

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
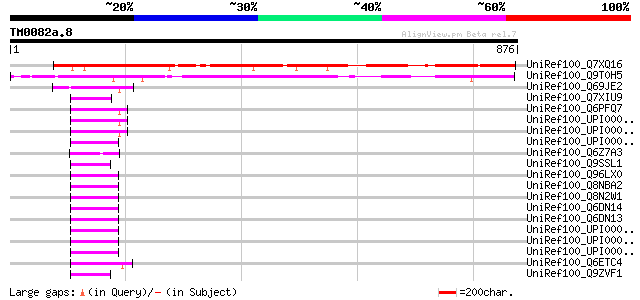
Query= TM0082a.8
(876 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XQ16 OSJNBb0065L13.3 protein [Oryza sativa] 722 0.0
UniRef100_Q9T0H5 Hypothetical protein T6G15.100 [Arabidopsis tha... 720 0.0
UniRef100_Q69JE2 Putative CLB1 protein [Oryza sativa] 58 1e-06
UniRef100_Q7XIU9 Zinc finger and C2 domain protein-like [Oryza s... 57 3e-06
UniRef100_Q6PFQ7 Rasa4 protein [Mus musculus] 56 4e-06
UniRef100_UPI00001D08E3 UPI00001D08E3 UniRef100 entry 55 9e-06
UniRef100_UPI00001D080C UPI00001D080C UniRef100 entry 55 9e-06
UniRef100_UPI00001CECA6 UPI00001CECA6 UniRef100 entry 53 5e-05
UniRef100_Q6Z7A3 Putative C2 domain-containing protein [Oryza sa... 53 5e-05
UniRef100_Q9SSL1 F15H11.6 protein [Arabidopsis thaliana] 53 5e-05
UniRef100_Q96LX0 Hypothetical protein FLJ33132 [Homo sapiens] 53 5e-05
UniRef100_Q8NBA2 Hypothetical protein FLJ34011 [Homo sapiens] 53 5e-05
UniRef100_Q8N2W1 MCTP1 protein [Homo sapiens] 53 5e-05
UniRef100_Q6DN14 MCTP1L [Homo sapiens] 53 5e-05
UniRef100_Q6DN13 MCTP1S [Homo sapiens] 53 5e-05
UniRef100_UPI000036D052 UPI000036D052 UniRef100 entry 52 6e-05
UniRef100_UPI000036D051 UPI000036D051 UniRef100 entry 52 6e-05
UniRef100_UPI000036D050 UPI000036D050 UniRef100 entry 52 6e-05
UniRef100_Q6ETC4 Putative CLB1 protein [Oryza sativa] 52 6e-05
UniRef100_Q9ZVF1 Hypothetical protein At2g01540 [Arabidopsis tha... 52 6e-05
>UniRef100_Q7XQ16 OSJNBb0065L13.3 protein [Oryza sativa]
Length = 888
Score = 722 bits (1863), Expect = 0.0
Identities = 421/866 (48%), Positives = 525/866 (60%), Gaps = 159/866 (18%)
Query: 77 ERAERPPFDINLAVILAGFAFEAYTTPPGT----------------SDPYVVFQMDSQTV 120
E RPPFD+NLAV+LAGFAFEAYT+PP SD ++ D Q V
Sbjct: 83 EDPPRPPFDLNLAVVLAGFAFEAYTSPPEDVGWREIDAAECQTVFLSDSFLREVYDGQLV 142
Query: 121 KSNIKWG-------------------TKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAP 161
+K G TKEPTWNEEFTFNI L LQVAAWDANLV P
Sbjct: 143 V-RLKKGVNLPVMDPWIVISVIDQRRTKEPTWNEEFTFNISLSRENLLQVAAWDANLVTP 201
Query: 162 HKRMGNAGVDLEWLCDGDVHEILVELEGMGGGGKVQLEVKYKTFDEIDDEKKWWKMPFVS 221
HKRMGNAG+ LE LCDG H + VELEG+GGGG + +EV+YK++D+I+ EK+WW++PFVS
Sbjct: 202 HKRMGNAGLCLESLCDGSNHNVTVELEGLGGGGTIDVEVRYKSYDDIEREKQWWRIPFVS 261
Query: 222 DFLKINGFDSALKKVTGSNTVQTRQFVEYAFGQLKSFNKAYLQKGQMSDTDND-----EY 276
DFL + SAL+ V GS ++ QFV+ AFGQL SF YL K D + E
Sbjct: 262 DFLVKSSLGSALRTVLGSESINASQFVQSAFGQLSSFTYTYLPKPPSLDIRAEAPKRAEQ 321
Query: 277 DTEGSGELNESPFVLEMPPREAGSSEASNEACSEQRNTEEFHSHESDTENGHTSEPSTQT 336
+ S NE LE ++ +S ++ CSE +T + E +S P+ +
Sbjct: 322 SVDNSAGSNE----LEQYKMDSKASGDDSDCCSEAESTATVVNTEG------SSSPNMKE 371
