

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
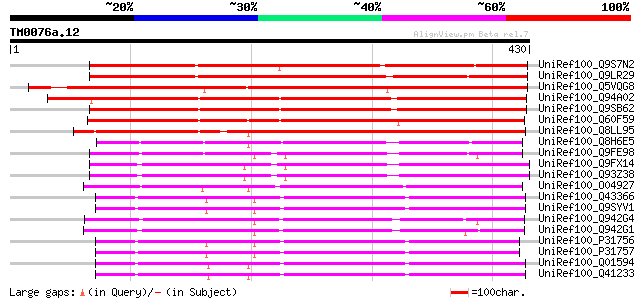
Query= TM0076a.12
(430 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9S7N2 Putative alliinase; 54807-57232 [Arabidopsis th... 377 e-103
UniRef100_Q9LR29 F26F24.17 [Arabidopsis thaliana] 365 1e-99
UniRef100_Q5VQG8 Putative alliinase [Oryza sativa] 350 6e-95
UniRef100_Q94A02 Putative alliin lyase [Arabidopsis thaliana] 344 2e-93
UniRef100_Q9SB62 Putative alliin lyase [Arabidopsis thaliana] 342 1e-92
UniRef100_Q60F59 Putative alliin lyase [Oryza sativa] 342 1e-92
UniRef100_Q8LL95 Putative alliin lyase [Aegilops tauschii] 322 2e-86
UniRef100_Q8H6E5 Hypothetical alliin lyase-like protein [Marchan... 276 8e-73
UniRef100_Q9FE98 Alliinase, putative; 28821-30567 [Arabidopsis t... 254 4e-66
UniRef100_Q9FX14 F12G12.12 protein [Arabidopsis thaliana] 253 6e-66
UniRef100_Q93Z38 At1g34060/F12G12_150 [Arabidopsis thaliana] 253 7e-66
UniRef100_O04927 Alliinase [Allium tuberosum] 243 6e-63
UniRef100_Q43366 Alliinase precursor [Allium cepa] 239 1e-61
UniRef100_Q9SYV1 Alliinase [Allium cepa] 239 1e-61
UniRef100_Q942G4 Putative alliinase [Oryza sativa] 237 4e-61
UniRef100_Q942G1 Putative alliinase [Oryza sativa] 234 3e-60
UniRef100_P31756 Alliin lyase precursor [Allium ascalonicum] 234 4e-60
UniRef100_P31757 Alliin lyase precursor [Allium cepa] 233 1e-59
UniRef100_Q01594 Alliin lyase 1 precursor [Allium sativum] 229 1e-58
UniRef100_Q41233 Alliin lyase 2 precursor [Allium sativum] 228 3e-58
>UniRef100_Q9S7N2 Putative alliinase; 54807-57232 [Arabidopsis thaliana]
Length = 391
Score = 377 bits (968), Expect = e-103
Identities = 183/365 (50%), Positives = 254/365 (69%), Gaps = 7/365 (1%)
Query: 67 LNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVV 126
+NLD GDP + +YW+K + C V I+GC+LMSY SD +N+CW++ PEL+DAI+ LH VV
Sbjct: 25 VNLDHGDPTAYEEYWRKMGDRCTVTIRGCDLMSYFSDMTNLCWFLEPELEDAIKDLHGVV 84
Query: 127 RNAVTKDKYIVIGTGTTQLFMAALFALSPSMTTSHPINVVSATPYYSEYKDQLDLFCSER 186
NA T+D+YIV+GTG+TQL AA+ ALS S+ S P++VV+A P+YS Y ++ S
Sbjct: 85 GNAATEDRYIVVGTGSTQLCQAAVHALS-SLARSQPVSVVAAAPFYSTYVEETTYVRSGM 143
Query: 187 FQWGGDAGEYDKNETYIEVVNYPNNPDGTIKEPMVK--SIVEGKLVHDLAYYWPHYTPIT 244
++W GDA +DK YIE+V PNNPDGTI+E +V E K++HD AYYWPHYTPIT
Sbjct: 144 YKWEGDAWGFDKKGPYIELVTSPNNPDGTIRETVVNRPDDDEAKVIHDFAYYWPHYTPIT 203
Query: 245 HQANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIM 304
+ +HD+MLFT SK TGHAGSRIGWA+VKD EVAKKMV ++ ++SIGV+KESQ R AKI+
Sbjct: 204 RRQDHDIMLFTFSKITGHAGSRIGWALVKDKEVAKKMVEYIIVNSIGVSKESQVRTAKIL 263
Query: 305 GVICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKAYCNFANES 364
V+ + +S + FF+Y + +++ RWEKLR ++ FT+ KYP+A+CN+ +S
Sbjct: 264 NVL---KETCKSESESENFFKYGREMMKNRWEKLREVVKESDAFTLPKYPEAFCNYFGKS 320
Query: 365 FEALPAFVWLKCEEGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELIKR 424
E+ PAF WL +E + L + K+ R G R G+ KHVR+SM+ +D F ++R
Sbjct: 321 LESYPAFAWLGTKEETDLVSE-LRRHKVMSRAGERCGSDKKHVRVSMLSREDVFNVFLER 379
Query: 425 LSNAK 429
L+N K
Sbjct: 380 LANMK 384
>UniRef100_Q9LR29 F26F24.17 [Arabidopsis thaliana]
Length = 387
Score = 365 bits (937), Expect = 1e-99
Identities = 178/363 (49%), Positives = 246/363 (67%), Gaps = 7/363 (1%)
Query: 67 LNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVV 126
+NLD+GDP F +YW KK + C VVI +LMSY SDT NVCW++ PEL+ AI+ LH +
Sbjct: 25 INLDQGDPTAFQEYWMKKKDRCTVVIPAWDLMSYFSDTKNVCWFLEPELEKAIKALHGAI 84
Query: 127 RNAVTKDKYIVIGTGTTQLFMAALFALSPSMTTSHPINVVSATPYYSEYKDQLDLFCSER 186
NA T+++YIV+GTG++QL AALFALS S++ P+++V+A PYYS Y ++ S
Sbjct: 85 GNAATEERYIVVGTGSSQLCQAALFALS-SLSEVKPVSIVAAVPYYSTYVEEASYLQSTL 143
Query: 187 FQWGGDAGEYDKNETYIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAYYWPHYTPITHQ 246
++W GDA +DK YIE+V PNNPDG ++EP+V GK++HDLAYYWPHYTPIT +
Sbjct: 144 YKWEGDARTFDKKGPYIELVTSPNNPDGIMREPVVNRREGGKVIHDLAYYWPHYTPITRR 203
Query: 247 ANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIMGV 306
+HD+MLFT SK TGHAGSRIGWA+VKDIEVAKKMV ++ ++SIGV+KESQTRA I+
Sbjct: 204 QDHDLMLFTFSKITGHAGSRIGWALVKDIEVAKKMVHYLTINSIGVSKESQTRATTILNE 263
