

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0075.5
(667 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
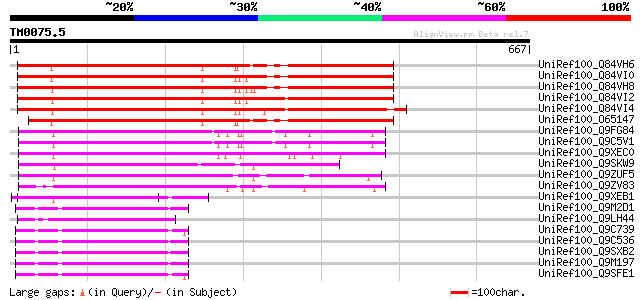
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84VH6 Gag-pol polyprotein [Glycine max] 466 e-129
UniRef100_Q84VI0 Gag-pol polyprotein [Glycine max] 456 e-126
UniRef100_Q84VH8 Gag-pol polyprotein [Glycine max] 451 e-125
UniRef100_Q84VI2 Gag-pol polyprotein [Glycine max] 447 e-124
UniRef100_Q84VI4 Gag-pol polyprotein [Glycine max] 443 e-123
UniRef100_O65147 Gag-pol polyprotein [Glycine max] 436 e-120
UniRef100_Q9FG84 Copia-like retroelement pol polyprotein [Arabid... 211 4e-53
UniRef100_Q9C5V1 Gag/pol polyprotein [Arabidopsis thaliana] 211 4e-53
UniRef100_Q9XEC0 Putative transposon protein [Arabidopsis thaliana] 202 3e-50
UniRef100_Q9SKW9 F5J5.1 [Arabidopsis thaliana] 185 3e-45
UniRef100_Q9ZUF5 Copia-like retroelement pol polyprotein [Arabid... 183 2e-44
UniRef100_Q9ZV83 Putative gag-protease polyprotein [Arabidopsis ... 168 5e-40
UniRef100_Q9XEB1 Putative transposon protein [Arabidopsis thaliana] 134 7e-30
UniRef100_Q9M2D1 Copia-type polyprotein [Arabidopsis thaliana] 124 1e-26
UniRef100_Q9LH44 Copia-like retrotransposable element [Arabidops... 122 3e-26
UniRef100_Q9C739 Copia-type polyprotein, putative [Arabidopsis t... 122 3e-26
UniRef100_Q9C536 Copia-type polyprotein, putative [Arabidopsis t... 122 3e-26
UniRef100_Q9SXB2 T28P6.8 protein [Arabidopsis thaliana] 122 3e-26
UniRef100_Q9M197 Copia-type reverse transcriptase-like protein [... 122 3e-26
UniRef100_Q9SFE1 T26F17.17 [Arabidopsis thaliana] 120 1e-25
>UniRef100_Q84VH6 Gag-pol polyprotein [Glycine max]
Length = 1577
Score = 466 bits (1198), Expect = e-129
Identities = 256/538 (47%), Positives = 340/538 (62%), Gaps = 74/538 (13%)
Query: 10 PPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIAST-----TELKPEDKWTKK 64
PPILDGTNY+YWKARM+ FLKS+DS WKA++KGW+HP + T ELKPE+ WTK+
Sbjct: 13 PPILDGTNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKE 72
Query: 65 EDDEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTT 124
ED+ ALGNSKALN +FNGVDKN+FRLINTCTVAK+AWEILKT HEGTSKV+MS+LQLL T
Sbjct: 73 EDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLAT 132
Query: 125 QFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEE 184
+FE +KM E+E I++FHM I ++AN+ ALGE M++EKL RKILRSLPKRFDMKVTAIEE
Sbjct: 133 KFENLKMKEEECIHDFHMTILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEE 192
Query: 185 AQDISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVS 244
AQDI N++VDELIGSLQTFE+ L+ R+EKK+K++ FVSN E +ED+ + DTD + AV
Sbjct: 193 AQDICNMRVDELIGSLQTFELGLSDRTEKKSKNLAFVSNDEGEEDEYDLDTDEGLTNAVV 252
Query: 245 LL----NKALKSLGRMSNTNVLDNVSDNVKNTEFQLKDKHENDTTKAI---------HVK 291
L NK L + R +V + D K +E+Q K + +K I H+K
Sbjct: 253 FLGKQFNKVLNRMDRRQKPHVRNISLDIRKGSEYQRKSDEKPSHSKGIQCRGCEGYGHIK 312
Query: 292 -----------------------------------ALIGKCYSDAESSDGDEEELVETYK 316
AL G+ S +SSD D E T+
Sbjct: 313 AECPTHLKKQRKGLSVCRSDDTESEQESDSDRDVNALTGRFESAEDSSDTDSE---ITFD 369
Query: 317 LLLAKWEESCMYGEKMRKEVKDLIAEKKQLQSNNSSLQEEVKTISKLREENEKLQITNAK 376
L + E C+ EK ++ ++ QL+ K I+ L E E + +K
Sbjct: 370 ELAIFYRELCIKSEK-------ILQQEAQLK----------KVIANLEAEKEAHEEEISK 412
Query: 377 LQEEVTLLNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGDMEGIGFSYKSANKSASSE- 435
L+ EV LNSKLE M KSI+M+NK +++L+Z+L++GK VG+ G+GF++KSA ++ +E
Sbjct: 413 LKGEVGFLNSKLENMTKSIKMLNKGSDMLDZVLQLGKKVGNQRGLGFNHKSAGRTTMTEF 472
Query: 436 KQTKQPMSDPMLHHSVRHVYPQFRKSKKSTWRCHHCGKLGHIRPYCYKLYGYPQSHDQ 493
K M H RH Q ++SK+ WRCH+CGK GHI+P+CY L+G+P Q
Sbjct: 473 VPAKNSTGATMSQHRSRHHGTQQKRSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQ 530
>UniRef100_Q84VI0 Gag-pol polyprotein [Glycine max]
Length = 1576
Score = 456 bits (1172), Expect = e-126
Identities = 255/534 (47%), Positives = 339/534 (62%), Gaps = 67/534 (12%)
Query: 10 PPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIAST-----TELKPEDKWTKK 64
PPILDGTNY+YWKARM+ FLKS+DS WKA++KGW+HP + T ELKPE+ WTK+
Sbjct: 13 PPILDGTNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKE 72
Query: 65 EDDEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAW-EILKTAHEGTSKVRMSKLQLLT 123
ED+ ALGNSKALN +FNGVDKN+FRLINTCTVAK+A EILKT HEGTSKV+MS+LQLL
Sbjct: 73 EDELALGNSKALNALFNGVDKNIFRLINTCTVAKDACGEILKTTHEGTSKVKMSRLQLLA 132
Query: 124 TQFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIE 183
