

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
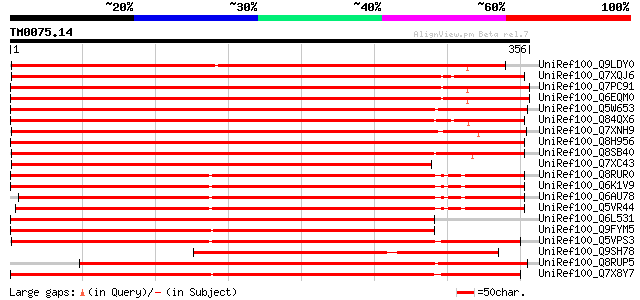
Query= TM0075.14
(356 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LDY0 Gb|AAF34840.1 [Arabidopsis thaliana] 407 e-112
UniRef100_Q7XQJ6 OSJNBa0017B10.12 protein [Oryza sativa] 403 e-111
UniRef100_Q7PC91 Putative transposase [Oryza sativa] 393 e-108
UniRef100_Q6EQM0 Ribosomal protein-like [Oryza sativa] 393 e-108
UniRef100_Q5W653 Hypothetical protein OSJNBb0052F16.9 [Oryza sat... 390 e-107
UniRef100_Q84QX6 Hypothetical protein OSJNBa0093I13.9 [Oryza sat... 387 e-106
UniRef100_Q7XNH9 OSJNBb0032D24.14 protein [Oryza sativa] 387 e-106
UniRef100_Q8H956 B protein [Oryza sativa] 384 e-105
UniRef100_Q8SB40 Putative ribosomal protein [Oryza sativa] 384 e-105
UniRef100_Q7XC43 Hypothetical protein [Oryza sativa] 382 e-104
UniRef100_Q8RUR0 OSJNBa0026J14.30 protein [Oryza sativa] 372 e-101
UniRef100_Q6K1V9 Ribosomal protein-like [Oryza sativa] 372 e-101
UniRef100_Q6AU78 Hypothetical protein OSJNBa0014C03.5 [Oryza sat... 363 3e-99
UniRef100_Q5VR44 Ribosomal protein-like [Oryza sativa] 363 3e-99
UniRef100_Q6L531 Hypothetical protein OJ1005_B11.14 [Oryza sativa] 360 4e-98
UniRef100_Q9FYM5 F21J9.3 [Arabidopsis thaliana] 360 4e-98
UniRef100_Q5VPS3 Ribosomal protein-like [Oryza sativa] 355 8e-97
UniRef100_Q9SH78 Hypothetical protein At2g07520 [Arabidopsis tha... 351 2e-95
UniRef100_Q8RUP5 Simiar to ribosomal protein [Oryza sativa] 348 1e-94
UniRef100_Q7X8Y7 OSJNBa0085H03.2 protein [Oryza sativa] 348 1e-94
>UniRef100_Q9LDY0 Gb|AAF34840.1 [Arabidopsis thaliana]
Length = 415
Score = 407 bits (1047), Expect = e-112
Identities = 200/342 (58%), Positives = 252/342 (73%), Gaps = 4/342 (1%)
Query: 2 FRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADAV 61
FRRR+RM+K +FLRIV LSS +F R DA + G SP+ KCT A+R+LAYG A+DAV
Sbjct: 75 FRRRFRMKKPLFLRIVDRLSSELMFFQHRRDATGRFGHSPIQKCTAAIRLLAYGYASDAV 134
Query: 62 DEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSIDC 121
DEY+++G TTA+ CL F GII + EYLRAPT +L+R+L + + RGFPGMIGS+DC
Sbjct: 135 DEYLRMGETTAMSCLENFTKGIISFFGDEYLRAPTATNLRRLLNIGKIRGFPGMIGSLDC 194
Query: 122 MHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPV 181
MHWEWKNCP AW+GQ+TRG G T++LEA+AS DLWIWH FFG PGTLNDIN+LDRSP+
Sbjct: 195 MHWEWKNCPTAWKGQYTRGS-GKPTIVLEAIASQDLWIWHVFFGPPGTLNDINILDRSPI 253
Query: 182 FDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQERC 241
FDD+ QG+AP V + VN R Y++AYYL DGIYP + TF++SIRLPQ+ LFA QE
Sbjct: 254 FDDILQGRAPNVKYKVNGREYHLAYYLTDGIYPKWATFIQSIRLPQNRKATLFATHQEAD 313
Query: 242 RKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDFE 301
RKD+ERAFGVLQARF II+ PA +WD +G IM++CIILHNMIVEDERD + ++ E
Sbjct: 314 RKDVERAFGVLQARFHIIKNPALVWDKEKIGNIMKACIILHNMIVEDERDGYNIQFDVSE 373
Query: 302 QSGEGGSSTPQ---PYSTEVLPAFANHVRARSELRDSNVHHE 340
G+ TPQ YST + N + R++LRD N+H +
Sbjct: 374 FLQVEGNQTPQVDLSYSTGMPLNIENMMGMRNQLRDQNMHQQ 415
>UniRef100_Q7XQJ6 OSJNBa0017B10.12 protein [Oryza sativa]
Length = 454
Score = 403 bits (1036), Expect = e-111
Identities = 206/353 (58%), Positives = 244/353 (68%), Gaps = 3/353 (0%)
Query: 2 FRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADAV 61
FR RYRM+KH+FL IV LS F QR DA K G SPL KCT A+RMLAYG +AD +
Sbjct: 97 FRWRYRMKKHLFLHIVQTLSHWSPVFQQRKDAFGKVGFSPLLKCTAALRMLAYGTSADIL 156
Query: 62 DEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSIDC 121
DE ++I TT +E L FC GII + +YLR PT ED+QR+L V E RGFPGM+GSIDC