Query: 307 ICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKAYCNFANESFE 366
+ + + FFEY ++ RWE+LR +E FT+ YP+ +CNF ++
Sbjct: 264 LTKTCR-----TQSESFFEYGYEKMKSRWERLREVVESGDAFTLPNYPQDFCNFFGKTLS 318
Query: 367 ALPAFVWLKCEEGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELIKRLS 426
PAF WL +E + + L + K+ RGG R G ++VR+SM+ DD+F ++RL+
Sbjct: 319 TSPAFAWLGYKEE-RDLGSLLKEKKVLTRGGDRCGCNKRYVRVSMLSRDDDFDVSLQRLA 377
Query: 427 NAK 429
K
Sbjct: 378 TIK 380
>UniRef100_Q5VQG8 Putative alliinase [Oryza sativa]
Length = 507
Score = 350 bits (897), Expect = 6e-95
Identities = 181/424 (42%), Positives = 258/424 (60%), Gaps = 24/424 (5%)
Query: 16 HSSHLTTRFMSTKQKQRARGFFQEHTLYPSMVVAKAASPINKNGFTSPSSTLNLDKGDPL 75
H + L TR +RAR A + +P + S +NLD GDP
Sbjct: 98 HDAGLATRSSDAAVHRRAR-------------TASSMAPSTGKPAVTTDSVINLDHGDPT 144
Query: 76 VFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVVRNAVTKDKY 135
+F ++W++ + VVI G + MSY SD +NVCW++ PEL + RLH VV NA +
Sbjct: 145 MFEEFWRETGDAAEVVIPGWQTMSYFSDVTNVCWFLEPELDRQVRRLHRVVGNAAVDGYH 204
Query: 136 IVIGTGTTQLFMAALFALSPSMTTS---HPINVVSATPYYSEYKDQLDLFCSERFQWGGD 192
+++GTG+TQLFMAAL+AL+P + PI+VVS PYYS Y D S F+W GD
Sbjct: 205 VLVGTGSTQLFMAALYALAPDAAAAAAGEPISVVSTAPYYSSYPAVTDFLRSGLFRWAGD 264
Query: 193 AGEYDKNETYIEVVNYPNNPDGTIKEPMVKSIV-EGKLVHDLAYYWPHYTPITHQANHDV 251
A + K ++YIE+V PNNPDG I+E ++ G+ VHDLAYYWP YTPIT +A+HD+
Sbjct: 265 ADAF-KGDSYIELVCSPNNPDGAIREAVLDPKTGNGRTVHDLAYYWPQYTPITKRASHDI 323
Query: 252 MLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIMGVICDGY 311
MLFT+SK TGHAG+RIGWA+VKD +A+KM FV++++IGV+K+SQ RAAK++ + DGY
Sbjct: 324 MLFTVSKSTGHAGTRIGWALVKDRAIARKMTKFVELNTIGVSKDSQMRAAKVLAAVSDGY 383
Query: 312 -----QKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKAYCNFANESFE 366
Q E++ F++ + + ERW LR + +F++ + +CNF E+
Sbjct: 384 ERRPEQTKETMTTPLRLFDFGRRKMVERWSMLRAAAAASGIFSLPEETSGFCNFTKETAA 443
Query: 367 ALPAFVWLKCE-EGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELIKRL 425
PAF WL+C+ E +E+ +L KI R G +FGA +++VR+SM+ DD F I RL
Sbjct: 444 TNPAFAWLRCDREDVEDCAGFLRGHKILTRSGAQFGADARYVRVSMLDRDDAFDIFINRL 503
Query: 426 SNAK 429
S+ K
Sbjct: 504 SSLK 507
>UniRef100_Q94A02 Putative alliin lyase [Arabidopsis thaliana]
Length = 440
Score = 344 bits (883), Expect = 2e-93
Identities = 182/409 (44%), Positives = 256/409 (62%), Gaps = 18/409 (4%)
Query: 32 RARGFFQEHTLYPSMVVAKAASPINKNGFTSPSST-----------LNLDKGDPLVFGKY 80
R RG + T Y S+ + + + +S S +NL GDP V+ +Y
Sbjct: 33 RERGDSWDRTAYVSIWPVVSTTASESSSLSSASCNYSKIEEDDDRIINLKFGDPTVYERY 92
Query: 81 WKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVVRNAVTKDKYIVIGT 140
W++ E +VI G + +SY SD +N+CW++ PEL I R+H VV NAVT+D++IV+GT
Sbjct: 93 WQENGEVTTMVIPGWQSLSYFSDENNLCWFLEPELAKEIVRVHKVVGNAVTQDRFIVVGT 152
Query: 141 GTTQLFMAALFALSPSMTTSHPINVVSATPYYSEYKDQLDLFCSERFQWGGDAGEYDKNE 200
G+TQL+ AAL+ALSP S PINVVSATPYYS Y D S ++WGGDA Y ++
Sbjct: 153 GSTQLYQAALYALSPH-DDSGPINVVSATPYYSTYPLITDCLKSGLYRWGGDAKTYKEDG 211
Query: 201 TYIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAYYWPHYTPITHQANHDVMLFTLSKCT 260
YIE+V PNNPDG ++E +V S EG L+HDLAYYWP YTPIT A+HDVMLFT SK T
Sbjct: 212 PYIELVTSPNNPDGFLRESVVNS-TEGILIHDLAYYWPQYTPITSPADHDVMLFTASKST 270
Query: 261 GHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIMGVICDGYQKFESIEPD 320
GHAG RIGWA+VKD E A+KM+ ++++++IGV+K+SQ R AK++ V+ D
Sbjct: 271 GHAGIRIGWALVKDRETARKMIEYIELNTIGVSKDSQLRVAKVLKVVSDSCGNVTG---- 326
Query: 321 QLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKAYCNFANESFEALPAFVWLKCEEGI 380
+ FF++ + + ERW+ L+++ + K F+V + CNF FE PAF W KCEEGI
Sbjct: 327 KSFFDHSYDAMYERWKLLKQAAKDTKRFSVPDFVSQRCNFFGRVFEPQPAFAWFKCEEGI 386
Query: 381 ENAENYL-GKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELIKRLSNA 428
+ E +L + KI + G FG +VR+SM+ D F + R++++
Sbjct: 387 VDCEKFLREEKKILTKSGKYFGDELSNVRISMLDRDTNFNIFLHRITSS 435
>UniRef100_Q9SB62 Putative alliin lyase [Arabidopsis thaliana]
Length = 454
Score = 342 bits (877), Expect = 1e-92
Identities = 173/363 (47%), Positives = 242/363 (66%), Gaps = 7/363 (1%)
Query: 67 LNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVV 126
