

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0082b.11
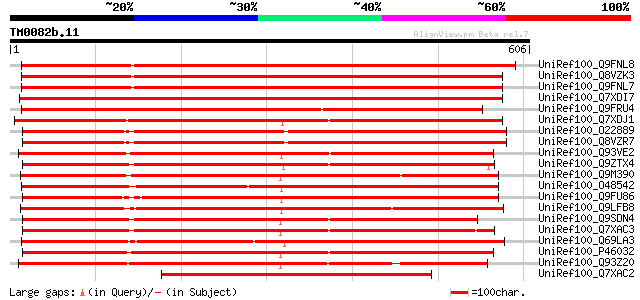
(606 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FNL8 Peptide transporter [Arabidopsis thaliana] 689 0.0
UniRef100_Q8VZK3 Putative peptide transporter protein [Arabidops... 687 0.0
UniRef100_Q9FNL7 Peptide transporter [Arabidopsis thaliana] 687 0.0
UniRef100_Q7XDI7 Putative peptide transport protein [Oryza sativa] 682 0.0
UniRef100_Q9FRU4 Putative nitrate transporter NRT1-3 [Glycine max] 673 0.0
UniRef100_Q7XDJ1 Putative peptide transport protein [Oryza sativa] 647 0.0
UniRef100_O22889 Putative PTR2 family peptide transporter [Arabi... 594 e-168
UniRef100_Q8VZR7 Putative PTR2 family peptide transporter protei... 591 e-167
UniRef100_Q93VE2 Putative peptide transporter 1 [Oryza sativa] 536 e-151
UniRef100_Q9ZTX4 LeOPT1 [Lycopersicon esculentum] 520 e-146
UniRef100_Q9M390 Putative peptide transport protein [Arabidopsis... 513 e-144
UniRef100_O48542 Peptide transporter [Hordeum vulgare] 511 e-143
UniRef100_Q9FU86 Putative peptide transport protein [Oryza sativa] 508 e-142
UniRef100_Q9LFB8 Oligopeptide transporter-like protein [Arabidop... 500 e-140
UniRef100_Q9SDN4 Amino acid/peptide transporter [Prunus dulcis] 495 e-138
UniRef100_Q7XAC3 Peptide transporter 1 [Vicia faba] 493 e-138
UniRef100_Q69LA3 Putative peptide transport protein [Oryza sativa] 476 e-132
UniRef100_P46032 Peptide transporter PTR2-B [Arabidopsis thaliana] 473 e-132
UniRef100_Q93Z20 At1g62200/F19K23_13 [Arabidopsis thaliana] 472 e-131
UniRef100_Q7XAC2 Peptide transporter 2 [Vicia faba] 469 e-131
>UniRef100_Q9FNL8 Peptide transporter [Arabidopsis thaliana]
Length = 586
Score = 689 bits (1777), Expect = 0.0
Identities = 341/579 (58%), Positives = 437/579 (74%), Gaps = 5/579 (0%)
Query: 14 EDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGT 73
+DYT+DGTVDL+G V RS+TG WKAC+F+V YE+FERMAYYGISSNLV+++ +KLH+GT
Sbjct: 8 DDYTKDGTVDLRGNRVRRSQTGRWKACSFVVVYEVFERMAYYGISSNLVIYMTTKLHQGT 67
Query: 74 VASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRP 133
V SSNNV+NW G W TP++GAY+ADA+ GRY TF+I+S IYLLGM LLTL+VSLP L+P
Sbjct: 68 VKSSNNVTNWVGTSWLTPILGAYVADAHFGRYITFVISSAIYLLGMALLTLSVSLPGLKP 127
Query: 134 PPCAQG-VENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTY 192
P C+ VEN C +AS +F+ ALY +A+G GGTKPNIST+GADQFDE++PK+ +
Sbjct: 128 PKCSTANVEN--CEKASVIQLAVFFGALYTLAIGTGGTKPNISTIGADQFDEFDPKDKIH 185
Query: 193 KLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHK 252
K SFFNWW+FSI G F+TTVLVY+QDN+ W++GYGL T+GLAFSI +FL+GT YRHK
Sbjct: 186 KHSFFNWWMFSIFFGTFFATTVLVYVQDNVGWAIGYGLSTLGLAFSIFIFLLGTRLYRHK 245
Query: 253 LPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKA 312
LP GSP T+M +V VA+ K + + D + +EL EYA+ I TS+LRFL++A
Sbjct: 246 LPMGSPFTKMARVIVASLRKAREPMSSDSTRFYELPPMEYASKRAFPIHSTSSLRFLNRA 305
Query: 313 AVKTGKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTLDRSMG 372
++KTG T WRLCT+T+VEETKQM KM+P+L T +P ML Q TLFIKQGTTLDR +
Sbjct: 306 SLKTGSTHKWRLCTITEVEETKQMLKMLPVLFVTFVPSMMLAQIMTLFIKQGTTLDRRLT 365
Query: 373 PNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLMYVLIM 432
NF IPPA L SML SI IYDRVFV +R+ T NPRGIT+LQR+G G+++++LIM
Sbjct: 366 NNFSIPPASLLGFTTFSMLVSIVIYDRVFVKFMRKLTGNPRGITLLQRMGIGMILHILIM 425
Query: 433 VIAWLTERKRLRVAREKHLLGQHDI-IPLSIFILIPQYALTGVAENFAEIAKMDFFYYQA 491
+IA +TER RL+VA E L Q + IPLSIF L+PQY L G+A+ F EIAK++FFY QA
Sbjct: 426 IIASITERYRLKVAAEHGLTHQTAVPIPLSIFTLLPQYVLMGLADAFIEIAKLEFFYDQA 485
Query: 492 PEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYYYAFMV 551
PE MKSLG SY++TS+A+G F+SS LLS+V+ ITKK +GWI +NLN SRLD YY F
Sbjct: 486 PESMKSLGTSYTSTSMAVGYFMSSILLSSVSQITKKQG-RGWIQNNLNESRLDNYYMFFA 544
Query: 552 ILSFLNFLCFLVTAKFFDYNVDVTHEKSGSELNPASLDN 590
+L+ LNF+ FLV +F++Y DVT + + P +DN
Sbjct: 545 VLNLLNFILFLVVIRFYEYRADVTQSANVEQKEPNMVDN 583
>UniRef100_Q8VZK3 Putative peptide transporter protein [Arabidopsis thaliana]
Length = 582
Score = 687 bits (1773), Expect = 0.0
Identities = 332/564 (58%), Positives = 431/564 (75%), Gaps = 5/564 (0%)
Query: 14 EDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGT 73
+DYT+DGTVDL+G PV RS G WKAC+F+V YE+FERMAYYGISSNL +++ +KLH+GT
Sbjct: 8 DDYTKDGTVDLQGNPVRRSIRGRWKACSFVVVYEVFERMAYYGISSNLFIYMTTKLHQGT 67
Query: 74 VASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRP 133
V SSNNV+NW G W TP++GAY+ DA LGRY TF+I+ IY GM +LTL+V++P ++P
Sbjct: 68 VKSSNNVTNWVGTSWLTPILGAYVGDALLGRYITFVISCAIYFSGMMVLTLSVTIPGIKP 127
Query: 134 PPCAQ-GVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTY 192
P C+ VEN C +AS +F+ ALY +A+G GGTKPNIST+GADQFD ++PKE T
Sbjct: 128 PECSTTNVEN--CEKASVLQLAVFFGALYTLAIGTGGTKPNISTIGADQFDVFDPKEKTQ 185
Query: 193 KLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHK 252
KLSFFNWW+FSI G LF+ TVLVY+QDN+ W+LGYGLPT+GLA SI +FL+GTPFYRHK
Sbjct: 186 KLSFFNWWMFSIFFGTLFANTVLVYVQDNVGWTLGYGLPTLGLAISITIFLLGTPFYRHK 245
Query: 253 LPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKA 312
LP+GSP T+M +V VA+ K A + HD HEL EY I T +LRFLD+A
Sbjct: 246 LPTGSPFTKMARVIVASFRKANAPMTHDITSFHELPSLEYERKGAFPIHPTPSLRFLDRA 305
Query: 313 AVKTGKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTLDRSMG 372
++KTG W LCT T+VEETKQM +M+P+L T +P ML Q +TLF+KQGTTLDR +
Sbjct: 306 SLKTGTNHKWNLCTTTEVEETKQMLRMLPVLFITFVPSMMLAQINTLFVKQGTTLDRKVT 365
Query: 373 PNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLMYVLIM 432
+F IPPA LS + +SML SI +YDRVFV + R++T NPRGIT+LQR+G GL+ ++LIM
Sbjct: 366 GSFSIPPASLSGFVTLSMLISIVLYDRVFVKITRKFTGNPRGITLLQRMGIGLIFHILIM 425
Query: 433 VIAWLTERKRLRVAREKHLLGQHDI-IPLSIFILIPQYALTGVAENFAEIAKMDFFYYQA 491
++A +TER RL+VA + L+ Q + +PL+IF L+PQ+ L G+A++F E+AK++FFY QA
