

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0081b.1
(360 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
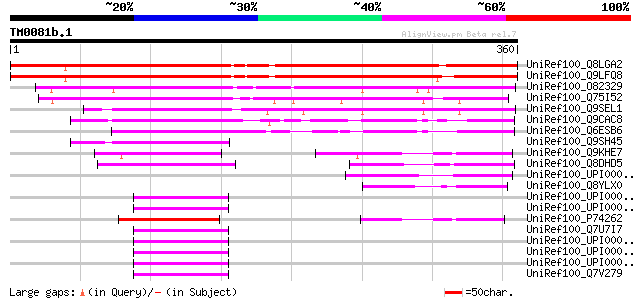
UniRef100_Q8LGA2 Seed maturation-like protein [Arabidopsis thali... 361 2e-98
UniRef100_Q9LFQ8 Seed maturation-like protein [Arabidopsis thali... 360 3e-98
UniRef100_O82329 Hypothetical protein At2g14910 [Arabidopsis tha... 199 1e-49
UniRef100_Q75I52 Expressed protein [Oryza sativa] 194 4e-48
UniRef100_Q9SEL1 Seed maturation protein PM23 [Glycine max] 157 6e-37
UniRef100_Q9CAC8 Hypothetical protein F24D7.19 [Arabidopsis thal... 100 1e-19
UniRef100_Q6ESB6 Seed maturation-like protein [Oryza sativa] 91 4e-17
UniRef100_Q9SH45 F2K11.3 [Arabidopsis thaliana] 62 2e-08
UniRef100_Q9KHE7 Hypothetical protein [Anabaena sp.] 61 5e-08
UniRef100_Q8DHD5 Tll2024 protein [Synechococcus elongatus] 54 9e-06
UniRef100_UPI00002D5DAB UPI00002D5DAB UniRef100 entry 52 3e-05
UniRef100_Q8YLX0 All5176 protein [Anabaena sp.] 51 4e-05
UniRef100_UPI00002DF66A UPI00002DF66A UniRef100 entry 50 7e-05
UniRef100_UPI00002641F6 UPI00002641F6 UniRef100 entry 50 7e-05
UniRef100_P74262 Slr1674 protein [Synechocystis sp.] 50 7e-05
UniRef100_Q7U7I7 Hypothetical protein [Synechococcus sp.] 50 7e-05
UniRef100_UPI00002F4274 UPI00002F4274 UniRef100 entry 50 1e-04
UniRef100_UPI00002A30AC UPI00002A30AC UniRef100 entry 50 1e-04
UniRef100_UPI000025E7FA UPI000025E7FA UniRef100 entry 50 1e-04
UniRef100_Q7V279 Hypothetical protein [Prochlorococcus marinus s... 50 1e-04
>UniRef100_Q8LGA2 Seed maturation-like protein [Arabidopsis thaliana]
Length = 355
Score = 361 bits (926), Expect = 2e-98
Identities = 209/366 (57%), Positives = 260/366 (70%), Gaps = 17/366 (4%)
Query: 1 MAALSWRPF-ILSRLTDLSPNPLHPPKPPPLFLRRRRCF-----LTSCYADGFSSSSSSS 54
MAA S R F +LSR+TDLS L +PPP R + ++S S S
Sbjct: 1 MAAASARAFFMLSRVTDLSKKKLILHQPPPSSSPHRLPYAPNRAVSSSAVISCLSGGGVS 60
Query: 55 SSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLLPS 114
S D VSTR+S DRGF VIAN++ RI+PLD SVISKG+SD+A+DSMKQTIS+MLGLLPS
Sbjct: 61 SDDSYVSTRRSKLDRGFAVIANLVNRIQPLDTSVISKGLSDSAKDSMKQTISSMLGLLPS 120
Query: 115 DHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLDIASPCGARDSDCEKRSEI 174
D FSV+VT+S+ PL+RLL+SSIITGYTLWNAEYR+SL RN DI P R + E +S
Sbjct: 121 DQFSVSVTISEQPLYRLLISSIITGYTLWNAEYRVSLRRNFDI--PIDPRKEE-EDQSSK 177
Query: 175 LEVKGGGEDGGEIEVASDLGLKDLENCSSSPRVFGDLPPQALNYIQQLQSELTNVKEELN 234
V+ G E G ++ DLG E SP+VFGDL P+AL+YIQ LQSEL+++KEEL+
Sbjct: 178 DNVRFGSEKG----MSEDLGNCVEEFERLSPQVFGDLSPEALSYIQLLQSELSSMKEELD 233
Query: 235 ARKQEMMQLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVD 294
++K++ +++E ++G RN+LL+YLRSLDPEMVTELS+ SS EVE+I++QLVQN+L R F D
Sbjct: 234 SQKKKALRIECEKGNRNDLLDYLRSLDPEMVTELSQLSSPEVEEIVNQLVQNVLERLFED 293
Query: 295 DASGSFMEQSVEGNIDNHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQLS 354
+ +FM+ D GD V TSRDYLAKLLFWCMLLGHHLRGLENRL LS
Sbjct: 294 QTTSNFMQNPG----IRTTDGGDGTGRKVDTSRDYLAKLLFWCMLLGHHLRGLENRLHLS 349