++L GDP V+ +YW++ E +VI G + +SY SD +N+CW++ PEL I R+H VV
Sbjct: 93 ISLCSGDPTVYERYWQENGEVTTMVIPGWQSLSYFSDENNLCWFLEPELAKEIVRVHKVV 152
Query: 127 RNAVTKDKYIVIGTGTTQLFMAALFALSPSMTTSHPINVVSATPYYSEYKDQLDLFCSER 186
NAVT+D++IV+GTG+TQL+ AAL+ALSP S PINVVSATPYYS Y D S
Sbjct: 153 GNAVTQDRFIVVGTGSTQLYQAALYALSPH-DDSGPINVVSATPYYSTYPLITDCLKSGL 211
Query: 187 FQWGGDAGEYDKNETYIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAYYWPHYTPITHQ 246
++WGGDA Y ++ YIE+V PNNPDG ++E +V S EG L+HDLAYYWP YTPIT
Sbjct: 212 YRWGGDAKTYKEDGPYIELVTSPNNPDGFLRESVVNS-TEGILIHDLAYYWPQYTPITSP 270
Query: 247 ANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIMGV 306
A+HDVMLFT SK TGHAG RIGWA+VKD E A+KM+ ++++++IGV+K+SQ R AK++ V
Sbjct: 271 ADHDVMLFTASKSTGHAGIRIGWALVKDRETARKMIEYIELNTIGVSKDSQLRVAKVLKV 330
Query: 307 ICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKAYCNFANESFE 366
+ D + FF++ + + ERW+ L+++ + K F+V + CNF FE
Sbjct: 331 VSDSCGNVTG----KSFFDHSYDAMYERWKLLKQAAKDTKRFSVPDFVSQRCNFFGRVFE 386
Query: 367 ALPAFVWLKCEEGIENAENYL-GKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELIKRL 425
PAF W KCEEGI + E +L + KI + G FG +VR+SM+ D F + R+
Sbjct: 387 PQPAFAWFKCEEGIVDCEKFLREEKKILTKSGKYFGDELSNVRISMLDRDTNFNIFLHRI 446
Query: 426 SNA 428
+++
Sbjct: 447 TSS 449
>UniRef100_Q60F59 Putative alliin lyase [Oryza sativa]
Length = 441
Score = 342 bits (877), Expect = 1e-92
Identities = 172/368 (46%), Positives = 243/368 (65%), Gaps = 9/368 (2%)
Query: 65 STLNLDKGDPLVFGKYWKKKS-EECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLH 123
S +NLD GDP ++ +W+ + E +VI G + MSY SD ++CW++ P L+ + RLH
Sbjct: 74 SIINLDHGDPTMYEAFWRGGAGERATIVIPGWQTMSYFSDVGSLCWFLEPGLEREVRRLH 133
Query: 124 HVVRNAVTKDKYIVIGTGTTQLFMAALFALSPSMTTSHPINVVSATPYYSEYKDQLDLFC 183
+V NAV ++++GTG+TQLF AAL+ALSP S P+NVVS PYYS Y D
Sbjct: 134 RLVGNAVADGYHVLVGTGSTQLFQAALYALSPP-GPSAPMNVVSPAPYYSSYPAVTDFLK 192
Query: 184 SERFQWGGDAGEYDKNETYIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAYYWPHYTPI 243
S ++W GDA +D +TY+E+V P+NPDG I+E ++KS +G VHDLAYYWP YTPI
Sbjct: 193 SGLYRWAGDAKMFD-GDTYVELVCSPSNPDGGIREAVLKS-GDGVAVHDLAYYWPQYTPI 250
Query: 244 THQANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKI 303
T A HD+MLFT+SKCTGHAG+R+GWA+VKD VA+KM F+++++IGV+K+SQ RAAKI
Sbjct: 251 TSAAAHDIMLFTVSKCTGHAGTRLGWALVKDRAVAQKMSKFIELNTIGVSKDSQLRAAKI 310
Query: 304 MGVICDGYQKFESIEPD----QLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKAYCN 359
+ I DGY + + D F + + + RW KLR ++ +FT+ +C
Sbjct: 311 LKAITDGYDRAPAAGDDDDDSSRLFHFARRKMVSRWAKLRAAVAASGIFTLPDELPGHCT 370
Query: 360 FANESFEALPAFVWLKC-EEGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGTDDEF 418
FANE+ A P F WL+C +EG+++ E YL + KI RGG +FGA + VR+SM+ TD+ F
Sbjct: 371 FANETVSAYPPFAWLRCGKEGVDDLEGYLRERKIISRGGGKFGADGRVVRISMLDTDEAF 430
Query: 419 TELIKRLS 426
+ RL+
Sbjct: 431 AIFVDRLA 438
>UniRef100_Q8LL95 Putative alliin lyase [Aegilops tauschii]
Length = 427
Score = 322 bits (824), Expect = 2e-86
Identities = 166/378 (43%), Positives = 236/378 (61%), Gaps = 11/378 (2%)
Query: 54 PINKNGFTSPSSTLNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLP 113
P K SP + +NLD GDP ++ +W++ E VI G +SY S+ +CWY+ P
Sbjct: 55 PDAKGDALSPDAVINLD-GDPTMYEAFWREAGERAAAVIPGWWGVSYFSNAGGLCWYLEP 113
Query: 114 ELKDAIERLHHVVRNAVTKDKYIVIGTGTTQLFMAALFALSPSMTTSHPINVVSATPYYS 173
EL+ + R+H +V NA+ ++V+GTGTTQLF AA++AL+P+ +T P+ VVS PYYS
Sbjct: 114 ELEREVRRVHRLVGNAMADGYHLVVGTGTTQLFQAAMYALAPT-STDRPVGVVSPAPYYS 172
Query: 174 EYKDQLDLFCSERFQWGGDAGEY--DKNETYIEVVNYPNNPDGTIKEPMVKS-IVEGKLV 230
DL S ++W GDA + D IE+V PNNPDG I+E ++ S +GK +
Sbjct: 173 T-----DLLLSGYYRWAGDANAFHGDHGGENIELVVSPNNPDGAIREAVLSSDSSKGKAI 227
Query: 231 HDLAYYWPHYTPITHQANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSI 290
HDL YYWP YTPIT A HD+MLFT+SK +GH G+R+GWA+VKD +VAKKMV FV S+I
Sbjct: 228 HDLVYYWPQYTPITSPAVHDIMLFTMSKLSGHGGTRLGWALVKDSDVAKKMVYFVYGSTI 287
Query: 291 GVAKESQTRAAKIMGVICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTV 350
GV+K+SQ RAAKI+GV+ D Y+ F++ + ERW LR ++ F++
Sbjct: 288 GVSKDSQLRAAKILGVVSDAYEPGPGDGTQLRLFDFARRRTGERWRALRAAVAATGTFSL 347