Sbjct: 426 IVASVTERYRLKVAADHGLIHQTGVKLPLTIFALLPQFVLMGMADSFLEVAKLEFFYDQA 485
Query: 492 PEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYYYAFMV 551
PE MKSLG SYSTTS+A+G F+SSFLLSTV++ITKK +GWIL+NLN SRLDYYY F
Sbjct: 486 PESMKSLGTSYSTTSLAIGNFMSSFLLSTVSEITKKRG-RGWILNNLNESRLDYYYLFFA 544
Query: 552 ILSFLNFLCFLVTAKFFDYNVDVT 575
+L+ +NF+ FLV KF+ Y +VT
Sbjct: 545 VLNLVNFILFLVVVKFYVYRAEVT 568
>UniRef100_Q9FNL7 Peptide transporter [Arabidopsis thaliana]
Length = 582
Score = 687 bits (1772), Expect = 0.0
Identities = 332/564 (58%), Positives = 431/564 (75%), Gaps = 5/564 (0%)
Query: 14 EDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGT 73
+DYT+DGTVDL+G PV RS G WKAC+F+V YE+FERMAYYGISSNL +++ +KLH+GT
Sbjct: 8 DDYTKDGTVDLQGNPVRRSIRGRWKACSFVVVYEVFERMAYYGISSNLFIYMTTKLHQGT 67
Query: 74 VASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRP 133
V SSNNV+NW G W TP++GAY+ DA LGRY TF+I+ IY GM +LTL+V++P ++P
Sbjct: 68 VKSSNNVTNWVGTSWLTPILGAYVGDALLGRYITFVISCAIYFSGMMVLTLSVTIPGIKP 127
Query: 134 PPCAQ-GVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTY 192
P C+ VEN C +AS +F+ ALY +A+G GGTKPNIST+GADQFD ++PKE T
Sbjct: 128 PECSTTNVEN--CEKASVLQLAVFFGALYTLAIGTGGTKPNISTIGADQFDVFDPKEKTQ 185
Query: 193 KLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHK 252
KLSFFNWW+FSI G LF+ TVLVY+QDN+ W+LGYGLPT+GLA SI +FL+GTPFYRHK
Sbjct: 186 KLSFFNWWMFSIFFGTLFANTVLVYVQDNVGWTLGYGLPTLGLAISITIFLLGTPFYRHK 245
Query: 253 LPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKA 312
LP+GSP T+M +V VA+ K A + HD HEL EY I T +LRFLD+A
Sbjct: 246 LPTGSPFTKMARVIVASFRKANAPMTHDITSFHELPSLEYERKGAFPIHPTPSLRFLDRA 305
Query: 313 AVKTGKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTLDRSMG 372
++KTG W LCT T+VEETKQM +M+P+L T +P ML Q +TLF+KQGTTLDR +
Sbjct: 306 SLKTGTNHKWNLCTTTEVEETKQMLRMLPVLFITFVPSMMLAQINTLFVKQGTTLDRKVT 365
Query: 373 PNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLMYVLIM 432
+F IPPA LS + +SML SI +YDRVFV + R++T NPRGIT+LQR+G GL+ ++LIM
Sbjct: 366 GSFSIPPASLSGFVTLSMLISIVLYDRVFVKITRKFTGNPRGITLLQRMGIGLIFHILIM 425
Query: 433 VIAWLTERKRLRVAREKHLLGQHDI-IPLSIFILIPQYALTGVAENFAEIAKMDFFYYQA 491
++A +TER RL+VA + L+ Q + +PL+IF L+PQ+ L G+A++F E+AK++FFY QA
Sbjct: 426 IVASVTERYRLKVAADHGLIHQTGVKLPLTIFALLPQFVLMGMADSFLEVAKLEFFYDQA 485
Query: 492 PEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYYYAFMV 551
PE MKSLG SYSTTS+A+G F+SSFLLSTV++ITKK +GWIL+NLN SRLDYYY F
Sbjct: 486 PESMKSLGTSYSTTSLAIGNFMSSFLLSTVSEITKKRG-RGWILNNLNESRLDYYYLFFA 544
Query: 552 ILSFLNFLCFLVTAKFFDYNVDVT 575
+L+ +NF+ FLV KF+ Y +VT
Sbjct: 545 VLNLVNFVLFLVVVKFYVYRAEVT 568
>UniRef100_Q7XDI7 Putative peptide transport protein [Oryza sativa]
Length = 608
Score = 682 bits (1760), Expect = 0.0
Identities = 336/568 (59%), Positives = 429/568 (75%), Gaps = 4/568 (0%)
Query: 12 GKEDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHE 71
G +DYTQDGTVDL G PVLRSK G WKAC F+V YE+FERMAYYGISSNLV++L +KLH+
Sbjct: 12 GDDDYTQDGTVDLHGNPVLRSKRGGWKACGFVVVYEVFERMAYYGISSNLVLYLTTKLHQ 71
Query: 72 GTVASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPAL 131
GTV+S+NNV+NW G +W TP++GAYIADA+LGRY TF+IAS IYL+GM LLTL VS+P+L
Sbjct: 72 GTVSSANNVTNWVGTIWMTPILGAYIADAHLGRYRTFMIASLIYLIGMSLLTLAVSVPSL 131
Query: 132 RPPPCAQGVENQDCPQ-ASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKEN 190
+PP C G + C + AS G+F+LALYI+A+G GGTKPNIST+GADQFD++ P+E
Sbjct: 132 KPPKCGAGTADPGCSEKASSLQLGVFFLALYILAVGTGGTKPNISTIGADQFDDHHPRER 191
Query: 191 TYKLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYR 250
+KLSFFNWW+FSI G LF+ TVLVY+QDN+ W++GY LPT+GLA SI +F GTPFYR
Sbjct: 192 RHKLSFFNWWMFSIFFGTLFANTVLVYLQDNVGWTVGYALPTLGLAVSIAIFTAGTPFYR 251
Query: 251 HKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLD 310
HK SGS RM +V VAA K +P D ++LHEL E YA + +T L+ L
Sbjct: 252 HKPTSGSSFARMARVIVAAIRKLAVALPDDARELHELDDEYYAKKKTTPLPYTPYLKILS 311
Query: 311 KAAVKTGKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTLDRS 370
KAAVKT TS W L TVTQVEETKQ+ KM+P+L T +P M+ Q +TLF+KQGTTLDR
Sbjct: 312 KAAVKTSTTSRWSLSTVTQVEETKQILKMLPVLAVTFVPAAMMAQVNTLFVKQGTTLDRR 371
Query: 371 M-GPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLMYV 429
+ G FEIPPA L A + ISML S+ +YDRVF+P++ R T NPRGIT+LQR+G GL++++
Sbjct: 372 VGGGGFEIPPASLQAFVTISMLVSVVLYDRVFMPLMARATGNPRGITLLQRMGVGLVIHI 431
Query: 430 LIMVIAWLTERKRLRVAREKHLL-GQHDIIPLSIFILIPQYALTGVAENFAEIAKMDFFY 488
IM IA +TER RL VARE + + IPL+IF+L+PQ+ L GVA+ F E+AK++FFY
Sbjct: 432 AIMGIASVTERHRLAVAREHGIADSKGTTIPLTIFVLLPQFVLMGVADAFLEVAKIEFFY 491
Query: 489 YQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKH-SHQGWILDNLNISRLDYYY 547
QAPEGMKSLG SY+ TS+ +G FLSS LLSTVA +T++H GWI +NLN SRLD+YY
Sbjct: 492 DQAPEGMKSLGTSYAMTSLGVGNFLSSLLLSTVAHVTRRHGGGGGWIQNNLNASRLDHYY 551
Query: 548 AFMVILSFLNFLCFLVTAKFFDYNVDVT 575
AF +L+ +N + F + + + YN +V+
Sbjct: 552 AFFAVLNCVNLVFFFLVCRLYVYNAEVS 579
>UniRef100_Q9FRU4 Putative nitrate transporter NRT1-3 [Glycine max]
Length = 574
Score = 673 bits (1736), Expect = 0.0
Identities = 326/540 (60%), Positives = 419/540 (77%), Gaps = 3/540 (0%)
Query: 14 EDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGT 73
EDYTQDGTV++KG+P+LRSK+G WKAC+F+V YE+FERMAYYGISSNL+++L +KLH+GT
Sbjct: 11 EDYTQDGTVNIKGKPILRSKSGGWKACSFVVVYEVFERMAYYGISSNLILYLTTKLHQGT 70
Query: 74 VASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRP 133
V+S+NNV+NW G +W TP++GAYIADA+LGRYWTF+IAS +YL GM LLTL VSLP+L+P
Sbjct: 71 VSSANNVTNWVGTIWMTPILGAYIADAFLGRYWTFVIASTVYLSGMSLLTLAVSLPSLKP 130
Query: 134 PPCAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTYK 193
P C + + C +AS +FY ALY +A+G GGTKPNIST+GADQFD++ PKE +K