Query: 337 SGEELSNQLFWRDFTNVINVNVVQKLGLTVPGKFKWDGLEFLDKIGSQSQNIAEATYIQS 396
+ E FW+ FT+V+N NV+Q G ++P + DG + L +G +S IAE Y++S
Sbjct: 372 TDE-----YFWKAFTSVLNQNVLQNFGFSLPEVKQLDGFDLLSSLGLKSSEIAEKEYLES 426
Query: 397 GLAMPRGIDDTEDKTSGKPAIAA-----------------IQSALPEVKKATESLMRQTD 439
GLA +T+ K AI +Q+ P+V K + ++ QT+
Sbjct: 427 GLATVDASISEGHETTPKDAIDVDKEDGTIPIKENLPKEEVQAPFPDVSKVSRDVLSQTE 486
Query: 440 SILGGLMLLAATVSKMKDEGSSSEERKIEEDSAKVGGNDIQCLPKIPSSENGSV-----L 494
+ILG LMLL+ ++S E ++ +EDS K + QC +++ +V
Sbjct: 487 NILGALMLLSRSLSPQDKESVMVDDGSNKEDSVK----EEQCASDYTDNDDDAVSTEVFT 542
Query: 495 DDKKAEEMRELFSTAETAMEAWAMLATSLGHASFIKSEFEKICFLDNAPTDTQ-----VA 549
D +KAE+ + LF +AETAMEAWAMLATSLG SFIKS+FEKICFLDN TDTQ VA
Sbjct: 543 DAQKAEDRQRLFESAETAMEAWAMLATSLGRNSFIKSDFEKICFLDNVSTDTQLKSLKVA 602
Query: 550 IWRDSMRRRLVVAFRGTEQSQWKDLITDLMIVPAGLASSSFSSAQASILDPKGVFVGVRY 609
IWRD RRRLVVAFRGTEQS+WKDL+TDLM+VPAGL
Sbjct: 603 IWRDCSRRRLVVAFRGTEQSKWKDLLTDLMLVPAGL------------------------ 638
Query: 610 PIARLNPERIGGDFKQEVQVHSGFLSAYDSVRTRIISLIRLAIGYVDDH-FEPLHKWHIY 668
NPER+GGDFKQE+QVHSGFLSAYDSVR RII+L++ A+GY D+ E + KWH+Y
Sbjct: 639 -----NPERLGGDFKQEIQVHSGFLSAYDSVRNRIIALVKYAVGYQDEEDGENIPKWHVY 693
Query: 669 VTGHSLGGALATLLALELSSNQLTNMVDYVEEPKTHHIRQIMIANYGRISAMALSCPIFD 728
VTGHSLGGALATLLALELSS S MA S
Sbjct: 694 VTGHSLGGALATLLALELSS-----------------------------SLMAKS----- 719
Query: 729 NNAHLRGAISITMYNFGSPRVGNKRFAEVYNEKIKDSWRVVNHRDIIPTVPRLMGYCHVN 788
G I +TMYNFGSPRVGN+RFAEVYN K+KDSWRVVNHRDIIPTVPRLMGYCHV
Sbjct: 720 ------GVIFVTMYNFGSPRVGNRRFAEVYNAKVKDSWRVVNHRDIIPTVPRLMGYCHVE 773
Query: 789 QPVFLAAGVLRNSLENKDILGDGYEGDVLGESTPDVIVSEFMKGEKELIEKLLQTEINIF 848
PV+L G L+++L +++ + D EGD +GE TPDV+VSEFMKGEK+L+EKLLQTEIN+
Sbjct: 774 APVYLKFGDLKDALVDEETIDD--EGDSIGEYTPDVLVSEFMKGEKQLVEKLLQTEINLL 831
Query: 849 RSIRDGTAFMQHMEDFYYVTLLEVQC 874
RSIRDG+A MQHMEDFYYVTLLE C
Sbjct: 832 RSIRDGSALMQHMEDFYYVTLLEGYC 857
>UniRef100_Q9T0H5 Hypothetical protein T6G15.100 [Arabidopsis thaliana]
Length = 805
Score = 720 bits (1858), Expect = 0.0
Identities = 415/914 (45%), Positives = 539/914 (58%), Gaps = 178/914 (19%)
Query: 1 MASLQLRYLLSPPSLNFPSQHSSKPRFSRSFPPHFPGNLRAFSLAWRKQRRKRVFSTCCS 60
M SLQL + S H +P+ + P F N R F+ + R R
Sbjct: 1 MTSLQLHF----------SVHFLRPKHRLRYSPIF--NTRTFNFPGKITFRLRAKPNSLR 48
Query: 61 SKTGSQVERISVQEDDERAERPPFDINLAVILAGFAFEAYTTPPGTSDPYVVFQMDSQTV 120
S+ + +S+ E DER+ PFDINLAVILAGFAFE+Y +PPGTSDPYVV +D Q
Sbjct: 49 CFAQSETKEVSLPEKDERS---PFDINLAVILAGFAFESYASPPGTSDPYVVMDLDGQVA 105
Query: 121 KSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRMGNAGVDLEWLCD--- 177
KS KWGTKEP WNE+F FNIKLPP K +++AAWDANLV PHKRMGN+ ++LE +CD
