

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0081b.3
(322 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
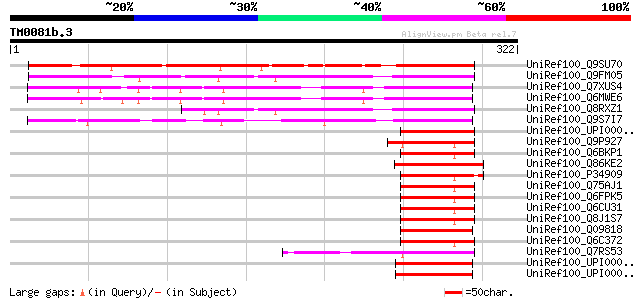
UniRef100_Q9SU70 Hypothetical protein T17F15.60 [Arabidopsis tha... 219 8e-56
UniRef100_Q9FM05 Arabidopsis thaliana genomic DNA, chromosome 5,... 191 3e-47
UniRef100_Q7XUS4 OSJNBa0060P14.3 protein [Oryza sativa] 165 2e-39
UniRef100_Q6MWE6 B1358B12.22 protein [Oryza sativa] 155 1e-36
UniRef100_Q8RXZ1 Hypothetical protein At5g62910 [Arabidopsis tha... 155 1e-36
UniRef100_Q9S7I7 Hypothetical protein F9E10.28 [Arabidopsis thal... 108 1e-22
UniRef100_UPI000042D574 UPI000042D574 UniRef100 entry 69 2e-10
UniRef100_Q9P927 NOT4p [Candida albicans] 67 5e-10
UniRef100_Q6BKP1 Similar to CA0449|CaMOT2.3 Candida albicans CaM... 67 6e-10
UniRef100_Q86KE2 Similar to Dictyostelium discoideum (Slime mold... 67 8e-10
UniRef100_P34909 General negative regulator of transcription sub... 67 8e-10
UniRef100_Q75AJ1 ADL064Wp [Ashbya gossypii] 66 1e-09
UniRef100_Q6FPK5 Similar to sp|P34909 Saccharomyces cerevisiae Y... 65 3e-09
UniRef100_Q6CU31 Similarity [Kluyveromyces lactis] 64 7e-09
UniRef100_Q8J1S7 CCr4/NOT complex/transcription factor subunit [... 64 7e-09
UniRef100_Q09818 Putative general negative regulator of transcri... 64 7e-09
UniRef100_Q6C372 Similar to KLLA0C08041g Kluyveromyces lactis [Y... 63 1e-08
UniRef100_Q7RS53 Hypothetical protein [Plasmodium yoelii yoelii] 62 3e-08
UniRef100_UPI00003661E0 UPI00003661E0 UniRef100 entry 61 4e-08
UniRef100_UPI00003661DF UPI00003661DF UniRef100 entry 61 4e-08
>UniRef100_Q9SU70 Hypothetical protein T17F15.60 [Arabidopsis thaliana]
Length = 319
Score = 219 bits (558), Expect = 8e-56
Identities = 137/289 (47%), Positives = 175/289 (60%), Gaps = 26/289 (8%)
Query: 13 KKKRTNGSAKLKQIKLDVRREQWLSRVKKGCNVDSNGRMDNCPSSKHTASE--ENRSSCK 70
KKKRTN SAKLKQ KL +RREQWLS+V + + G + S++ SE + R +
Sbjct: 22 KKKRTNRSAKLKQSKLGLRREQWLSQVA----MINKGDKEEMESNRRIGSEKPDQRDLPR 77
Query: 71 EMTRKGEDIEGTCIQDSDSRSTINSPDRSTYSHDESSGKDFSGSSSNSISTSCSGNDSEE 130
+ ED GT +S S NSP+ + +S +FS SSS+ SCSGN +EE
Sbjct: 78 PVENLDEDNNGTHRHESFIESLSNSPN--SILSGMNSIPNFSSSSSSGSGGSCSGNITEE 135
Query: 131 ED--DDGCLDDWEAVADALYANDKSHSVVS--ESPAEHEAECRYTVLEDDKNPRVDFSKA 186
ED DDGCLDDWEA+ADAL A+D+ H + ES EHE + + + L + + +KA
Sbjct: 136 EDADDDGCLDDWEAIADALAADDEKHEKENPPESCQEHE-DIKQSALIGGADVVLRDAKA 194
Query: 187 DFKSEVPESKPNRRAWKPDDTLRPRCLPNLSKQRNSPLNSNWRGSRKTVPWAWQTIISQP 246
D S+ + K N+ AW+PDD LRP+ LPNL KQR+ P+ N S TV P