Sbjct: 131 PQCFEK-DVTKCAKASTLQLAVFYGALYTLAVGTGGTKPNISTIGADQFDDFHPKEKLHK 189
Query: 194 LSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHKL 253
LSFFNWW+FSI G LF+ +VLVYIQDN+ W+LGY LPT+GL SI++F+ GTPFY+ +
Sbjct: 190 LSFFNWWMFSIFFGTLFANSVLVYIQDNVGWTLGYALPTLGLLVSIMIFVAGTPFYQAQS 249
Query: 254 PSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKAA 313
+GS TRM +V VAA K K VP D K+L+EL E YA RIDHT TL+FLDKA
Sbjct: 250 AAGSTFTRMARVIVAACRKSKVPVPSDSKELYELDKEGYAKKGSYRIDHTPTLKFLDKAC 309
Query: 314 VKT-GKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTLDRSMG 372
VKT TSPW LCTVTQV ETKQM +M+PIL+ T +P TM+ Q +TLF+KQG R
Sbjct: 310 VKTDSNTSPWMLCTVTQVGETKQMIRMIPILVATFVPSTMMAQINTLFVKQG-YYSRQTP 368
Query: 373 PNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLMYVLIM 432
+IP L+A + +S+L I +YDR FV +++R+TKNPRGIT+LQR+G GL+++ LIM
Sbjct: 369 WKLQIPQQSLAAFVTVSLLVCIVLYDRFFVKIMQRFTKNPRGITLLQRMGIGLVIHTLIM 428
Query: 433 VIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAKMDFFYYQAP 492
+IA TE RL+VARE ++ +PLSIFIL+PQ+ L G A+ F E+AK++FFY Q+P
Sbjct: 429 IIASGTESYRLKVAREHGVVESGGQVPLSIFILLPQFILMGTADAFLEVAKIEFFYDQSP 488
Query: 493 EGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYYYAFMVI 552
E MKS+G SYSTT++ LG F+SSFLLSTV++ITKK+ H+GWIL+NLN S LDYYYAF I
Sbjct: 489 EHMKSIGTSYSTTTLGLGNFISSFLLSTVSNITKKNGHKGWILNNLNESHLDYYYAFFAI 548
>UniRef100_Q7XDJ1 Putative peptide transport protein [Oryza sativa]
Length = 607
Score = 647 bits (1670), Expect = 0.0
Identities = 316/579 (54%), Positives = 417/579 (71%), Gaps = 11/579 (1%)
Query: 6 EKGLPSGKEDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFL 65
E G +G ++YT+DG+VDL+G PVLRSK G WKAC+FIV YELFERMAYYGI+SNLV++L
Sbjct: 2 ENGAGAGDDEYTRDGSVDLRGNPVLRSKRGGWKACSFIVVYELFERMAYYGIASNLVIYL 61
Query: 66 GSKLHEGTVASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLT 125
KLH+GTV ++NNV+NW+G V+ TPL+GA +ADA+LGRYWTF+ S +YL+GM LLTL
Sbjct: 62 TEKLHQGTVEAANNVTNWSGTVFITPLIGAVVADAWLGRYWTFVAGSAVYLMGMLLLTLA 121
Query: 126 VSLPALRPPPCAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEY 185
VS+PAL+PPPC G CP+AS G+++ LY IALG GGTKPNIST+GADQFD++
Sbjct: 122 VSVPALKPPPC-DGGGGAACPRASALQLGVYFGGLYTIALGHGGTKPNISTIGADQFDDF 180
Query: 186 EPKENTYKLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVG 245
P E +KLSFFNWW+F+I +G+LFSTTVLVY+QDN+SW++GYG+PT+GL S+ VFL G
Sbjct: 181 HPPEKLHKLSFFNWWMFTIFLGILFSTTVLVYLQDNVSWTVGYGIPTLGLMVSVAVFLSG 240
Query: 246 TPFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTST 305
TP YRHK+P GSPL M +V AA KW+ +P D K+LHEL +E Y R+D T +
Sbjct: 241 TPLYRHKVPQGSPLATMGRVVAAAVWKWRVPLPADSKELHELELEHYTTRRGFRMDATVS 300
Query: 306 LRFLDKAAVKTG--------KTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAH 357
+ FL+KAAVK G + W LCTVTQVEETKQ+ K+VP+L T ++PCT++ QA
Sbjct: 301 MAFLNKAAVKPGEGGGGSVARLPGWTLCTVTQVEETKQIVKLVPLLATMVVPCTLVAQAG 360
Query: 358 TLFIKQGTTLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITM 417
TLF+KQG TLDR +G F +PPA L A + +ML I +YDR VP +RR TKNPRGIT+
Sbjct: 361 TLFVKQGVTLDRRIG-KFHVPPASLGAFVTATMLICIVLYDRFLVPAVRRRTKNPRGITL 419
Query: 418 LQRLGAGLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAEN 477
LQR+ G+L+ ++ MV+ + E +RL AR L+ +P++IFIL+PQ+ L GVA+
Sbjct: 420 LQRISLGMLLQIVTMVVTSVVESQRLGYARRHGLVATGGQLPVTIFILLPQFVLLGVADA 479
Query: 478 FAEIAKMDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADIT-KKHSHQGWILD 536
F + +++FFY QAPE MKSLG + S T+ G LSS +L+ V +T W+ +
Sbjct: 480 FLVVGQIEFFYDQAPESMKSLGTAMSLTAYGAGNLLSSAILAAVERVTGGGKGRTPWVTN 539
Query: 537 NLNISRLDYYYAFMVILSFLNFLCFLVTAKFFDYNVDVT 575
NLN SRLDYYYAF+ L+ N L F+V + + Y V+ T
Sbjct: 540 NLNASRLDYYYAFLATLAAANLLAFVVLSCKYSYRVEST 578
>UniRef100_O22889 Putative PTR2 family peptide transporter [Arabidopsis thaliana]
Length = 583
Score = 594 bits (1531), Expect = e-168
Identities = 296/567 (52%), Positives = 401/567 (70%), Gaps = 10/567 (1%)
Query: 16 YTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGTVA 75
YTQDGTVDL+GRPVL SKTG W+AC+F++GYE FERMA+YGI+SNLV +L +LHE T++
Sbjct: 7 YTQDGTVDLQGRPVLASKTGRWRACSFLLGYEAFERMAFYGIASNLVNYLTKRLHEDTIS 66
Query: 76 SSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRPPP 135
S NV+NW+G VW TP+ GAYIAD+Y+GR+WTF +S IY+LGM LLT+ V++ +LRP
Sbjct: 67 SVRNVNNWSGAVWITPIAGAYIADSYIGRFWTFTASSLIYVLGMILLTMAVTVKSLRPT- 125
Query: 136 CAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTYKLS 195
C GV C +AS FY++LY IA+GAGGTKPNIST GADQFD Y +E K+S
Sbjct: 126 CENGV----CNKASSLQVTFFYISLYTIAIGAGGTKPNISTFGADQFDSYSIEEKKQKVS 181
Query: 196 FFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHKLPS 255
FFNWW+FS +G LF+T LVYIQ+NL W LGYG+PTVGL S++VF +GTPFYRHK+
Sbjct: 182 FFNWWMFSSFLGALFATLGLVYIQENLGWGLGYGIPTVGLLVSLVVFYIGTPFYRHKVIK 241
Query: 256 GSPLTR-MLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKAAV 314
L + ++QV +AA K P D +L+EL Y +N ++++ HT RFLDKAA+
Sbjct: 242 TDNLAKDLVQVPIAAFKNRKLQCPDDHLELYELDSHYYKSNGKHQVHHTPVFRFLDKAAI 301
Query: 315 KTGKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTLDRSMGPN 374
KT P CTVT+VE K++ ++ I + TLIP T+ Q +TLF+KQGTTLDR +G N
Sbjct: 302 KTSSRVP---CTVTKVEVAKRVLGLIFIWLVTLIPSTLWAQVNTLFVKQGTTLDRKIGSN 358
Query: 375 FEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLMYVLIMVI 434
F+IP A L + + +SML S+P+YD+ FVP +R+ T NPRGIT+LQRLG G + ++ + I
Sbjct: 359 FQIPAASLGSFVTLSMLLSVPMYDQSFVPFMRKKTGNPRGITLLQRLGVGFAIQIVAIAI 418
Query: 435 AWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAKMDFFYYQAPEG 494
A E KR+RV +E H+ ++P+SIF L+PQY+L G+ + F I ++FFY Q+PE
Sbjct: 419 ASAVEVKRMRVIKEFHITSPTQVVPMSIFWLLPQYSLLGIGDVFNAIGLLEFFYDQSPEE 478
Query: 495 MKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYYYAFMVILS 554
M+SLG ++ T+ + LG FL+SFL++ + IT K + WI +NLN SRLDYYY F+V++S
