNSFHVTCAKLIPECRW 668
Query: 432 DQEKYLLLCPVHSNAKFPHEKSRPKKQATQEHPASAHLSSQLSNPLG----------AFH 481
D K+++LCP+ ++ K P E++ K + + P L SQ G FH
Sbjct: 669 DNVKFVMLCPLDASIKLPCEEANSKDRKCKRTPKEP-LHSQPKQVSGKANIRELHIKQFH 727
Query: 482 DDGKKVVFCGSALSDEEKGSIENLRVMAVVVSGRGEVLLINFASKVGATVTKFWTSDVTH 541
KK+V S L+ EEK ++ FA G T++K W S VTH
Sbjct: 728 GFSKKLVLSCSGLTVEEK------------------TVIAEFAELSGVTISKNWDSTVTH 769
Query: 542 VIAATDANGACSRTLKVLMAILNGRWVLKIDWIKACMKVMSLVEEEPYEISLDNQGCNGG 601
VIA+ + NGAC RTLK +MAIL G+W+L IDWIKACMK V EEPYEI++D G G
Sbjct: 770 VIASINENGACKRTLKFMMAILEGKWILTIDWIKACMKNTKYVSEEPYEITMDVHGIREG 829
Query: 602 PKAGRLSALANEPKLFSGLKFYFSGDYVSSYKEDLEDLVEVGGGAVL----TSKDKLEAK 657
P GR AL +PKLF+GLKFY GD+ +YK L+DL+ GG +L S D EA
Sbjct: 830 PYLGRQRALKKKPKLFTGLKFYIMGDFELAYKGYLQDLIVAAGGTILRRRPVSSDDNEA- 888
Query: 658 RHEFEVTSNLLIVYNLDPPQGSKLGDEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIA 717
+ ++V++++P + TL QR ++AE LA + ++ +W+L+SIA
Sbjct: 889 --------STIVVFSVEP-------SKKKTLTQRRSDAEALAKSARARAASSSWVLDSIA 933
Query: 718 ACKL 721
C++
Sbjct: 934 GCQI 937
Score = 84.3 bits (207), Expect = 1e-14
Identities = 52/169 (30%), Positives = 91/169 (53%), Gaps = 18/169 (10%)
Query: 20 HFQKLALELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSGSECAVCKTKYAQTDIRNV 79
H +++ ELKCP+CLSL+ V L C+H+FC++C+V S + C VCK Y + +IR
Sbjct: 6 HLERMGRELKCPICLSLYNSAVSLSCNHVFCNACIVKSMKMDATCPVCKIPYHRREIRGA 65
Query: 80 PFVENMVAIYRSL-DASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQSSPN 138
P ++++V+IY+++ DAS + Q + S +Q RD+S ++KA +
Sbjct: 66 PHMDSLVSIYKNMEDASGIKLFVSQNNPSPSDKEKQ---VRDAS---VEKAS-------D 112
Query: 139 SNGFGVGENRKSMITMHVKPEELEMSSGGRAGFRNDVKPYPMQRSRVEI 187
N G + R S + K +E+++ + G +KP + RV++
Sbjct: 113 KNRQGSRKGRASKRNEYGKTKEIDVDAPGPI----VMKPSSQTKKRVQL 157
>UniRef100_O49549 Hypothetical protein F7J7.10 [Arabidopsis thaliana]
Length = 1331
Score = 305 bits (780), Expect = 4e-81
Identities = 195/569 (34%), Positives = 271/569 (47%), Gaps = 94/569 (16%)
Query: 212 KGSDNDCSELDSDHPFNPEILENSSLK---RASTGKGNLKERKSQFRSESSASETDKPTR 268
KG + HP E SLK R S +LK+ + + ++S + +
Sbjct: 794 KGDQDQAHGPSDTHPEKRSPTEKPSLKKRGRKSNASSSLKDLSGKTQKKTSEKKLKLDSH 853
Query: 269 DLKRKKYLTKGDDHIQHVSTHHSKLVDSHCGLDLKSGKEPGELLPANIPIDLNPSTS--- 325
+ K G+ + DS GK+ + +N S+S
Sbjct: 854 MISSKATQPHGNGILTAGLNQGGDKQDSRNNRKSTVGKDDHTMQVIEKCSTINKSSSGGS 913
Query: 326 --------------ICSFCQSSETSEATGPMLHYANGKSVIGDAAMQPNVIHVHRCCVDW 371
C+FCQ SE +EA+G M HY G+ V D VIHVH+ C +W
Sbjct: 914 AHLRRCNGSLTKKFTCAFCQCSEDTEASGEMTHYYRGEPVSADFNGGSKVIHVHKNCAEW 973
Query: 372 APQVYFVDETCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAMDVSTCRW 431
AP VYF D T NL E+ R ++ CS CGLKGAALGCY KSCK ++HV CA + CRW
Sbjct: 974 APNVYFNDLTIVNLDVELTRSRRISCSCCGLKGAALGCYNKSCKNSFHVTCAKLIPECRW 1033
Query: 432 D-------------------------QEKYLLLCPVHSNAKFPHEKSRPKKQATQEHPAS 466
D Q K+++LCP+ ++ K P E++ K + + P
Sbjct: 1034 DNVSVAYIVEALYSPLHSSFNTFSYQQVKFVMLCPLDASIKLPCEEANSKDRKCKRTPKE 1093