Sbjct: 106 KSKTKWGTKEPKWNEDFVFNIKLPPAKKIEIAAWDANLVTPHKRMGNSEINLESVCDAVL 165
Query: 178 ------GDVHEILVELEGMGGGGKVQLEVKYKTFDEIDDEKKWWKMPFVSDFLKIN---- 227
G++H++LVEL+G+GGGGKVQLE+KYK F E+++EKKWW+ PFVS+FL+ N
Sbjct: 166 FCFASEGNLHKVLVELDGIGGGGKVQLEIKYKGFGEVEEEKKWWRFPFVSEFLQRNEIKS 225
Query: 228 ---------GFDSALKKVTGSNTVQTRQFVEYAFGQLKSFNKAYLQKGQMSDTDNDEYDT 278
+S LK + S V RQFVEYAFGQLKS N A L+ ++ +N D+
Sbjct: 226 VLKNFVDSEAVESVLKNLVDSEAVPARQFVEYAFGQLKSLNDAPLKNTEL--LNNTAEDS 283
Query: 279 EGSGELNESPFVLEMPPREAGSSEASNEACSEQRNTEEFHSHESDTE-NGHTSE-PSTQT 336
EG A S ++S++ S ++ S + D + +GH +E
Sbjct: 284 EG-----------------ASSEDSSDQHRSTNLSSSGKLSKDKDGDGDGHGNELEDDNE 326
Query: 337 SGEELSNQLFWRDFTNVINVNVVQKLGLTVPGKFKWDGLEFLDKIGSQSQNIAEATYIQS 396
SG S FW + +++ N+VQKLGL P K KW+G E L+ G QS+ AEA YI+S
Sbjct: 327 SGSIQSESNFWDNIPDIVGQNIVQKLGLPSPEKLKWNGTELLENFGLQSRKTAEAGYIES 386
Query: 397 GLAMPRGIDDTEDKTSGKPAIAAIQSALPEVKKATESLMRQTDSILGGLMLLAATVSKMK 456
GLA + ++K G+ AI A +S+L ++K AT+ L++Q D++ G LM+L A V +
Sbjct: 387 GLATADTREADDEKEDGQVAINASKSSLADMKNATQELLKQADNVFGALMVLKAVVPHLS 446
Query: 457 DEGSSSEERKIEEDSAKVGGNDI---QCLPKIPSSENGSVLDDKKAEEMRELFSTAETAM 513
+ S E+ IE++ + +D+ KI N D+K AEEM+ LFS+AE+AM
Sbjct: 447 KD-SVGSEKVIEKNGSSSVTDDVSGSSKTEKISGLVNVDGADEKNAEEMKTLFSSAESAM 505
Query: 514 EAWAMLATSLGHASFIKSEFEKICFLDNAPTDTQVAIWRDSMRRRLVVAFRGTEQSQWKD 573
EAWAMLAT+LGH SFIKSEFEK+CFL+N TDTQVAIWRD+ R+R+V+AFRGTEQ
Sbjct: 506 EAWAMLATALGHPSFIKSEFEKLCFLENDITDTQVAIWRDARRKRVVIAFRGTEQ----- 560
Query: 574 LITDLMIVPAGLASSSFSSAQASILDPKGVFVGVRYPIARLNPERIGGDFKQEVQVHSGF 633
S F SA S+
Sbjct: 561 ------------VHSGFLSAYDSV------------------------------------ 572
Query: 634 LSAYDSVRTRIISLIRLAIGYVDDHFEPLHKWHIYVTGHSLGGALATLLALELSSNQLTN 693
R RIISL+++ IGY+DD E KWH+YVTGHSLGGALATLLALELSS+QL
Sbjct: 573 -------RIRIISLLKMTIGYIDDVTEREDKWHVYVTGHSLGGALATLLALELSSSQLAK 625
Query: 694 MVDYVEEPKTHHIRQIMIANYGRISAMALSCPIFDNNAHLRGAISITMYNFGSPRVGNKR 753
RGAI++TMYNFGSPRVGNK+
Sbjct: 626 ----------------------------------------RGAITVTMYNFGSPRVGNKQ 645
Query: 754 FAEVYNEKIKDSWRVVNHRDIIPTVPRLMGYCHVNQPVFLAAG----------------V 797
FAE+YN+K+KDSWRVVNHRDIIPTVPRLMGYCHV PV+L+AG V
Sbjct: 646 FAEIYNQKVKDSWRVVNHRDIIPTVPRLMGYCHVAHPVYLSAGDVYELCKLYNMIFQYKV 705
Query: 798 LRNSLENKDILGDGYEGDVLGESTPDVIVSEFMKGEKELIEKLLQTEINIFRSIRDGTAF 857
++ E+ + DGY +V+GE+TPD++VS FMKGEKEL+EK+LQTEI IF ++RDG+A
Sbjct: 706 IKMLQEDIEFQKDGYHAEVIGEATPDILVSRFMKGEKELVEKILQTEIKIFNALRDGSAL 765
Query: 858 MQHMEDFYYVTLLE 871
MQHMEDFYY+TLLE
Sbjct: 766 MQHMEDFYYITLLE 779
>UniRef100_Q69JE2 Putative CLB1 protein [Oryza sativa]
Length = 539
Score = 57.8 bits (138), Expect = 1e-06