Sbjct: 479 MQSLGTTFFTSGIGLGNFLNSFLVTMIDKITSKGGGKSWIGNNLNDSRLDYYYGFLVVIS 538
Query: 555 FLNFLCFLVTAKFFDY-NVDVTHEKSG 580
+N F+ A + Y + D T E SG
Sbjct: 539 IVNMGLFVWAASKYVYKSDDDTKEFSG 565
>UniRef100_Q8VZR7 Putative PTR2 family peptide transporter protein [Arabidopsis
thaliana]
Length = 583
Score = 591 bits (1524), Expect = e-167
Identities = 295/567 (52%), Positives = 400/567 (70%), Gaps = 10/567 (1%)
Query: 16 YTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGTVA 75
YTQDGTVDL+GRPVL SKTG W+AC+F++GYE FERMA+YGI+SNLV +L +LHE T++
Sbjct: 7 YTQDGTVDLQGRPVLASKTGRWRACSFLLGYEAFERMAFYGIASNLVNYLTKRLHEDTIS 66
Query: 76 SSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRPPP 135
S NV+NW+G VW TP+ GAYIAD+Y+GR+WTF +S IY+LGM LLT+ V++ +LRP
Sbjct: 67 SVRNVNNWSGAVWITPIAGAYIADSYIGRFWTFTASSLIYVLGMILLTMAVTVKSLRPT- 125
Query: 136 CAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTYKLS 195
C GV C +AS FY++LY IA+GAGGTKPNIST GADQFD Y +E K+S
Sbjct: 126 CENGV----CNKASSLQVTFFYISLYTIAIGAGGTKPNISTFGADQFDSYSIEEKKQKVS 181
Query: 196 FFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHKLPS 255
FFNWW+FS +G LF+T LVYIQ+NL W LGYG+PTVGL S++VF +GTPFYRHK+
Sbjct: 182 FFNWWMFSSFLGALFATLGLVYIQENLGWGLGYGIPTVGLLVSLVVFYIGTPFYRHKVIK 241
Query: 256 GSPLTR-MLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKAAV 314
L + ++QV +AA K P D +L+EL Y +N ++++ HT RFLDKAA+
Sbjct: 242 TDNLAKDLVQVPIAAFKNRKLQCPDDHLELYELDSHYYKSNGKHQVHHTPVFRFLDKAAI 301
Query: 315 KTGKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTLDRSMGPN 374
KT P CTVT+VE K++ ++ I + TLIP T+ Q +TLF+KQGTTLDR +G N
Sbjct: 302 KTSSRVP---CTVTKVEVAKRVLGLIFIWLVTLIPSTLWAQVNTLFVKQGTTLDRKIGSN 358
Query: 375 FEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLMYVLIMVI 434
F+IP A L + + +SML S+P+YD+ FVP +R+ T NPRGIT+LQRLG G + ++ + I
Sbjct: 359 FQIPAASLGSFVTLSMLLSVPMYDQSFVPFMRKKTGNPRGITLLQRLGVGFAIQIVAIAI 418
Query: 435 AWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAKMDFFYYQAPEG 494
A E KR+RV +E H+ ++P+SIF L+PQY+L G+ + F I ++FFY Q+P
Sbjct: 419 ASAVEVKRMRVIKEFHITSPTQVVPMSIFWLLPQYSLLGIGDVFNAIGLLEFFYDQSPGE 478
Query: 495 MKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYYYAFMVILS 554
M+SLG ++ T+ + LG FL+SFL++ + IT K + WI +NLN SRLDYYY F+V++S
Sbjct: 479 MQSLGTTFFTSGIGLGNFLNSFLVTMIDKITSKGGGKSWIGNNLNDSRLDYYYGFLVVIS 538
Query: 555 FLNFLCFLVTAKFFDY-NVDVTHEKSG 580
+N F+ A + Y + D T E SG
Sbjct: 539 IVNMGLFVWAASKYVYKSDDDTKEFSG 565
>UniRef100_Q93VE2 Putative peptide transporter 1 [Oryza sativa]
Length = 593
Score = 536 bits (1381), Expect = e-151
Identities = 262/562 (46%), Positives = 371/562 (65%), Gaps = 12/562 (2%)
Query: 11 SGKEDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLH 70
S + YT DG+VD G PV++ +TG W+AC FI+G E ER+AYYGIS+NLV +L KLH
Sbjct: 28 SNQLTYTGDGSVDFSGNPVVKERTGRWRACPFILGNECCERLAYYGISTNLVTYLTKKLH 87
Query: 71 EGTVASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPA 130
+G ++++NV+ W G + TPL+GA +ADAY GRYWT S IY +GM +LTL+ S+P
Sbjct: 88 DGNASAASNVTAWQGTCYLTPLIGAILADAYWGRYWTIATFSTIYFIGMAVLTLSASVPT 147
Query: 131 LRPPPCAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKEN 190
PPPC E CP A+P +F+L LY+IALG GG KP +S+ GADQFD+ +P E
Sbjct: 148 FMPPPC----EGSFCPPANPLQYTVFFLGLYLIALGTGGIKPCVSSFGADQFDDTDPVER 203
Query: 191 TYKLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYR 250
K SFFNW+ FSI +G L S++ LV++QDN+ W +G+G+PT+ + +I+ F GT YR
Sbjct: 204 IQKGSFFNWFYFSINIGALISSSFLVWVQDNIGWGIGFGIPTIFMGLAIISFFSGTSLYR 263
Query: 251 HKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLD 310
+ P GSP+TR+ QV VA+ KW HVP D +L+EL A +++HT LR LD
Sbjct: 264 FQKPGGSPITRVCQVVVASFRKWNVHVPEDSSRLYELPDGASAIEGSRQLEHTDELRCLD 323
Query: 311 KAAVKT-------GKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQ 363
KAA T T+PWR+CTVTQVEE K + +M P+ TT++ + Q T+F++Q
Sbjct: 324 KAATITDLDVKADSFTNPWRICTVTQVEELKILVRMFPVWATTIVFSAVYAQMSTMFVEQ 383
Query: 364 GTTLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGA 423
G LD S+GP F+IPPA LS ++S++ +P+YD + VP+ RR+T NPRG T LQR+G
Sbjct: 384 GMMLDTSVGP-FKIPPASLSTFDVVSVIIWVPLYDSILVPIARRFTGNPRGFTELQRMGI 442
Query: 424 GLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAK 483
GL++ + M A + E KRL +AR +HL+ Q+ +PL+I IPQY L G +E F +
Sbjct: 443 GLVISIFSMAAAAVLEIKRLDIARAEHLVDQNVPVPLNICWQIPQYFLVGASEVFTFVGS 502
Query: 484 MDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRL 543
++FFY Q+P+ M+SL + + ALG +LS+F+L+ VA T + + GWI DNLN L
Sbjct: 503 LEFFYDQSPDAMRSLCSALQLVTTALGNYLSAFILTLVAYFTTRGGNPGWIPDNLNQGHL 562
Query: 544 DYYYAFMVILSFLNFLCFLVTA 565
DY++ + LSFLNF+ +++ A
Sbjct: 563 DYFFWLLAGLSFLNFVIYVICA 584
>UniRef100_Q9ZTX4 LeOPT1 [Lycopersicon esculentum]
Length = 580
Score = 520 bits (1340), Expect = e-146
Identities = 265/563 (47%), Positives = 366/563 (64%), Gaps = 17/563 (3%)
Query: 16 YTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGTVA 75
YT+DG+VD+KG PVL+S+TG+W+AC FI+G E ER+AYYGI++NLV +L KLHEG V+
Sbjct: 20 YTRDGSVDIKGNPVLKSETGNWRACPFILGNECCERLAYYGIAANLVTYLTKKLHEGNVS 79
Query: 76 SSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRPPP 135
++ NV+ W G + TPL+GA +ADAY GRYWT S IY +GMC LTL+ S+PA +PP
Sbjct: 80 AARNVTTWQGTCYITPLIGAVLADAYWGRYWTIATFSTIYFIGMCTLTLSASVPAFKPPQ 139
Query: 136 CAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTYKLS 195
C V CP ASP IF+ LY+IALG GG KP +S+ GADQFD+ +PKE K S
Sbjct: 140 CVGSV----CPSASPAQYAIFFFGLYLIALGTGGIKPCVSSFGADQFDDTDPKERVKKGS 195
Query: 196 FFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHKLPS 255
FFNW+ FSI +G L S++++V+IQ+N W LG+G+P V + +I F GTP YR + P
Sbjct: 196 FFNWFYFSINIGALISSSLIVWIQENAGWGLGFGIPAVFMGIAIASFFFGTPLYRFQKPG 255