Query: 467 AHLSSQLSNPLGA----------FHDDGKKVVFCGSALSDEEKGSIENLRVMAVVVSGRG 516
L SQ G FH KK+V S L+ EEK
Sbjct: 1094 P-LHSQPKQVSGKANIRELHIKQFHGFSKKLVLSCSGLTVEEK----------------- 1135
Query: 517 EVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILNGRWVLKIDWIKA 576
++ FA G T++K W S VTHVIA+ + NGAC RTLK +MAIL G+W+L IDWIKA
Sbjct: 1136 -TVIAEFAELSGVTISKNWDSTVTHVIASINENGACKRTLKFMMAILEGKWILTIDWIKA 1194
Query: 577 CMKVMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKLFSGLKFYFSGDYVSSYKEDL 636
CMK V EEPYEI++D G GP GR AL +PKLF+GLKFY GD+ +YK L
Sbjct: 1195 CMKNTKYVSEEPYEITMDVHGIREGPYLGRQRALKKKPKLFTGLKFYIMGDFELAYKGYL 1254
Query: 637 EDLVEVGGGAVL----TSKDKLEAKRHEFEVTSNLLIVYNLDPPQGSKLGDEVSTLWQRL 692
+DL+ GG +L S D EA + ++V++++P + TL QR
Sbjct: 1255 QDLIVAAGGTILRRRPVSSDDNEA---------STIVVFSVEP-------SKKKTLTQRR 1298
Query: 693 NEAEDLAANTGSQVIGHTWILESIAACKL 721
++AE LA + ++ +W+L+SIA C++
Sbjct: 1299 SDAEALAKSARARAASSSWVLDSIAGCQI 1327
Score = 84.3 bits (207), Expect = 1e-14
Identities = 52/169 (30%), Positives = 91/169 (53%), Gaps = 18/169 (10%)
Query: 20 HFQKLALELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSGSECAVCKTKYAQTDIRNV 79
H +++ ELKCP+CLSL+ V L C+H+FC++C+V S + C VCK Y + +IR
Sbjct: 371 HLERMGRELKCPICLSLYNSAVSLSCNHVFCNACIVKSMKMDATCPVCKIPYHRREIRGA 430
Query: 80 PFVENMVAIYRSL-DASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQSSPN 138
P ++++V+IY+++ DAS + Q + S +Q RD+S ++KA +
Sbjct: 431 PHMDSLVSIYKNMEDASGIKLFVSQNNPSPSDKEKQ---VRDAS---VEKAS-------D 477
Query: 139 SNGFGVGENRKSMITMHVKPEELEMSSGGRAGFRNDVKPYPMQRSRVEI 187
N G + R S + K +E+++ + G +KP + RV++
Sbjct: 478 KNRQGSRKGRASKRNEYGKTKEIDVDAPGPI----VMKPSSQTKKRVQL 522
>UniRef100_Q9SUA7 Hypothetical protein T13K14.230 [Arabidopsis thaliana]
Length = 1495
Score = 281 bits (720), Expect = 4e-74
Identities = 175/492 (35%), Positives = 233/492 (46%), Gaps = 74/492 (15%)
Query: 212 KGSDNDCSELDSDHPFNPEILENSSLK---RASTGKGNLKERKSQFRSESSASETDKPTR 268
KG + HP E SLK R S +LK+ + + ++S + +
Sbjct: 1011 KGDQDQAHGPSDTHPEKRSPTEKPSLKKRGRKSNASSSLKDLSGKTQKKTSEKKLKLDSH 1070
Query: 269 DLKRKKYLTKGDDHIQHVSTHHSKLVDSHCGLDLKSGKEPGELLPANIPIDLNPSTS--- 325
+ K G+ + DS GK+ + +N S+S
Sbjct: 1071 MISSKATQPHGNGILTAGLNQGGDKQDSRNNRKSTVGKDDHTMQVIEKCSTINKSSSGGS 1130
Query: 326 --------------ICSFCQSSETSEATGPMLHYANGKSVIGDAAMQPNVIHVHRCCVDW 371
C+FCQ SE +EA+G M HY G+ V D VIHVH+ C +W
Sbjct: 1131 AHLRRCNGSLTKKFTCAFCQCSEDTEASGEMTHYYRGEPVSADFNGGSKVIHVHKNCAEW 1190
Query: 372 APQVYFVDETCKNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPCAMDVSTCRW 431
AP VYF D T NL E+ R ++ CS CGLKGAALGCY KSCK ++HV CA + CRW
Sbjct: 1191 APNVYFNDLTIVNLDVELTRSRRISCSCCGLKGAALGCYNKSCKNSFHVTCAKLIPECRW 1250
Query: 432 D-------------------------QEKYLLLCPVHSNAKFPHEKSRPKKQATQEHPAS 466
D Q K+++LCP+ ++ K P E++ K + + P
Sbjct: 1251 DNVSVAYIVEALYSPLHSSFNTFSYQQVKFVMLCPLDASIKLPCEEANSKDRKCKRTPKE 1310
Query: 467 AHLSSQLSNPLGA----------FHDDGKKVVFCGSALSDEEKGSIENLRVMAVVVSGRG 516
L SQ G FH KK+V S L+ EEK
Sbjct: 1311 P-LHSQPKQVSGKANIRELHIKQFHGFSKKLVLSCSGLTVEEK----------------- 1352