Sbjct: 157 DENLQIAETTVIESLVNFCKGIIDCFGPKYLRRPTAEDIQRLLHVGEARGFPGMLGSIDC 216
Query: 122 MHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPV 181
MHWEWKNCP AW+GQ+TRGD G TV+LEAVASHDLWIWHAFFG G+ NDINVL++S +
Sbjct: 217 MHWEWKNCPVAWKGQYTRGDHGVPTVMLEAVASHDLWIWHAFFGVAGSNNDINVLNQSSL 276
Query: 182 FDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQERC 241
F QG AP V F VN Y+M YYLADGIYP + FVK+ +LPQ E KLFAQ QE
Sbjct: 277 FTAQRQGIAPDVHFTVNGNEYDMGYYLADGIYPEWAAFVKTFKLPQYEKHKLFAQKQEST 336
Query: 242 RKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDFE 301
RKDIE AFGVLQ+RF II PAR+W LG IM +CIILHNMIVEDERD++ R DF
Sbjct: 337 RKDIECAFGVLQSRFAIICTPARMWKKPTLGEIMYACIILHNMIVEDERDSYEGR-ADFN 395
Query: 302 QSGEGGSSTP-QPYSTEVLPAFANHVRARSELRDSNVHHELQADLVKHIWTKF 353
+G S P Y + F + +R+ ++HH L+ DL +HIW KF
Sbjct: 396 YE-QGSSPVPLNGYGQGPIHGFDRVLEIGVAIRNKDMHHRLKNDLTEHIWNKF 447
>UniRef100_Q7PC91 Putative transposase [Oryza sativa]
Length = 482
Score = 393 bits (1009), Expect = e-108
Identities = 197/358 (55%), Positives = 250/358 (69%), Gaps = 3/358 (0%)
Query: 1 MFRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADA 60
+FRRR+RM + +FLRIV L YFTQRVDA ++G+SPL KCT A+R LA G AD
Sbjct: 120 IFRRRFRMSRPLFLRIVEALGQWSVYFTQRVDAVNRKGLSPLQKCTAAIRQLATGSGADE 179
Query: 61 VDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSID 120
+DEY+KIG TTA+E ++ F G+ ++ YLR PT ED +R+LQ+ E+RGFPGM GSID
Sbjct: 180 LDEYLKIGETTAMEAMKNFVKGLQDVFGERYLRRPTMEDTERLLQLGEKRGFPGMFGSID 239
Query: 121 CMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSP 180
CMHW W+ CP AW+GQFTRGD+ T+ILEAVASHDLWIWHAFFG G+ NDINVL++S
Sbjct: 240 CMHWHWERCPVAWKGQFTRGDQKVPTLILEAVASHDLWIWHAFFGAAGSNNDINVLNQST 299
Query: 181 VFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQER 240
VF +G+AP V ++VN YN Y+LADGIYP + FVKSIRLP +E +KL+A +QE
Sbjct: 300 VFIKELKGQAPRVQYMVNGNQYNTGYFLADGIYPEWAVFVKSIRLPNTEKEKLYADMQEG 359
Query: 241 CRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDF 300
RKDIERAFGVLQ RF I++ PARL+D L ++ +CIILHNMIVEDE++T D
Sbjct: 360 ARKDIERAFGVLQRRFCILKRPARLYDRGVLRDVVLACIILHNMIVEDEKETRIIE-EDL 418
Query: 301 EQSGEGGSSTPQ--PYSTEVLPAFANHVRARSELRDSNVHHELQADLVKHIWTKFANA 356
+ + SST Q +S E F + +RD H+ L+ DLV+HIW KF A
Sbjct: 419 DLNVPPSSSTVQEPEFSPEQNTPFDRVLEKDISIRDRAAHNRLKKDLVEHIWNKFGGA 476
>UniRef100_Q6EQM0 Ribosomal protein-like [Oryza sativa]
Length = 447
Score = 393 bits (1009), Expect = e-108
Identities = 197/358 (55%), Positives = 250/358 (69%), Gaps = 3/358 (0%)
Query: 1 MFRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADA 60
+FRRR+RM + +FLRIV L YFTQRVDA ++G+SPL KCT A+R LA G AD
Sbjct: 85 IFRRRFRMSRPLFLRIVEALGQWSVYFTQRVDAVNRKGLSPLQKCTAAIRQLATGSGADE 144
Query: 61 VDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSID 120
+DEY+KIG TTA+E ++ F G+ ++ YLR PT ED +R+LQ+ E+RGFPGM GSID
Sbjct: 145 LDEYLKIGETTAMEAMKNFVKGLQDVFGERYLRRPTMEDTERLLQLGEKRGFPGMFGSID 204
Query: 121 CMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSP 180
CMHW W+ CP AW+GQFTRGD+ T+ILEAVASHDLWIWHAFFG G+ NDINVL++S
Sbjct: 205 CMHWHWERCPVAWKGQFTRGDQKVPTLILEAVASHDLWIWHAFFGAAGSNNDINVLNQST 264
Query: 181 VFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQER 240
VF +G+AP V ++VN YN Y+LADGIYP + FVKSIRLP +E +KL+A +QE
Sbjct: 265 VFIKELKGQAPRVQYMVNGNQYNTGYFLADGIYPEWAVFVKSIRLPNTEKEKLYADMQEG 324
Query: 241 CRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDF 300
RKDIERAFGVLQ RF I++ PARL+D L ++ +CIILHNMIVEDE++T D