Query: 256 GSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKAAVK 315
GSPLTRM QV VA KW VP D L+E + A ++ HT LR LDKAAV
Sbjct: 256 GSPLTRMCQVLVAVFHKWNLSVPDDSTLLYETPDKSSAIEGSRKLLHTDELRCLDKAAVV 315
Query: 316 TGK-------TSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTLD 368
+ ++ WRLCTVTQVEE K + +M PI T ++ + Q T+F++QG +D
Sbjct: 316 SDNELTTGDYSNAWRLCTVTQVEELKILIRMFPIWATGIVFSAVYAQMSTMFVEQGMVMD 375
Query: 369 RSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLMY 428
++G +F+IP A LS IS++ +P+YD++ VP+ RR+T RG + LQR+G GL +
Sbjct: 376 TAVG-SFKIPAASLSTFDTISVIVWVPVYDKILVPIARRFTGIERGFSELQRMGIGLFLS 434
Query: 429 VLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAKMDFFY 488
+L M A + E +RL++AR+ L+ + +PLSIF IPQY + G AE F I +++FFY
Sbjct: 435 MLCMSAAAIVEIRRLQLARDLGLVDEAVSVPLSIFWQIPQYFILGAAEIFTFIGQLEFFY 494
Query: 489 YQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYYYA 548
Q+P+ M+SL + S + ALG +LSSF+L+ V IT + GWI +NLN LDY++
Sbjct: 495 DQSPDAMRSLCSALSLLTTALGNYLSSFILTVVTSITTRGGKPGWIPNNLNGGHLDYFFW 554
Query: 549 FMVILSFLN-----FLCFLVTAK 566
+ LSF N FLC + +K
Sbjct: 555 LLAALSFFNLVIYVFLCQMYKSK 577
>UniRef100_Q9M390 Putative peptide transport protein [Arabidopsis thaliana]
Length = 570
Score = 513 bits (1321), Expect = e-144
Identities = 265/565 (46%), Positives = 363/565 (63%), Gaps = 13/565 (2%)
Query: 13 KEDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEG 72
K+ YTQDGTVD+ P + KTG+WKAC FI+G E ER+AYYG+ +NLV +L S+L++G
Sbjct: 4 KDVYTQDGTVDIHKNPANKEKTGNWKACRFILGNECCERLAYYGMGTNLVNYLESRLNQG 63
Query: 73 TVASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALR 132
++NNV+NW+G + TPL+GA+IADAYLGRYWT IY+ GM LLTL+ S+P L+
Sbjct: 64 NATAANNVTNWSGTCYITPLIGAFIADAYLGRYWTIATFVFIYVSGMTLLTLSASVPGLK 123
Query: 133 PPPCAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTY 192
P C N D + +F++ALY+IALG GG KP +S+ GADQFDE + E
Sbjct: 124 PGNC-----NADTCHPNSSQTAVFFVALYMIALGTGGIKPCVSSFGADQFDENDENEKIK 178
Query: 193 KLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHK 252
K SFFNW+ FSI VG L + TVLV+IQ N+ W G+G+PTV + ++ F G+ FYR +
Sbjct: 179 KSSFFNWFYFSINVGALIAATVLVWIQMNVGWGWGFGVPTVAMVIAVCFFFFGSRFYRLQ 238
Query: 253 LPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKA 312
P GSPLTR+ QV VAA K VP D L E + +E ++ HT L+F DKA
Sbjct: 239 RPGGSPLTRIFQVIVAAFRKISVKVPEDKSLLFETADDESNIKGSRKLVHTDNLKFFDKA 298
Query: 313 AV-------KTGKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGT 365
AV K G+ +PWRLC+VTQVEE K + ++P+ T ++ T+ Q T+F+ QG
Sbjct: 299 AVESQSDSIKDGEVNPWRLCSVTQVEELKSIITLLPVWATGIVFATVYSQMSTMFVLQGN 358
Query: 366 TLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGL 425
T+D+ MG NFEIP A LS +S+L P+YD+ +P+ R++T+N RG T LQR+G GL
Sbjct: 359 TMDQHMGKNFEIPSASLSLFDTVSVLFWTPVYDQFIIPLARKFTRNERGFTQLQRMGIGL 418
Query: 426 LMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAKMD 485
++ + M+ A + E RL + + Q I +SIF IPQY L G AE F I +++
Sbjct: 419 VVSIFAMITAGVLEVVRLDYVKTHNAYDQKQ-IHMSIFWQIPQYLLIGCAEVFTFIGQLE 477
Query: 486 FFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDY 545
FFY QAP+ M+SL + S T+VALG +LS+ L++ V ITKK+ GWI DNLN LDY
Sbjct: 478 FFYDQAPDAMRSLCSALSLTTVALGNYLSTVLVTVVMKITKKNGKPGWIPDNLNRGHLDY 537
Query: 546 YYAFMVILSFLNFLCFLVTAKFFDY 570
++ + LSFLNFL +L +K + Y
Sbjct: 538 FFYLLATLSFLNFLVYLWISKRYKY 562
>UniRef100_O48542 Peptide transporter [Hordeum vulgare]
Length = 579
Score = 511 bits (1316), Expect = e-143
Identities = 256/563 (45%), Positives = 367/563 (64%), Gaps = 13/563 (2%)
Query: 14 EDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGT 73
E YTQDGTVD+KG P L+ TG+W+AC +I+ E ER+AYYG+S+NLV F+ ++
Sbjct: 7 EMYTQDGTVDIKGNPALKKDTGNWRACPYILANECCERLAYYGMSTNLVNFMKDRMGMAN 66
Query: 74 VASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRP 133
A++NNV+NW G + TPL+GA++ADAYLGR+WT IY+ G+ LLT+ S+ L P
Sbjct: 67 AAAANNVTNWGGTCYITPLIGAFLADAYLGRFWTIASFMIIYIFGLGLLTMATSVHGLVP 126
Query: 134 PPCAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTYK 193
++GV C +P ++ALY+IALG GG KP +S+ GADQFDE++ E K
Sbjct: 127 ACASKGV----C-DPTPGQSAAVFIALYLIALGTGGIKPCVSSFGADQFDEHDDVERKSK 181
Query: 194 LSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHKL 253
SFFNW+ FSI +G L +++VLVY+Q ++ WS G+G+P V +A ++ F VGT YRH+
Sbjct: 182 SSFFNWFYFSINIGALVASSVLVYVQTHVGWSWGFGIPAVVMAIAVGSFFVGTSLYRHQR 241
Query: 254 PSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKAA 313
P GSPLTR+ QV VAA K V D L+E + +E +++HT RFLDKAA
Sbjct: 242 PGGSPLTRIAQVLVAATRKLGVAV--DGSALYETADKESGIEGSRKLEHTRQFRFLDKAA 299
Query: 314 VKT------GKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTL 367
V+T SPWRLCTVTQVEE K + +++PI + ++ T+ Q T+F+ QG TL
Sbjct: 300 VETHADRTAAAPSPWRLCTVTQVEELKSVVRLLPIWASGIVFATVYGQMSTMFVLQGNTL 359
Query: 368 DRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLM 427
D SMGP F+IP A LS +S++ +P+YDR+ VP +R T PRG T LQR+G GL++
Sbjct: 360 DASMGPKFKIPSASLSIFDTLSVIAWVPVYDRILVPAVRSVTGRPRGFTQLQRMGIGLVV 419
Query: 428 YVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAKMDFF 487
+ M+ A + E RLR + L G+ D++P+SIF +PQY + G AE F + +++FF
Sbjct: 420 SMFAMLAAGVLELVRLRTIAQHGLYGEKDVVPISIFWQVPQYFIIGCAEVFTFVGQLEFF 479
Query: 488 YYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYYY 547
Y QAP+ M+S+ + S T+VALG +LS+ L++ VA +T + QGWI DNLN+ LDY++
Sbjct: 480 YDQAPDAMRSMCSALSLTTVALGNYLSTLLVTVVAKVTTRGGKQGWIPDNLNVGHLDYFF 539
Query: 548 AFMVILSFLNFLCFLVTAKFFDY 570
+ LS +NF +L+ A ++ Y
Sbjct: 540 WLLAALSLVNFAVYLLIASWYTY 562
>UniRef100_Q9FU86 Putative peptide transport protein [Oryza sativa]
Length = 580
Score = 508 bits (1307), Expect = e-142
Identities = 252/561 (44%), Positives = 362/561 (63%), Gaps = 12/561 (2%)