Query: 517 EVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILNGRWVLKIDWIKA 576
++ FA G T++K W S VTHVIA+ + NGAC RTLK +MAIL G+W+L IDWIKA
Sbjct: 1353 -TVIAEFAELSGVTISKNWDSTVTHVIASINENGACKRTLKFMMAILEGKWILTIDWIKA 1411
Query: 577 CMKVMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKLFSGLKFYFSGDYVSSYKEDL 636
CMK V EEPYEI++D G GP GR AL +PKLF+GLKFY GD+ +YK L
Sbjct: 1412 CMKNTKYVSEEPYEITMDVHGIREGPYLGRQRALKKKPKLFTGLKFYIMGDFELAYKGYL 1471
Query: 637 EDLVEVGGGAVL 648
+DL+ GG +L
Sbjct: 1472 QDLIVAAGGTIL 1483
Score = 84.3 bits (207), Expect = 1e-14
Identities = 52/169 (30%), Positives = 91/169 (53%), Gaps = 18/169 (10%)
Query: 20 HFQKLALELKCPLCLSLFEKPVLLPCDHLFCDSCLVDSSLSGSECAVCKTKYAQTDIRNV 79
H +++ ELKCP+CLSL+ V L C+H+FC++C+V S + C VCK Y + +IR
Sbjct: 588 HLERMGRELKCPICLSLYNSAVSLSCNHVFCNACIVKSMKMDATCPVCKIPYHRREIRGA 647
Query: 80 PFVENMVAIYRSL-DASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQSSPN 138
P ++++V+IY+++ DAS + Q + S +Q RD+S ++KA +
Sbjct: 648 PHMDSLVSIYKNMEDASGIKLFVSQNNPSPSDKEKQ---VRDAS---VEKAS-------D 694
Query: 139 SNGFGVGENRKSMITMHVKPEELEMSSGGRAGFRNDVKPYPMQRSRVEI 187
N G + R S + K +E+++ + G +KP + RV++
Sbjct: 695 KNRQGSRKGRASKRNEYGKTKEIDVDAPGPI----VMKPSSQTKKRVQL 739
>UniRef100_UPI00003AE4EC UPI00003AE4EC UniRef100 entry
Length = 770
Score = 93.6 bits (231), Expect = 2e-17
Identities = 55/149 (36%), Positives = 82/149 (54%), Gaps = 10/149 (6%)
Query: 505 LRVMAVVVSGRGEVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILN 564
L ++ +S + + LL A+ + A + S VTHV+ + T+K +MA+L+
Sbjct: 565 LGILGSGLSSKQQKLLNKLATVLKARRCTEFNSTVTHVVVP---DVPMPSTVKCMMAVLS 621
Query: 565 GRWVLKIDWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKLFSGLKFYF 624
G WVLK +W++AC++ +EE YEI GGP+ GRL+ PKLF G FYF
Sbjct: 622 GCWVLKFEWVQACLQSTVREQEEKYEIQ-------GGPQRGRLNREQLLPKLFDGCYFYF 674
Query: 625 SGDYVSSYKEDLEDLVEVGGGAVLTSKDK 653
G + S K DL +LV+ GGG +L + K
Sbjct: 675 LGSFNSHQKSDLVELVKAGGGQILVRQPK 703
Score = 41.6 bits (96), Expect = 0.084
Identities = 34/126 (26%), Positives = 53/126 (41%), Gaps = 17/126 (13%)
Query: 5 GKPNNKSKLMNPWM---LHFQKLALELKCPLCLSLFEKPVLLP-CDHLFCDSCLVDSSLS 60
G+P + + PW ++L L C C + +PV L C+H+FC SC+ D
Sbjct: 14 GQPGPAASMARPWAHTRAALERLERALSCSRCAGVLREPVSLGGCEHVFCLSCMGDH--V 71
Query: 61 GSECAVCKTKYAQTDIRNVPFVENMV----AIYRSLDASFCASMLQQRSNDDSRVLQQCQ 116
G C VC D++ +++MV + LDA SM +S C
Sbjct: 72 GKGCPVCHVPAWVQDMQINRQLDDMVQLCGKLRHLLDAGTSDSM-------ESTPTPMCS 124
Query: 117 TFRDSS 122
F ++S
Sbjct: 125 DFGENS 130
>UniRef100_Q5ZKJ1 Hypothetical protein [Gallus gallus]
Length = 750
Score = 93.6 bits (231), Expect = 2e-17
Identities = 55/149 (36%), Positives = 82/149 (54%), Gaps = 10/149 (6%)
Query: 505 LRVMAVVVSGRGEVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILN 564
L ++ +S + + LL A+ + A + S VTHV+ + T+K +MA+L+
Sbjct: 544 LGILGSGLSSKQQKLLNKLATVLKARRCTEFNSTVTHVVVP---DVPMPSTVKCMMAVLS 600
Query: 565 GRWVLKIDWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKLFSGLKFYF 624
G WVLK +W++AC++ +EE YEI GGP+ GRL+ PKLF G FYF