Query: 355 CVVGLL 360
CVVGLL
Sbjct: 350 CVVGLL 355
>UniRef100_Q9LFQ8 Seed maturation-like protein [Arabidopsis thaliana]
Length = 355
Score = 360 bits (924), Expect = 3e-98
Identities = 210/370 (56%), Positives = 263/370 (70%), Gaps = 25/370 (6%)
Query: 1 MAALSWRPF-ILSRLTDLSPNPLHPPKPPPLFLRRRRCF-----LTSCYADGFSSSSSSS 54
MAA S R F +LSR+TDLS L +PPP R + ++S S S
Sbjct: 1 MAAASARAFFMLSRVTDLSKKKLILHQPPPSSSPHRLPYAPNRAVSSSAVISCLSGGGVS 60
Query: 55 SSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLLPS 114
S D VSTR+S DRGF VIAN++ RI+PLD SVISKG+SD+A+DSMKQTIS+MLGLLPS
Sbjct: 61 SDDSYVSTRRSKLDRGFAVIANLVNRIQPLDTSVISKGLSDSAKDSMKQTISSMLGLLPS 120
Query: 115 DHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLDIASPCGARDSDCEKRSEI 174
D FSV+VT+S+ PL+RLL+SSIITGYTLWNAEYR+SL RN DI P R + E +S
Sbjct: 121 DQFSVSVTISEQPLYRLLISSIITGYTLWNAEYRVSLRRNFDI--PIDPRKEE-EDQSSK 177
Query: 175 LEVKGGGEDGGEIEVASDLGLKDLENCSSSPRVFGDLPPQALNYIQQLQSELTNVKEELN 234
V+ G E G ++ DLG E SP+VFGDL P+AL+YIQ LQSEL+++KEEL+
Sbjct: 178 DNVRFGSEKG----MSEDLGNCVEEFERLSPQVFGDLSPEALSYIQLLQSELSSMKEELD 233
Query: 235 ARKQEMMQLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVD 294
++K++ +++E ++G RN+LL+YLRSLDPEMVTELS+ SS EVE+I++QLVQN+L R F D
Sbjct: 234 SQKKKALRIECEKGNRNDLLDYLRSLDPEMVTELSQLSSPEVEEIVNQLVQNVLERLFED 293
Query: 295 DASGSFME----QSVEGNIDNHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENR 350
+ +FM+ ++ EG GD V TSRDYLAKLLFWCMLLGHHLRGLENR
Sbjct: 294 QTTSNFMQNPGIRTTEG--------GDGTGRKVDTSRDYLAKLLFWCMLLGHHLRGLENR 345
Query: 351 LQLSCVVGLL 360
L LSCVVGLL
Sbjct: 346 LHLSCVVGLL 355
>UniRef100_O82329 Hypothetical protein At2g14910 [Arabidopsis thaliana]
Length = 386
Score = 199 bits (505), Expect = 1e-49
Identities = 135/366 (36%), Positives = 203/366 (54%), Gaps = 29/366 (7%)
Query: 19 PNPLHPPKPP-------PLFLRRRRCFLTSCYADGFSSSSSSSSSDDVVSTRKSTFDRGF 71
P LH P P P F RR R + + S++ SS+ DD S T
Sbjct: 14 PQLLHKPTKPLPFLFLLPRFNRRFRSLTITSSSTTSSNNFSSNCGDDGFSLDDFTLHSDS 73
Query: 72 T-----VIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKH 126
V++++++ IEPLD S+I K V D+MK+TIS MLGLLPSD F V +
Sbjct: 74 RSPKKCVLSDLIQEIEPLDVSLIQKDVPVTTLDAMKRTISGMLGLLPSDRFQVHIESLWE 133
Query: 127 PLHRLLVSSIITGYTLWNAEYRMSLTRNLDIASPCGARDSDCEKRSEILEVKGGGEDGGE 186
PL +LLVSS++TGYTL NAEYR+ L +NLD++ G DS + +E +++G D
Sbjct: 134 PLSKLLVSSMMTGYTLRNAEYRLFLEKNLDMSG--GGLDSHASENTE-YDMEGTFPDEDH 190
Query: 187 IEVASDLGLKDLENCSSSPRVFGDLPPQALNYIQQLQSELTNVKEELNARKQEMMQLEYD 246
+ D ++L + G + +A YI +LQS+L++VK+EL +++ L+
Sbjct: 191 VSSKRDSRTQNLSE-TIDEEGLGRVSSEAQEYILRLQSQLSSVKKELQEMRRKNAALQMQ 249
Query: 247 RGI---RNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNIL--------SRFFVDD 295
+ + +N+LL+YLRSL PE V ELS P++ EV++ IH +V +L S+F +
Sbjct: 250 QFVGEEKNDLLDYLRSLQPEKVAELSEPAAPEVKETIHSVVHGLLATLSPKMHSKFPASE 309
Query: 296 A--SGSFMEQSVEGNIDNHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQL 353
+ + +S E + + +F + +RDYLA+LLFWCMLLGH+LRGLE R++L