Sbjct: 325 ARKDIERAFGVLQRRFCILKRPARLYDRGVLRDVVLACIILHNMIVEDEKETRIIE-EDL 383
Query: 301 EQSGEGGSSTPQ--PYSTEVLPAFANHVRARSELRDSNVHHELQADLVKHIWTKFANA 356
+ + SST Q +S E F + +RD H+ L+ DLV+HIW KF A
Sbjct: 384 DLNVPPSSSTVQEPEFSPEQNTPFDRVLEKDISIRDRAAHNRLKKDLVEHIWNKFGGA 441
>UniRef100_Q5W653 Hypothetical protein OSJNBb0052F16.9 [Oryza sativa]
Length = 444
Score = 390 bits (1003), Expect = e-107
Identities = 192/355 (54%), Positives = 245/355 (68%), Gaps = 1/355 (0%)
Query: 1 MFRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADA 60
MFR R+RM K +FLRIV LS YFTQR D + +SPL KCT A+RMLAYG ADA
Sbjct: 87 MFRTRFRMHKPLFLRIVEALSQWSPYFTQRGDCSGHTSLSPLQKCTAALRMLAYGTPADA 146
Query: 61 VDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSID 120
+DEY+KIG +TALECL F G+I ++ YLR PT+ED++ IL V+E RGFPGM+GSID
Sbjct: 147 LDEYLKIGKSTALECLEMFSRGVIVVFGGTYLRRPTREDVEHILHVNESRGFPGMLGSID 206
Query: 121 CMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSP 180
CMHW W++CP+AW GQFTRGD T+ILEAVASHDLWIWHAFFG G+ NDINVL++SP
Sbjct: 207 CMHWRWESCPRAWRGQFTRGDYKVPTIILEAVASHDLWIWHAFFGVAGSNNDINVLNQSP 266
Query: 181 VFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQER 240
+F D +G+AP V + VN YN YYL DGIYP + F K+I LPQ E KL+A+ QE
Sbjct: 267 LFLDTVRGEAPRVHYYVNGEEYNHGYYLDDGIYPEWAVFQKTIPLPQIEKHKLYAEHQEG 326
Query: 241 CRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDF 300
RKD+ERAFGVLQARF I+R A+ W +G M +C+ILHNMIVEDE + A D
Sbjct: 327 ARKDVERAFGVLQARFNIVRRSAKKWKRKSIGNTMLACVILHNMIVEDEGED-AICDLDL 385
Query: 301 EQSGEGGSSTPQPYSTEVLPAFANHVRARSELRDSNVHHELQADLVKHIWTKFAN 355
+ P ++ P F + + ++ +R ++H +L+ DL++HIW +F N
Sbjct: 386 NRIPRTSIVLPPEVTSGGNPCFRDVLSRKAAIRARSMHTQLKTDLIEHIWNRFRN 440
>UniRef100_Q84QX6 Hypothetical protein OSJNBa0093I13.9 [Oryza sativa]
Length = 626
Score = 387 bits (994), Expect = e-106
Identities = 196/356 (55%), Positives = 253/356 (71%), Gaps = 5/356 (1%)
Query: 1 MFRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADA 60
MFRRR+RM+K +FLRIV+ LS YFT R+DA + G SPL KCT A+RMLAYG AD
Sbjct: 270 MFRRRFRMRKPLFLRIVSALSEWSPYFTNRLDATGRAGHSPLQKCTAAIRMLAYGTPADQ 329
Query: 61 VDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSID 120
+DE +KIG TALECL +F G+I ++ EYLRAP ++++R+LQV++ RGFPGM+G+ID
Sbjct: 330 LDEVLKIGPNTALECLGKFAEGVIEIFRKEYLRAPRSDEVERLLQVADSRGFPGMLGNID 389
Query: 121 CMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSP 180
CMHW WKNCP +W GQFTRGDKG T+ILEAVAS DL IWH FF G+ NDINVL++SP
Sbjct: 390 CMHWAWKNCPVSWCGQFTRGDKGVPTMILEAVASKDLRIWHDFFATAGSNNDINVLNKSP 449
Query: 181 VFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQER 240
+F + +G+AP V F VN YN YYLADGIYP + TFVK+I+LPQ++ KL+A +E
Sbjct: 450 LFIEALRGEAPRVQFSVNGNQYNTWYYLADGIYPEWATFVKTIQLPQTDEHKLYAAREEG 509
Query: 241 CRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDF 300
RKD+ERAFGVLQ+RF I+ AR+W D+ IM +C+IL NMIVEDE++ A+ D
Sbjct: 510 TRKDVERAFGVLQSRFNIVCRLARMWRQGDVINIMEACVILRNMIVEDEQE-MAEIPLDL 568
Query: 301 EQSGEGGSSTPQP---YSTEVLPAFANHVRARSELRDSNVHHELQADLVKHIWTKF 353
++ G S P S++ P FA +R S +RD H +L+ DLV HIW +F
Sbjct: 569 NEN-PGASFVLPPEVRNSSDPNPCFAAVLRRNSSIRDRAKHMQLKKDLVAHIWQRF 623
>UniRef100_Q7XNH9 OSJNBb0032D24.14 protein [Oryza sativa]
Length = 450
Score = 387 bits (993), Expect = e-106
Identities = 191/355 (53%), Positives = 240/355 (66%), Gaps = 5/355 (1%)
Query: 2 FRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADAV 61
FRRR+RM++H+FLRIV L YF R DA K G+SPL KCT AMRMLAYG AD +