Sbjct: 601 GCWVLKFEWVQACLQSTVREQEEKYEIQ-------GGPQRGRLNREQLLPKLFDGCYFYF 653
Query: 625 SGDYVSSYKEDLEDLVEVGGGAVLTSKDK 653
G + S K DL +LV+ GGG +L + K
Sbjct: 654 LGSFNSHQKSDLVELVKAGGGQILVRQPK 682
Score = 37.0 bits (84), Expect = 2.1
Identities = 32/118 (27%), Positives = 49/118 (41%), Gaps = 17/118 (14%)
Query: 13 LMNPWM---LHFQKLALELKCPLCLSLFEKPVLLP-CDHLFCDSCLVDSSLSGSECAVCK 68
+ PW ++L L C C + +PV L C+H+FC SC+ D G C VC
Sbjct: 1 MARPWAHTRAALERLERALSCSRCAGVLREPVSLGGCEHVFCLSCMGDH--VGKGCPVCH 58
Query: 69 TKYAQTDIRNVPFVENMV----AIYRSLDASFCASMLQQRSNDDSRVLQQCQTFRDSS 122
D++ +++MV + LDA SM +S C F ++S
Sbjct: 59 VPAWVQDMQINRQLDDMVQLCGKLRHLLDAGTSDSM-------ESTPTPMCSDFGENS 109
>UniRef100_Q6T8H0 Breast cancer 1 [Tetraodon nigroviridis]
Length = 1267
Score = 92.8 bits (229), Expect = 3e-17
Identities = 80/272 (29%), Positives = 121/272 (44%), Gaps = 38/272 (13%)
Query: 462 EHPASAHLSSQLSNPLGAFHDDGKKVVFCGSALS---DEEKGSIENLRVMA---VVVSGR 515
+H + L+S++SN +G K + GS D+E + E R +A +V SG
Sbjct: 1011 KHSSITELNSRISNTVG-LSSAAKTLKXDGSPSDGHEDKENNTPERARSLARMLLVTSGL 1069
Query: 516 G---EVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILNGRWVLKID 572
G ++ + FA ++GATV T +VTHV+ TD C RTLK + I +WV+
Sbjct: 1070 GPSQQITVKKFAKRIGATVVSQVTPEVTHVVMHTDEQLVCERTLKYFLGIAGRKWVVSFQ 1129
Query: 573 WIKACMKVMSLVEEEPYEISLD--NQGCNGGPKAGRLSALANEPKLFSGLKFYFSGDYVS 630
WI C+K L+ E +E+ D N + GP R A A+ L G F G +
Sbjct: 1130 WISECIKQKKLLNETLFEVRGDVVNGFDHQGPMKAR--ATADNNLLMKGYSICFQGPFTD 1187
Query: 631 SYKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNLLIVYNLDPPQGSKLGDEVSTLWQ 690
++E +VE G V+ L+ KR TS+ LIV GS+ VS
Sbjct: 1188 MTTAEMELMVEXCGATVVQDPLLLDGKR-----TSHQLIVVQ----SGSESSRSVS---- 1234
Query: 691 RLNEAEDLAANTGSQVIGHTWILESIAACKLQ 722
+ V+ W+L+S+A +Q
Sbjct: 1235 -----------GKATVVTRGWLLDSVATYTIQ 1255
Score = 51.2 bits (121), Expect = 1e-04
Identities = 21/64 (32%), Positives = 37/64 (57%), Gaps = 4/64 (6%)
Query: 28 LKCPLCLSLFEKPVLLPCDHLFCDSCLV----DSSLSGSECAVCKTKYAQTDIRNVPFVE 83
L+CP+CL L +PV CDH FC C++ ++ + + C VCK+K + ++ P +
Sbjct: 20 LQCPICLDLMSEPVSTKCDHQFCRFCMLKLLSNTKQNKANCPVCKSKITKRSLQESPGFQ 79
Query: 84 NMVA 87
+V+
Sbjct: 80 RLVS 83
>UniRef100_Q6J724 BRCA1-associated RING domain protein 1 beta isoform [Rattus
norvegicus]
Length = 602
Score = 90.5 bits (223), Expect = 2e-16
Identities = 61/213 (28%), Positives = 97/213 (44%), Gaps = 16/213 (7%)
Query: 512 VSGRGEVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILNGRWVLKI 571
+S + + LL + + A + + VTHVI + TLK ++ ILNG WVLK
Sbjct: 402 LSSQQQKLLSKLETVLKAKKCAEFDNTVTHVIVPDEE---AQSTLKCMLGILNGCWVLKF 458
Query: 572 DWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKLFSGLKFYFSGDYVSS 631
DW+KAC+ +EE YE+ GGP+ RL+ PKLF G F+ G++
Sbjct: 459 DWVKACLDSQEREQEEKYEVP-------GGPQRSRLNREQLLPKLFDGCYFFLGGNFKHH 511
Query: 632 YKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNL-LIVYNLDPPQGSKLGDEVSTLWQ 690
KEDL L+ GG +L+ K K ++ +VT + + Y+ P + +
Sbjct: 512 PKEDLLKLIAAAGGRILSRKPKPDS-----DVTQTINTVAYHAKPDSDQRFCTQYIVYED 566