Sbjct: 310 VPPTETVKAKSDEDCAELVENTSLQFQPLISLTRDYLARLLFWCMLLGHYLRGLEYRMEL 369
Query: 354 SCVVGL 359
V+ L
Sbjct: 370 MEVLSL 375
>UniRef100_Q75I52 Expressed protein [Oryza sativa]
Length = 405
Score = 194 bits (492), Expect = 4e-48
Identities = 141/368 (38%), Positives = 198/368 (53%), Gaps = 45/368 (12%)
Query: 21 PLHPPKPPP---LFLRRRRCFLTSCYADGFSSSSSSSSSDDVVSTRKSTFDRGFTVIANM 77
PL PP PPP LF R G ++S+S + R + +++ N+
Sbjct: 29 PLPPPPPPPQQQLFQAALRLPRRRLAGVGVVAASASPFDELYARGRPAHGSSKKSILWNL 88
Query: 78 LRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSII 137
++ IEPLD SVI K V D+MK+TIS MLGLLPSD F V V +P +LLVSSI+
Sbjct: 89 IQDIEPLDLSVIQKDVPPETVDAMKRTISGMLGLLPSDQFRVVVEALWNPFFKLLVSSIM 148
Query: 138 TGYTLWNAEYRMSLTRNLDIASPCGARDSDCEKRSEILEVKGGGEDGGE----IEVASDL 193
TGYTL NAEYR+S RNL+++ DS+ + R +I E + G ++ +
Sbjct: 149 TGYTLRNAEYRLSFERNLELSE----EDSEGQNR-DISEDNHHNINLGSPVTIFRLSEED 203
Query: 194 GLKDLEN------CSSSPRVFGDLPPQALNYIQQLQSELTNVKEELN--ARKQEMMQLEY 245
L+D E C + G+L PQA +YI QLQS L +K+EL+ RK +Q++
Sbjct: 204 MLQDTEKNDEELPCETVGEDLGNLTPQAEDYIIQLQSRLDAMKKELHDLRRKNSALQMQQ 263
Query: 246 DRG-IRNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFF----------VD 294
G +N+LL+YLRSL PE V ELS +S V++ IH +V +L+ +
Sbjct: 264 FVGEEKNDLLDYLRSLTPEKVAELSESTSPGVQEAIHSVVHGLLATLSPKIHSKAPPPLG 323
Query: 295 DASGSFMEQSVEGNIDNHPDNGDE--------FSDTVGTSRDYLAKLLFWCMLLGHHLRG 346
+ASG + N+ D+ E F + RDYLA+LLFWCMLLGH++RG
Sbjct: 324 NASGGVL------NLGGEDDDCAELVENASLPFQPLISVPRDYLARLLFWCMLLGHYIRG 377
Query: 347 LENRLQLS 354
LE RL+L+
Sbjct: 378 LEYRLELA 385
>UniRef100_Q9SEL1 Seed maturation protein PM23 [Glycine max]
Length = 404
Score = 157 bits (396), Expect = 6e-37
Identities = 113/327 (34%), Positives = 175/327 (52%), Gaps = 32/327 (9%)
Query: 53 SSSSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLL 112
++SS D S K + V+ +++ IEPLD S I K V D+MK+TIS MLGLL
Sbjct: 75 AASSHDFASNSKKS------VLTELIQEIEPLDVSHIQKDVPPTTADAMKRTISGMLGLL 128
Query: 113 PSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLDIASPCGARDSDCEK-R 171
PSD F V + PL +LL+SS++TGYTL N EYR+ L +NLD+ + D EK +
Sbjct: 129 PSDQFHVVIEALWEPLSKLLISSMMTGYTLRNVEYRLCLEKNLDMF------EGDIEKPK 182
Query: 172 SEILEVKGGG---EDGGEIEVASDLGLKD-LENCSSSPRV--FGDLPPQALNYIQQLQSE 225
+E ++V G + IE + L +E + G++ +A YI LQS
Sbjct: 183 AESMKVDLQGLMHDSVNAIEFGKNKNLSSKVEKLHEEVDIQELGEISAEAQQYIFNLQSR 242
Query: 226 LTNVKEELNARKQEMMQLEYDRGI---RNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQ 282
L+++K+EL+ K++ L+ + + +N+LL+YLRSL PE V +LS +S E++D I
Sbjct: 243 LSSMKKELHEVKRKSAALQMQQFVGEEKNDLLDYLRSLQPEQVAQLSEFTSPELKDTILS 302
Query: 283 LVQNILSRFF--VDDASGSFMEQSVEGNIDNHPDNGDE--------FSDTVGTSRDYLAK 332
+V +L+ + + E + G + ++ E F + +RDYLA+
Sbjct: 303 VVHGLLATLSPKMHSKPSTMSENTTVGATNAGSEDCAEVLENSALQFQPVISLTRDYLAR 362
Query: 333 LLFWCMLLGHHLRGLENRLQLSCVVGL 359
LLFWCML L GL +L+ ++ L
Sbjct: 363 LLFWCMLWDTILEGLSVDWKLTDLLSL 389
>UniRef100_Q9CAC8 Hypothetical protein F24D7.19 [Arabidopsis thaliana]