T+FE +KM E+E I++FHM I ++AN+ ALGE M++EKL RKILRSLPKRFDMKVTAIE
Sbjct: 133 TKFENLKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIE 192
Query: 184 EAQDISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAV 243
EAQDI N++VDELIGSLQTFE+ L+ R+EKK+K++ FVSN E +ED+ + DTD + AV
Sbjct: 193 EAQDICNMRVDELIGSLQTFELGLSDRNEKKSKNLAFVSNDEGEEDEYDLDTDEGLTNAV 252
Query: 244 SLL----NKALKSLGRMSNTNVLDNVSDNVKNTEFQLKDKHENDTTKAI---------HV 290
LL NK L + R +V + D K +E+ K + +K I H+
Sbjct: 253 GLLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGSEYHKKSDEKPSHSKGIQCHGCEGYGHI 312
Query: 291 KAL-----------IGKCYS-DAES------------------SDGDEEELVETYKLLLA 320
KA + C S D ES SD D ++ T+ L
Sbjct: 313 KAECPTHLKKQRKGLSVCRSDDTESEQESDSDRDVNALTGRFESDEDSSDIEITFDELAI 372
Query: 321 KWEESCMYGEKMRKEVKDLIAEKKQLQSNNSSLQEEVKTISKLREENEKLQITNAKLQEE 380
+ + C+ EK ++ ++ QL+ K I+ L E E + ++L+ E
Sbjct: 373 SYRKLCIKSEK-------ILQQEAQLK----------KVIANLEAEKEAHEEEISELKGE 415
Query: 381 VTLLNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGDMEGIGFSYKSANKSASSE-KQTK 439
V LNSKLE M KSI+M+NK +++L+E+L++GK VG+ G+GF++KSA + +E K
Sbjct: 416 VGFLNSKLENMTKSIKMLNKGSDMLDEVLQLGKNVGNQRGLGFNHKSACRITMTEFVPAK 475
Query: 440 QPMSDPMLHHSVRHVYPQFRKSKKSTWRCHHCGKLGHIRPYCYKLYGYPQSHDQ 493
M H RH Q +KSK+ WRCH+CGK GHI+P+CY L+G+P Q
Sbjct: 476 NSTGATMSQHRSRHHGTQQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQ 529
>UniRef100_Q84VH8 Gag-pol polyprotein [Glycine max]
Length = 1576
Score = 451 bits (1160), Expect = e-125
Identities = 252/534 (47%), Positives = 338/534 (63%), Gaps = 67/534 (12%)
Query: 10 PPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTT-----ELKPEDKWTKK 64
PPILDG+NY+YWKARM+ FLKS+DS WKA++KGW+HP + T ELKPE+ WTK+
Sbjct: 13 PPILDGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKE 72
Query: 65 EDDEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTT 124
ED+ ALGNSKALN +FNGVDKN+FRLINTCTVAK+AWEILK HEGTSKV+MS+LQLL T
Sbjct: 73 EDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKMSRLQLLAT 132
Query: 125 QFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEE 184
+FE +KM E+E I++FHM I ++AN+ ALGE +++EKL RKILRSLPKRFDMKVTAIEE
Sbjct: 133 KFENLKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEE 192
Query: 185 AQDISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVS 244
AQDI N++VDELIGSLQTFE+ L+ R+EKK+K++ FVSN E +ED+ + DTD + AV
Sbjct: 193 AQDICNMRVDELIGSLQTFELGLSDRAEKKSKNLAFVSNDEGEEDEYDLDTDEGLTNAVV 252
Query: 245 LL----NKALKSLGRMSNTNVLDNVSDNVKNTEFQLKDKHENDTTKAI------------ 288
LL NK L + + +V + D K +++Q + + +K I
Sbjct: 253 LLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGSKYQKRSDVKPSHSKGIQCHGCEGYGHII 312
Query: 289 -----HVKAL---IGKCYSDAES---SDGDEE--------ELVE---------TYKLLLA 320
H+K + C SD ES SD D + E E T+ L A
Sbjct: 313 AECPTHLKKHRKGLSVCQSDTESEQESDSDRDVNALTGIFETAEDSSDTDSEITFDELAA 372
Query: 321 KWEESCMYGEKMRKEVKDLIAEKKQLQSNNSSLQEEVKTISKLREENEKLQITNAKLQEE 380
+ + C+ EK ++ ++ QL+ K I+ L E E + ++L+ E
Sbjct: 373 SYRKLCIKSEK-------ILQQEAQLK----------KVIADLEAEKEAHEEEISELKGE 415
Query: 381 VTLLNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGDMEGIGFSYKSANKSASSE-KQTK 439
V LNSKLE MKKSI+M+NK ++ L+E+L +GK G+ G+GF+ K A ++ +E K
Sbjct: 416 VGFLNSKLETMKKSIKMLNKGSDTLDEVLLLGKNAGNQRGLGFNPKFAGRTTMTEFVPAK 475
Query: 440 QPMSDPMLHHSVRHVYPQFRKSKKSTWRCHHCGKLGHIRPYCYKLYGYPQSHDQ 493
M H RH Q +KSK+ WRCH+CGK GHI+P+CY L+G+P Q
Sbjct: 476 NRTGTTMSQHLSRHHGTQQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQ 529
>UniRef100_Q84VI2 Gag-pol polyprotein [Glycine max]
Length = 1576
Score = 447 bits (1150), Expect = e-124
Identities = 245/520 (47%), Positives = 338/520 (64%), Gaps = 39/520 (7%)
Query: 10 PPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTT-----ELKPEDKWTKK 64
PPILDG+NY+YWKARM+ FLKS+DS WKA++KGW+HP + T ELKPE+ WTK+
Sbjct: 13 PPILDGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKE 72
Query: 65 EDDEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTT 124
ED+ ALGNSKALN +FNGVDKN+FRLINTCTVAK+A EILK+ HEGTSKV+MS+LQLL T
Sbjct: 73 EDELALGNSKALNALFNGVDKNIFRLINTCTVAKDACEILKSTHEGTSKVKMSRLQLLAT 132
Query: 125 QFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEE 184
+FE +KM E+E I++FHM I ++AN+ ALGE +++EKL RKILRSLPKRFDMKVTAIEE
Sbjct: 133 KFENLKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEE 192
Query: 185 AQDISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVS 244