Query: 691 RLNEAEDLAANTGSQVIGHTWILESIAACKLQP 723
N + + TW++ + A +L P
Sbjct: 567 LFNCHPERVRQGKVWMAPSTWLISCVMAFELLP 599
>UniRef100_Q9QZH2 BRCA1-associated RING domain protein 1 [Rattus norvegicus]
Length = 768
Score = 90.5 bits (223), Expect = 2e-16
Identities = 61/213 (28%), Positives = 97/213 (44%), Gaps = 16/213 (7%)
Query: 512 VSGRGEVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILNGRWVLKI 571
+S + + LL + + A + + VTHVI + TLK ++ ILNG WVLK
Sbjct: 568 LSSQQQKLLSKLETVLKAKKCAEFDNTVTHVIVPDEE---AQSTLKCMLGILNGCWVLKF 624
Query: 572 DWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKLFSGLKFYFSGDYVSS 631
DW+KAC+ +EE YE+ GGP+ RL+ PKLF G F+ G++
Sbjct: 625 DWVKACLDSQEREQEEKYEVP-------GGPQRSRLNREQLLPKLFDGCYFFLGGNFKHH 677
Query: 632 YKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNL-LIVYNLDPPQGSKLGDEVSTLWQ 690
KEDL L+ GG +L+ K K ++ +VT + + Y+ P + +
Sbjct: 678 PKEDLLKLIAAAGGRILSRKPKPDS-----DVTQTINTVAYHAKPDSDQRFCTQYIVYED 732
Query: 691 RLNEAEDLAANTGSQVIGHTWILESIAACKLQP 723
N + + TW++ + A +L P
Sbjct: 733 LFNCHPERVRQGKVWMAPSTWLISCVMAFELLP 765
Score = 47.4 bits (111), Expect = 0.002
Identities = 36/136 (26%), Positives = 62/136 (45%), Gaps = 14/136 (10%)
Query: 28 LKCPLCLSLFEKPVLLP-CDHLFCDSCLVDSSLSGSECAVCKTKYAQTDIRNVPFVENMV 86
L+C C ++ +PV L C+H+FC C+ S GS C VC T D++ +++M+
Sbjct: 42 LRCSRCANILREPVCLGGCEHIFCSGCI--SDCVGSGCPVCHTPAWILDLKINRQLDSMI 99
Query: 87 AIYRSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQS--SPNSNGFGV 144
+Y L ++L DS+ T R S + + ++ N + SP S
Sbjct: 100 QLYSKLQ-----NLLHDNKGSDSK----DDTSRASLFGDAERKKNSVKMWFSPRSKKIRC 150
Query: 145 GENRKSMITMHVKPEE 160
N+ S+ T K ++
Sbjct: 151 VVNKVSVQTQPQKAKD 166
>UniRef100_O70445 BRCA1-associated RING domain protein 1 [Mus musculus]
Length = 765
Score = 86.7 bits (213), Expect = 2e-15
Identities = 64/238 (26%), Positives = 103/238 (42%), Gaps = 34/238 (14%)
Query: 487 VVFCGSALSDEEKGSIENLRVMAVVVSGRGEVLLINFASKVGATVTKFWTSDVTHVIAAT 546
+VF GS LS +++ + L + + A + S VTHVI
Sbjct: 558 LVFIGSGLSSQQQKMLSKLETV------------------LKAKKCMEFDSTVTHVIVPD 599
Query: 547 DANGACSRTLKVLMAILNGRWVLKIDWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGR 606
+ TLK ++ IL+G W+LK DW+KAC+ +EE YE+ GGP+ R
Sbjct: 600 EE---AQSTLKCMLGILSGCWILKFDWVKACLDSKVREQEEKYEVP-------GGPQRSR 649
Query: 607 LSALANEPKLFSGLKFYFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSN 666
L+ PKLF G F+ G++ ++DL L+ GG VL+ K K ++ +VT
Sbjct: 650 LNREQLLPKLFDGCYFFLGGNFKHHPRDDLLKLIAAAGGKVLSRKPKPDS-----DVTQT 704
Query: 667 L-LIVYNLDPPQGSKLGDEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAACKLQP 723
+ + Y+ P + + N + + TW++ I A +L P
Sbjct: 705 INTVAYHAKPESDQRFCTQYIVYEDLFNCHPERVRQGKVWMAPSTWLISCIMAFELLP 762
Score = 44.3 bits (103), Expect = 0.013
Identities = 39/132 (29%), Positives = 57/132 (42%), Gaps = 33/132 (25%)
Query: 28 LKCPLCLSLFEKPVLLP-CDHLFCDSCLVDSSLSGSECAVCKTKYAQTDIRNVPFVENMV 86
L+C C ++ ++PV L C+H+FC C+ S GS C VC T D++
Sbjct: 42 LRCSRCANILKEPVCLGGCEHIFCSGCI--SDCVGSGCPVCYTPAWILDLK--------- 90