Length = 340
Score = 99.8 bits (247), Expect = 1e-19
Identities = 84/318 (26%), Positives = 151/318 (47%), Gaps = 55/318 (17%)
Query: 44 ADGFSSSSSSSSSDDVVSTRKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMKQ 103
A G SS SS+ SS TR+ R ++ ++ ++P + K ++M+Q
Sbjct: 61 AYGSSSDSSADSSTPPNGTRQPKSRRD--ILLEYVQNVKPEFMEMFVKRAPKHVVEAMRQ 118
Query: 104 TISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLDIASPCGA 163
T++ M+G LP F+VTVT L +L++S ++TGY NA+YR+ L ++L+ +
Sbjct: 119 TVTNMIGTLPPQFFAVTVTSVAENLAQLMMSVLMTGYMFRNAQYRLELQQSLEQVALPEP 178
Query: 164 RDSDCEKRSEILEVKGGGED---GGEIEVASDLGLKDLENCSSSPRVFGDLPPQALNYIQ 220
RD KGG ED G + V+ + + N S ++ A YI+
Sbjct: 179 RDQ-----------KGGDEDYAPGTQKNVSGE--VIRWNNVSGPEKI------DAKKYIE 219
Query: 221 QLQSELTNVKEELNARKQEMMQLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSVEVEDII 280
L++E+ + ++ + +N +LEYL+SL+P+ + EL+ + +V +
Sbjct: 220 LLEAEIEELNRQVGRKSANQ---------QNEILEYLKSLEPQNLKELTSTAGEDVAVAM 270
Query: 281 HQLVQNILSRFFVDDASGSFMEQSVEGNIDNHPDNGDEFSDTVGTSRDYLAKLLFWCMLL 340
+ V+ +L+ V D + ++ N+ TS LAKLL+W M++
Sbjct: 271 NTFVKRLLA---VSDPN------QMKTNVTE-------------TSAADLAKLLYWLMVV 308
Query: 341 GHHLRGLENRLQLSCVVG 358
G+ +R +E R + V+G
Sbjct: 309 GYSIRNIEVRFDMERVLG 326
>UniRef100_Q6ESB6 Seed maturation-like protein [Oryza sativa]
Length = 336
Score = 91.3 bits (225), Expect = 4e-17
Identities = 74/288 (25%), Positives = 136/288 (46%), Gaps = 52/288 (18%)
Query: 73 VIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKHPLHRLL 132
++ ++ ++P + K D+M+QT++ M+G LP F+VTVT L +L+
Sbjct: 85 ILLEYVKNVQPEFMELFIKRAPPQVVDAMRQTVTNMIGTLPPQFFAVTVTTVAENLAQLM 144
Query: 133 VSSIITGYTLWNAEYRMSLTRNLD-IASPCGARDSDCEKRSEILEVKGGGEDGGEIEVAS 191
S ++TGY NA+YR+ L ++L+ IA P ++D + + K GE I
Sbjct: 145 YSVLMTGYMFRNAQYRLELQQSLEQIALPEPKEENDSADYAPGTQKKVTGE---VIRWNK 201
Query: 192 DLGLKDLENCSSSPRVFGDLPPQALNYIQQLQSELTNVKEELNARKQEMMQLEYDRGIRN 251
G + ++ A+ YI+ L++E+ + ++ ARK N
Sbjct: 202 TTGPEKID---------------AVKYIELLEAEIDELSHQV-ARKSSQGS--------N 237
Query: 252 NLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVDDASGSFMEQSVEGNIDN 311
LLEYL++L+P+ + EL+ + +V ++ ++ +L+ V D +
Sbjct: 238 ELLEYLKTLEPQNLKELASSAGEDVVFAMNAFIKRLLA---VSDPA-------------- 280
Query: 312 HPDNGDEFSDTVG-TSRDYLAKLLFWCMLLGHHLRGLENRLQLSCVVG 358
+ TV TS + LA L+FW M++G+ +R +E R + V+G
Sbjct: 281 ------QMKTTVSETSANQLANLMFWLMIVGYSMRNIEVRFDMERVLG 322
Score = 33.9 bits (76), Expect = 7.2
Identities = 26/104 (25%), Positives = 49/104 (47%), Gaps = 23/104 (22%)
Query: 250 RNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVDDASGSFMEQSVEGNI 309
R+ LLEY++++ PE + + + +V D + Q V N++ G+ Q
Sbjct: 83 RDILLEYVKNVQPEFMELFIKRAPPQVVDAMRQTVTNMI---------GTLPPQF----- 128
Query: 310 DNHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQL 353
F+ TV T + LA+L++ ++ G+ R + RL+L
Sbjct: 129 ---------FAVTVTTVAENLAQLMYSVLMTGYMFRNAQYRLEL 163
>UniRef100_Q9SH45 F2K11.3 [Arabidopsis thaliana]