AQDI N++VDELIGSLQTFE+ L+ R+EKK+K++ FVSN E +ED+ + DTD + AV
Sbjct: 193 AQDICNMRVDELIGSLQTFELGLSDRAEKKSKNLAFVSNDEGEEDEYDLDTDEGLTNAVV 252
Query: 245 LL----NKALKSLGRMSNTNVLDNVSDNVKNTEFQLKDKHENDTTKAI------------ 288
LL NK L + + +V + D K +++Q + + +K I
Sbjct: 253 LLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGSKYQKRSDVKPSHSKGIQCHGCEGYGHII 312
Query: 289 -----HVKAL---IGKCYSDAES-----SDGDEEELVETYKLLLAKWE-ESCMYGEKMRK 334
H+K + C SD ES SD D L+ ++ + +S + +++
Sbjct: 313 AECPTHLKKHRKGLSVCQSDTESEQESDSDRDVNALIGIFETAEDSSDTDSEITFDELAA 372
Query: 335 EVKDLIAEKKQLQSNNSSLQEEVKTISKLREENEKLQITNAKLQEEVTLLNSKLEGMKKS 394
+ L + +++ + L+ K I+ L E E + ++L+ EV LNSKLE M KS
Sbjct: 373 SYRKLCIKSEKILQQEAQLK---KVIADLEAEKEAHKEEISELKGEVGFLNSKLENMTKS 429
Query: 395 IRMMNKSTNVLEEILEVGKTVGDMEGIGFSYKSANKSASSE-KQTKQPMSDPMLHHSVRH 453
I+M+NK ++ L+E+L +GK G+ G+GF+ KSA ++ +E K M H RH
Sbjct: 430 IKMLNKGSDTLDEVLLLGKNAGNQRGLGFNPKSAGRTTMTEFVPAKNRTGATMSQHRSRH 489
Query: 454 VYPQFRKSKKSTWRCHHCGKLGHIRPYCYKLYGYPQSHDQ 493
Q +KSK+ WRCH+CGK GHI+P+CY L+G+P Q
Sbjct: 490 HGMQQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQ 529
>UniRef100_Q84VI4 Gag-pol polyprotein [Glycine max]
Length = 1574
Score = 443 bits (1139), Expect = e-123
Identities = 247/538 (45%), Positives = 341/538 (62%), Gaps = 49/538 (9%)
Query: 10 PPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTT-----ELKPEDKWTKK 64
PPILDG+NY+YWKARM+ FLKS+DS WKA++KGW+HP + T ELKPE+ WTK+
Sbjct: 13 PPILDGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKE 72
Query: 65 EDDEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTT 124
ED+ ALGNSKALN +FNGVDKN+FRLINTCTVAK+AWEILK HEGTSKV++S+LQLL T
Sbjct: 73 EDELALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKISRLQLLAT 132
Query: 125 QFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEE 184
+FE +KM E+E I++FHM I ++AN+ ALGE +++EKL RKILRSLPKRFDMKVTAIEE
Sbjct: 133 KFENLKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEE 192
Query: 185 AQDISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVS 244
AQDI N++VDELIGSLQTFE+ L+ R+EKK+K++ FVSN E +ED+ + +TD + AV
Sbjct: 193 AQDICNMRVDELIGSLQTFELGLSDRAEKKSKNLAFVSNDEGEEDEYDLNTDEGLTNAVV 252
Query: 245 LL----NKALKSLGRMSNTNVLDNVSDNVKNTEFQLKDKHENDTTKAI------------ 288
LL NK L + + +V + D K +++Q K + +K I
Sbjct: 253 LLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGSKYQKKSDVKPSHSKGIQCHGCEGYGHII 312
Query: 289 -----HVKAL---IGKCYSDAES-SDGDEEELVETYKLLLAKWEESC-----MYGEKMRK 334
H+K + C SD ES + D + V + E+S + +++
Sbjct: 313 AECPTHLKKHRKGLSVCQSDTESEQESDSDRDVNALTGIFETAEDSSDTDSEITFDELAT 372
Query: 335 EVKDLIAEKKQLQSNNSSLQEEVKTISKLREENEKLQITNAKLQEEVTLLNSKLEGMKKS 394
+ L + +++ + L+ K I+ L E E + ++L+ EV LNSKLE M KS
Sbjct: 373 SYRKLCIKSEKILQQEAQLK---KVIADLEAEKEAHKEEISELKGEVGFLNSKLENMTKS 429
Query: 395 IRMMNKSTNVLEEILEVGKTVGDMEGIGFSYKSANKSASSE-KQTKQPMSDPMLHHSVRH 453
I+M+NK ++ L+E+L +GK G+ G+GF+ KSA ++ +E K M H RH
Sbjct: 430 IKMLNKGSDTLDEVLLLGKNAGNQRGLGFNPKSAGRTTMTEFVPAKNRTGATMSQHRSRH 489
Query: 454 VYPQFRKSKKSTWRCHHCGKLGHIRPYCYKLYGYPQSHDQPRTNPQVAPTRKE--WKP 509
Q +KSK+ WRCH+CGK GHI+P+CY L+ P Q + +RK+ W P
Sbjct: 490 HGMQQKKSKRKKWRCHYCGKYGHIKPFCYHLH--------PHHGTQSSNSRKKMMWVP 539
>UniRef100_O65147 Gag-pol polyprotein [Glycine max]
Length = 1550
Score = 436 bits (1120), Expect = e-120
Identities = 243/523 (46%), Positives = 326/523 (61%), Gaps = 74/523 (14%)
Query: 25 MMVFLKSMDSIAWKAIVKGWKHPVIAST-----TELKPEDKWTKKEDDEALGNSKALNVI 79
M+ FLKS+DS WKA++KGW+HP + T ELKPE+ WTK+ED+ ALGNSKALN +
Sbjct: 1 MVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKEEDELALGNSKALNAL 60
Query: 80 FNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFETMKMNEDESIYE 139
FNGVDKN+FRLINTCTVAK+AWEILKT HEGTSKV+MS+LQLL T+FE +KM E+E I+E
Sbjct: 61 FNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLATKFENLKMKEEECIHE 120
Query: 140 FHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDISNIKVDELIGS 199
FHM I ++AN+ ALGE M++EKL RKILRSLPKRFDMKVTAIEEAQDI N++VDELIGS
Sbjct: 121 FHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQDICNMRVDELIGS 180
Query: 200 LQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVSLL----NKALKSLGR 255
LQTFE+ L+ R+EKK+K++ FVSN E +ED+ + DTD + AV LL NK L + R
Sbjct: 181 LQTFELGLSDRTEKKSKNLAFVSNDEGEEDEYDLDTDEGLTNAVVLLGKQFNKVLNRMDR 240
Query: 256 MSNTNVLDNVSDNVKNTEFQLKDKHENDTTKAI---------HVK--------------- 291
+V + D K +E+Q + + +K I H+K