Identities = 43/151 (28%), Positives = 66/151 (43%), Gaps = 13/151 (8%)
Query: 75 DDERAERPPFDINLAVILAGFAFEAYTTPPGTSDPYVVFQMDSQTV---KSNIKWGTKEP 131
D +A + P I L +L G SDPYV +M + K+ +K P
Sbjct: 250 DPSKASKKPVGILLVKVLRAQNLRKKDLL-GKSDPYVKLKMSDDKLPSKKTTVKRSNLNP 308
Query: 132 TWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRMGNAGVDLEWLCDGDVHEILVEL---- 187
WNE+F F + P + L++ +D V H++MG + L+ L + + V L
Sbjct: 309 EWNEDFKFVVTDPETQALEINVFDWEQVGKHEKMGMNNILLKELPADETKVMTVNLLKTM 368
Query: 188 -----EGMGGGGKVQLEVKYKTFDEIDDEKK 213
+ G++ LEV YK F E D EK+
Sbjct: 369 DPNDVQNEKSRGQLTLEVTYKPFKEEDMEKE 399
>UniRef100_Q7XIU9 Zinc finger and C2 domain protein-like [Oryza sativa]
Length = 161
Score = 56.6 bits (135), Expect = 3e-06
Identities = 27/71 (38%), Positives = 42/71 (59%), Gaps = 1/71 (1%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDPYVV +D+Q +K+ + T P WNEE T ++ P P+Q+ +D + + +
Sbjct: 24 GGSDPYVVLHLDNQKLKTGVVKKTTNPVWNEELTLAVR-NPETPIQLEVFDKDTFSKDDQ 82
Query: 165 MGNAGVDLEWL 175
MG+A D+E L
Sbjct: 83 MGDAEFDIEAL 93
>UniRef100_Q6PFQ7 Rasa4 protein [Mus musculus]
Length = 802
Score = 56.2 bits (134), Expect = 4e-06
Identities = 32/107 (29%), Positives = 55/107 (50%), Gaps = 8/107 (7%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDP+V + +T ++++ + P WNE F F ++ ++ L V AWD +LV+ +
Sbjct: 152 GASDPFVRVHYNGRTQETSVVKKSCYPRWNETFDFELEKGASEALLVEAWDWDLVSRNDF 211
Query: 165 MGNAGVDLEWLCDGDVHEILVEL--------EGMGGGGKVQLEVKYK 203
+G V+++ LC E L +G G G +QLEV+ +
Sbjct: 212 LGKVAVNVQRLCSAQQEEGWFRLQPDQSKSRQGKGNLGSLQLEVRLR 258
>UniRef100_UPI00001D08E3 UPI00001D08E3 UniRef100 entry
Length = 784
Score = 55.1 bits (131), Expect = 9e-06
Identities = 32/107 (29%), Positives = 54/107 (49%), Gaps = 8/107 (7%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDP+V + +T ++++ + P WNE F F ++ + L V AWD +LV+ +
Sbjct: 152 GASDPFVRVHYNGRTQETSVVKKSCYPRWNETFEFELEKGATEALLVEAWDWDLVSRNDF 211
Query: 165 MGNAGVDLEWLCDGDVHEILVEL--------EGMGGGGKVQLEVKYK 203
+G V+++ LC E L +G G G +QLEV+ +
Sbjct: 212 LGKVVVNVQTLCSAQQEEGWFRLQPDQSKSRQGKGNLGSLQLEVRLR 258
>UniRef100_UPI00001D080C UPI00001D080C UniRef100 entry
Length = 803
Score = 55.1 bits (131), Expect = 9e-06
Identities = 32/107 (29%), Positives = 54/107 (49%), Gaps = 8/107 (7%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDP+V + +T ++++ + P WNE F F ++ + L V AWD +LV+ +
Sbjct: 152 GASDPFVRVHYNGRTQETSVVKKSCYPRWNETFEFELEKGATEALLVEAWDWDLVSRNDF 211
Query: 165 MGNAGVDLEWLCDGDVHEILVEL--------EGMGGGGKVQLEVKYK 203
+G V+++ LC E L +G G G +QLEV+ +
Sbjct: 212 LGKVVVNVQTLCSAQQEEGWFRLQPDQSKSRQGKGNLGSLQLEVRLR 258
>UniRef100_UPI00001CECA6 UPI00001CECA6 UniRef100 entry
Length = 546
Score = 52.8 bits (125), Expect = 5e-05