Query: 16 YTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGTVA 75
YTQDGTVD+KG P + TG+W+AC +I+ E ER+AYYG+S+NLV ++ ++L + +
Sbjct: 9 YTQDGTVDVKGNPATKKNTGNWRACPYILANECCERLAYYGMSTNLVNYMKTRLGQESAI 68
Query: 76 SSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRPPP 135
++NNV+NW+G + TPL+GA++ADAY+GR+WT IY+LG+ LLT+ S+ L P
Sbjct: 69 AANNVTNWSGTCYITPLLGAFLADAYMGRFWTIASFMIIYILGLALLTMASSVKGL-VPA 127
Query: 136 CAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTYKLS 195
C G C T G+ +LALY+IALG GG KP +S+ GADQFDE + E K S
Sbjct: 128 CDGGA----CHPTEAQT-GVVFLALYLIALGTGGIKPCVSSFGADQFDENDEGEKRSKSS 182
Query: 196 FFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHKLPS 255
FFNW+ FSI +G L +++VLVY+Q ++ W G+G+P V +A ++ F VGTP YRH+ P
Sbjct: 183 FFNWFYFSINIGALVASSVLVYVQTHVGWGWGFGIPAVVMAVAVASFFVGTPLYRHQRPG 242
Query: 256 GSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKAAVK 315
GSPLTR+ QV VA+ KW VP D +LHE E +++HT LD+AAV+
Sbjct: 243 GSPLTRIAQVLVASARKWGVEVPADGSRLHETLDRESGIEGSRKLEHTGQFACLDRAAVE 302
Query: 316 T------GKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTLDR 369
T S WRLCTVTQVEE K + +++PI + ++ T+ Q T+F+ QG TLD
Sbjct: 303 TPEDRSAANASAWRLCTVTQVEELKSVVRLLPIWASGIVFATVYGQMSTMFVLQGNTLDA 362
Query: 370 SMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLMYV 429
SMGP+F IP A LS +S++ +P+YDR+ VP +R T PRG T LQR+G GL++ V
Sbjct: 363 SMGPHFSIPAASLSIFDTLSVIVWVPVYDRLIVPAVRAVTGRPRGFTQLQRMGIGLVISV 422
Query: 430 LIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAKMDFFYY 489
M+ A + + RLR L G D++P+SIF +PQY + G AE F + +++FFY
Sbjct: 423 FSMLAAGVLDVVRLRAIARHGLYGDKDVVPISIFWQVPQYFIIGAAEVFTFVGQLEFFYD 482
Query: 490 QAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYYYAF 549
QAP+ M+S+ + S T+VALG +LS+ L++ V +T ++ GWI DNLN LDY++
Sbjct: 483 QAPDAMRSMCSALSLTTVALGNYLSTLLVTIVTHVTTRNGAVGWIPDNLNRGHLDYFFWL 542
Query: 550 MVILSFLNFLCFLVTAKFFDY 570
+ +LS +NF +LV A ++ Y
Sbjct: 543 LAVLSLINFGVYLVIASWYTY 563
>UniRef100_Q9LFB8 Oligopeptide transporter-like protein [Arabidopsis thaliana]
Length = 570
Score = 500 bits (1288), Expect = e-140
Identities = 258/571 (45%), Positives = 368/571 (64%), Gaps = 14/571 (2%)
Query: 13 KEDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEG 72
K+ YT+DGT+D+ +P ++KTG+WKAC FI+G E ER+AYYG+S+NL+ +L +++
Sbjct: 5 KDIYTKDGTLDIHKKPANKNKTGTWKACRFILGTECCERLAYYGMSTNLINYLEKQMNME 64
Query: 73 TVASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALR 132
V++S +VSNW+G + TPL+GA+IADAYLGRYWT IY+ GM LLT++ S+P L
Sbjct: 65 NVSASKSVSNWSGTCYATPLIGAFIADAYLGRYWTIASFVVIYIAGMTLLTISASVPGLT 124
Query: 133 PPPCAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTY 192
P + C A+ I ++ALY+IALG GG KP +S+ GADQFD+ + KE
Sbjct: 125 PT-----CSGETC-HATAGQTAITFIALYLIALGTGGIKPCVSSFGADQFDDTDEKEKES 178
Query: 193 KLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHK 252
K SFFNW+ F I VG + +++VLV+IQ N+ W G G+PTV +A +++ F G+ FYR +
Sbjct: 179 KSSFFNWFYFVINVGAMIASSVLVWIQMNVGWGWGLGVPTVAMAIAVVFFFAGSNFYRLQ 238
Query: 253 LPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKA 312
P GSPLTRMLQV VA+ K K +P D L+E E + +++HT L F DKA
Sbjct: 239 KPGGSPLTRMLQVIVASCRKSKVKIPEDESLLYENQDAESSIIGSRKLEHTKILTFFDKA 298
Query: 313 AVKT-------GKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGT 365
AV+T K+S W+LCTVTQVEE K + +++PI T ++ ++ Q T+F+ QG
Sbjct: 299 AVETESDNKGAAKSSSWKLCTVTQVEELKALIRLLPIWATGIVFASVYSQMGTVFVLQGN 358
Query: 366 TLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGL 425
TLD+ MGPNF+IP A LS +S+L P+YD++ VP R+YT + RG T LQR+G GL
Sbjct: 359 TLDQHMGPNFKIPSASLSLFDTLSVLFWAPVYDKLIVPFARKYTGHERGFTQLQRIGIGL 418
Query: 426 LMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAKMD 485
++ + MV A + E RL + H L + IP++IF +PQY L G AE F I +++
Sbjct: 419 VISIFSMVSAGILEVARLNYV-QTHNLYNEETIPMTIFWQVPQYFLVGCAEVFTFIGQLE 477
Query: 486 FFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDY 545
FFY QAP+ M+SL + S T++A G +LS+FL++ V +T+ GWI NLN LDY
Sbjct: 478 FFYDQAPDAMRSLCSALSLTAIAFGNYLSTFLVTLVTKVTRSGGRPGWIAKNLNNGHLDY 537
Query: 546 YYAFMVILSFLNFLCFLVTAKFFDYNVDVTH 576
++ + LSFLNFL +L AK++ Y H
Sbjct: 538 FFWLLAGLSFLNFLVYLWIAKWYTYKKTTGH 568
>UniRef100_Q9SDN4 Amino acid/peptide transporter [Prunus dulcis]
Length = 559
Score = 495 bits (1274), Expect = e-138
Identities = 253/538 (47%), Positives = 347/538 (64%), Gaps = 12/538 (2%)
Query: 16 YTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGTVA 75
YT DG+VD+ G+PVL+ TG+W AC FI+G E ER+A+YGIS+NLV +L KLHEG V+
Sbjct: 24 YTGDGSVDITGKPVLKQSTGNWXACPFILGTECCERLAFYGISTNLVTYLTHKLHEGNVS 83
Query: 76 SSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRPPP 135
++ NV+ W+G + TPL+GA +ADAY GRYWT I S IY +GMC LT++ S+PAL+PP
Sbjct: 84 AARNVTTWSGTCYLTPLIGAVLADAYWGRYWTIAIFSTIYFIGMCTLTISASVPALKPPQ 143
Query: 136 CAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTYKLS 195
C V CP ASP G+F+ LY+IAL GG KP +S+ GADQFD+ + +E K S
Sbjct: 144 CVDSV----CPSASPAQYGVFFFGLYLIALRTGGIKPCVSSFGADQFDDTDSRERVKKGS 199
Query: 196 FFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHKLPS 255
FFNW+ FSI +G L S+T++V++QDN W LG+G+P + + +I+ F GTP YR + P
Sbjct: 200 FFNWFYFSINIGALVSSTLIVWVQDNAGWGLGFGIPALFMGIAIVSFFSGTPLYRFQKPG 259
Query: 256 GSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKAAV- 314
GSPLTRM QV VA+ KW VP D L+E + A +++H+ L LDKAAV
Sbjct: 260 GSPLTRMCQVLVASFRKWNLDVPRDSSLLYETQDKGSAIKGSRKLEHSDELNCLDKAAVI 319
Query: 315 -----KTGK-TSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTLD 368
KTG ++PWR+CTVTQVEE K + +M PI T ++ + Q T+F++QG +D
Sbjct: 320 SETETKTGDFSNPWRICTVTQVEELKILIRMFPIWATGIVFSAVYAQMATMFVEQGMMMD 379
Query: 369 RSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLMY 428