Sbjct: 195 D--SQRRKQKSNQ-AWRPDDKLRPQGLPNLEKQRSFPV-MNLHFSSVTVV---------P 241
Query: 247 SQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRKLYD 295
S CPICYEDLD+TDS+FLPC CGF LCLFCHK I + D RCP CRK Y+
Sbjct: 242 SSCPICYEDLDLTDSNFLPCPCGFRLCLFCHKTICDGDGRCPGCRKPYE 290
>UniRef100_Q9FM05 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MQB2
[Arabidopsis thaliana]
Length = 327
Score = 191 bits (484), Expect = 3e-47
Identities = 124/301 (41%), Positives = 153/301 (50%), Gaps = 40/301 (13%)
Query: 13 KKKRTNGSAKLKQIKLDVRREQWLSRVKKGCNVDSNGRMDNCPSSKHTASEENRSSCKEM 72
KKKR N SAK+KQ KL +RREQWLS+V R N H S +
Sbjct: 20 KKKRNNKSAKMKQNKLGLRREQWLSQVAVSNKEVKEERSVNRSQKPHHESSDK------- 72
Query: 73 TRKGEDIEG---TCIQDSDSRSTINSPDRSTYSHDESSGKDFSGSSSNSISTSCSGNDSE 129
R+ ED G +S S NS TYS SG+ S SS+S S CSGN +E
Sbjct: 73 VRREEDNNGGNNLLHHESFMESPSNSSVGGTYSSTNFSGR--SSRSSSSSSGFCSGNITE 130
Query: 130 EE----DDDGCLDDWEAVADALYANDKSHSVVSESPAEHEAE-----------CRYTVLE 174
EE DDDGC+DDWEAVADAL A ++ P E E C ++ +
Sbjct: 131 EENVDDDDDGCVDDWEAVADALAAEEEIEK--KSRPLESVKEQVSVGQSASNVCDSSISD 188
Query: 175 DDKNPRVDFSKADFKSEVPESKPNRRAWKPDDTLRPRCLPNLSKQRNSPLNSNWRGSRKT 234
V+ K + + + RAW+ DD LRP+ LPNL+KQ + P S
Sbjct: 189 ASDVVGVEDPKQECLRVSSRKQTSNRAWRLDDDLRPQGLPNLAKQLSFPELDKRFSS--- 245
Query: 235 VPWAWQTIISQPSQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRKLY 294
++ PS CPICYEDLD+TDS+FLPC CGF LCLFCHK I + D RCP CRK Y
Sbjct: 246 --------VAIPSSCPICYEDLDLTDSNFLPCPCGFRLCLFCHKTICDGDGRCPGCRKPY 297
Query: 295 D 295
+
Sbjct: 298 E 298
>UniRef100_Q7XUS4 OSJNBa0060P14.3 protein [Oryza sativa]
Length = 327
Score = 165 bits (417), Expect = 2e-39
Identities = 128/320 (40%), Positives = 155/320 (48%), Gaps = 60/320 (18%)
Query: 12 SKKKRTN-GSAKLKQIKLDVRREQWLSRVKKG----CNVDSNGRMDNCPS----SKHTAS 62
+KKKR N SAKLKQ KLD RREQWLS+VK G + G N S S H
Sbjct: 16 AKKKRGNRSSAKLKQCKLDARREQWLSQVKDGKEAKASTSPTGTEPNAGSMTVPSPHPPL 75
Query: 63 EENRSSCKEMTRKGEDIE------GTCIQDSDSRSTINSPDRSTYSHDESS--------- 107
R + KG D E G Q+ S S ++SP S S + S
Sbjct: 76 PRRRLDVRS---KGGDPEEDREERGAARQELGS-SYLDSPVHSPSSDNSGSVGGMHRKHY 131
Query: 108 --GKDFSGSSSNSISTSCSGNDSEEEDDD--------GCLDDWEAVADA-LYANDKSHSV 156
G + SSS+S+ +S S + SE EDDD G LDDWEAVADA D HS
Sbjct: 132 NNGGGLNLSSSSSVWSS-SRSVSEAEDDDTGGPEEENGVLDDWEAVADADALTVDDCHSH 190
Query: 157 VSESPAEHEAECRYTVLEDDKNPRVDFSKADFKSEVPESKPNRRAWKPDDTLRPRCLPNL 216
S A ++ R D + +AW PDD RP+ LP++
Sbjct: 191 QSSGHVAPPAAPNVCTAPANQTGRQDPIQ------------RTKAWAPDDIFRPQSLPSI 238
Query: 217 SKQRNSP--LNSNWRGSRKTVPWAWQTIISQPSQCPICYEDLDVTDSSFLPCSCGFHLCL 274