Sbjct: 95 FRRRFRMRRHLFLRIVQALGVWSPYFRLRRDAFGKVGLSPLQKCTAAMRMLAYGTPADLM 154
Query: 62 DEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSIDC 121
DE + +TA+EC+ F G+ L+ +YLR PT ED+QR+LQ E GFPGM+GSIDC
Sbjct: 155 DETFGVAESTAMECMINFVQGVRHLFGEQYLRRPTVEDIQRLLQFGEAHGFPGMLGSIDC 214
Query: 122 MHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPV 181
MHWEW++CP AW+GQFTRGD G T++LEAVAS DLWIWHAFFG G+ NDINVLD+SP+
Sbjct: 215 MHWEWQSCPVAWKGQFTRGDYGVPTIMLEAVASLDLWIWHAFFGAAGSNNDINVLDQSPL 274
Query: 182 FDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQERC 241
F ++ QG+AP V F +N YNM YYL D IYP + F KSI P+S KL+AQ QE
Sbjct: 275 FTEMIQGRAPPVQFTINGTQYNMGYYLTDRIYPEWAAFAKSITRPRSAKHKLYAQRQESA 334
Query: 242 RKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDFE 301
RKD+ERAFGVLQ R+ IIR PAR+W+ +L IM +CIILHNMIVEDER ++ +
Sbjct: 335 RKDVERAFGVLQKRWAIIRHPARIWEREELADIMYACIILHNMIVEDERGSYD---IPDD 391
Query: 302 QSGEGGSSTPQPYSTEVLP--AFANHVRARSELRDSNVHHELQADLVKHIWTKFA 354
+ E G PQ + P F + + D H L+ DL++H+W KF+
Sbjct: 392 NTYEQGQYYPQMTGLDHGPIYGFQEVLEQNKAIHDRQTHRRLKGDLIEHVWQKFS 446
>UniRef100_Q8H956 B protein [Oryza sativa]
Length = 455
Score = 384 bits (987), Expect = e-105
Identities = 189/354 (53%), Positives = 244/354 (68%), Gaps = 1/354 (0%)
Query: 1 MFRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADA 60
+FRRR+RM + +FLRIV L +YFTQRVDAA ++G+SPL KCT A+R LA G AD
Sbjct: 86 IFRRRFRMYRPLFLRIVDALGQWSDYFTQRVDAAGRQGLSPLQKCTAAIRQLATGSGADE 145
Query: 61 VDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSID 120
+DEY+KIG TTA++ ++ F GI ++ YLR PT ED +R+L++ E+RGFPGM GSID
Sbjct: 146 LDEYLKIGETTAMDAMKNFVKGIREVFGERYLRRPTVEDTERLLELGERRGFPGMFGSID 205
Query: 121 CMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSP 180
CMHW+W+ CP AW+GQFTRGD+ T+ILEAVASHDLWIWHAFFG G+ NDINVL RS
Sbjct: 206 CMHWQWERCPTAWKGQFTRGDQKVPTLILEAVASHDLWIWHAFFGVAGSNNDINVLSRST 265
Query: 181 VFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQER 240
VF + +G+AP V ++VN YN Y+LADGIYP + F KS RLP +E +KL+AQ QE
Sbjct: 266 VFINELKGQAPRVQYMVNGNQYNEGYFLADGIYPEWKVFAKSYRLPITEKEKLYAQHQEG 325
Query: 241 CRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDT-FAQRWTD 299
RKDIERAFGVLQ RF I++ PARL+D L ++ CIILHNMIVEDE++ + D
Sbjct: 326 ARKDIERAFGVLQRRFCILKRPARLYDRGVLRDVVLGCIILHNMIVEDEKEARLIEENLD 385
Query: 300 FEQSGEGGSSTPQPYSTEVLPAFANHVRARSELRDSNVHHELQADLVKHIWTKF 353
+ + +S + + + +RD H L+ DLV+HIW KF
Sbjct: 386 LNEPASSSTVQAPEFSPDQHVPLERILEKDTSMRDRLAHRRLKNDLVEHIWNKF 439
>UniRef100_Q8SB40 Putative ribosomal protein [Oryza sativa]
Length = 915
Score = 384 bits (986), Expect = e-105
Identities = 193/354 (54%), Positives = 246/354 (68%), Gaps = 3/354 (0%)
Query: 2 FRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADAV 61
FRRR+RM K +FLRIV LS+ D +FTQRVDA ++ SPL KCT A+RML YG ADA+
Sbjct: 558 FRRRFRMNKPLFLRIVNALSNWDQFFTQRVDATGRDSHSPLQKCTAAIRMLGYGTPADAL 617
Query: 62 DEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSIDC 121
DE +KI +T+LECL +F GII + EYLR PT ++L++ILQ +E RGFPGMIGSIDC
Sbjct: 618 DEVLKIAASTSLECLGKFAVGIIECFGSEYLRPPTSDELEKILQENEARGFPGMIGSIDC 677
Query: 122 MHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPV 181
MHW+WKNCPK W G F G KG T+ILEAVAS DL IWHAFFG G+ NDI VL++SP+
Sbjct: 678 MHWQWKNCPKGWAGMFINGFKGKPTMILEAVASRDLRIWHAFFGNAGSQNDIQVLNKSPL 737
Query: 182 FDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQERC 241
F +G+AP VS+ VN Y+ YYLADGIYP + FVK+IR PQ+E KL+AQ QE