S+G +F IPPA LS+ +IS++ +PIYDR VP+ R++T RG + LQR+G GL +
Sbjct: 380 TSVG-SFTIPPASLSSFDVISVIFWVPIYDRFIVPIARKFTGKERGFSELQRMGIGLFLS 438
Query: 429 VLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAKMDFFY 488
VL M A + E KRL++A E L+ + +PLSIF IPQY L G AE F I +++FFY
Sbjct: 439 VLCMSAAAVVEMKRLQLATELGLVDKEVAVPLSIFWQIPQYFLLGAAEIFTFIGQLEFFY 498
Query: 489 YQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYY 546
Q+ + M+SL + S + +G F V T + GWI DNLN LDY+
Sbjct: 499 DQSSDAMRSLCSALSASDDFIGKLSELFDSDIVTYFTTQGGKAGWIPDNLNDGHLDYF 556
>UniRef100_Q7XAC3 Peptide transporter 1 [Vicia faba]
Length = 584
Score = 493 bits (1270), Expect = e-138
Identities = 259/559 (46%), Positives = 363/559 (64%), Gaps = 14/559 (2%)
Query: 16 YTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVV-FLGSKLHEGTV 74
YT DG+VD KGRPVL+ TG+WKAC FI+G E ER+AYYGI++NLV L +KLHEG V
Sbjct: 24 YTGDGSVDFKGRPVLKKNTGNWKACPFILGNECCERLAYYGIATNLVKPILLAKLHEGNV 83
Query: 75 ASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRPP 134
+++ NV+ W G + PL+GA +AD+Y GRYWT I S IY +GM LTL+ S+PAL+P
Sbjct: 84 SAARNVTTWQGTCYLAPLIGAVLADSYWGRYWTIAIFSMIYFIGMGTLTLSASIPALKPA 143
Query: 135 PCAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTYKL 194
C V CP A+P +F++ LY+IALG GG KP +S+ GADQFD+ + +E K
Sbjct: 144 ECLGAV----CPPATPAQYAVFFIGLYLIALGTGGIKPCVSSFGADQFDDTDSRERVKKG 199
Query: 195 SFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHKLP 254
SFFNW+ FSI +G L S++ +V+IQ+N W LG+G+P + + +I F +GTP YR + P
Sbjct: 200 SFFNWFYFSINIGALISSSFIVWIQENAGWGLGFGIPALFMGLAIGSFFLGTPLYRFQKP 259
Query: 255 SGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKAAV 314
GSPLTRM QV A+ K VP D L+E + A ++ H+ LR LD+AAV
Sbjct: 260 GGSPLTRMCQVVAASFRKRNLTVPEDSSLLYETPDKSSAIEGSRKLQHSDELRCLDRAAV 319
Query: 315 ------KTGKTSP-WRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTL 367
K G S WRLCTVTQVEE K + +M P+ T ++ + Q T+F++QGT +
Sbjct: 320 ISDDERKRGDYSNLWRLCTVTQVEELKILIRMFPVWATGIVFSAVYAQMSTMFVEQGTMM 379
Query: 368 DRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLM 427
D S+G +F+IP A LS +IS++ +P+YDR VP+ R++T RG + LQR+G GL +
Sbjct: 380 DTSVG-SFKIPAASLSTFDVISVIFWVPVYDRFIVPIARKFTGKERGFSELQRMGIGLFI 438
Query: 428 YVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAKMDFF 487
VL M A + E KRL++A+E L+ + +PL+IF+ IPQY L G AE F + +++FF
Sbjct: 439 SVLCMSAAAIVEIKRLQLAKELDLVDKAVPVPLTIFLQIPQYFLLGAAEVFTFVGQLEFF 498
Query: 488 YYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYYY 547
Y Q+P+ M+SL + S + +LG +LSSF+L+ V T + + GWI DNLN LD Y+
Sbjct: 499 YDQSPDAMRSLCSALSLLTTSLGNYLSSFILTVVLYFTTRGGNPGWIPDNLNKGHLD-YF 557
Query: 548 AFMVILSFLNFLCFLVTAK 566
+ + LSFLN ++V AK
Sbjct: 558 SGLAGLSFLNMFLYIVAAK 576
>UniRef100_Q69LA3 Putative peptide transport protein [Oryza sativa]
Length = 572
Score = 476 bits (1224), Expect = e-132
Identities = 253/571 (44%), Positives = 359/571 (62%), Gaps = 12/571 (2%)
Query: 14 EDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGT 73
E T DGT D G+P +RSK+G+W+AC FI+G E ER+AYYG+S+NLV ++ +L +G
Sbjct: 2 EATTTDGTTDHAGKPAVRSKSGTWRACPFILGNECCERLAYYGMSANLVNYMVDRLRQGN 61
Query: 74 VASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRP 133
++ +V+NW+G + PLVGA++ADAYLGRY T +Y++G+ LLT++ S+P ++P
Sbjct: 62 AGAAASVNNWSGTCYVMPLVGAFLADAYLGRYRTIAAFMALYIVGLALLTMSASVPGMKP 121
Query: 134 PPCAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTYK 193
P CA + C SP F++ALY+IALG GG KP +S+ GADQFD+ +P+E+ K
Sbjct: 122 PNCAT-ISASSCGP-SPGQSAAFFVALYLIALGTGGIKPCVSSFGADQFDDADPREHRSK 179
Query: 194 LSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHKL 253
SFFNW+ SI VG L +++VLV++Q N+ W G+G+P V +A ++ FL+G+ YRH+
Sbjct: 180 ASFFNWFYMSINVGALVASSVLVWVQMNVGWGWGFGIPAVAMAVAVASFLMGSSLYRHQK 239
Query: 254 PSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKAA 313
P GSPLTRMLQV VAA K + +P D L L + R+ HT R+LD+AA
Sbjct: 240 PGGSPLTRMLQVVVAAARKSRVALPADAAAL--LYEGDKLACGTRRLAHTEQFRWLDRAA 297
Query: 314 VKTGKT-------SPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTT 366
V T T S WRLC VTQVEE K + +++P+ + ++ + Q T+F+ QG T
Sbjct: 298 VVTPTTDKDDDTGSRWRLCPVTQVEELKAVVRLLPVWASGIVMSAVYGQMSTMFVLQGNT 357
Query: 367 LDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLL 426
LD MG F+IP A LS +++L +P+YDR+ VP RR+T +PRG T LQR+G GLL
Sbjct: 358 LDPRMGATFKIPSASLSIFDTLAVLAWVPVYDRLIVPAARRFTGHPRGFTQLQRMGIGLL 417
Query: 427 MYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAKMDF 486
+ V MV A + E RLRVA +L +P+SIF + QY + G AE FA I ++DF
Sbjct: 418 ISVFSMVAAGVLEVVRLRVAAAHGMLDSTSYLPISIFWQV-QYFIIGAAEVFAFIGQIDF 476
Query: 487 FYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYY 546
FY QAP+ M+S + S TS ALG +LS+ L+ V + + GWI DNLN LDY+
Sbjct: 477 FYDQAPDDMRSTCTALSLTSSALGNYLSTLLVVIVTAASTRGGGLGWIPDNLNRGHLDYF 536
Query: 547 YAFMVILSFLNFLCFLVTAKFFDYNVDVTHE 577
+ + LS +NFL +L A ++ T E
Sbjct: 537 FWLLAALSAVNFLVYLWIANWYRCKTITTTE 567
>UniRef100_P46032 Peptide transporter PTR2-B [Arabidopsis thaliana]
Length = 585
Score = 473 bits (1216), Expect = e-132
Identities = 242/557 (43%), Positives = 351/557 (62%), Gaps = 12/557 (2%)
Query: 16 YTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLHEGTVA 75
Y +DG+VD G P L+ KTG+WKAC FI+G E ER+AYYGI+ NL+ +L +KLH+G V+
Sbjct: 24 YAEDGSVDFNGNPPLKEKTGNWKACPFILGNECCERLAYYGIAGNLITYLTTKLHQGNVS 83
Query: 76 SSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPALRPPP 135
++ NV+ W G + TPL+GA +ADAY GRYWT S IY +GM LTL+ S+PAL+P
Sbjct: 84 AATNVTTWQGTCYLTPLIGAVLADAYWGRYWTIACFSGIYFIGMSALTLSASVPALKPAE 143
Query: 136 CAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKENTYKLS 195
C CP A+P +F+ LY+IALG GG KP +S+ GADQFD+ + +E K S
Sbjct: 144 CI----GDFCPSATPAQYAMFFGGLYLIALGTGGIKPCVSSFGADQFDDTDSRERVRKAS 199