Length = 222
Score = 62.0 bits (149), Expect = 2e-08
Identities = 39/114 (34%), Positives = 64/114 (55%), Gaps = 5/114 (4%)
Query: 44 ADGFSSSSSSSSSDDVVSTR-KSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMK 102
A G SS SS+ SS TR KS D ++ ++ ++P + K ++M+
Sbjct: 61 AYGSSSDSSADSSTPPNGTRPKSRRD----ILLEYVQNVKPEFMEMFVKRAPKHVVEAMR 116
Query: 103 QTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNLD 156
QT++ M+G LP F+VTVT L +L++S ++TGY NA+YR+ L ++L+
Sbjct: 117 QTVTNMIGTLPPQFFAVTVTSVAENLAQLMMSVLMTGYMFRNAQYRLELQQSLE 170
>UniRef100_Q9KHE7 Hypothetical protein [Anabaena sp.]
Length = 157
Score = 60.8 bits (146), Expect = 5e-08
Identities = 38/94 (40%), Positives = 53/94 (55%), Gaps = 4/94 (4%)
Query: 61 STRKSTFDRGFTVIANML----RRIEPLDNSVISKGVSDAARDSMKQTISTMLGLLPSDH 116
S R S F + AN+L + + P + +SK S M++ I +LG LP +H
Sbjct: 48 SNRVSEFFNSDSETANLLWQYVKSLSPETVTQLSKPTSPEVFQVMERNIIGLLGNLPPEH 107
Query: 117 FSVTVTVSKHPLHRLLVSSIITGYTLWNAEYRMS 150
F VT+T S+ L RLL S++I+GY L NAE RMS
Sbjct: 108 FGVTITTSREHLGRLLASAMISGYFLRNAEQRMS 141
Score = 56.6 bits (135), Expect = 1e-06
Identities = 42/143 (29%), Positives = 67/143 (46%), Gaps = 26/143 (18%)
Query: 218 YIQQLQSELTNVKEELNARKQEMMQLEY---DRGIRNNLLEYLRSLDPEMVTELSRPSSV 274
Y++ L S + +E Q E+ D N L +Y++SL PE VT+LS+P+S
Sbjct: 27 YLKPLSSHPPVLLQEEKVSNQSNRVSEFFNSDSETANLLWQYVKSLSPETVTQLSKPTSP 86
Query: 275 EVEDIIHQLVQNILSRFFVDDASGSFMEQSVEGNIDNHPDNGDEFSDTVGTSRDYLAKLL 334
EV + ME+++ G + N P + F T+ TSR++L +LL
Sbjct: 87 EVFQV---------------------MERNIIGLLGNLPP--EHFGVTITTSREHLGRLL 123
Query: 335 FWCMLLGHHLRGLENRLQLSCVV 357
M+ G+ LR E R+ V+
Sbjct: 124 ASAMISGYFLRNAEQRMSFETVL 146
>UniRef100_Q8DHD5 Tll2024 protein [Synechococcus elongatus]
Length = 110
Score = 53.5 bits (127), Expect = 9e-06
Identities = 37/117 (31%), Positives = 57/117 (48%), Gaps = 23/117 (19%)
Query: 242 QLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVDDASGSFM 301
Q D G N LL+YL+S PE++T ++R S E+ II
Sbjct: 12 QSSTDDGQPNLLLKYLQSQSPEVLTRIARSVSPEIRQII--------------------- 50
Query: 302 EQSVEGNIDNHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQLSCVVG 358
Q+V+G + P ++F+ + T RD LA LL M+ G+ LR +E R++L +G
Sbjct: 51 SQNVQGLVGGLP--SEDFNVQIATDRDNLAGLLASAMMTGYFLRQMEQRMELEMSLG 105
Score = 47.4 bits (111), Expect = 6e-04
Identities = 31/99 (31%), Positives = 51/99 (51%), Gaps = 1/99 (1%)
Query: 63 RKSTFDRGFTVIANMLRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVT 122
+ ST D ++ L+ P + I++ VS R + Q + ++G LPS+ F+V +
Sbjct: 12 QSSTDDGQPNLLLKYLQSQSPEVLTRIARSVSPEIRQIISQNVQGLVGGLPSEDFNVQIA 71
Query: 123 VSKHPLHRLLVSSIITGYTLWNAEYRMSLTRNL-DIASP 160
+ L LL S+++TGY L E RM L +L D+ P
Sbjct: 72 TDRDNLAGLLASAMMTGYFLRQMEQRMELEMSLGDLPMP 110
>UniRef100_UPI00002D5DAB UPI00002D5DAB UniRef100 entry
Length = 111
Score = 51.6 bits (122), Expect = 3e-05
Identities = 34/119 (28%), Positives = 57/119 (47%), Gaps = 23/119 (19%)
Query: 239 EMMQLEYDRGIRNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVDDASG 298