Sbjct: 241 RQKPHVRNIPFDIRKGSEYQKRSDEKPSHSKGIQCHGCEGYGHIKAECPTHLKKQRKGLS 300
Query: 292 --------------------ALIGKCYSDAESSDGDEEELVETYKLLLAKWEESCMYGEK 331
AL G+ S +SSD D E T+ L + E C+ EK
Sbjct: 301 VCRSDDTESEQESDSDRDVNALTGRFESAEDSSDTDSE---ITFDELAISYRELCIKSEK 357
Query: 332 MRKEVKDLIAEKKQLQSNNSSLQEEVKTISKLREENEKLQITNAKLQEEVTLLNSKLEGM 391
++ ++ QL+ K I+ L E E + ++L+ E+ LNSKLE M
Sbjct: 358 -------ILQQEAQLK----------KVIANLEAEKEAHEDEISELKGEIGFLNSKLENM 400
Query: 392 KKSIRMMNKSTNVLEEILEVGKTVGDMEGIGFSYKSANKSASSE-KQTKQPMSDPMLHHS 450
KSI+M+NK +++L+E+L++GK VG+ G+GF++KSA ++ +E K M H
Sbjct: 401 TKSIKMLNKGSDLLDEVLQLGKNVGNQRGLGFNHKSAGRTTMTEFVPAKNSTGATMSQHR 460
Query: 451 VRHVYPQFRKSKKSTWRCHHCGKLGHIRPYCYKLYGYPQSHDQ 493
RH Q +KSK+ WRCH+CGK GHI+P+CY L+G+P Q
Sbjct: 461 SRHHGTQQKKSKRKKWRCHYCGKYGHIKPFCYHLHGHPHHGTQ 503
>UniRef100_Q9FG84 Copia-like retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1013
Score = 211 bits (538), Expect = 4e-53
Identities = 155/557 (27%), Positives = 259/557 (45%), Gaps = 90/557 (16%)
Query: 12 ILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTE---LKPEDKWTKKEDDE 68
+L+ NY +WK +M ++ + AW A GWK PV+ LK ED+WT E+ +
Sbjct: 15 MLEKGNYGHWKVKMRALIRGLGKEAWIATSVGWKAPVVKGENGEDVLKTEDQWTDAEEAK 74
Query: 69 ALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFET 128
A NS+AL++IFN V++N F+ I C AKEAW+ L A+EGTS V+ S++ +L +QFE
Sbjct: 75 ATANSRALSLIFNSVNQNQFKRIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQFEN 134
Query: 129 MKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDI 188
+ M+E E+I EF +I +A+ LG+ ++KL +K+LR LP RF+ K TA+ + D
Sbjct: 135 LTMDESENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSLDT 194
Query: 189 SNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVSLLNK 248
I +E++G LQ +E+ + +K + ++E++E Q KD+ + +A+ S K
Sbjct: 195 DTIDFEEVVGMLQAYELEITSGKGGYSKGVALAVSSEKNEIQELKDSMSMMAKNFSRAMK 254
Query: 249 ALKSLGRMSNTNVLDNVSD-----NVKNTEFQLKD-------KHENDTTKAIHVKA---- 292
++ G N D D N K +E Q + K E + K +K
Sbjct: 255 RVEKRGFARNQG-SDRDRDRDRDRNSKRSEIQCHECQGYGHIKAECPSLKRKDLKCSECR 313
Query: 293 --------LIGK---------CYSDAESSDGDEEELVETYKLLLAKWEESCMYGEKMRKE 335
IG SD++S D D EE V+ + + E+ + + E
Sbjct: 314 GIGHTKFDCIGSKSKPDRSYIAESDSDSDDEDSEEDVKGFVSFVGIIEDDNVSSDSSDSE 373
Query: 336 VKDLIAEKKQLQSNNSS---------------------------LQEEVKTISKLREENE 368
V EK+++ +++ S L+E+VK ++ +
Sbjct: 374 VG---CEKEEISADDESDVEMDVDGEFRKLYENWLVLSKEKVIWLEEKVKVQEQIEQLKG 430
Query: 369 KLQITNAKLQEEVTL-----------LNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGD 417
+L + N +++ E+ L L+ L +K I M+NK T L+ IL G+
Sbjct: 431 ELAVAN-QIKSEMILKYSAKEEKNRELSQDLSDTRKKIHMLNKGTKDLDSILAAGRVGKS 489
Query: 418 MEGIGF-SYKSANKSASSEKQTKQPMSDPMLHHSVRHVYPQFRKSKKST----------W 466
G+G+ S+ K+ + P + S + P RK + +
Sbjct: 490 NFGLGYHGGGSSTKTNFVRSKAAAPTQSQSVFRSKSNSVPARRKYQNQNHYHSQRTVTGY 549
Query: 467 RCHHCGKLGHIRPYCYK 483
C++CG+ GHI+ YCY+
Sbjct: 550 ECYYCGRHGHIQRYCYR 566
>UniRef100_Q9C5V1 Gag/pol polyprotein [Arabidopsis thaliana]
Length = 1643
Score = 211 bits (538), Expect = 4e-53
Identities = 155/557 (27%), Positives = 259/557 (45%), Gaps = 90/557 (16%)
Query: 12 ILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTE---LKPEDKWTKKEDDE 68
+L+ NY +WK +M ++ + AW A GWK PV+ LK ED+WT E+ +
Sbjct: 15 MLEKGNYGHWKVKMRALIRGLGKEAWIATSVGWKAPVVKGENGEDVLKTEDQWTDAEEAK 74
Query: 69 ALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFET 128
A NS+AL++IFN V++N F+ I C AKEAW+ L A+EGTS V+ S++ +L +QFE
Sbjct: 75 ATANSRALSLIFNSVNQNQFKRIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQFEN 134
Query: 129 MKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDI 188
+ M+E E+I EF +I +A+ LG+ ++KL +K+LR LP RF+ K TA+ + D
Sbjct: 135 LTMDESENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSLDT 194
Query: 189 SNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVSLLNK 248
I +E++G LQ +E+ + +K + ++E++E Q KD+ + +A+ S K
Sbjct: 195 DTIDFEEVVGMLQAYELEITSGKGGYSKGVALAVSSEKNEIQELKDSMSMMAKNFSRAMK 254
Query: 249 ALKSLGRMSNTNVLDNVSD-----NVKNTEFQLKD-------KHENDTTKAIHVKA---- 292
++ G N D D N K +E Q + K E + K +K
Sbjct: 255 RVEKRGFARNQG-SDRDRDRDRDRNSKRSEIQCHECQGYGHIKAECPSLKRKDLKCSECR 313
Query: 293 --------LIGK---------CYSDAESSDGDEEELVETYKLLLAKWEESCMYGEKMRKE 335
IG SD++S D D EE V+ + + E+ + + E
Sbjct: 314 GIGHTKFDCIGSKSKPDRSYIAESDSDSDDEDSEEDVKGFVSFVGIIEDDNVSSDSSDSE 373
Query: 336 VKDLIAEKKQLQSNNSS---------------------------LQEEVKTISKLREENE 368