Sbjct: 738 FIHAIKGEAPRVSYTVNGTQYDTGYYLADGIYPEWAAFVKTIRKPQTEKHKLYAQRQEGA 797
Query: 242 RKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDFE 301
RKD+E AFGVLQ+RF I+ PARLW D+ IM++C+ILHNMIVEDE+D + D
Sbjct: 798 RKDVECAFGVLQSRFDIVNRPARLWKRNDVVNIMQACVILHNMIVEDEKD-LVKIPLDLN 856
Query: 302 QSGEGGSSTPQPYST--EVLPAFANHVRARSELRDSNVHHELQADLVKHIWTKF 353
++ P T P F + + S +R ++ H +L+ DLV+HIW ++
Sbjct: 857 ENPSATIVLPPEVQTNDNPNPCFVDVLNRNSAIRAASTHRQLKNDLVEHIWQRY 910
>UniRef100_Q7XC43 Hypothetical protein [Oryza sativa]
Length = 513
Score = 382 bits (980), Expect = e-104
Identities = 183/288 (63%), Positives = 219/288 (75%)
Query: 2 FRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADAV 61
FRRR+RM++HVFL IV +L +YFT RVD G SPL KCT A+RMLAYG AAD +
Sbjct: 84 FRRRFRMRRHVFLHIVDELGKWSSYFTHRVDCTGCLGHSPLQKCTAAIRMLAYGTAADTL 143
Query: 62 DEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSIDC 121
DEY+K+ +TALECL F G++ ++ YLR PT EDL+R+LQV E RGFPGM+GSIDC
Sbjct: 144 DEYLKVPQSTALECLENFVEGVVEVFSSRYLRRPTAEDLERLLQVGESRGFPGMLGSIDC 203
Query: 122 MHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPV 181
MHW WKNCP AW+GQ+TRGD+ T+ILEAVAS+DL IWHAFFG PG+ NDINVL++SP+
Sbjct: 204 MHWRWKNCPTAWKGQYTRGDQKYPTIILEAVASYDLHIWHAFFGIPGSNNDINVLNQSPL 263
Query: 182 FDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQERC 241
F + +G+AP + FIVN YN YYLADGIYP + FVKSIR PQ E KLFA+ QE
Sbjct: 264 FIEAIKGEAPQIQFIVNGTQYNTGYYLADGIYPEWAAFVKSIRSPQLEKHKLFAREQEGK 323
Query: 242 RKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDE 289
RKDIERAFGVLQARF I+ PAR W L IM++C+ILHNMIVEDE
Sbjct: 324 RKDIERAFGVLQARFNIVHRPARSWSQKVLRKIMQACVILHNMIVEDE 371
>UniRef100_Q8RUR0 OSJNBa0026J14.30 protein [Oryza sativa]
Length = 626
Score = 372 bits (954), Expect = e-101
Identities = 183/353 (51%), Positives = 243/353 (67%), Gaps = 8/353 (2%)
Query: 1 MFRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADA 60
+FRRR RM + +FLRI+ + D+YF Q+ +AA G S K T AMR LAYG+AADA
Sbjct: 278 LFRRRNRMSRPLFLRIMNAIEDHDDYFVQKRNAAGLIGFSCHQKVTAAMRQLAYGIAADA 337
Query: 61 VDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSID 120
+DEY+ I +TA+E LRRF ++ ++EHEYLR+P + D R+L++ E RGFPGM+GSID
Sbjct: 338 LDEYLGIAESTAIESLRRFVKAVVQVFEHEYLRSPNENDTTRLLELGEDRGFPGMLGSID 397
Query: 121 CMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSP 180
CMHW+WKNCP G + +G T+ILEAVAS DLWIWHAFFG PG+ NDINVL RSP
Sbjct: 398 CMHWKWKNCPTELHGMY-QGHVHEPTIILEAVASKDLWIWHAFFGMPGSHNDINVLHRSP 456
Query: 181 VFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQER 240
+F + +GKAP V++ +N Y M YYLADGIYPS+ TFVK+I P K FA+ QE
Sbjct: 457 LFAKLAEGKAPEVNYSINGHDYMMGYYLADGIYPSWATFVKTIPEPHGNKRKYFAKAQEA 516
Query: 241 CRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDF 300
RKD+ERAFGVLQARF I+R PAR WD LG IM++C+I+HNMI+EDE + W
Sbjct: 517 VRKDVERAFGVLQARFAIVRGPARHWDEKTLGYIMKACVIMHNMIIEDEGEV---DWE-- 571
Query: 301 EQSGEGGSSTPQPYSTEVLPAFANHVRARSELRDSNVHHELQADLVKHIWTKF 353
E+ EGG + S + +P + ++ ++RD H++L+ DLV+H+W +
Sbjct: 572 ERFPEGGENV--RVSHDEIPDLDDFIQMHKKIRDDETHYQLREDLVEHLWQHY 622
>UniRef100_Q6K1V9 Ribosomal protein-like [Oryza sativa]
Length = 916
Score = 372 bits (954), Expect = e-101
Identities = 183/353 (51%), Positives = 243/353 (67%), Gaps = 8/353 (2%)
Query: 1 MFRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADA 60
+FRRR RM + +FLRI+ + D+YF Q+ +AA G S K T AMR LAYG+AADA
Sbjct: 568 LFRRRNRMSRPLFLRIMNAIEDHDDYFVQKRNAAGLIGFSCHQKVTAAMRQLAYGIAADA 627