Query: 351 AKYPKAYCNFANESFEALPAFVWLKC-EEGIENAENYLGKLKIRGRGGVRFGAGSKHVRL 409
YCNF ++ A PAF WL+C ++G+E+ +L I RGG +FG ++ VR+
Sbjct: 348 PDEITGYCNFTKQTVAAYPAFAWLRCHKDGVEDCAEFLRGHGIVARGGEQFGGDARCVRV 407
Query: 410 SMIGTDDEFTELIKRLSN 427
+M+ D F LI+RLS+
Sbjct: 408 NMLDRDAVFNVLIQRLSS 425
>UniRef100_Q8H6E5 Hypothetical alliin lyase-like protein [Marchantia polymorpha]
Length = 390
Score = 276 bits (706), Expect = 8e-73
Identities = 150/355 (42%), Positives = 211/355 (59%), Gaps = 16/355 (4%)
Query: 73 DPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVVRNAVTK 132
DP +F YW++ + C I G + MSY +++ +++ EL I LH V+ NAVT+
Sbjct: 40 DPTMFENYWREHGDSCTTTILGWQRMSYYANSKQF-FFVQTELDQVIRSLHDVIGNAVTE 98
Query: 133 DKYIVIGTGTTQLFMAALFALSPSMTTSHPINVVSATPYYSEYKDQLDLFCSERFQWGGD 192
++IVIG G+TQLF AAL+ALSP + P VVS P+YS Y D S +QW GD
Sbjct: 99 GRHIVIGVGSTQLFQAALYALSPPDRAT-PTKVVSVAPFYSSYPTITDYLKSSLYQWAGD 157
Query: 193 AGEY--DKNETYIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAYYWPHYTPITHQANHD 250
A Y +++YIE+V PNNP G +++ +V G VHDLAYYWPH+TPIT A+HD
Sbjct: 158 ASVYRPQSDDSYIELVTTPNNPTGEVRQSVVSGGA-GFPVHDLAYYWPHHTPITSPADHD 216
Query: 251 VMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIMGVICDG 310
+MLFT+SK TGHAG+RIGWA+VKD++VA+KM F+++++IGV+K++Q RAA I+
Sbjct: 217 LMLFTVSKSTGHAGTRIGWALVKDLKVAQKMAKFIELNTIGVSKDAQIRAAHILRARIGA 276
Query: 311 YQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKAYCNFANESFEALPA 370
Q F + + RW++LR + F+V ++ A+C F E PA
Sbjct: 277 KQ----------LFHFAAAEMSYRWQRLRSVLTKGTRFSVPEFQPAWCTFFGEIRSPAPA 326
Query: 371 FVWLKCEEGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELIKRL 425
F WL+C E + E +L I R G FG+ + VRLSM+ F LI RL
Sbjct: 327 FAWLRCNEEAD-CERFLRHSGILTRSGGYFGSNTSFVRLSMLDRRASFEILIDRL 380
>UniRef100_Q9FE98 Alliinase, putative; 28821-30567 [Arabidopsis thaliana]
Length = 457
Score = 254 bits (648), Expect = 4e-66
Identities = 142/371 (38%), Positives = 222/371 (59%), Gaps = 29/371 (7%)
Query: 67 LNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVV 126
++ + GDPL +W +K+EE VV G MSY + + +M EL+ I +LH+VV
Sbjct: 90 VDANSGDPLFLEPFWIRKAEESAVVESGWHRMSYTFNGYGL--FMSAELEKIIRKLHNVV 147
Query: 127 RNAVTKDKYIVIGTGTTQLFMAALFALSPSMTTSHPINVVSATPYYSEYKDQLDLFCSER 186
NAVT +++I+ G G TQL A++ ALS + + S P +V++ PYY+ YK Q D F S
Sbjct: 148 GNAVTDNRFIIFGAGATQLLAASVHALSQTNSLS-PSRLVTSVPYYNLYKQQADFFNSTN 206
Query: 187 FQWGGDAGEYDKNET------YIEVVNYPNNPDGTIKEPMVKSIVEG---KLVHDLAYYW 237
++ GDA + ++E IE+V PNNPDG +K +++++G K +HD AYYW
Sbjct: 207 LKFEGDASAWKRSERNDDIKQVIEIVTSPNNPDGKLK----RAVLDGPNVKYIHDYAYYW 262
Query: 238 PHYTPITHQANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQ 297
P+++PIT QA+ D+ LF+LSK TGHAGSR GWA+VK+ V +KM +++ +SS+GV++++Q
Sbjct: 263 PYFSPITRQADEDLSLFSLSKTTGHAGSRFGWALVKEKTVYEKMKIYISLSSMGVSRDTQ 322
Query: 298 TRAAKIMGVIC-DGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKA 356
RA +++ V+ DG + F + L++RWE L + + F++
Sbjct: 323 LRALQLLKVVIGDGGNE---------IFRFGYGTLKKRWEILNKIFSMSTRFSLETIKPE 373
Query: 357 YCNFANESFEALPAFVWLKCEEGIENAENY--LGKLKIRGRGGVRFGAGSKHVRLSMIGT 414
YCN+ + E P++ W+KCE E+ + Y KI GR G FG+ + VRLS+I +
Sbjct: 374 YCNYFKKVREFTPSYAWVKCERP-EDTDCYEIFKAAKITGRNGEMFGSDERFVRLSLIRS 432
Query: 415 DDEFTELIKRL 425
D+F +LI L
Sbjct: 433 QDDFDQLIAML 443
>UniRef100_Q9FX14 F12G12.12 protein [Arabidopsis thaliana]
Length = 463
Score = 253 bits (647), Expect = 6e-66
Identities = 137/376 (36%), Positives = 217/376 (57%), Gaps = 28/376 (7%)
Query: 67 LNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVV 126
++ + GDPL +W +++E +++ G MSY+ + Y+ EL+ I +LH VV
Sbjct: 91 VDANSGDPLFLEPFWMRQAERSAILVSGWHRMSYIYEDGT---YVSRELEKVIRKLHSVV 147
Query: 127 RNAVTKDKYIVIGTGTTQLFMAALFALS-PSMTTSHPINVVSATPYYSEYKDQLDLFCSE 185
NAVT +++++ G+GTTQL AA+ ALS + + S P ++++ PYY+ YKDQ + F S
Sbjct: 148 GNAVTDNRFVIFGSGTTQLLAAAVHALSLTNSSVSSPARLLTSIPYYAMYKDQAEFFDSA 207
Query: 186 RFQWGGDA------GEYDKNETYIEVVNYPNNPDGTIKEPMVKSIVEG---KLVHDLAYY 236
++ G+A G D IEVV PNNPDG +K +++++G K +HD AYY
Sbjct: 208 HLKFEGNASAWKQSGRNDNITQVIEVVTSPNNPDGKLK----RAVLDGPNVKTLHDYAYY 263