V EK+++ +++ S L+E+VK ++ +
Sbjct: 374 VG---CEKEEISADDESDVEMDVDGEFRKLYENWLVLSKEKVIWLEEKVKVQEQIEQLKG 430
Query: 369 KLQITNAKLQEEVTL-----------LNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGD 417
+L + N +++ E+ L L+ L +K I M+NK T L+ IL G+
Sbjct: 431 ELAVAN-QIKSEMILKYSAKEEKNRELSQDLSDTRKKIHMLNKGTKDLDSILAAGRVGKS 489
Query: 418 MEGIGF-SYKSANKSASSEKQTKQPMSDPMLHHSVRHVYPQFRKSKKST----------W 466
G+G+ S+ K+ + P + S + P RK + +
Sbjct: 490 NFGLGYHGGGSSTKTNFVRSKAAAPTQSQSVFRSKSNSVPARRKYQNQNHYHSQRTVTGY 549
Query: 467 RCHHCGKLGHIRPYCYK 483
C++CG+ GHI+ YCY+
Sbjct: 550 ECYYCGRHGHIQRYCYR 566
>UniRef100_Q9XEC0 Putative transposon protein [Arabidopsis thaliana]
Length = 1008
Score = 202 bits (514), Expect = 3e-50
Identities = 154/547 (28%), Positives = 264/547 (48%), Gaps = 75/547 (13%)
Query: 12 ILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTE---LKPEDKWTKKEDDE 68
+L+ NY +WK +M ++ + AW A GWK PVI LK +D+W E+ +
Sbjct: 15 MLEKGNYGHWKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTKDQWNDAEEAK 74
Query: 69 ALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFET 128
A NS+AL++IFN V++N F+ I C AKEAW+ L A+EGTS V+ S++ +L +QFE
Sbjct: 75 AKANSRALSLIFNFVNQNQFKRIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQFEN 134
Query: 129 MKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDI 188
+ M E E+I EF +I +A+ LG+ ++KL +K+LR LP RF+ K TA+ + D
Sbjct: 135 LSMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSLDT 194
Query: 189 SNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVSLLNK 248
+I +E++G LQ +E+ + +K + ++ +++E Q KDT + +A+ S +
Sbjct: 195 DSIDFEEVVGMLQAYELEITSGKGGYSKGLALAASAKKNEIQELKDTMSMMAKDFSRAMR 254
Query: 249 AL--KSLGRMSNTNVLDNVS---DNVKNTEFQ-------------LKDKHENDTTKAIHV 290
+ K GR T+ + S D ++ E Q KD ++ H
Sbjct: 255 RVEKKGFGRNQGTDRYRDRSSKRDEIQCHECQGYGHIKAECPSLKRKDLKCSECNGLGHT 314
Query: 291 K-ALIG-------KCYSDAE--SSDGDEEELVETYKLLLAKWEESCMYGE-KMRKEVKDL 339
K +G C S++E S+DGD E+ ++ + + EE + + E +D
Sbjct: 315 KFDCVGSKSKPDKSCSSESESDSNDGDSEDYIKGFVSFVGIIEEKDESSDSEADGEDEDN 374
Query: 340 IAEKKQLQSNNSSLQEEVK-------TISKLR----EENEKLQITNAKLQEEVTLLNSK- 387
A++ + ++ EE + +SK + EE K+Q KL+ E+T N K
Sbjct: 375 SADEDSDIEKDVNINEEFRKLYDSWLMLSKEKVAWLEEKLKVQELTEKLKGELTAANQKN 434
Query: 388 --------------------LEGMKKSIRMMNKSTNVLEEILEVGKTVGDMEGIGF---- 423
L +K+I M+N T L+ IL G+ G+G+
Sbjct: 435 SELIQKCSVAEEKNRELSQELSDTRKNIHMLNSGTKDLDSILAAGRVGKSNFGLGYNGAG 494
Query: 424 -----SYKSANKSASSEKQTK-QPMSDPMLHHSVRHVYPQFRKSKKST-WRCHHCGKLGH 476
++ + +A ++ QT + D + V + ++ + T + C++CG+ GH
Sbjct: 495 SGTKTNFVRSEAAAPTKSQTGFRSNYDAVPARRVYQNHDHYQSRRTVTGYECYYCGRHGH 554
Query: 477 IRPYCYK 483
I+ YCY+
Sbjct: 555 IQRYCYR 561
>UniRef100_Q9SKW9 F5J5.1 [Arabidopsis thaliana]
Length = 1463
Score = 185 bits (470), Expect = 3e-45
Identities = 122/424 (28%), Positives = 208/424 (48%), Gaps = 20/424 (4%)
Query: 12 ILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELK---PEDKWTKKEDDE 68
+LD Y YWK RM ++ AW A+ +GW+ P + K P+ WT +E +
Sbjct: 15 LLDTKRYGYWKVRMTQIIRGQGEDAWTAVEEGWEPPFDLTEDGFKITKPKANWTAEEKLQ 74
Query: 69 ALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFET 128
+ N++A+N I NG+D++ F+LI C AK+AW+ L+ +HEGTS V+ ++L + TQFE
Sbjct: 75 SKFNARAMNAIVNGIDEDEFKLIQGCKSAKQAWDTLQKSHEGTSSVKRTRLDHIATQFEY 134
Query: 129 MKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDI 188
+KM E+I +F +I LAN LG+ ++KL +K+LR LP +F + A +
Sbjct: 135 LKMEPYETIVKFSSKISALANEAEVLGKTYKDQKLVKKLLRCLPPKFPAHKAVMRVAGNT 194
Query: 189 SNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVSLLNK 248
I +L+G L++ EM + K +K+I F ++ ++ Q+ KD A +A K
Sbjct: 195 DKISFVDLVGMLKSEEMEPDQDKVKPSKNIAFNADQGSEQFQQIKDGMALLARN---FGK 251
Query: 249 ALKSLGRMSNTNVLDNVSDNVKNTEFQLKDKHENDTTKAIHVKALIGKCYSDAESSDGDE 308
ALK + R N + + + + + + +D K + +CY D ES D E
Sbjct: 252 ALKRVERGQNRDSTSWSNKDGETSRGRFSRSENDDLGKKKEI-----QCYDDPESDDEGE 306
Query: 309 EEL-------VETYKLLLAKWEESCMYGEKMRKEVKDLIAEKKQLQSNN--SSLQEEVKT 359
E L +++ + C + E + L QL+ S + + +
Sbjct: 307 ELLNFVAFMASSDSSKVMSDTDSDCDQEVNPKDEYRVLYDSWMQLKDKQKLSGITVDENS 366
Query: 360 ISKLREENEKLQITNAKLQEEVTLLNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGDME 419
+++ + LQ ++ LL +L K IRM+NK + L++IL +G+T
Sbjct: 367 QDYYQKKFDWLQEECHMERDRAKLLERELNDKHKQIRMLNKGSESLDKILAMGRTDSQPR 426
Query: 420 GIGF 423
G+G+
Sbjct: 427 GLGY 430
>UniRef100_Q9ZUF5 Copia-like retroelement pol polyprotein [Arabidopsis thaliana]
Length = 916