S+Q + P + + W G+ A Q +S P CPIC EDLD+TDSSF PC C F LCL
Sbjct: 239 SRQVSFPASIGNGWMGA------AQQANLSTPLTCPICCEDLDLTDSSFCPCPCKFCLCL 292
Query: 275 FCHKKILEADARCPSCRKLY 294
FCH KILEAD RCP CRK Y
Sbjct: 293 FCHNKILEADGRCPGCRKEY 312
>UniRef100_Q6MWE6 B1358B12.22 protein [Oryza sativa]
Length = 341
Score = 155 bits (393), Expect = 1e-36
Identities = 125/332 (37%), Positives = 158/332 (46%), Gaps = 70/332 (21%)
Query: 12 SKKKRTN-GSAKLKQIKLDVRREQWLSRVKKGCN-------VDSNGRMDNCPSSKHTASE 63
+KKKR N SAKLKQ KLD RREQWLS+ + + +G+ +S T +E
Sbjct: 16 AKKKRGNRSSAKLKQCKLDARREQWLSQDCSNSDPPAFWIGIVKDGKEAKASTSP-TGTE 74
Query: 64 ENRSSCK-------------EMTRKGEDIE------GTCIQDSDSRSTINSPDRSTYSHD 104
N S ++ KG D E G Q+ S S ++SP S S +
Sbjct: 75 PNAGSMTVPSPHPPLPRRRLDVRSKGGDPEEDREERGAARQELGS-SYLDSPVHSPSSDN 133
Query: 105 ESS-----------GKDFSGSSSNSISTSCSGNDSEEEDDD--------GCLDDWEAVAD 145
S G + SSS+S+ +S S + SE EDDD G LDDWEAVAD
Sbjct: 134 SGSVGGMHRKHYNNGGGLNLSSSSSVWSS-SRSVSEAEDDDTGGPEEENGVLDDWEAVAD 192
Query: 146 A-LYANDKSHSVVSESPAEHEAECRYTVLEDDKNPRVDFSKADFKSEVPESKPNRRAWKP 204
A D HS S A ++ R D + +AW P
Sbjct: 193 ADALTVDDCHSHQSSGHVAPPAAPNVCTAPANQTGRQDPIQ------------RTKAWAP 240
Query: 205 DDTLRPRCLPNLSKQRNSP--LNSNWRGSRKTVPWAWQTIISQPSQCPICYEDLDVTDSS 262
DD RP+ LP++S+Q + P + + W G+ A Q +S P CPIC EDLD+TDSS
Sbjct: 241 DDIFRPQSLPSISRQVSFPASIGNGWMGA------AQQANLSTPLTCPICCEDLDLTDSS 294
Query: 263 FLPCSCGFHLCLFCHKKILEADARCPSCRKLY 294
F PC C F LCLFCH KILEAD RCP CRK Y
Sbjct: 295 FCPCPCKFCLCLFCHNKILEADGRCPGCRKEY 326
>UniRef100_Q8RXZ1 Hypothetical protein At5g62910 [Arabidopsis thaliana]
Length = 222
Score = 155 bits (392), Expect = 1e-36
Identities = 90/204 (44%), Positives = 113/204 (55%), Gaps = 31/204 (15%)
Query: 110 DFSGSSSNSISTS---CSGNDSEEE----DDDGCLDDWEAVADALYANDKSHSVVSESPA 162
+FSG SS S S+S CSGN +EEE DDDGC+DDWEAVADAL A ++ P
Sbjct: 3 NFSGRSSRSSSSSSGFCSGNITEEENVDDDDDGCVDDWEAVADALAAEEEIEK--KSRPL 60
Query: 163 EHEAE-----------CRYTVLEDDKNPRVDFSKADFKSEVPESKPNRRAWKPDDTLRPR 211
E E C ++ + V+ K + + + RAW+ DD LRP+
Sbjct: 61 ESVKEQVSVGQSASNVCDSSISDASDVVGVEDPKQECLRVSSRKQTSNRAWRLDDDLRPQ 120
Query: 212 CLPNLSKQRNSPLNSNWRGSRKTVPWAWQTIISQPSQCPICYEDLDVTDSSFLPCSCGFH 271
LPNL+KQ + P S ++ PS CPICYEDLD+TDS+FLPC CGF
Sbjct: 121 GLPNLAKQLSFPELDKRFSS-----------VAIPSSCPICYEDLDLTDSNFLPCPCGFR 169
Query: 272 LCLFCHKKILEADARCPSCRKLYD 295
LCLFCHK I + D RCP CRK Y+
Sbjct: 170 LCLFCHKTICDGDGRCPGCRKPYE 193
>UniRef100_Q9S7I7 Hypothetical protein F9E10.28 [Arabidopsis thaliana]
Length = 289
Score = 108 bits (271), Expect = 1e-22