Identities = 28/84 (33%), Positives = 41/84 (48%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDPYV F++ Q KS I T P W E+F F++ + + AWD +
Sbjct: 222 GLSDPYVKFRLGHQKYKSKIMPKTLNPQWREQFDFHLYEERGGVMDITAWDKDAGKRDDF 281
Query: 165 MGNAGVDLEWLCDGDVHEILVELE 188
+G VDL L H++ ++LE
Sbjct: 282 IGRCQVDLSSLSREQTHKLELQLE 305
Score = 39.3 bits (90), Expect = 0.52
Identities = 24/98 (24%), Positives = 50/98 (50%), Gaps = 3/98 (3%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDP+ V ++++ + ++ + P WN+ FTFNIK + L+V +D +
Sbjct: 378 GKSDPFCVVELNNDRLLTHTVYKNLNPEWNKVFTFNIK-DIHSVLEVTVYDEDRDRSADF 436
Query: 165 MGNAGVDLEWLCDGDVHEILVELEGMGG--GGKVQLEV 200
+G + L + +G+ +++ + + G G + LE+
Sbjct: 437 LGRVAIPLLSIQNGEQKAYVLKNKQLTGPTKGVIHLEI 474
>UniRef100_Q6Z7A3 Putative C2 domain-containing protein [Oryza sativa]
Length = 1111
Score = 52.8 bits (125), Expect = 5e-05
Identities = 29/88 (32%), Positives = 47/88 (52%), Gaps = 4/88 (4%)
Query: 104 PGTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHK 163
PG DPYVVF + + S++K+ T EP WNE F FN P L+V D+ PH
Sbjct: 648 PGLPDPYVVFTCNGKRKTSSVKFQTSEPKWNEIFEFNAMDDPPSRLEVVVHDSE--GPHN 705
Query: 164 RMGNAGVDLEWLCD--GDVHEILVELEG 189
++ ++ +L + D+ ++ + L+G
Sbjct: 706 KIPIGQTEVNFLKNNLSDLGDMWLPLDG 733
Score = 36.2 bits (82), Expect = 4.4
Identities = 23/65 (35%), Positives = 31/65 (47%), Gaps = 2/65 (3%)
Query: 77 ERAERPPFDINLAVILAGFAFEAYTTPPGTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEE 136
ER P + + V+ A AY T G SDPYV QM + K+ + P W+EE
Sbjct: 14 ERPGSAPMRLCVHVLEARGLQAAYLT--GHSDPYVRLQMGRRRAKTTVVKRCLSPLWDEE 71
Query: 137 FTFNI 141
F F +
Sbjct: 72 FGFAV 76
>UniRef100_Q9SSL1 F15H11.6 protein [Arabidopsis thaliana]
Length = 165
Score = 52.8 bits (125), Expect = 5e-05
Identities = 23/68 (33%), Positives = 41/68 (59%), Gaps = 1/68 (1%)
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
+SDP+VV M SQ +K+ + P WNEE T ++ P++P+ + +D + H +M
Sbjct: 25 SSDPFVVITMGSQKLKTRVVENNCNPEWNEELTLALR-HPDEPVNLIVYDKDTFTSHDKM 83
Query: 166 GNAGVDLE 173
G+A +D++
Sbjct: 84 GDAKIDIK 91
>UniRef100_Q96LX0 Hypothetical protein FLJ33132 [Homo sapiens]
Length = 692
Score = 52.8 bits (125), Expect = 5e-05
Identities = 28/84 (33%), Positives = 41/84 (48%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDPYV F++ Q KS I T P W E+F F++ + + AWD +
Sbjct: 222 GLSDPYVKFRLGHQKYKSKIMPKTLNPQWREQFDFHLYEERGGVIDITAWDKDAGKRDDF 281
Query: 165 MGNAGVDLEWLCDGDVHEILVELE 188
+G VDL L H++ ++LE
Sbjct: 282 IGRCQVDLSALSREQTHKLELQLE 305
Score = 38.9 bits (89), Expect = 0.68
Identities = 24/98 (24%), Positives = 50/98 (50%), Gaps = 3/98 (3%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDP+ V ++++ + ++ + P WN+ FTFNIK + L+V +D +