Query: 87 AIYRSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQSSPNSNGFGVGE 146
I R LD SM+Q S ++ + N DN S++S FG E
Sbjct: 91 -INRQLD-----SMIQLSSK-----------LQNLLHDNKDSKDNTSRASL----FGDAE 129
Query: 147 NRKSMITMHVKP 158
+K+ I M P
Sbjct: 130 RKKNSIKMWFSP 141
>UniRef100_Q99728 BRCA1-associated RING domain protein 1 [Homo sapiens]
Length = 777
Score = 86.3 bits (212), Expect = 3e-15
Identities = 59/167 (35%), Positives = 83/167 (49%), Gaps = 28/167 (16%)
Query: 487 VVFCGSALSDEEKGSIENLRVMAVVVSGRGEVLLINFASKVGATVTKFWTSDVTHVIAAT 546
+V GS LS E++ + L AV++ + T+F S VTHV+
Sbjct: 570 LVLIGSGLSSEQQKMLSEL---AVILKAK--------------KYTEF-DSTVTHVVVPG 611
Query: 547 DANGACSRTLKVLMAILNGRWVLKIDWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGR 606
DA TLK ++ ILNG W+LK +W+KAC++ +EE YEI GP+ R
Sbjct: 612 DA---VQSTLKCMLGILNGCWILKFEWVKACLRRKVCEQEEKYEIP-------EGPRRSR 661
Query: 607 LSALANEPKLFSGLKFYFSGDYVSSYKEDLEDLVEVGGGAVLTSKDK 653
L+ PKLF G FY G + K++L LV GGG +L+ K K
Sbjct: 662 LNREQLLPKLFDGCYFYLWGTFKHHPKDNLIKLVTAGGGQILSRKPK 708
Score = 43.9 bits (102), Expect = 0.017
Identities = 33/132 (25%), Positives = 55/132 (41%), Gaps = 31/132 (23%)
Query: 28 LKCPLCLSLFEKPVLLP-CDHLFCDSCLVDSSLSGSECAVCKTKYAQTDIRNVPFVENMV 86
L+C C ++ +PV L C+H+FC +C+ S G+ C VC T D++
Sbjct: 48 LRCSRCTNILREPVCLGGCEHIFCSNCV--SDCIGTGCPVCYTPAWIQDLK--------- 96
Query: 87 AIYRSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQSSPNSNGFGVGE 146
I R LD+ ++Q C R+ + N + + + P + F
Sbjct: 97 -INRQLDS----------------MIQLCSKLRNLLHDN--ELSDLKEDKPRKSLFNDAG 137
Query: 147 NRKSMITMHVKP 158
N+K+ I M P
Sbjct: 138 NKKNSIKMWFSP 149
>UniRef100_UPI00003699A5 UPI00003699A5 UniRef100 entry
Length = 218
Score = 85.5 bits (210), Expect = 5e-15
Identities = 52/142 (36%), Positives = 74/142 (51%), Gaps = 10/142 (7%)
Query: 512 VSGRGEVLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILNGRWVLKI 571
+S + +L A+ + A + S VTHV+ DA TLK ++ ILNG W+LK
Sbjct: 18 LSSEQQKMLSELAAILKAKKYTEFDSTVTHVVVPGDA---VQSTLKCMLGILNGCWILKF 74
Query: 572 DWIKACMKVMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKLFSGLKFYFSGDYVSS 631
+W+KAC++ +EE YEI GP+ RL+ PKLF G FY G +
Sbjct: 75 EWVKACLRRKVCEQEEKYEIP-------EGPRRSRLNREQLLPKLFDGCYFYLWGTFKHH 127
Query: 632 YKEDLEDLVEVGGGAVLTSKDK 653
K++L LV GGG +L+ K K
Sbjct: 128 PKDNLIKLVTAGGGQILSRKPK 149
>UniRef100_Q9LIM2 Similarity to 26S proteasome subunit 4 [Arabidopsis thaliana]
Length = 1964
Score = 84.0 bits (206), Expect = 1e-14
Identities = 39/88 (44%), Positives = 52/88 (58%), Gaps = 2/88 (2%)
Query: 364 VHRCCVDWAPQVYFVDETC-KNLKAEVARGAKLKCSTCGLKGAALGCYVKSCKRTYHVPC 422
VH+ C W+P+VYF C KN++A + RG LKC+ C GA GC V C RTYH+PC
Sbjct: 560 VHQNCAVWSPEVYFAGVGCLKNIRAALFRGRSLKCTRCDRPGATTGCRVDRCPRTYHLPC 619
Query: 423 AMDVSTCRWDQEKYLLLCPVHSNAKFPH 450
A + C +D K+L+ C H + PH
Sbjct: 620 AR-ANGCIFDHRKFLIACTDHRHHFQPH 646
>UniRef100_Q90X95 BRCA1-associated RING domain protein [Xenopus laevis]
Length = 772
Score = 82.0 bits (201), Expect = 6e-14