Query: 237 WPHYTPITHQANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKES 296
WPH++PITH + D+ LF+LSK TGHAGSR GW +VKD + +KM F++++S+GV+KE+
Sbjct: 264 WPHFSPITHPVDEDLSLFSLSKTTGHAGSRFGWGLVKDKAIYEKMDRFIRLTSMGVSKET 323
Query: 297 QTRAAKIMGVIC-DGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPK 355
Q +++ V+ DG + F + +++RWE L + + F++
Sbjct: 324 QLHVLQLLKVVVGDGGNE---------IFSFGYGTVKKRWETLNKIFSMSTRFSLQTIKP 374
Query: 356 AYCNFANESFEALPAFVWLKCEEGIE-NAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGT 414
YCN+ + E P++ W+KCE + N KI GR G FG+ + VRLS+I +
Sbjct: 375 EYCNYFKKVREFTPSYAWVKCERPEDTNCYEIFRAAKITGRNGNVFGSEERFVRLSLIRS 434
Query: 415 DDEFTELIKRLSNAKY 430
D+F +LI L Y
Sbjct: 435 QDDFDQLIAMLKKLVY 450
>UniRef100_Q93Z38 At1g34060/F12G12_150 [Arabidopsis thaliana]
Length = 463
Score = 253 bits (646), Expect = 7e-66
Identities = 137/376 (36%), Positives = 217/376 (57%), Gaps = 28/376 (7%)
Query: 67 LNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVV 126
++ + GDPL +W +++E +++ G MSY+ + Y+ EL+ I +LH VV
Sbjct: 91 VDANSGDPLSLEPFWMRQAERSAILVSGWHRMSYIYEDGT---YVSRELEKVIRKLHSVV 147
Query: 127 RNAVTKDKYIVIGTGTTQLFMAALFALS-PSMTTSHPINVVSATPYYSEYKDQLDLFCSE 185
NAVT +++++ G+GTTQL AA+ ALS + + S P ++++ PYY+ YKDQ + F S
Sbjct: 148 GNAVTDNRFVIFGSGTTQLLAAAVHALSLTNSSVSSPARLLTSIPYYAMYKDQAEFFDSA 207
Query: 186 RFQWGGDA------GEYDKNETYIEVVNYPNNPDGTIKEPMVKSIVEG---KLVHDLAYY 236
++ G+A G D IEVV PNNPDG +K +++++G K +HD AYY
Sbjct: 208 HLKFEGNASAWKQSGRNDNITQVIEVVTSPNNPDGKLK----RAVLDGPNVKTLHDYAYY 263
Query: 237 WPHYTPITHQANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKES 296
WPH++PITH + D+ LF+LSK TGHAGSR GW +VKD + +KM F++++S+GV+KE+
Sbjct: 264 WPHFSPITHPVDEDLSLFSLSKTTGHAGSRFGWGLVKDKAIYEKMDRFIRLTSMGVSKET 323
Query: 297 QTRAAKIMGVIC-DGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPK 355
Q +++ V+ DG + F + +++RWE L + + F++
Sbjct: 324 QLHVLQLLKVVVGDGGNE---------IFSFGYGTVKKRWETLNKIFSMSTRFSLQTIKP 374
Query: 356 AYCNFANESFEALPAFVWLKCEEGIE-NAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGT 414
YCN+ + E P++ W+KCE + N KI GR G FG+ + VRLS+I +
Sbjct: 375 EYCNYFKKVREFTPSYAWVKCERPEDTNCYEIFRAAKITGRNGNVFGSEERFVRLSLIRS 434
Query: 415 DDEFTELIKRLSNAKY 430
D+F +LI L Y
Sbjct: 435 QDDFDQLIAMLKKLVY 450
>UniRef100_O04927 Alliinase [Allium tuberosum]
Length = 476
Score = 243 bits (621), Expect = 6e-63
Identities = 136/373 (36%), Positives = 216/373 (57%), Gaps = 15/373 (4%)
Query: 62 SPSSTLNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIER 121
+P+ + ++ GD L +YWK E V++ G MSY + +M ELK I
Sbjct: 82 TPNCSADVASGDALFLEEYWKDHKENTAVLVSGWHRMSYFFPEKDSD-FMSAELKRTITE 140
Query: 122 LHHVVRNAVTKDKYIVIGTGTTQLFMAALFALSPSMT---TSHPINVVSATPYYSEYKDQ 178
LH +V NA TK K+IV G G TQL + +SP+++ T+ P VV+ PYY+ ++DQ
Sbjct: 141 LHEIVGNAETKGKHIVFGVGVTQLLHGLVLTISPNISNCPTAGPAKVVARAPYYAVFRDQ 200
Query: 179 LDLFCSERFQWGGDAGEY--DKNET-YIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAY 235
F ++ ++W G+A Y D N +IE+V PNNP+G +++ M IV ++D+ Y
Sbjct: 201 TSYFDNKGYEWKGNAANYVNDPNPNQFIELVTSPNNPEGNLRKAM---IVGSTAIYDMVY 257
Query: 236 YWPHYTPITHQANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKE 295
YWPH+TPIT++A+ D+MLFT+SK TGH+GSR GWAI+KD VA K+V F+ ++ G ++E
Sbjct: 258 YWPHFTPITYKADEDIMLFTMSKYTGHSGSRFGWAIIKDENVAIKLVEFMSKNTEGTSRE 317
Query: 296 SQTRAAKIMGVICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSI-ELRKVFTVAKYP 354
+Q R ++ + + + D FF + LR+RWEK+ + + K F+
Sbjct: 318 TQLRTLILLKEVIAMIKTHKGTPKDINFFGF--QHLRQRWEKVTELLDQSNKRFSYQHLN 375
Query: 355 KA-YCNFANESFEALPAFVWLKCE-EGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMI 412
++ +CN+ P++ W++C G EN + I + G RF AGS++VRLS+I
Sbjct: 376 QSEHCNYMRMKRPPSPSYAWVRCNWPGEENCSEVFKEGGIITQDGPRFEAGSQYVRLSLI 435
Query: 413 GTDDEFTELIKRL 425
T+D+F +++ L
Sbjct: 436 KTNDDFDQMMDHL 448
>UniRef100_Q43366 Alliinase precursor [Allium cepa]
Length = 479
Score = 239 bits (609), Expect = 1e-61
Identities = 131/364 (35%), Positives = 209/364 (56%), Gaps = 15/364 (4%)
Query: 72 GDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVVRNAVT 131
GD L +YW++ E V++ G MSY + V ++ EL+ I+ LH +V NA