Sbjct: 378 GKSDPFCVVELNNDRLLTHTVYKNLNPEWNKVFTFNIK-DIHSVLEVTVYDEDRDRSADF 436
Query: 165 MGNAGVDLEWLCDGDVHEILVELEGMGG--GGKVQLEV 200
+G + L + +G+ +++ + + G G + LE+
Sbjct: 437 LGKVAIPLLSIQNGEQKAYVLKNKQLTGPTKGVIYLEI 474
>UniRef100_Q8NBA2 Hypothetical protein FLJ34011 [Homo sapiens]
Length = 515
Score = 52.8 bits (125), Expect = 5e-05
Identities = 28/84 (33%), Positives = 41/84 (48%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDPYV F++ Q KS I T P W E+F F++ + + AWD +
Sbjct: 5 GLSDPYVKFRLGHQKYKSKIMPKTLNPQWREQFDFHLYEERGGVIDITAWDKDAGKRDDF 64
Query: 165 MGNAGVDLEWLCDGDVHEILVELE 188
+G VDL L H++ ++LE
Sbjct: 65 IGRCQVDLSALSREQTHKLELQLE 88
Score = 38.9 bits (89), Expect = 0.68
Identities = 24/98 (24%), Positives = 50/98 (50%), Gaps = 3/98 (3%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDP+ V ++++ + ++ + P WN+ FTFNIK + L+V +D +
Sbjct: 161 GKSDPFCVVELNNDRLLTHTVYKNLNPEWNKVFTFNIK-DIHSVLEVTVYDEDRDRSADF 219
Query: 165 MGNAGVDLEWLCDGDVHEILVELEGMGG--GGKVQLEV 200
+G + L + +G+ +++ + + G G + LE+
Sbjct: 220 LGKVAIPLLSIQNGEQKAYVLKNKQLTGPTKGVIYLEI 257
>UniRef100_Q8N2W1 MCTP1 protein [Homo sapiens]
Length = 600
Score = 52.8 bits (125), Expect = 5e-05
Identities = 28/84 (33%), Positives = 41/84 (48%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDPYV F++ Q KS I T P W E+F F++ + + AWD +
Sbjct: 268 GLSDPYVKFRLGHQKYKSKIMPKTLNPQWREQFDFHLYEERGGVIDITAWDKDAGKRDDF 327
Query: 165 MGNAGVDLEWLCDGDVHEILVELE 188
+G VDL L H++ ++LE
Sbjct: 328 IGRCQVDLSALSREQTHKLELQLE 351
Score = 38.9 bits (89), Expect = 0.68
Identities = 24/98 (24%), Positives = 50/98 (50%), Gaps = 3/98 (3%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDP+ V ++++ + ++ + P WN+ FTFNIK + L+V +D +
Sbjct: 424 GKSDPFCVVELNNDRLLTHTVYKNLNPEWNKVFTFNIK-DIHSVLEVTVYDEDRDRSADF 482
Query: 165 MGNAGVDLEWLCDGDVHEILVELEGMGG--GGKVQLEV 200
+G + L + +G+ +++ + + G G + LE+
Sbjct: 483 LGKVAIPLLSIQNGEQKAYVLKNKQLTGPTKGVIYLEI 520
>UniRef100_Q6DN14 MCTP1L [Homo sapiens]
Length = 999
Score = 52.8 bits (125), Expect = 5e-05
Identities = 28/84 (33%), Positives = 41/84 (48%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDPYV F++ Q KS I T P W E+F F++ + + AWD +
Sbjct: 489 GLSDPYVKFRLGHQKYKSKIMPKTLNPQWREQFDFHLYEERGGVIDITAWDKDAGKRDDF 548
Query: 165 MGNAGVDLEWLCDGDVHEILVELE 188
+G VDL L H++ ++LE
Sbjct: 549 IGRCQVDLSALSREQTHKLELQLE 572
Score = 38.9 bits (89), Expect = 0.68
Identities = 24/98 (24%), Positives = 50/98 (50%), Gaps = 3/98 (3%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDP+ V ++++ + ++ + P WN+ FTFNIK + L+V +D +
Sbjct: 645 GKSDPFCVVELNNDRLLTHTVYKNLNPEWNKVFTFNIK-DIHSVLEVTVYDEDRDRSADF 703
Query: 165 MGNAGVDLEWLCDGDVHEILVELEGMGG--GGKVQLEV 200