Query: 61 VDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSID 120
+DEY+ I +TA+E LRRF ++ ++EHEYLR+P + D R+L++ E RGFPGM+GSID
Sbjct: 628 LDEYLGIAESTAIESLRRFVKAVVQVFEHEYLRSPNENDTTRLLELGEDRGFPGMLGSID 687
Query: 121 CMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSP 180
CMHW+WKNCP G + +G T+ILEAVAS DLWIWHAFFG PG+ NDINVL RSP
Sbjct: 688 CMHWKWKNCPTELHGMY-QGHVHEPTIILEAVASKDLWIWHAFFGMPGSHNDINVLHRSP 746
Query: 181 VFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQER 240
+F + +GKAP V++ +N Y M YYLADGIYPS+ TFVK+I P K FA+ QE
Sbjct: 747 LFAKLAEGKAPEVNYSINGHDYMMGYYLADGIYPSWATFVKTIPEPHGNKRKYFAKAQEA 806
Query: 241 CRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDF 300
RKD+ERAFGVLQARF I+R PAR WD LG IM++C+I+HNMI+EDE + W
Sbjct: 807 VRKDVERAFGVLQARFAIVRGPARHWDEKTLGYIMKACVIMHNMIIEDEGEV---DWE-- 861
Query: 301 EQSGEGGSSTPQPYSTEVLPAFANHVRARSELRDSNVHHELQADLVKHIWTKF 353
E+ EGG + S + +P + ++ ++RD H++L+ DLV+H+W +
Sbjct: 862 ERFPEGGENV--RVSHDEIPDLDDFIQMHKKIRDDETHYQLREDLVEHLWQHY 912
>UniRef100_Q6AU78 Hypothetical protein OSJNBa0014C03.5 [Oryza sativa]
Length = 446
Score = 363 bits (933), Expect = 3e-99
Identities = 179/347 (51%), Positives = 239/347 (68%), Gaps = 8/347 (2%)
Query: 7 RMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADAVDEYIK 66
RM++ +FLRI+ + D+YF Q+ +AA G S K T AMR LAYG+AADA+DEY+
Sbjct: 104 RMRRPLFLRIMNAVEDHDDYFVQKRNAAGLIGFSCHQKVTAAMRQLAYGIAADALDEYLG 163
Query: 67 IGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSIDCMHWEW 126
I +TA+E LRRF ++ ++EHEYLR+P + D R+L++ E RGFPGM+GSIDCMHW+W
Sbjct: 164 IAESTAIESLRRFVKAVVQVFEHEYLRSPNENDTTRLLELGEDRGFPGMLGSIDCMHWKW 223
Query: 127 KNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDDVE 186
KNCP G + +G T+ILEAVAS DLWIWHAFFG PG+ NDINVL RSP+F +
Sbjct: 224 KNCPTELHGMY-QGHVHEPTIILEAVASKDLWIWHAFFGMPGSHNDINVLHRSPLFAKLA 282
Query: 187 QGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQERCRKDIE 246
+GKAP V++ +N Y M YYLADGIYPS+ TFVK+I P K FA+ QE RKD+E
Sbjct: 283 EGKAPEVNYSINGHDYMMGYYLADGIYPSWATFVKTIPEPHGNKRKYFAKAQEAIRKDVE 342
Query: 247 RAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDFEQSGEG 306
RAFGVLQARF I+R PAR WD LG IM++C+I+HNMI+EDE + W E+ EG
Sbjct: 343 RAFGVLQARFAIVRGPARHWDEKTLGYIMKACVIMHNMIIEDEGEV---DWE--ERFPEG 397
Query: 307 GSSTPQPYSTEVLPAFANHVRARSELRDSNVHHELQADLVKHIWTKF 353
G + S + +P + ++ ++RD H++L+ DLV+H+W +
Sbjct: 398 GENV--RVSHDEIPDLDDFIQMHKKIRDDETHYQLREDLVEHLWQHY 442
>UniRef100_Q5VR44 Ribosomal protein-like [Oryza sativa]
Length = 428
Score = 363 bits (933), Expect = 3e-99
Identities = 179/349 (51%), Positives = 240/349 (68%), Gaps = 8/349 (2%)
Query: 5 RYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADAVDEY 64
R RM + +FLRI+ + D+YF Q+ +AA G S K T AMR LAYG+AADA+DEY
Sbjct: 84 RNRMSRPLFLRIMNAVEDHDDYFVQKRNAAGLIGFSCHQKVTAAMRQLAYGIAADALDEY 143
Query: 65 IKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSIDCMHW 124
+ I +TA+E LRRF ++ ++EHEYLR+P + D R+L++ E RGFPGM+GSIDCMHW
Sbjct: 144 LGIAESTAIESLRRFVKAVVQVFEHEYLRSPNENDTTRLLELGEDRGFPGMLGSIDCMHW 203
Query: 125 EWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDD 184
+WKNCP G + +G T+ILEAVAS DLWIWHAFFG PG+ NDINVL RSP+F
Sbjct: 204 KWKNCPTELHGMY-QGHVHEPTIILEAVASKDLWIWHAFFGMPGSHNDINVLHRSPLFAK 262
Query: 185 VEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQERCRKD 244
+ +GKAP V++ +N+ Y M YYLADGIYPS+ TFVK+I P K FA+ QE RKD