E + E D N+L++YL+ P+++ ++R +S E+++II VQ +L
Sbjct: 5 EFLTSETDAISGNDLIKYLQEQSPDVLQRVARSASPEIQEIIRHNVQGLLGLL------- 57
Query: 299 SFMEQSVEGNIDNHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQLSCVV 357
G++F + TSRD LA LL M+ G+ LR +E R++L V
Sbjct: 58 ----------------PGEQFEVKIQTSRDNLAGLLASAMMTGYFLRQMEQRMELEGAV 100
Score = 44.7 bits (104), Expect = 0.004
Identities = 21/63 (33%), Positives = 36/63 (56%)
Query: 89 ISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYR 148
+++ S ++ ++ + +LGLLP + F V + S+ L LL S+++TGY L E R
Sbjct: 34 VARSASPEIQEIIRHNVQGLLGLLPGEQFEVKIQTSRDNLAGLLASAMMTGYFLRQMEQR 93
Query: 149 MSL 151
M L
Sbjct: 94 MEL 96
>UniRef100_Q8YLX0 All5176 protein [Anabaena sp.]
Length = 135
Score = 51.2 bits (121), Expect = 4e-05
Identities = 34/103 (33%), Positives = 55/103 (53%), Gaps = 23/103 (22%)
Query: 251 NNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVDDASGSFMEQSVEGNID 310
N L EY++S+ P+ VT+LS+P S EV +I + V L GN+
Sbjct: 22 NGLWEYVQSMSPQTVTQLSKPGSREVLQLIQRAVVATL------------------GNLP 63
Query: 311 NHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQL 353
+ ++F+ + TSR+ L++LL M+ G+ LR +E RL+L
Sbjct: 64 H-----EQFNTNITTSREELSQLLGAAMVDGYFLRNVEQRLEL 101
Score = 41.6 bits (96), Expect = 0.034
Identities = 21/77 (27%), Positives = 42/77 (54%)
Query: 78 LRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSII 137
++ + P + +SK S +++ + LG LP + F+ +T S+ L +LL ++++
Sbjct: 28 VQSMSPQTVTQLSKPGSREVLQLIQRAVVATLGNLPHEQFNTNITTSREELSQLLGAAMV 87
Query: 138 TGYTLWNAEYRMSLTRN 154
GY L N E R+ L ++
Sbjct: 88 DGYFLRNVEQRLELEKS 104
>UniRef100_UPI00002DF66A UPI00002DF66A UniRef100 entry
Length = 86
Score = 50.4 bits (119), Expect = 7e-05
Identities = 23/67 (34%), Positives = 39/67 (57%)
Query: 89 ISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYR 148
++K S+ +D ++ + +LG+LP +HF V VT ++ L +L S+++TGY L E R
Sbjct: 4 VAKSASNDIQDIIRHNVQGLLGMLPGEHFEVKVTANRDNLANMLASAMMTGYFLRQMEQR 63
Query: 149 MSLTRNL 155
L L
Sbjct: 64 KELEETL 70
>UniRef100_UPI00002641F6 UPI00002641F6 UniRef100 entry
Length = 117
Score = 50.4 bits (119), Expect = 7e-05
Identities = 23/67 (34%), Positives = 39/67 (57%)
Query: 89 ISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYR 148
++K S+ +D ++ + +LG+LP +HF V VT ++ L +L S+++TGY L E R
Sbjct: 35 VAKSASNDIQDIIRHNVQGLLGMLPGEHFEVKVTANRDNLANMLASAMMTGYFLRQMEQR 94
Query: 149 MSLTRNL 155
L L
Sbjct: 95 KELEETL 101
Score = 43.1 bits (100), Expect = 0.012
Identities = 27/103 (26%), Positives = 48/103 (46%), Gaps = 23/103 (22%)
Query: 251 NNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVDDASGSFMEQSVEGNID 310
N+L++YL+ P+ + +++ +S +++DII VQ +L
Sbjct: 18 NSLIQYLQEQTPDTLQRVAKSASNDIQDIIRHNVQGLLGML------------------- 58
Query: 311 NHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQL 353
G+ F V +RD LA +L M+ G+ LR +E R +L
Sbjct: 59 ----PGEHFEVKVTANRDNLANMLASAMMTGYFLRQMEQRKEL 97
>UniRef100_P74262 Slr1674 protein [Synechocystis sp.]