Sbjct: 98 GDGLFLEEYWQQHKENSAVLVSGWHRMSYFFNP--VSNFISFELEKTIKELHEIVGNAAA 155
Query: 132 KDKYIVIGTGTTQLFMAALFALSPSMTTSH---PINVVSATPYYSEYKDQLDLFCSERFQ 188
KD+YIV G G TQL + +LSP+MT + VV+ PYY +++Q F + ++
Sbjct: 156 KDRYIVFGVGVTQLIHGLVISLSPNMTATPCAPQSKVVAHAPYYPVFREQTKYFDKKGYE 215
Query: 189 WGGDAGEYDKNET---YIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAYYWPHYTPITH 245
W G+A +Y T +IE+V PNNP+G ++ ++K K ++D+ YYWPHYTPI +
Sbjct: 216 WKGNAADYVNTSTPEQFIEMVTSPNNPEGLLRHEVIKGC---KSIYDMVYYWPHYTPIKY 272
Query: 246 QANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIMG 305
+A+ D+MLFT+SK TGH+GSR GWA++KD V K++ ++ ++ G ++E+Q R+ KI+
Sbjct: 273 KADEDIMLFTMSKYTGHSGSRFGWALIKDETVYNKLLNYMTKNTEGTSRETQLRSLKILK 332
Query: 306 VICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKA-YCNFANES 364
+ + + D F + K LRERW + ++ F+ K P++ YCN+
Sbjct: 333 EVIAMVKTQKGTMRDLNTFGFQK--LRERWVNITSLLDKSDRFSYQKLPQSEYCNYFRRM 390
Query: 365 FEALPAFVWLKCE-EGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELIK 423
P++ W+KCE E ++ +I + G F AGS++VRLS+I T D+F +L+
Sbjct: 391 RPPSPSYAWVKCEWEEDKDCYQTFQNGRINTQNGEGFEAGSRYVRLSLIKTKDDFDQLMY 450
Query: 424 RLSN 427
L N
Sbjct: 451 YLKN 454
>UniRef100_Q9SYV1 Alliinase [Allium cepa]
Length = 479
Score = 239 bits (609), Expect = 1e-61
Identities = 131/364 (35%), Positives = 209/364 (56%), Gaps = 15/364 (4%)
Query: 72 GDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVVRNAVT 131
GD L +YW++ E V++ G MSY + V ++ EL+ I+ LH +V NA
Sbjct: 98 GDGLFLEEYWQQHKENSAVLVSGWHRMSYFFNP--VSNFISFELEKTIKELHEIVGNAAA 155
Query: 132 KDKYIVIGTGTTQLFMAALFALSPSMTTSH---PINVVSATPYYSEYKDQLDLFCSERFQ 188
KD+YIV G G TQL + +LSP+MT + VV+ PYY +++Q F + ++
Sbjct: 156 KDRYIVFGVGVTQLIHGLVISLSPNMTATPCAPQSKVVAHAPYYPVFREQTKYFDKKGYE 215
Query: 189 WGGDAGEYDKNET---YIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAYYWPHYTPITH 245
W G+A +Y T +IE+V PNNP+G ++ ++K K ++D+ YYWPHYTPI +
Sbjct: 216 WKGNAADYVNTSTPEQFIEMVTSPNNPEGLLRHEVIKGC---KSIYDMVYYWPHYTPIKY 272
Query: 246 QANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIMG 305
+A+ D+MLFT+SK TGH+GSR GWA++KD V K++ ++ ++ G ++E+Q R+ KI+
Sbjct: 273 KADEDIMLFTMSKYTGHSGSRFGWALIKDETVYNKLLNYMTKNTEGTSRETQLRSLKILK 332
Query: 306 VICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKA-YCNFANES 364
+ + + D F + K LRERW + ++ F+ K P++ YCN+
Sbjct: 333 EVIAMVKTQKGTMRDLNTFGFQK--LRERWVNITSLLDKSDRFSYQKLPQSEYCNYFRRM 390
Query: 365 FEALPAFVWLKCE-EGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELIK 423
P++ W+KCE E ++ +I + G F AGS++VRLS+I T D+F +L+
Sbjct: 391 RPPSPSYAWVKCEWEEDKDCYQTFQNGRINTQNGEGFEAGSRYVRLSLIKTKDDFDQLMY 450
Query: 424 RLSN 427
L N
Sbjct: 451 YLKN 454
>UniRef100_Q942G4 Putative alliinase [Oryza sativa]
Length = 485
Score = 237 bits (605), Expect = 4e-61
Identities = 135/374 (36%), Positives = 201/374 (53%), Gaps = 23/374 (6%)
Query: 63 PSSTLNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERL 122
P+ + GDPL YWK+ + VV G +SY++ ++ EL I RL
Sbjct: 107 PNCAADGQGGDPLFLEPYWKRHAAASAVVFSGWHRLSYITTDGHL---KSVELDRQIRRL 163
Query: 123 HHVVRNAVTKDKYIVIGTGTTQLFMAALFALSPS-MTTSHPINVVSATPYYSEYKDQLDL 181
H V NAV DKY+V GTG+T L A ++ALSP S P +VV+ PY++ YK Q +
Sbjct: 164 HRAVGNAVVDDKYLVFGTGSTHLINALVYALSPEGNAASPPASVVATVPYFAMYKSQTVM 223
Query: 182 FCSERFQWGGDAGEYDKNET-------YIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLA 234
F ++W G + N + +IE V PNNPD T+ EP++ + D A
Sbjct: 224 FDGREYRWDGTTAAWANNNSSRNPTRGFIEFVTSPNNPDSTLHEPILAG---SSAIVDHA 280
Query: 235 YYWPHYTPITHQANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAK 294
YYWPH T I A+ DVMLFT SK +GHAGSR GWA+V+D +VA + + +++ S++G ++
Sbjct: 281 YYWPHLTHIPAPADEDVMLFTTSKLSGHAGSRFGWALVRDEKVASRAISYIEESTVGTSR 340
Query: 295 ESQTRAAKIMGVICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYP 354
E+Q R KI+ VI E I F + +++ RW +L + ++ K P
Sbjct: 341 ETQLRVLKILKVILANLHGKEDI------FAFGYDVMSSRWRRLNAVVSRSTRISLQKMP 394
Query: 355 KAYCNFANESFEALPAFVWLKCEEGIENAENY--LGKLKIRGRGGVRFGAGSKHVRLSMI 412
YC + N E PA+ W+KC E +E+ + Y L I G AG+++ RLS+I
Sbjct: 395 PQYCTYFNRIKEPSPAYAWVKC-EWVEDDDCYETLLAAGINSLTGTVNEAGTRYTRLSLI 453