Score = 183 bits (464), Expect = 2e-44
Identities = 141/491 (28%), Positives = 234/491 (46%), Gaps = 41/491 (8%)
Query: 12 ILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTE---LKPEDKWTKKEDDE 68
IL+ NY +WK +M ++ + AW A GWK PVI LK ED+W E+ +
Sbjct: 27 ILEKGNYGHWKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTEDQWNDAEEAK 86
Query: 69 ALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFET 128
A NS+AL++IFN V++N F+ I C AKEAW+ L A+EGTS V+ S++ +L +QFE
Sbjct: 87 ATANSRALSLIFNSVNQNQFKQIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQFEN 146
Query: 129 MKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDI 188
+ M E E+I EF +I +A+ LG+ ++KL +K+LR LP RF+ K TA+ + D
Sbjct: 147 LTMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSLDT 206
Query: 189 SNIKVDELIGSLQTFEMSL-NGRSEKKAKSITFVSNTEEDEDQREKDTDANIA-EAVSLL 246
++I +E++G Q +E+ + +G+ S +D E +I + V
Sbjct: 207 NSIDFEEVVGMFQAYELEITSGKGGYGHIKAECPSLKRKDLKCSECKGLGHIKFDCVGSK 266
Query: 247 NKALKSLGRMSNTNVLDNVS-DNVKN-TEFQ--LKDKHENDTTKAIHVKALIGKCYSDAE 302
+K +S S ++ D S D +K F +++K E+ ++A G+ ++
Sbjct: 267 SKPDRSCSSESESDSNDGDSEDYIKGFVSFVGIIEEKDESSDSEA------DGEDEDNSA 320
Query: 303 SSDGDEEELV---ETYKLLLAKWEESCMYGEKMRKEVKDLIAEKKQLQSNNSSLQEEVKT 359
D D E+ V E ++ L W + KE + EK ++Q L+ E+
Sbjct: 321 DEDSDIEKDVKINEEFRKLYDSW-------LMLSKEKVAWLEEKLKVQELTEKLKGELTA 373
Query: 360 ISKLREE-NEKLQITNAKLQEEVTLLNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGDM 418
++ E +K + K +E L+ +L +K I M+N T L+ IL G+
Sbjct: 374 ANQKNSELTQKCSVAEEKNRE----LSQELSDTRKKIHMLNSGTKDLDSILAAGRVGKSN 429
Query: 419 EGIGFS-YKSANKSASSEKQTKQPMSDPMLHHSVRHVYPQFR----------KSKKSTWR 467
G+G++ S K+ + P S P R + + +
Sbjct: 430 FGLGYNGVGSGTKTNFVRSEAGAPTKSQTGFRSNYDAVPARRMYQNHDHYHSRRTVTGYE 489
Query: 468 CHHCGKLGHIR 478
C++CG+ GH R
Sbjct: 490 CYYCGRHGHFR 500
>UniRef100_Q9ZV83 Putative gag-protease polyprotein [Arabidopsis thaliana]
Length = 627
Score = 168 bits (425), Expect = 5e-40
Identities = 144/558 (25%), Positives = 232/558 (40%), Gaps = 111/558 (19%)
Query: 12 ILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDDEALG 71
+LD Y YWK M ++ + G+K KP+ WT +E ++
Sbjct: 15 LLDTKRYGYWKVCMTQIIRGQED--------GFKIT--------KPKANWTAEEKLQSKF 58
Query: 72 NSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFETMKM 131
N++A+ IFNGVD++ F+LI C AK+AW+ L+ +HEGTS V+ ++L + TQFE +KM
Sbjct: 59 NARAMKAIFNGVDEDEFKLIQGCKSAKQAWDTLQKSHEGTSSVKRTRLDHIATQFEYLKM 118
Query: 132 NEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDISNI 191
DE I +F +I LAN +G+ ++KL +K+LR LP +F + A + I
Sbjct: 119 EPDEKIVKFSSKISALANEAEVMGKTYKDQKLVKKLLRCLPPKFAAHKAVMRVAGNTDKI 178
Query: 192 KVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDANIAEAVSLLNKALK 251
+L+G L+ EM + K +K+I F ++ ++ Q KD A +A K ++
Sbjct: 179 SFVDLVGMLKLEEMKADQDKVKPSKNIAFNADQGSEQFQEIKDGMALLARNFGKALKRVE 238
Query: 252 SLGRMSNTNVLDNVSDNV-KNTEFQLKD-------KHENDTTKAIHVKALIGKC------ 297
G S + +D++ K E Q + K E TK +K L KC
Sbjct: 239 IDGERSRGRFSRSENDDLRKKKEIQCYECGGFGHIKPECPITKRKEMKCL--KCKGVGHT 296
Query: 298 -----------------YSDAESSDGDEEEL----------------------------V 312
+SD+ES D EE L
Sbjct: 297 KFECPNKSKLKEKSLISFSDSESDDEGEELLNFVAFMASSDSSKFMSDTDSDCDEELNPK 356
Query: 313 ETYKLLLAKWEESCMYGEKMRKEVKDLIAEKKQLQSNNSSLQ-EEVKTISKLREENEKLQ 371
+ Y++L W + + K+ L+ EK L++ +++ E+ + +S + +
Sbjct: 357 DKYRVLYDSWVQ-------LSKDKLKLVKEKLTLEAKLANVSTEDKQKLSGITVDGNSQD 409
Query: 372 ITNAKL----------QEEVTLLNSKLEGMKKSIRMMNKSTNVLEEILEVGKTVGDMEGI 421
KL ++ LL +L K IRM+NK L++IL +G+T G+
Sbjct: 410 YYQKKLDCLQKECHRERDRTKLLERELNDKYKQIRMLNKGLESLDKILAMGRTDSQQRGL 469
Query: 422 GFSYKSANKSASSEKQTKQPMSDPMLHHSVRHVYPQFRKSKKSTWR-------------- 467
G+ + + VR Y + +K KS
Sbjct: 470 GYQGYTGKIKKEEGRVINFVSGGSTSETVVRQSYIEPKKQVKSHVEIKRESVVRTRMVGV 529
Query: 468 --CHHCGKLGHIRPYCYK 483
C HCGK H+R CYK
Sbjct: 530 ICCDHCGKRFHMREQCYK 547
>UniRef100_Q9XEB1 Putative transposon protein [Arabidopsis thaliana]
Length = 590
Score = 134 bits (338), Expect = 7e-30
Identities = 75/258 (29%), Positives = 135/258 (52%), Gaps = 8/258 (3%)
Query: 3 ARRSIYMP-PI-LDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTE---LKP 57
A+R I +P P+ LD +Y YWK + ++S+D AW A+ GW P K
Sbjct: 336 AQRFIAIPKPLKLDAEHYGYWKVLIKRSIQSIDMDAWFAVEDGWMPPTTKDAKRDIVSKS 395
Query: 58 EDKWTKKEDDEALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMS 117
+W E A NS+AL+VIF + +N F + C AKE WEIL+ + E T+ V+ +