Length = 116
Score = 50.4 bits (119), Expect = 7e-05
Identities = 26/72 (36%), Positives = 44/72 (61%)
Query: 78 LRRIEPLDNSVISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSII 137
++ + P + +S+ S M++ I +LG LP +HF VT++ S+ L RLL S+++
Sbjct: 27 VQELSPETIAQLSRPDSQEVFQVMERNIIGLLGNLPPEHFGVTISTSRENLGRLLASAMM 86
Query: 138 TGYTLWNAEYRM 149
+GY L NAE R+
Sbjct: 87 SGYFLRNAEQRL 98
Score = 48.9 bits (115), Expect = 2e-04
Identities = 32/102 (31%), Positives = 50/102 (48%), Gaps = 23/102 (22%)
Query: 250 RNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVDDASGSFMEQSVEGNI 309
+++L Y++ L PE + +LSRP S EV + ME+++ G +
Sbjct: 20 KDSLWTYVQELSPETIAQLSRPDSQEVFQV---------------------MERNIIGLL 58
Query: 310 DNHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRL 351
N P + F T+ TSR+ L +LL M+ G+ LR E RL
Sbjct: 59 GNLPP--EHFGVTISTSRENLGRLLASAMMSGYFLRNAEQRL 98
>UniRef100_Q7U7I7 Hypothetical protein [Synechococcus sp.]
Length = 117
Score = 50.4 bits (119), Expect = 7e-05
Identities = 23/67 (34%), Positives = 39/67 (57%)
Query: 89 ISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYR 148
++K S+ +D ++ + +LG+LP +HF V VT ++ L +L S+++TGY L E R
Sbjct: 35 VAKSASNDIQDIIRHNVQGLLGMLPGEHFEVKVTANRDNLANMLASAMMTGYFLRQMEQR 94
Query: 149 MSLTRNL 155
L L
Sbjct: 95 KELEETL 101
Score = 43.1 bits (100), Expect = 0.012
Identities = 27/103 (26%), Positives = 48/103 (46%), Gaps = 23/103 (22%)
Query: 251 NNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVDDASGSFMEQSVEGNID 310
N+L++YL+ P+ + +++ +S +++DII VQ +L
Sbjct: 18 NSLIQYLQEQTPDTLQRVAKSASNDIQDIIRHNVQGLLGML------------------- 58
Query: 311 NHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQL 353
G+ F V +RD LA +L M+ G+ LR +E R +L
Sbjct: 59 ----PGEHFEVKVTANRDNLANMLASAMMTGYFLRQMEQRKEL 97
>UniRef100_UPI00002F4274 UPI00002F4274 UniRef100 entry
Length = 112
Score = 50.1 bits (118), Expect = 1e-04
Identities = 24/67 (35%), Positives = 40/67 (58%)
Query: 89 ISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYR 148
++K S+ ++ ++ + +LG+LPSD F V +T SK + LL S+++TGY L E R
Sbjct: 35 VAKSASEDIQEIIRHNVQGLLGMLPSDQFDVKITSSKDNIANLLSSAMMTGYFLRQMEQR 94
Query: 149 MSLTRNL 155
L + L
Sbjct: 95 KELEQTL 101
Score = 44.7 bits (104), Expect = 0.004
Identities = 27/108 (25%), Positives = 51/108 (47%), Gaps = 23/108 (21%)
Query: 246 DRGIRNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVDDASGSFMEQSV 305
D N+L++YL+ PE++ +++ +S ++++II VQ +L
Sbjct: 13 DPNDENDLIQYLQKQSPEVLQRVAKSASEDIQEIIRHNVQGLLGML-------------- 58
Query: 306 EGNIDNHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQL 353
D+F + +S+D +A LL M+ G+ LR +E R +L
Sbjct: 59 ---------PSDQFDVKITSSKDNIANLLSSAMMTGYFLRQMEQRKEL 97
>UniRef100_UPI00002A30AC UPI00002A30AC UniRef100 entry
Length = 112
Score = 50.1 bits (118), Expect = 1e-04
Identities = 24/67 (35%), Positives = 40/67 (58%)
Query: 89 ISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYR 148
++K S+ ++ ++ + +LG+LPSD F V +T SK + LL S+++TGY L E R
Sbjct: 35 VAKSASEDIQEIIRHNVQGLLGMLPSDQFDVKITSSKDNIANLLSSAMMTGYFLRQMEQR 94
Query: 149 MSLTRNL 155