Identities = 90/289 (31%), Positives = 119/289 (41%), Gaps = 57/289 (19%)
Query: 12 SKKKRTNGSAKLKQIKLDVRREQWLSRVKKGCNVDSN--GRMDNCPSSKHTASEENRSSC 69
SKK++ N KLKQ K+D RR+QW+S+ KK NVD GR K T + R
Sbjct: 20 SKKRKANWFCKLKQWKIDARRKQWISQWKKA-NVDEEEIGRRLRSLLEKLTDQKAWRIDY 78
Query: 70 KEMTRKGEDIEGTCIQDSDSRSTINSPDRSTYSHDESSGKDFSGSSSNSISTSCSGNDSE 129
+ D+E T S +SP D SG F CS +E
Sbjct: 79 DDDEDDEIDLERTS-------SFASSPTSVLKRKDSVSGDCFC----------CSKQMTE 121
Query: 130 EEDD--DGCLDDWEAVADALYANDKSHSVVSESPAEHEAECRYTVLEDDKNPRVDFSKAD 187
EE++ D D+W+ DAL + E+D N D
Sbjct: 122 EEEEVFDDAYDNWDGFKDALNS-----------------------FENDNNESSRLVTED 158
Query: 188 FKSEVPESKPN--RRAWKPDDTLRPRCLPNLSKQRNSPLNSNWRGSRKTVPWAWQTIISQ 245
F+ E + P+ +R K P + + N SN R
Sbjct: 159 FEQEEEDLIPDTSQRMNKCKQEAAPGNQTTIHRNSNKKKRSNSEKQRG----------DG 208
Query: 246 PSQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRKLY 294
+CPIC E +D TD F PC+CGF +CLFCH KI E +ARCP+CRK Y
Sbjct: 209 DEECPICSELMDATDLEFEPCTCGFRICLFCHNKISENEARCPACRKDY 257
>UniRef100_UPI000042D574 UPI000042D574 UniRef100 entry
Length = 873
Score = 68.9 bits (167), Expect = 2e-10
Identities = 25/47 (53%), Positives = 33/47 (70%)
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADARCPSCRKLYD 295
C +C E LD++D +F PC CG +C FC+ K+L DARCP CR+ YD
Sbjct: 64 CLLCAEPLDLSDLNFKPCQCGLQICQFCYNKLLSTDARCPGCRRTYD 110
>UniRef100_Q9P927 NOT4p [Candida albicans]
Length = 576
Score = 67.4 bits (163), Expect = 5e-10
Identities = 27/60 (45%), Positives = 39/60 (65%), Gaps = 5/60 (8%)
Query: 241 TIISQPSQ--CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKIL---EADARCPSCRKLYD 295
T IS + CP+C E++D++D +F PC CG+ +C FC+ I E + RCP CR+LYD
Sbjct: 7 TFISDDEEEYCPLCVEEMDISDKNFKPCPCGYQICQFCYNNIRQNPELNGRCPGCRRLYD 66
>UniRef100_Q6BKP1 Similar to CA0449|CaMOT2.3 Candida albicans CaMOT2.3
transcriptional repressor [Debaryomyces hansenii]
Length = 652
Score = 67.0 bits (162), Expect = 6e-10
Identities = 24/50 (48%), Positives = 35/50 (70%), Gaps = 3/50 (6%)
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKIL---EADARCPSCRKLYD 295
CP+C E++D++D +F PC CG+ +C FC+ I E + RCP CR+LYD
Sbjct: 18 CPLCVEEMDISDKNFKPCPCGYQICQFCYNNIRQNPELNGRCPGCRRLYD 67
>UniRef100_Q86KE2 Similar to Dictyostelium discoideum (Slime mold). MkpA protein
[Dictyostelium discoideum]
Length = 1348
Score = 66.6 bits (161), Expect = 8e-10
Identities = 25/58 (43%), Positives = 40/58 (68%), Gaps = 1/58 (1%)
Query: 245 QPSQCPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILEADA-RCPSCRKLYDHVDGNV 301
+ + CP+C ++L D F PC CG+ +C+FC ++I E++ RCP+CRK YD V+ +V