Sbjct: 396 RTEWIADEKTAANHNSQALSVIFGSLLRNKFTQVQGCLSAKEVWEILQVSFECTNNVKRT 455
Query: 118 KLQLLTTQFETMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDM 177
+L +L ++FE + M +ES+ +F+ ++ + LG+ ++K+ +K LRSLP +F
Sbjct: 456 RLDMLASEFENLTMEAEESVDDFNGKLSSITQEAVVLGKTYKDKKMVKKFLRSLPDKFQS 515
Query: 178 KVTAIEEAQDISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQREKDTDA 237
+AI+ + + +K D+++G +Q ++ + E T+ E+D+ E+D
Sbjct: 516 HKSAIDVSLNSDQLKFDQVVGMMQAYD---TDKEEILNSYATYFGAIEDDDHTVEEDAQM 572
Query: 238 NIAEAVSLLNKALKSLGR 255
+++ L+ + G+
Sbjct: 573 GTIKSLILIQSDSEKEGK 590
Score = 58.5 bits (140), Expect = 6e-07
Identities = 43/183 (23%), Positives = 84/183 (45%), Gaps = 29/183 (15%)
Query: 11 PILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDDEA- 69
PI + NY +W+ +M ++ W+ + +G P + + PE K + A
Sbjct: 8 PIFNKENYGFWRIKMKTIFQTKK--LWEIVDEGVPKP--PAEGDHSPEAVQQKTRCEAAS 63
Query: 70 LGNSKALNVIFNGVDKNMF-RLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFET 128
L + AL ++ V ++F R+ + + WE +E
Sbjct: 64 LKDLTALQILQTAVSDSIFPRIAPASSALGKPWE-----------------------YEN 100
Query: 129 MKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDI 188
+KM E ++I F ++ ++ N GE S+ ++ +KIL SLPKRFD+ V +++ +D+
Sbjct: 101 LKMKESDNINTFMTKLIEMGNQLRVHGEEKSDYQIVQKILISLPKRFDIIVAMMKQTKDL 160
Query: 189 SNI 191
+++
Sbjct: 161 TSL 163
>UniRef100_Q9M2D1 Copia-type polyprotein [Arabidopsis thaliana]
Length = 1352
Score = 124 bits (310), Expect = 1e-26
Identities = 69/223 (30%), Positives = 122/223 (53%), Gaps = 7/223 (3%)
Query: 8 YMPPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDD 67
+ P+L +NYD W RM L + D W+ + KG+ P + +D D
Sbjct: 8 FQVPVLTKSNYDNWSLRMKAILGAHD--VWEIVEKGFIEPENEGSLSQTQKDGLR----D 61
Query: 68 EALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFE 127
+ KAL +I+ G+D++ F + T AKEAWE L+T+++G +V+ +LQ L +FE
Sbjct: 62 SRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFE 121
Query: 128 TMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQD 187
++M E E + ++ R+ + N+ GE + + ++ K+LRSL +F+ VT IEE +D
Sbjct: 122 ALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKD 181
Query: 188 ISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQ 230
+ + +++L+GSLQ +E + E A+ + + T+E+ Q
Sbjct: 182 LEAMTIEQLLGSLQAYE-EKKKKKEDIAEQVLNMQITKEENGQ 223
>UniRef100_Q9LH44 Copia-like retrotransposable element [Arabidopsis thaliana]
Length = 1499
Score = 122 bits (307), Expect = 3e-26
Identities = 66/203 (32%), Positives = 116/203 (56%), Gaps = 7/203 (3%)
Query: 11 PILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDDEAL 70
PI +G +Y +WK +M+ LK+ W I G + S + + T++ DD+ +
Sbjct: 10 PIFNGESYGFWKIKMITILKTRK--LWDVIENG-----VTSNSSPETSPALTRERDDQVM 62
Query: 71 GNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFETMK 130
+ AL ++ + V ++F I + A EAW L+ +G+S+V+M LQ L ++E +K
Sbjct: 63 KDMMALQILQSAVSDSIFPRIAPASSATEAWNALEMEFQGSSQVKMINLQTLRREYENLK 122
Query: 131 MNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQDISN 190
M E E+I +F ++ +L+N GE S+ ++ +KIL S+P++FD V +E+ +D+S
Sbjct: 123 MEEGETINDFTTKLINLSNQLRVHGEEKSDYQVVQKILISVPQQFDSIVGVLEQTKDLST 182
Query: 191 IKVDELIGSLQTFEMSLNGRSEK 213
+ V ELIG+L+ E LN R ++
Sbjct: 183 LSVTELIGTLKAHERRLNLREDR 205
>UniRef100_Q9C739 Copia-type polyprotein, putative [Arabidopsis thaliana]
Length = 1352
Score = 122 bits (307), Expect = 3e-26
Identities = 72/226 (31%), Positives = 123/226 (53%), Gaps = 13/226 (5%)
Query: 8 YMPPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDD 67
+ P+L +NYD W RM L + D W+ + KG+ P + +D D
Sbjct: 8 FQVPVLTKSNYDNWSLRMKAILGAHD--VWEIVEKGFIEPENEGSLSQTQKDGLR----D 61
Query: 68 EALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFE 127
+ KAL +I+ G+D++ F + T AKEAWE L+T+++G +V+ +LQ L +FE
Sbjct: 62 SRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFE 121
Query: 128 TMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQD 187
++M E E + ++ R+ + N+ GE + + ++ K+LRSL +F+ VT IEE +D
Sbjct: 122 ALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKD 181
Query: 188 ISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSN---TEEDEDQ 230
+ + +++L+GSLQ +E + +KK I V N T+E+ Q
Sbjct: 182 LEAMTIEQLLGSLQAYE----EKKKKKEDIIEQVLNMQITKEENGQ 223
>UniRef100_Q9C536 Copia-type polyprotein, putative [Arabidopsis thaliana]
Length = 1320
Score = 122 bits (306), Expect = 3e-26
Identities = 68/223 (30%), Positives = 121/223 (53%), Gaps = 7/223 (3%)