L + L
Sbjct: 95 KELEQTL 101
Score = 44.7 bits (104), Expect = 0.004
Identities = 27/108 (25%), Positives = 51/108 (47%), Gaps = 23/108 (21%)
Query: 246 DRGIRNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVDDASGSFMEQSV 305
D N+L++YL+ PE++ +++ +S ++++II VQ +L
Sbjct: 13 DPNDENDLIQYLQKQSPEVLQRVAKSASEDIQEIIRHNVQGLLGML-------------- 58
Query: 306 EGNIDNHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQL 353
D+F + +S+D +A LL M+ G+ LR +E R +L
Sbjct: 59 ---------PSDQFDVKITSSKDNIANLLSSAMMTGYFLRQMEQRKEL 97
>UniRef100_UPI000025E7FA UPI000025E7FA UniRef100 entry
Length = 111
Score = 50.1 bits (118), Expect = 1e-04
Identities = 24/67 (35%), Positives = 40/67 (58%)
Query: 89 ISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYR 148
++K S+ ++ ++ + +LG+LPSD F V +T SK + LL S+++TGY L E R
Sbjct: 35 VAKSASEDIQEIIRHNVQGLLGMLPSDQFDVKITSSKDNIANLLSSAMMTGYFLRQMEQR 94
Query: 149 MSLTRNL 155
L + L
Sbjct: 95 KELEQTL 101
Score = 44.7 bits (104), Expect = 0.004
Identities = 27/108 (25%), Positives = 51/108 (47%), Gaps = 23/108 (21%)
Query: 246 DRGIRNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVDDASGSFMEQSV 305
D N+L++YL+ PE++ +++ +S ++++II VQ +L
Sbjct: 13 DPNDENDLIQYLQKQSPEVLQRVAKSASEDIQEIIRHNVQGLLGML-------------- 58
Query: 306 EGNIDNHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQL 353
D+F + +S+D +A LL M+ G+ LR +E R +L
Sbjct: 59 ---------PSDQFDVKITSSKDNIANLLSSAMMTGYFLRQMEQRKEL 97
>UniRef100_Q7V279 Hypothetical protein [Prochlorococcus marinus subsp. pastoris]
Length = 112
Score = 50.1 bits (118), Expect = 1e-04
Identities = 24/67 (35%), Positives = 40/67 (58%)
Query: 89 ISKGVSDAARDSMKQTISTMLGLLPSDHFSVTVTVSKHPLHRLLVSSIITGYTLWNAEYR 148
++K S+ ++ ++ + +LG+LPSD F V +T SK + LL S+++TGY L E R
Sbjct: 35 VAKSASEDIQEIIRHNVQGLLGMLPSDQFDVKITSSKDNIANLLSSAMMTGYFLRQMEQR 94
Query: 149 MSLTRNL 155
L + L
Sbjct: 95 KELEQTL 101
Score = 44.7 bits (104), Expect = 0.004
Identities = 27/108 (25%), Positives = 51/108 (47%), Gaps = 23/108 (21%)
Query: 246 DRGIRNNLLEYLRSLDPEMVTELSRPSSVEVEDIIHQLVQNILSRFFVDDASGSFMEQSV 305
D N+L++YL+ PE++ +++ +S ++++II VQ +L
Sbjct: 13 DPNDENDLIQYLQKQSPEVMQRVAKSASEDIQEIIRHNVQGLLGML-------------- 58
Query: 306 EGNIDNHPDNGDEFSDTVGTSRDYLAKLLFWCMLLGHHLRGLENRLQL 353
D+F + +S+D +A LL M+ G+ LR +E R +L
Sbjct: 59 ---------PSDQFDVKITSSKDNIANLLSSAMMTGYFLRQMEQRKEL 97
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.135 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 593,123,088
Number of Sequences: 2790947
Number of extensions: 24157604
Number of successful extensions: 110044
Number of sequences better than 10.0: 98
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 61
Number of HSP's that attempted gapping in prelim test: 109865
Number of HSP's gapped (non-prelim): 171
length of query: 360
length of database: 848,049,833
effective HSP length: 128
effective length of query: 232
effective length of database: 490,808,617
effective search space: 113867599144
effective search space used: 113867599144
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0081b.1