Sbjct: 4 EDNSCPLCLDELSKADRKFRPCPCGYQICVFCFERIRESEQNRCPACRKTYDSVNFSV 61
>UniRef100_P34909 General negative regulator of transcription subunit 4
[Saccharomyces cerevisiae]
Length = 587
Score = 66.6 bits (161), Expect = 8e-10
Identities = 27/56 (48%), Positives = 37/56 (65%), Gaps = 5/56 (8%)
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKIL---EADARCPSCRKLYDHVDGNV 301
CP+C E +D+TD +F PC CG+ +C FC+ I E + RCP+CR+ YD D NV
Sbjct: 33 CPLCIEPMDITDKNFFPCPCGYQICQFCYNNIRQNPELNGRCPACRRKYD--DENV 86
>UniRef100_Q75AJ1 ADL064Wp [Ashbya gossypii]
Length = 646
Score = 66.2 bits (160), Expect = 1e-09
Identities = 25/50 (50%), Positives = 34/50 (68%), Gaps = 3/50 (6%)
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKIL---EADARCPSCRKLYD 295
CP+C E LD+TD +F PC CG+ +C FC+ I E + RCP+CR+ YD
Sbjct: 33 CPLCMEPLDITDKNFKPCPCGYQICQFCYNNIRQNPELNGRCPACRRKYD 82
>UniRef100_Q6FPK5 Similar to sp|P34909 Saccharomyces cerevisiae YER068w MOT2
transcriptional repressor [Candida glabrata]
Length = 620
Score = 64.7 bits (156), Expect = 3e-09
Identities = 23/50 (46%), Positives = 34/50 (68%), Gaps = 3/50 (6%)
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKIL---EADARCPSCRKLYD 295
CP+C E +D+TD +F PC CG+ +C FC+ I E + RCP+CR+ +D
Sbjct: 33 CPLCIEPMDITDKNFFPCPCGYQICQFCYNNIRQNPELNGRCPACRRKFD 82
>UniRef100_Q6CU31 Similarity [Kluyveromyces lactis]
Length = 574
Score = 63.5 bits (153), Expect = 7e-09
Identities = 25/50 (50%), Positives = 32/50 (64%), Gaps = 3/50 (6%)
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKIL---EADARCPSCRKLYD 295
CP+C E LDV D F PC CG+ +C FC+ I E + RCP+CR+ YD
Sbjct: 38 CPLCIEPLDVMDKHFKPCPCGYQICQFCYNNIRQNPELNGRCPACRRKYD 87
>UniRef100_Q8J1S7 CCr4/NOT complex/transcription factor subunit [Kluyveromyces
lactis]
Length = 574
Score = 63.5 bits (153), Expect = 7e-09
Identities = 25/50 (50%), Positives = 32/50 (64%), Gaps = 3/50 (6%)
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKIL---EADARCPSCRKLYD 295
CP+C E LDV D F PC CG+ +C FC+ I E + RCP+CR+ YD
Sbjct: 38 CPLCIEPLDVMDKHFKPCPCGYQICQFCYNNIRQNPELNGRCPACRRKYD 87
>UniRef100_Q09818 Putative general negative regulator of transcription C16C9.04c
[Schizosaccharomyces pombe]
Length = 489
Score = 63.5 bits (153), Expect = 7e-09
Identities = 23/47 (48%), Positives = 34/47 (71%), Gaps = 1/47 (2%)
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILE-ADARCPSCRKLY 294
CP+C E++D++D +F PC CG+ +C FC I E + RCP+CR+LY
Sbjct: 18 CPLCMEEIDISDKNFKPCQCGYRVCRFCWHHIKEDLNGRCPACRRLY 64