+G + L + +G+ +++ + + G G + LE+
Sbjct: 704 LGKVAIPLLSIQNGEQKAYVLKNKQLTGPTKGVIYLEI 741
>UniRef100_Q6DN13 MCTP1S [Homo sapiens]
Length = 778
Score = 52.8 bits (125), Expect = 5e-05
Identities = 28/84 (33%), Positives = 41/84 (48%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDPYV F++ Q KS I T P W E+F F++ + + AWD +
Sbjct: 268 GLSDPYVKFRLGHQKYKSKIMPKTLNPQWREQFDFHLYEERGGVIDITAWDKDAGKRDDF 327
Query: 165 MGNAGVDLEWLCDGDVHEILVELE 188
+G VDL L H++ ++LE
Sbjct: 328 IGRCQVDLSALSREQTHKLELQLE 351
Score = 38.9 bits (89), Expect = 0.68
Identities = 24/98 (24%), Positives = 50/98 (50%), Gaps = 3/98 (3%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDP+ V ++++ + ++ + P WN+ FTFNIK + L+V +D +
Sbjct: 424 GKSDPFCVVELNNDRLLTHTVYKNLNPEWNKVFTFNIK-DIHSVLEVTVYDEDRDRSADF 482
Query: 165 MGNAGVDLEWLCDGDVHEILVELEGMGG--GGKVQLEV 200
+G + L + +G+ +++ + + G G + LE+
Sbjct: 483 LGKVAIPLLSIQNGEQKAYVLKNKQLTGPTKGVIYLEI 520
>UniRef100_UPI000036D052 UPI000036D052 UniRef100 entry
Length = 515
Score = 52.4 bits (124), Expect = 6e-05
Identities = 28/84 (33%), Positives = 41/84 (48%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDPYV F++ Q KS I T P W E+F F++ + + AWD +
Sbjct: 5 GLSDPYVKFRLGHQKYKSKIMPKTLNPQWREQFDFHLYEERGGIIDITAWDKDAGKRDDF 64
Query: 165 MGNAGVDLEWLCDGDVHEILVELE 188
+G VDL L H++ ++LE
Sbjct: 65 IGRCQVDLSALSREQTHKLELQLE 88
Score = 38.9 bits (89), Expect = 0.68
Identities = 24/98 (24%), Positives = 50/98 (50%), Gaps = 3/98 (3%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDP+ V ++++ + ++ + P WN+ FTFNIK + L+V +D +
Sbjct: 161 GKSDPFCVVELNNDRLLTHTVYKNLNPEWNKVFTFNIK-DIHSVLEVTVYDEDRDRSADF 219
Query: 165 MGNAGVDLEWLCDGDVHEILVELEGMGG--GGKVQLEV 200
+G + L + +G+ +++ + + G G + LE+
Sbjct: 220 LGKVAIPLLSIQNGEQKAYVLKNKQLTGPTKGVIYLEI 257
>UniRef100_UPI000036D051 UPI000036D051 UniRef100 entry
Length = 457
Score = 52.4 bits (124), Expect = 6e-05
Identities = 28/84 (33%), Positives = 41/84 (48%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDPYV F++ Q KS I T P W E+F F++ + + AWD +
Sbjct: 134 GLSDPYVKFRLGHQKYKSKIMPKTLNPQWREQFDFHLYEERGGIIDITAWDKDAGKRDDF 193
Query: 165 MGNAGVDLEWLCDGDVHEILVELE 188
+G VDL L H++ ++LE
Sbjct: 194 IGRCQVDLSALSREQTHKLELQLE 217
Score = 38.9 bits (89), Expect = 0.68
Identities = 24/98 (24%), Positives = 50/98 (50%), Gaps = 3/98 (3%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDP+ V ++++ + ++ + P WN+ FTFNIK + L+V +D +
Sbjct: 290 GKSDPFCVVELNNDRLLTHTVYKNLNPEWNKVFTFNIK-DIHSVLEVTVYDEDRDRSADF 348
Query: 165 MGNAGVDLEWLCDGDVHEILVELEGMGG--GGKVQLEV 200
+G + L + +G+ +++ + + G G + LE+
Sbjct: 349 LGKVAIPLLSIQNGEQKAYVLKNKQLTGPTKGVIYLEI 386