Query: 413 GTDDEFTELIKRLS 426
T D+F L++R++
Sbjct: 454 KTQDDFDMLLERIT 467
>UniRef100_Q942G1 Putative alliinase [Oryza sativa]
Length = 484
Score = 234 bits (598), Expect = 3e-60
Identities = 131/374 (35%), Positives = 200/374 (53%), Gaps = 22/374 (5%)
Query: 62 SPSSTLNLDKGDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIER 121
+P+ T + D G+PL YW++ + VV G +SY++ + EL I
Sbjct: 106 TPNCTADADSGNPLFLEPYWRRHAAASAVVFSGWHRLSYITTGGR---FHSVELDRHIRL 162
Query: 122 LHHVVRNAVTKDKYIVIGTGTTQLFMAALFALSPSMTT-SHPINVVSATPYYSEYKDQLD 180
LH V NAV DKY+V G G+ QL A ++ALSP S P +VV+ PYY YK Q D
Sbjct: 163 LHRAVGNAVVDDKYLVFGAGSMQLINALVYALSPDGNADSPPASVVATVPYYPAYKSQTD 222
Query: 181 LFCSERFQWGGDAGEYDKNET------YIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLA 234
+F ++W G + N + +IE V PNNPD +++P++ + D A
Sbjct: 223 MFDGREYRWDGTTATWSNNGSRNSTKGFIEFVTSPNNPDTALRKPVLAG---SSAIVDHA 279
Query: 235 YYWPHYTPITHQANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAK 294
YYWPH T I A+ DVMLFT SK +GHAGSR GWA+++D +VAK+ + +V+ S +G ++
Sbjct: 280 YYWPHLTHIPAPADEDVMLFTASKLSGHAGSRFGWALIRDEKVAKRALSYVEQSIMGASR 339
Query: 295 ESQTRAAKIMGVICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYP 354
++Q R KI+ VI E I F + +++R RW +L + ++ K P
Sbjct: 340 DTQLRMLKILKVILANLHGKEDI------FAFGYDVMRSRWRRLNAVVSRSTRISLQKIP 393
Query: 355 KAYCNFANESFEALPAFVWLKC--EEGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMI 412
YC + N E PA+ W+KC EE ++ E L I R G A +++ R+S++
Sbjct: 394 PQYCTYFNRIKEPSPAYAWVKCEWEEDVDCYETLLA-AGIISRSGTLSEAEARYTRMSLL 452
Query: 413 GTDDEFTELIKRLS 426
D+F L++R++
Sbjct: 453 KAQDDFDVLLERIT 466
>UniRef100_P31756 Alliin lyase precursor [Allium ascalonicum]
Length = 447
Score = 234 bits (597), Expect = 4e-60
Identities = 128/359 (35%), Positives = 206/359 (56%), Gaps = 15/359 (4%)
Query: 72 GDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVVRNAVT 131
GD L +YW++ E V++ G SY + V ++ EL+ I+ LH +V NA
Sbjct: 66 GDGLFLEEYWQQHKENSAVLVSGWHRTSYFFNP--VSNFISFELEKTIKELHEIVGNAAA 123
Query: 132 KDKYIVIGTGTTQLFMAALFALSPSMTTSH---PINVVSATPYYSEYKDQLDLFCSERFQ 188
KD+YIV G G TQL + +LSP+MT + VV+ PYY +++Q F + ++
Sbjct: 124 KDRYIVFGVGVTQLIHGLVISLSPNMTATPCAPQSKVVAHAPYYPVFREQTKYFDKKGYE 183
Query: 189 WGGDAGEYDKNET---YIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAYYWPHYTPITH 245
W G+A +Y T +IE+V PNNP+G ++ ++K K ++D+ YYWPHYTPI +
Sbjct: 184 WKGNAADYVNTSTPEQFIEMVTSPNNPEGLLRHEVIKGC---KSIYDMVYYWPHYTPIKY 240
Query: 246 QANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIMG 305
+A+ D+MLFT+SK TGH+GSR GWA++KD V K++ ++ ++ G ++E+Q R+ KI+
Sbjct: 241 KADEDIMLFTMSKYTGHSGSRFGWALIKDETVYNKLLNYMTKNTEGTSRETQLRSLKILK 300
Query: 306 VICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKA-YCNFANES 364
+ + + D F + K LRERW + ++ F+ K P++ YCN+
Sbjct: 301 EVTAMIKTQKGTMRDLNTFGFQK--LRERWVNITALLDKSDRFSYQKLPQSEYCNYFRRM 358
Query: 365 FEALPAFVWLKCE-EGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELI 422
P++ W+KCE E ++ +I + G F AGS++VRLS+I T D+F +L+
Sbjct: 359 RPPSPSYAWVKCEWEEDKDCYQTFQNGRINTQSGEGFEAGSRYVRLSLIKTKDDFDQLM 417
>UniRef100_P31757 Alliin lyase precursor [Allium cepa]
Length = 479
Score = 233 bits (593), Expect = 1e-59
Identities = 128/359 (35%), Positives = 205/359 (56%), Gaps = 15/359 (4%)
Query: 72 GDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVVRNAVT 131
GD L +YW++ E V++ G MSY + V ++ EL+ I+ LH +V NA
Sbjct: 98 GDGLFLEEYWQQHKENSAVLVSGWHRMSYFFNP--VSNFISFELEKTIKELHEIVGNAAA 155
Query: 132 KDKYIVIGTGTTQLFMAALFALSPSMTTSH---PINVVSATPYYSEYKDQLDLFCSERFQ 188
KD+YIV G G TQL + +LSP+MT + VV+ PYY +++Q F + ++
Sbjct: 156 KDRYIVFGVGVTQLIHGLVISLSPNMTATPCAPQSKVVAHAPYYPVFREQTKYFDKKGYE 215
Query: 189 WGGDAGEYDKNET---YIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAYYWPHYTPITH 245
W G+A +Y T +IE+V PNNP+G ++ ++K K ++ + YYWPHYTPI +
Sbjct: 216 WKGNAADYVNTSTPEQFIEMVTSPNNPEGLLRHEVIKGC---KSIYYMVYYWPHYTPIKY 272
Query: 246 QANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIMG 305
+A+ D+MLFT+SK TGH+GSR GWA++KD V K++ ++ ++ G ++E+Q R+ KI+
Sbjct: 273 KADEDIMLFTMSKYTGHSGSRFGWALIKDETVYNKLLNYMTKNTEGTSRETQLRSLKILK 332