Identities = 59/206 (28%), Positives = 96/206 (45%), Gaps = 18/206 (8%)
Query: 520 LINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILNGRWVLKIDWIKACMK 579
L A + A++ + S VTHVI + + RT+K +M IL G W L+ W+KAC++
Sbjct: 581 LTKLAKMLKASICAEYDSTVTHVIVSDEP---VLRTMKCMMGILAGCWTLQFSWVKACLQ 637
Query: 580 VMSLVEEEPYEISLDNQGCNGGPKAGRLSALANEPKLFSGLKFYFSGDYVSSYKEDLEDL 639
+ EE YEI GP RL+ P+L G FYF G + KEDL +L
Sbjct: 638 SCNREPEESYEIP-------HGPHRARLNGQQMLPQLLDGCHFYFLGCFTEHRKEDLIEL 690
Query: 640 VEVGGGAVLTSKDKLEAKRHEFEVTSNL-LIVYNLDPPQGSKLGDEVSTLWQRLNEAEDL 698
V+ GG +L + K ++ +VT + + Y+ + + + ++ R ++
Sbjct: 691 VKAAGGQILIRQPKPDS-----DVTQTINTVAYHAEADSDQRFCTQY-IVFDRASKYRPE 744
Query: 699 AANTGSQVIG-HTWILESIAACKLQP 723
G +WI+E I + +L P
Sbjct: 745 RVRQGKVWFAPSSWIIECITSFQLLP 770
Score = 40.0 bits (92), Expect = 0.25
Identities = 24/96 (25%), Positives = 51/96 (53%), Gaps = 9/96 (9%)
Query: 22 QKLALELKCPLCLSLFEKPVLLP-CDHLFCDSCLVDSSLSGSECAVCKTKYAQTDIRNVP 80
+++ +L C C+ + + PV L C+H+FC +C+ ++ G+EC +C T D++
Sbjct: 36 REMERKLCCSKCMFILKDPVCLGGCEHVFCWTCVNENL--GNECPLCNTPAWVRDVQVNR 93
Query: 81 FVENMVAIYRSLDASFCASMLQQRSNDDSRVLQQCQ 116
++NM+ + L ++L D+++ + CQ
Sbjct: 94 QLDNMIQLCAKL-----RNLLSMGKTDENK-MDLCQ 123
>UniRef100_P48754 Breast cancer type 1 susceptibility protein homolog [Mus musculus]
Length = 1812
Score = 78.2 bits (191), Expect = 8e-13
Identities = 62/240 (25%), Positives = 111/240 (45%), Gaps = 28/240 (11%)
Query: 485 KKVVFCGSALSDEEKGSIENL-RVMAVVVSGRG--EVLLIN-FASKVGATVTKFWTSDVT 540
K VV S + E S E R +++VVSG EV+ + FA K T+T T + T
Sbjct: 1569 KAVVGIVSKIKPELTSSEERADRDISMVVSGLTPKEVMTVQKFAEKYRLTLTDAITEETT 1628
Query: 541 HVIAATDANGACSRTLKVLMAILNGRWVLKIDWIKACMKVMSLVEEEPYEISLD--NQGC 598
HVI TDA C RTLK + I G+W++ W+ ++ L+ +E++ D
Sbjct: 1629 HVIIKTDAEFVCERTLKYFLGIAGGKWIVSYSWVVRSIQERRLLNVHEFEVTGDVVTGRN 1688
Query: 599 NGGPKAGRLSALANEPKLFSGLKFYFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKR 658
+ GP+ R S KLF GL+ Y + + K+DLE ++++ G +V+ L
Sbjct: 1689 HQGPRRSRES----REKLFKGLQVYCCDPFTNMPKDDLERMLQLCGASVVKELPSL---- 1740
Query: 659 HEFEVTSNLLIVYNLDPPQGSKLGDEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAA 718
+ ++L+++ + W + D+ ++++ W+L+S+++
Sbjct: 1741 -THDTGAHLVVIVQ-------------PSAWTEDSNCPDIGQLCKARLVMWDWVLDSLSS 1786
Score = 49.3 bits (116), Expect = 4e-04
Identities = 27/111 (24%), Positives = 50/111 (44%), Gaps = 13/111 (11%)
Query: 28 LKCPLCLSLFEKPVLLPCDHLFCDSC---LVDSSLSGSECAVCKTKYAQTDIRNVPFVEN 84
L+CP+CL L ++PV CDH+FC C L++ S+C +CK + + ++
Sbjct: 22 LECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQCPLCKNEITKRSLQGSTRFSQ 81
Query: 85 MVAIYRSLDASF----------CASMLQQRSNDDSRVLQQCQTFRDSSYSN 125
+ + A+F S ++R+N R+ ++ + Y N
Sbjct: 82 LAEELLRIMAAFELDTGMQLTNGFSFSKKRNNSCERLNEEASIIQSVGYRN 132
>UniRef100_UPI0000141712 UPI0000141712 UniRef100 entry
Length = 1822
Score = 77.8 bits (190), Expect = 1e-12
Identities = 58/220 (26%), Positives = 99/220 (44%), Gaps = 26/220 (11%)
Query: 508 MAVVVSGRGE---VLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILN 564