>UniRef100_UPI000036D050 UPI000036D050 UniRef100 entry
Length = 560
Score = 52.4 bits (124), Expect = 6e-05
Identities = 28/84 (33%), Positives = 41/84 (48%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDPYV F++ Q KS I T P W E+F F++ + + AWD +
Sbjct: 90 GLSDPYVKFRLGHQKYKSKIMPKTLNPQWREQFDFHLYEERGGIIDITAWDKDAGKRDDF 149
Query: 165 MGNAGVDLEWLCDGDVHEILVELE 188
+G VDL L H++ ++LE
Sbjct: 150 IGRCQVDLSALSREQTHKLELQLE 173
Score = 38.9 bits (89), Expect = 0.68
Identities = 24/98 (24%), Positives = 50/98 (50%), Gaps = 3/98 (3%)
Query: 105 GTSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKR 164
G SDP+ V ++++ + ++ + P WN+ FTFNIK + L+V +D +
Sbjct: 246 GKSDPFCVVELNNDRLLTHTVYKNLNPEWNKVFTFNIK-DIHSVLEVTVYDEDRDRSADF 304
Query: 165 MGNAGVDLEWLCDGDVHEILVELEGMGG--GGKVQLEV 200
+G + L + +G+ +++ + + G G + LE+
Sbjct: 305 LGKVAIPLLSIQNGEQKAYVLKNKQLTGPTKGVIYLEI 342
>UniRef100_Q6ETC4 Putative CLB1 protein [Oryza sativa]
Length = 538
Score = 52.4 bits (124), Expect = 6e-05
Identities = 30/119 (25%), Positives = 58/119 (48%), Gaps = 12/119 (10%)
Query: 105 GTSDPYVVFQMDSQTV---KSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAP 161
G SDPYV ++ + + K+++K P WNE+F +K P ++ L++ +D V
Sbjct: 279 GKSDPYVKLKLTEEKLPSKKTSVKRSNLNPEWNEDFKLVVKDPESQALELTVYDWEQVGK 338
Query: 162 HKRMGNAGVDLEWLCDGDVHEILVELEGMGGG---------GKVQLEVKYKTFDEIDDE 211
H ++G + + L+ L + + ++L G++ ++V YK F E D +
Sbjct: 339 HDKIGMSVIPLKELIPDEAKSLTLDLHKTMDANDPANDKFRGQLTVDVTYKPFKEGDSD 397
>UniRef100_Q9ZVF1 Hypothetical protein At2g01540 [Arabidopsis thaliana]
Length = 180
Score = 52.4 bits (124), Expect = 6e-05
Identities = 24/68 (35%), Positives = 40/68 (58%), Gaps = 1/68 (1%)
Query: 106 TSDPYVVFQMDSQTVKSNIKWGTKEPTWNEEFTFNIKLPPNKPLQVAAWDANLVAPHKRM 165
+SDPY+V + QT+K+ + P WNEE T IK PN P+++ +D + +M
Sbjct: 26 SSDPYIVLNVADQTLKTRVVKKNCNPVWNEEMTVAIK-DPNVPIRLTVFDWDKFTGDDKM 84
Query: 166 GNAGVDLE 173
G+A +D++
Sbjct: 85 GDANIDIQ 92
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,483,558,548
Number of Sequences: 2790947
Number of extensions: 64425161
Number of successful extensions: 193545
Number of sequences better than 10.0: 652
Number of HSP's better than 10.0 without gapping: 203
Number of HSP's successfully gapped in prelim test: 451
Number of HSP's that attempted gapping in prelim test: 192476
Number of HSP's gapped (non-prelim): 1332
length of query: 876
length of database: 848,049,833
effective HSP length: 136
effective length of query: 740
effective length of database: 468,481,041
effective search space: 346675970340
effective search space used: 346675970340
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 79 (35.0 bits)
Lotus: description of TM0082a.8