Query: 306 VICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKA-YCNFANES 364
+ + D F + K LRERW + ++ F+ K P++ YCN+
Sbjct: 333 EVIAMVKTQNGTMRDLNTFGFQK--LRERWVNITALLDKSDRFSYQKLPQSEYCNYFRRM 390
Query: 365 FEALPAFVWLKCE-EGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELI 422
P++ W+KCE E ++ +I + G F AGS++VRLS+I T D+F +L+
Sbjct: 391 RPPSPSYAWVKCEWEEDKDCYQTFQNGRINTQSGEGFEAGSRYVRLSLIKTKDDFDQLM 449
>UniRef100_Q01594 Alliin lyase 1 precursor [Allium sativum]
Length = 486
Score = 229 bits (584), Expect = 1e-58
Identities = 127/364 (34%), Positives = 204/364 (55%), Gaps = 15/364 (4%)
Query: 72 GDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVVRNAVT 131
GD L +YWK+ E V++ MSY + V ++ EL+ I+ LH VV NA
Sbjct: 102 GDGLFLEEYWKQHKEASAVLVSPWHRMSYFFNP--VSNFISFELEKTIKELHEVVGNAAA 159
Query: 132 KDKYIVIGTGTTQLFMAALFALSPSMTTSHPI---NVVSATPYYSEYKDQLDLFCSERFQ 188
KD+YIV G G TQL + +LSP+MT + VV+ P+Y +++Q F + +
Sbjct: 160 KDRYIVFGVGVTQLIHGLVISLSPNMTATPDAPESKVVAHAPFYPVFREQTKYFNKKGYV 219
Query: 189 WGGDAGEY---DKNETYIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAYYWPHYTPITH 245
W G+A Y E YIE+V PNNP+G ++ ++K K ++D+ YYWPHYTPI +
Sbjct: 220 WAGNAANYVNVSNPEQYIEMVTSPNNPEGLLRHAVIKGC---KSIYDMVYYWPHYTPIKY 276
Query: 246 QANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIMG 305
+A+ D++LFT+SK TGH+GSR GWA++KD V ++ ++ ++ G +E+Q R+ K++
Sbjct: 277 KADEDILLFTMSKFTGHSGSRFGWALIKDESVYNNLLNYMTKNTEGTPRETQLRSLKVLK 336
Query: 306 VICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKA-YCNFANES 364
+ + + D F + K LRERW + ++ F+ + P++ YCN+
Sbjct: 337 EVVAMVKTQKGTMRDLNTFGFKK--LRERWVNITALLDQSDRFSYQELPQSEYCNYFRRM 394
Query: 365 FEALPAFVWLKCE-EGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELIK 423
P++ W+KCE E ++ +I + GV F A S++VRLS+I T D+F +L+
Sbjct: 395 RPPSPSYAWVKCEWEEDKDCYQTFQNGRINTQNGVGFEASSRYVRLSLIKTQDDFDQLMY 454
Query: 424 RLSN 427
L +
Sbjct: 455 YLKD 458
>UniRef100_Q41233 Alliin lyase 2 precursor [Allium sativum]
Length = 473
Score = 228 bits (580), Expect = 3e-58
Identities = 127/364 (34%), Positives = 203/364 (54%), Gaps = 15/364 (4%)
Query: 72 GDPLVFGKYWKKKSEECMVVIKGCELMSYLSDTSNVCWYMLPELKDAIERLHHVVRNAVT 131
GD L +YWK+ E V++ MSY + V ++ EL+ I+ LH VV NA
Sbjct: 89 GDGLFLEEYWKQHKEASAVLVSPWHRMSYFFNP--VSNFISFELEKTIKELHEVVGNAAA 146
Query: 132 KDKYIVIGTGTTQLFMAALFALSPSMTTSHPI---NVVSATPYYSEYKDQLDLFCSERFQ 188
KD+YIV G G TQL + +LSP+MT + VV+ P+Y +++Q F + +
Sbjct: 147 KDRYIVFGVGVTQLIHGLVISLSPNMTATPDAPESKVVAHAPFYPVFREQTKYFDKKGYV 206
Query: 189 WGGDAGEY---DKNETYIEVVNYPNNPDGTIKEPMVKSIVEGKLVHDLAYYWPHYTPITH 245
W G+A Y E YIE+V PNNP+G ++ ++K K ++D+ YYWPHYTPI +
Sbjct: 207 WAGNAANYVNVSNPEQYIEMVTSPNNPEGLLRHAVIKGC---KSIYDMVYYWPHYTPIKY 263
Query: 246 QANHDVMLFTLSKCTGHAGSRIGWAIVKDIEVAKKMVMFVQMSSIGVAKESQTRAAKIMG 305
+A+ D++LFT+SK TGH+GSR GWA++KD V ++ ++ ++ G +E+Q R+ K++
Sbjct: 264 KADEDILLFTMSKFTGHSGSRFGWALIKDESVYNNLLNYMTKNTEGTPRETQLRSLKVLK 323
Query: 306 VICDGYQKFESIEPDQLFFEYCKNILRERWEKLRRSIELRKVFTVAKYPKA-YCNFANES 364
I + + D F + K LRERW + ++ F+ + P++ YCN+
Sbjct: 324 EIVAMVKTQKGTMRDLNTFGFKK--LRERWVNITALLDQSDRFSYQELPQSEYCNYFRRM 381
Query: 365 FEALPAFVWLKCE-EGIENAENYLGKLKIRGRGGVRFGAGSKHVRLSMIGTDDEFTELIK 423
P++ W+ CE E ++ +I + GV F A S++VRLS+I T D+F +L+
Sbjct: 382 RPPSPSYAWVNCEWEEDKDCYQTFQNGRINTQSGVGFEASSRYVRLSLIKTQDDFDQLMY 441
Query: 424 RLSN 427
L +
Sbjct: 442 YLKD 445
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.135 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 727,862,311
Number of Sequences: 2790947
Number of extensions: 30962563
Number of successful extensions: 71838
Number of sequences better than 10.0: 209
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 166
Number of HSP's that attempted gapping in prelim test: 71498
Number of HSP's gapped (non-prelim): 222
length of query: 430
length of database: 848,049,833
effective HSP length: 130
effective length of query: 300
effective length of database: 485,226,723
effective search space: 145568016900
effective search space used: 145568016900
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0076a.12