Query: 196 FFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYRHKLPS 255
FFNW+ FSI +G L S+++LV+IQ+N W LG+G+PTV + +I F GTP YR + P
Sbjct: 200 FFNWFYFSINIGALVSSSLLVWIQENRGWGLGFGIPTVFMGLAIASFFFGTPLYRFQKPG 259
Query: 256 GSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLDKAAV- 314
GSP+TR+ QV VA+ K VP D L+E + A +I+HT ++LDKAAV
Sbjct: 260 GSPITRISQVVVASFRKSSVKVPEDATLLYETQDKNSAIAGSRKIEHTDDCQYLDKAAVI 319
Query: 315 -----KTGK-TSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQGTTLD 368
K+G ++ WRLCTVTQVEE K + +M PI + +I + Q T+F++QG ++
Sbjct: 320 SEEESKSGDYSNSWRLCTVTQVEELKILIRMFPIWASGIIFSAVYAQMSTMFVQQGRAMN 379
Query: 369 RSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGAGLLMY 428
+G +F++PPA L S++ +P+YDR VP+ R++T +G T +QR+G GL +
Sbjct: 380 CKIG-SFQLPPAALGTFDTASVIIWVPLYDRFIVPLARKFTGVDKGFTEIQRMGIGLFVS 438
Query: 429 VLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAKMDFFY 488
VL M A + E RL +A + L+ +P+S+ IPQY + G AE F I +++FFY
Sbjct: 439 VLCMAAAAIVEIIRLHMANDLGLVESGAPVPISVLWQIPQYFILGAAEVFYFIGQLEFFY 498
Query: 489 YQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRLDYYYA 548
Q+P+ M+SL + + + ALG +LSS +L+ V T ++ +GWI DNLN LDY++
Sbjct: 499 DQSPDAMRSLCSALALLTNALGNYLSSLILTLVTYFTTRNGQEGWISDNLNSGHLDYFFW 558
Query: 549 FMVILSFLNFLCFLVTA 565
+ LS +N + +A
Sbjct: 559 LLAGLSLVNMAVYFFSA 575
>UniRef100_Q93Z20 At1g62200/F19K23_13 [Arabidopsis thaliana]
Length = 590
Score = 472 bits (1215), Expect = e-131
Identities = 244/554 (44%), Positives = 352/554 (63%), Gaps = 17/554 (3%)
Query: 11 SGKEDYTQDGTVDLKGRPVLRSKTGSWKACTFIVGYELFERMAYYGISSNLVVFLGSKLH 70
+G +DG++D+ G P + KTG+WKAC FI+G E ER+AYYGI+ NL+ + S+LH
Sbjct: 30 TGLSSTAEDGSIDIYGNPPSKKKTGNWKACPFILGNECCERLAYYGIAKNLITYYTSELH 89
Query: 71 EGTVASSNNVSNWAGVVWTTPLVGAYIADAYLGRYWTFIIASCIYLLGMCLLTLTVSLPA 130
E V+++++V W G + TPL+GA IAD+Y GRYWT S IY +GM LLTL+ SLP
Sbjct: 90 ESNVSAASDVMIWQGTCYITPLIGAVIADSYWGRYWTIASFSAIYFIGMALLTLSASLPV 149
Query: 131 LRPPPCAQGVENQDCPQASPWTRGIFYLALYIIALGAGGTKPNISTMGADQFDEYEPKEN 190
L+P CA GV C A+ +F+ LY+IALG GG KP +S+ GADQFD+ +P+E
Sbjct: 150 LKPAACA-GVAAALCSPATTVQYAVFFTGLYLIALGTGGIKPCVSSFGADQFDDTDPRER 208
Query: 191 TYKLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAFSILVFLVGTPFYR 250
K SFFNW+ FSI +G S+T+LV++Q+N+ W LG+ +PTV + SI F +GTP YR
Sbjct: 209 VRKASFFNWFYFSINIGSFISSTLLVWVQENVGWGLGFLIPTVFMGVSIASFFIGTPLYR 268
Query: 251 HKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSRNRIDHTSTLRFLD 310
+ P GSP+TR+ QV VAA K K ++P D L+E + +I HT +FLD
Sbjct: 269 FQKPGGSPITRVCQVLVAAYRKLKLNLPEDISFLYETREKNSMIAGSRKIQHTDGYKFLD 328
Query: 311 KAAV------KTGK-TSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQAHTLFIKQ 363
KAAV K+G ++PW+LCTVTQVEE K + +M PI + ++ + Q TLF++Q
Sbjct: 329 KAAVISEYESKSGAFSNPWKLCTVTQVEEVKTLIRMFPIWASGIVYSVLYSQISTLFVQQ 388
Query: 364 GTTLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGITMLQRLGA 423
G +++R + +FEIPPA + +L SIPIYDR VP +RR+T P+G+T LQR+G
Sbjct: 389 GRSMNRII-RSFEIPPASFGVFDTLIVLISIPIYDRFLVPFVRRFTGIPKGLTDLQRMGI 447
Query: 424 GLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAENFAEIAK 483
GL + VL + A + E RL++A+ D + +SIF IPQY L G+AE F I +
Sbjct: 448 GLFLSVLSIAAAAIVETVRLQLAQ--------DFVAMSIFWQIPQYILMGIAEVFFFIGR 499
Query: 484 MDFFYYQAPEGMKSLGISYSTTSVALGCFLSSFLLSTVADITKKHSHQGWILDNLNISRL 543
++FFY ++P+ M+S+ + + + A+G +LSS +L+ VA T GW+ D+LN L
Sbjct: 500 VEFFYDESPDAMRSVCSALALLNTAVGSYLSSLILTLVAYFTALGGKDGWVPDDLNKGHL 559
Query: 544 DYYYAFMVILSFLN 557
DY++ +V L +N
Sbjct: 560 DYFFWLLVSLGLVN 573
>UniRef100_Q7XAC2 Peptide transporter 2 [Vicia faba]
Length = 317
Score = 469 bits (1208), Expect = e-131
Identities = 226/316 (71%), Positives = 271/316 (85%), Gaps = 1/316 (0%)
Query: 178 GADQFDEYEPKENTYKLSFFNWWVFSILVGVLFSTTVLVYIQDNLSWSLGYGLPTVGLAF 237
GADQFDE+EPKE +YKLSFFNWW FSI VG LFS T L+YIQD + W++GYGLPT GL
Sbjct: 2 GADQFDEFEPKERSYKLSFFNWWFFSIFVGTLFSNTFLIYIQDRVGWAVGYGLPTAGLTI 61
Query: 238 SILVFLVGTPFYRHKLPSGSPLTRMLQVFVAAGIKWKAHVPHDPKQLHELSMEEYANNSR 297
S+LVFL+GTP YRHKLPSGSP+TRMLQVFVA+ K + P DPK+LHELS++EYA N R
Sbjct: 62 SVLVFLIGTPLYRHKLPSGSPITRMLQVFVASIRKVEGTSPDDPKELHELSIDEYAYNGR 121
Query: 298 NRIDHTSTLR-FLDKAAVKTGKTSPWRLCTVTQVEETKQMTKMVPILITTLIPCTMLIQA 356
NRIDH+S+LR FLDKAA+KT +TS W LCTVTQVEETKQMTKM+PILITT+IP T+++Q+
Sbjct: 122 NRIDHSSSLRSFLDKAAMKTSQTSSWMLCTVTQVEETKQMTKMIPILITTIIPSTLIVQS 181
Query: 357 HTLFIKQGTTLDRSMGPNFEIPPAGLSAILIISMLTSIPIYDRVFVPVIRRYTKNPRGIT 416
TLFIKQGTTLDRSMGP+F+IPPA L+A + I ML SI +YDR FVP++RRYTKNPRGIT
Sbjct: 182 TTLFIKQGTTLDRSMGPHFDIPPACLTAFITIFMLISIVVYDRAFVPMVRRYTKNPRGIT 241
Query: 417 MLQRLGAGLLMYVLIMVIAWLTERKRLRVAREKHLLGQHDIIPLSIFILIPQYALTGVAE 476
MLQRLG GL++++ IMV A L ERKRLRVARE +L G+HD IPL+IFIL+PQ+AL GVA+
Sbjct: 242 MLQRLGIGLVLHIAIMVTACLAERKRLRVARENNLFGRHDTIPLTIFILLPQFALGGVAD 301
Query: 477 NFAEIAKMDFFYYQAP 492
+F EIAK++FFY Q+P
Sbjct: 302 SFVEIAKLEFFYDQSP 317
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.138 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,036,432,037
Number of Sequences: 2790947
Number of extensions: 44980433
Number of successful extensions: 104868
Number of sequences better than 10.0: 643
Number of HSP's better than 10.0 without gapping: 423
Number of HSP's successfully gapped in prelim test: 220
Number of HSP's that attempted gapping in prelim test: 102419
Number of HSP's gapped (non-prelim): 984
length of query: 606
length of database: 848,049,833
effective HSP length: 133
effective length of query: 473
effective length of database: 476,853,882
effective search space: 225551886186
effective search space used: 225551886186
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0082b.11