Query: 8 YMPPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDD 67
+ P+L +NYD W RM L + D W+ + KG+ P + +D D
Sbjct: 8 FQVPVLTKSNYDNWSLRMKAILGAHD--VWEIVEKGFIEPENEGSLSQTQKDGLR----D 61
Query: 68 EALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFE 127
+ KAL +I+ G+D++ F + T AKEAWE L+T+++G +V+ +LQ L +FE
Sbjct: 62 SRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFE 121
Query: 128 TMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQD 187
++M E E + ++ R+ + N+ GE + + ++ K+LRSL +F+ VT IEE +D
Sbjct: 122 ALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKD 181
Query: 188 ISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQ 230
+ + +++L+GSLQ +E + E + + + T+E+ Q
Sbjct: 182 LEAMTIEQLLGSLQAYE-EKKKKKEDIVEQVLNMQITKEENGQ 223
>UniRef100_Q9SXB2 T28P6.8 protein [Arabidopsis thaliana]
Length = 1352
Score = 122 bits (306), Expect = 3e-26
Identities = 68/223 (30%), Positives = 121/223 (53%), Gaps = 7/223 (3%)
Query: 8 YMPPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDD 67
+ P+L +NYD W RM L + D W+ + KG+ P + +D D
Sbjct: 8 FQVPVLTKSNYDNWSLRMKAILGAHD--VWEIVEKGFIEPENEGSLSQTQKDGLR----D 61
Query: 68 EALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFE 127
+ KAL +I+ G+D++ F + T AKEAWE L+T+++G +V+ +LQ L +FE
Sbjct: 62 SRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFE 121
Query: 128 TMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQD 187
++M E E + ++ R+ + N+ GE + + ++ K+LRSL +F+ VT IEE +D
Sbjct: 122 ALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKD 181
Query: 188 ISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQ 230
+ + +++L+GSLQ +E + E + + + T+E+ Q
Sbjct: 182 LEAMTIEQLLGSLQAYE-EKKKKKEDIVEQVLNMQITKEENGQ 223
>UniRef100_Q9M197 Copia-type reverse transcriptase-like protein [Arabidopsis
thaliana]
Length = 1272
Score = 122 bits (306), Expect = 3e-26
Identities = 68/223 (30%), Positives = 121/223 (53%), Gaps = 7/223 (3%)
Query: 8 YMPPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDD 67
+ P+L +NYD W RM L + D W+ + KG+ P + +D D
Sbjct: 8 FQVPVLTKSNYDNWSLRMKAILGAHD--VWEIVEKGFIEPENEGSLSQTQKDGLR----D 61
Query: 68 EALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFE 127
+ KAL +I+ G+D++ F + T AKEAWE L+T+++G +V+ +LQ L +FE
Sbjct: 62 SRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFE 121
Query: 128 TMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQD 187
++M E E + ++ R+ + N+ GE + + ++ K+LRSL +F+ VT IEE +D
Sbjct: 122 ALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKD 181
Query: 188 ISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSNTEEDEDQ 230
+ + +++L+GSLQ +E + E + + + T+E+ Q
Sbjct: 182 LEAMTIEQLLGSLQAYE-EKKKKKEDIVEQVLNMQITKEENGQ 223
>UniRef100_Q9SFE1 T26F17.17 [Arabidopsis thaliana]
Length = 1291
Score = 120 bits (302), Expect = 1e-25
Identities = 70/226 (30%), Positives = 123/226 (53%), Gaps = 13/226 (5%)
Query: 8 YMPPILDGTNYDYWKARMMVFLKSMDSIAWKAIVKGWKHPVIASTTELKPEDKWTKKEDD 67
+ P+L +NYD W +M L + D W+ + KG+ P + +D D
Sbjct: 8 FQVPVLTKSNYDNWSLQMKAILGAHD--VWEIVEKGFIEPENEGSLSQTQKDGLR----D 61
Query: 68 EALGNSKALNVIFNGVDKNMFRLINTCTVAKEAWEILKTAHEGTSKVRMSKLQLLTTQFE 127
+ KAL +I+ G+D++ F + T AKEAWE L+T+++G +V+ +LQ L +FE
Sbjct: 62 SRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGVDQVKKVRLQTLRGEFE 121
Query: 128 TMKMNEDESIYEFHMRIRDLANSTFALGEPMSEEKLARKILRSLPKRFDMKVTAIEEAQD 187
++M E E + ++ R+ + N+ GE + + ++ K+LRSL +F+ VT IEE +D
Sbjct: 122 ALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKD 181
Query: 188 ISNIKVDELIGSLQTFEMSLNGRSEKKAKSITFVSN---TEEDEDQ 230
+ + +++L+GSLQ +E + +KK + V N T+E+ Q
Sbjct: 182 LEAMTIEQLLGSLQAYE----EKKKKKEDIVEQVLNMRITKEENGQ 223
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.311 0.127 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,066,817,532
Number of Sequences: 2790947
Number of extensions: 44895993
Number of successful extensions: 201517
Number of sequences better than 10.0: 5449
Number of HSP's better than 10.0 without gapping: 468
Number of HSP's successfully gapped in prelim test: 5234
Number of HSP's that attempted gapping in prelim test: 182959
Number of HSP's gapped (non-prelim): 16936
length of query: 667
length of database: 848,049,833
effective HSP length: 134
effective length of query: 533
effective length of database: 474,062,935
effective search space: 252675544355
effective search space used: 252675544355
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0075.5