M++VVSG +L+ FA K T+T T + THV+ TDA C RTLK + I
Sbjct: 1609 MSMVVSGLTPEEFMLVYKFARKHHITLTNLITEETTHVVMKTDAEFVCERTLKYFLGIAG 1668
Query: 565 GRWVLKIDWIKACMKVMSLVEEEPYEISLD--NQGCNGGPKAGRLSALANEPKLFSGLKF 622
G+WV+ W+ +K ++ E +E+ D N + GPK R S + K+F GL+
Sbjct: 1669 GKWVVSYFWVTQSIKERKMLNEHDFEVRGDVVNGRNHQGPKRARES---QDRKIFRGLEI 1725
Query: 623 YFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNLLIVYNLDPPQGSKLG 682
G + + + LE +V++ G +V+ + F + + + + + P
Sbjct: 1726 CCYGPFTNMPTDQLEWMVQLCGASVV-------KELSSFTLGTGVHPIVVVQP------- 1771
Query: 683 DEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAACKLQ 722
W N + + V+ W+L+S+A + Q
Sbjct: 1772 ----DAWTEDNGFHAIGQMCEAPVVTREWVLDSVALYQCQ 1807
Score = 50.4 bits (119), Expect = 2e-04
Identities = 32/125 (25%), Positives = 55/125 (43%), Gaps = 15/125 (12%)
Query: 28 LKCPLCLSLFEKPVLLPCDHLFCDSC---LVDSSLSGSECAVCKT----KYAQTDIRNVP 80
L+CP+CL L ++PV CDH+FC C L++ S+C +CK + Q R
Sbjct: 22 LECPICLELIKEPVSTKCDHIFCKFCMLKLLNQKKGPSQCPLCKNDITKRSLQESTRFSQ 81
Query: 81 FVENMVAIY------RSLDASFCASMLQQRSNDDSRVLQQCQTFRDSSYSNIKKADNFSQ 134
VE ++ I L+ + + ++ +N + + + Y N +A Q
Sbjct: 82 LVEELLKIICAFQLDTGLEYANSYNFAKKENNSPEHLKDEVSIIQSMGYRN--RAKRLLQ 139
Query: 135 SSPNS 139
S P +
Sbjct: 140 SEPEN 144
>UniRef100_UPI000014170B BRCA1 protein [Homo sapiens]
Length = 1567
Score = 77.8 bits (190), Expect = 1e-12
Identities = 58/220 (26%), Positives = 99/220 (44%), Gaps = 26/220 (11%)
Query: 508 MAVVVSGRGE---VLLINFASKVGATVTKFWTSDVTHVIAATDANGACSRTLKVLMAILN 564
M++VVSG +L+ FA K T+T T + THV+ TDA C RTLK + I
Sbjct: 1354 MSMVVSGLTPEEFMLVYKFARKHHITLTNLITEETTHVVMKTDAEFVCERTLKYFLGIAG 1413
Query: 565 GRWVLKIDWIKACMKVMSLVEEEPYEISLD--NQGCNGGPKAGRLSALANEPKLFSGLKF 622
G+WV+ W+ +K ++ E +E+ D N + GPK R S + K+F GL+
Sbjct: 1414 GKWVVSYFWVTQSIKERKMLNEHDFEVRGDVVNGRNHQGPKRARES---QDRKIFRGLEI 1470
Query: 623 YFSGDYVSSYKEDLEDLVEVGGGAVLTSKDKLEAKRHEFEVTSNLLIVYNLDPPQGSKLG 682
G + + + LE +V++ G +V+ + F + + + + + P
Sbjct: 1471 CCYGPFTNMPTDQLEWMVQLCGASVV-------KELSSFTLGTGVHPIVVVQP------- 1516
Query: 683 DEVSTLWQRLNEAEDLAANTGSQVIGHTWILESIAACKLQ 722
W N + + V+ W+L+S+A + Q
Sbjct: 1517 ----DAWTEDNGFHAIGQMCEAPVVTREWVLDSVALYQCQ 1552
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.132 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,229,135,601
Number of Sequences: 2790947
Number of extensions: 51926055
Number of successful extensions: 132868
Number of sequences better than 10.0: 2491
Number of HSP's better than 10.0 without gapping: 1491
Number of HSP's successfully gapped in prelim test: 1000
Number of HSP's that attempted gapping in prelim test: 130121
Number of HSP's gapped (non-prelim): 3341
length of query: 726
length of database: 848,049,833
effective HSP length: 135
effective length of query: 591
effective length of database: 471,271,988
effective search space: 278521744908
effective search space used: 278521744908
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 79 (35.0 bits)
Lotus: description of TM0082b.8