>UniRef100_Q6C372 Similar to KLLA0C08041g Kluyveromyces lactis [Yarrowia lipolytica]
Length = 495
Score = 62.8 bits (151), Expect = 1e-08
Identities = 22/50 (44%), Positives = 34/50 (68%), Gaps = 3/50 (6%)
Query: 249 CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKIL---EADARCPSCRKLYD 295
CP+C E++D++D +F PC CG+ +C FC+ I + + RCP CR+ YD
Sbjct: 17 CPLCVEEMDISDRNFKPCPCGYQICQFCYNNIRQNPQLNGRCPGCRRPYD 66
>UniRef100_Q7RS53 Hypothetical protein [Plasmodium yoelii yoelii]
Length = 1386
Score = 61.6 bits (148), Expect = 3e-08
Identities = 38/128 (29%), Positives = 64/128 (49%), Gaps = 15/128 (11%)
Query: 174 EDDKNPRVDFSKADFKSEVPESKPNRRAWKPDDTLRPRCLPNLSKQRNSPLNSNWRGSRK 233
EDD+ +DF + + + + + P D+++ +K+ ++ ++N +K
Sbjct: 49 EDDE---IDFEEDEGEEDGDQINATDINEYPSDSIKN------TKRNDTSSSNNKITKKK 99
Query: 234 TVPWAWQTIISQPSQ-----CPICYEDLDVTDSSFLPCSCGFHLCLFCHKKILE-ADARC 287
TV T IS CP+C E LD TD +F PC CG+ +CL+C I + +C
Sbjct: 100 TVANEKDTQISTIENNNNVICPLCVEQLDETDRNFFPCDCGYQICLWCLYYIRDHMSNKC 159
Query: 288 PSCRKLYD 295
P+CR+ YD
Sbjct: 160 PACRRSYD 167
>UniRef100_UPI00003661E0 UPI00003661E0 UniRef100 entry
Length = 646
Score = 61.2 bits (147), Expect = 4e-08
Identities = 22/50 (44%), Positives = 34/50 (68%), Gaps = 1/50 (2%)
Query: 246 PSQCPICYEDLDVTDSSFLPCSCGFHLCLFC-HKKILEADARCPSCRKLY 294
P +CP+C E L++ D +F PC+CG+ +C FC H+ + + CP+CRK Y
Sbjct: 11 PMECPLCMEPLEIDDVNFFPCTCGYQICRFCWHRIRTDENGLCPACRKPY 60
>UniRef100_UPI00003661DF UPI00003661DF UniRef100 entry
Length = 724
Score = 61.2 bits (147), Expect = 4e-08
Identities = 22/50 (44%), Positives = 34/50 (68%), Gaps = 1/50 (2%)
Query: 246 PSQCPICYEDLDVTDSSFLPCSCGFHLCLFC-HKKILEADARCPSCRKLY 294
P +CP+C E L++ D +F PC+CG+ +C FC H+ + + CP+CRK Y
Sbjct: 11 PMECPLCMEPLEIDDVNFFPCTCGYQICRFCWHRIRTDENGLCPACRKPY 60
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.129 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 580,288,018
Number of Sequences: 2790947
Number of extensions: 25101929
Number of successful extensions: 83072
Number of sequences better than 10.0: 1157
Number of HSP's better than 10.0 without gapping: 294
Number of HSP's successfully gapped in prelim test: 892
Number of HSP's that attempted gapping in prelim test: 77041
Number of HSP's gapped (non-prelim): 3710
length of query: 322
length of database: 848,049,833
effective HSP length: 127
effective length of query: 195
effective length of database: 493,599,564
effective search space: 96251914980
effective search space used: 96251914980
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0081b.3