Sbjct: 263 LAEGKAPEVNYSINRHDYMMGYYLADGIYPSWATFVKTIPEPHGNKRKYFAKAQEAVRKD 322
Query: 245 IERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDFEQSG 304
+ERAFGVLQARF I+R PAR W+ LG IM++C+I+HNMI+EDE + W E+
Sbjct: 323 VERAFGVLQARFAIVRGPARHWNEKTLGYIMKACVIMHNMIIEDEGEV---DWE--ERFP 377
Query: 305 EGGSSTPQPYSTEVLPAFANHVRARSELRDSNVHHELQADLVKHIWTKF 353
EGG + S + +P + ++ ++RD H++L+ DLV+H+W +
Sbjct: 378 EGGENV--RVSHDEIPDLDDFIQMHKKIRDDETHYQLREDLVEHLWQHY 424
>UniRef100_Q6L531 Hypothetical protein OJ1005_B11.14 [Oryza sativa]
Length = 443
Score = 360 bits (923), Expect = 4e-98
Identities = 173/291 (59%), Positives = 215/291 (73%)
Query: 1 MFRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADA 60
MFRRR+RM++ +FLRIV +LS YFTQRVDA + +SPL KC A+RML YG AAD
Sbjct: 88 MFRRRFRMRRPLFLRIVHELSEWSPYFTQRVDATGRNSLSPLQKCIAAIRMLGYGTAADQ 147
Query: 61 VDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSID 120
+DE ++I +T LECL +F G+I + EYLR P ++L++ILQ +E RGFPGM GSID
Sbjct: 148 LDEVLRIAASTCLECLGKFAEGVIEKFGDEYLRLPRADELEKILQENEARGFPGMYGSID 207
Query: 121 CMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSP 180
CMHW WKNCPK W G FT G+KG T+ILEAVA+ DL IWHAFFG G+ NDINVL++SP
Sbjct: 208 CMHWPWKNCPKGWAGMFTSGNKGVPTMILEAVATKDLRIWHAFFGTAGSQNDINVLNKSP 267
Query: 181 VFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQER 240
+F +G+APTV + VN Y+M YYLAD IYP + FVK++ PQ DKLFA QE
Sbjct: 268 LFIQAIKGEAPTVHYNVNGTQYDMGYYLADRIYPEWAVFVKTVTAPQLAEDKLFALKQEG 327
Query: 241 CRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERD 291
RKD+E AFGVLQ+RF IIR PARLW D+ IM++C+ILHNMIVEDE++
Sbjct: 328 ARKDVECAFGVLQSRFDIIRRPARLWKQGDVINIMQACVILHNMIVEDEKE 378
>UniRef100_Q9FYM5 F21J9.3 [Arabidopsis thaliana]
Length = 457
Score = 360 bits (923), Expect = 4e-98
Identities = 169/291 (58%), Positives = 215/291 (73%), Gaps = 1/291 (0%)
Query: 1 MFRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADA 60
MFRRR+RM + +FLRI + DNYF QR D K G+S L K T A RMLAYGV AD+
Sbjct: 1 MFRRRFRMSRSLFLRIYDAIQRHDNYFVQRRDRVGKLGLSGLQKMTAAFRMLAYGVPADS 60
Query: 61 VDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSID 120
DEYIKIG +TALE L+RFC I+ ++ YLR+P D+ R+L + E RGFP M+GS+D
Sbjct: 61 TDEYIKIGESTALESLKRFCRAIVEVFACRYLRSPDANDVARLLHIGESRGFPRMLGSLD 120
Query: 121 CMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSP 180
CMHW+WKNCP AW GQ+ G + T+ILEAVA +DLWIWHA+FG PG+ NDINVL+ S
Sbjct: 121 CMHWKWKNCPTAWGGQYA-GRSRSPTIILEAVADYDLWIWHAYFGLPGSNNDINVLEASH 179
Query: 181 VFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQER 240
+F ++ +G AP S+++N++PYNM+YYLADGIYP + T V++I P+ KLFA QE
Sbjct: 180 LFANLAEGTAPPASYVINEKPYNMSYYLADGIYPKWSTLVQTIHDPRGPKKKLFAMKQEA 239
Query: 241 CRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERD 291
CRKD+ERAFGVLQ+RF I+ P+RLW+ L IM SCII+HNMI+EDERD
Sbjct: 240 CRKDVERAFGVLQSRFAIVAGPSRLWNKTVLHDIMTSCIIMHNMIIEDERD 290
>UniRef100_Q5VPS3 Ribosomal protein-like [Oryza sativa]
Length = 436
Score = 355 bits (912), Expect = 8e-97
Identities = 176/349 (50%), Positives = 240/349 (68%), Gaps = 4/349 (1%)
Query: 2 FRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADAV 61
FRRR+RM++ +FLRIV ++++ + +FTQR +AA + G S L KCT A++MLAY AD +
Sbjct: 76 FRRRFRMRRELFLRIVDEVTAKNRFFTQRRNAAGQLGFSALHKCTVALKMLAYVGPADEL 135
Query: 62 DEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSIDC 121
D+++K+G +T L+ L F +I ++ E+LR P E+++ IL ++ RGFPGMIGSIDC
Sbjct: 136 DDHLKMGESTVLKTLMEFVMTMIKVFGKEFLRPPRSEEIEHILSINLARGFPGMIGSIDC 195
Query: 122 MHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPV 181
MHWEW +CP W+G + RG KG T+ILE VA+ DL IWHA+FG PG+ NDIN+L RS V
Sbjct: 196 MHWEWSSCPTGWQGMY-RGHKGKPTLILEVVATEDLRIWHAYFGLPGSHNDINILHRSNV 254
Query: 182 FDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQERC 241
FDDV +G+AP+V F +N Y++ YYLADGIYP + T VKSI LP S K+F++ QE C
Sbjct: 255 FDDVAKGRAPSVEFHINDNIYSLGYYLADGIYPDWATLVKSIALPVSNKQKVFSEQQESC 314
Query: 242 RKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDFE 301
RKD+ERAFGVLQA++KI+ PARLW L IMR+C+ILHNM+VEDER +
Sbjct: 315 RKDVERAFGVLQAKWKILHRPARLWSPKVLNSIMRACVILHNMVVEDERGV---QLPTVH 371
Query: 302 QSGEGGSSTPQPYSTEVLPAFANHVRARSELRDSNVHHELQADLVKHIW 350
S G S P +PA V A++ ++D LQ DL++H+W
Sbjct: 372 PSEWPGVSDPPINQDREIPAIEKLVDAQNIIQDRETAALLQNDLMEHMW 420
>UniRef100_Q9SH78 Hypothetical protein At2g07520 [Arabidopsis thaliana]
Length = 222
Score = 351 bits (900), Expect = 2e-95
Identities = 169/209 (80%), Positives = 181/209 (85%), Gaps = 6/209 (2%)
Query: 127 KNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSPVFDDVE 186
KNCP AWEGQFTRGDKGT+T+ EAVASHDLWIWH FFGC TLNDIN LDRSPVFDD+E
Sbjct: 16 KNCPIAWEGQFTRGDKGTSTISFEAVASHDLWIWHTFFGCASTLNDINFLDRSPVFDDIE 75
Query: 187 QGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQERCRKDIE 246
QG P V+F V QR YNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLF Q+QE CRKDIE
Sbjct: 76 QGNTPRVNFFVYQRQYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFVQLQEGCRKDIE 135
Query: 247 RAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDFEQSGEG 306
RAFGVL ARFKII W+IADL IIMRSCIILHNMIVE+ERDT+AQ WTD++QS E
Sbjct: 136 RAFGVLHARFKII------WNIADLAIIMRSCIILHNMIVENERDTYAQHWTDYDQSKES 189
Query: 307 GSSTPQPYSTEVLPAFANHVRARSELRDS 335
GSSTPQP+STEVL AFANHV ARS+LRDS
Sbjct: 190 GSSTPQPFSTEVLHAFANHVHARSKLRDS 218
>UniRef100_Q8RUP5 Simiar to ribosomal protein [Oryza sativa]
Length = 310
Score = 348 bits (893), Expect = 1e-94
Identities = 171/307 (55%), Positives = 217/307 (69%), Gaps = 1/307 (0%)
Query: 49 MRMLAYGVAADAVDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSE 108
MRMLAYG ADA+DEY+KIG T+LECL RF G+I ++ EYLR PT D++R+LQV+E
Sbjct: 1 MRMLAYGTPADALDEYLKIGKCTSLECLDRFTRGVIEVFGGEYLRRPTSSDVERLLQVNE 60
Query: 109 QRGFPGMIGSIDCMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPG 168
RGF GM+GSIDCMHW W+ CP AW GQFTRGD G T++LEAVAS DL IWHAFFG G
Sbjct: 61 SRGFSGMLGSIDCMHWRWEKCPLAWRGQFTRGDYGVPTIVLEAVASQDLHIWHAFFGVAG 120
Query: 169 TLNDINVLDRSPVFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQS 228
+ ND+NVL++SP+F D +G+AP V F VN Y YYLADGIYP + F+K+I LPQ+
Sbjct: 121 SNNDLNVLNQSPLFFDALKGEAPQVQFSVNGNEYGTGYYLADGIYPEWAAFMKTIPLPQT 180
Query: 229 EPDKLFAQVQERCRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVED 288
E KLFA+ QE RKD+ERAFGVLQ+RF I+R PARLW +G IM +C+ILHNMIVED
Sbjct: 181 EKHKLFAKHQEGARKDVERAFGVLQSRFTIVRRPARLWRRKSVGRIMLACVILHNMIVED 240
Query: 289 ERDTFAQRWTDFEQSGEGGSSTPQPYSTEVLPAFANHVRARSELRDSNVHHELQADLVKH 348
ER+ A D ++ + P + FA+ +R ++ +R H +L+ DLV+H
Sbjct: 241 EREE-ATIHIDLNENPGASFALPPEVNIGGNLCFADVMRKKATVRSRPQHTQLKNDLVEH 299
Query: 349 IWTKFAN 355
IW KF +
Sbjct: 300 IWHKFGD 306
>UniRef100_Q7X8Y7 OSJNBa0085H03.2 protein [Oryza sativa]
Length = 440
Score = 348 bits (893), Expect = 1e-94
Identities = 169/351 (48%), Positives = 230/351 (65%), Gaps = 6/351 (1%)
Query: 1 MFRRRYRMQKHVFLRIVADLSSSDNYFTQRVDAAKKEGISPLAKCTTAMRMLAYGVAADA 60
+FRRR+RM + +FLRIVA + + D+YF QR +A G + L K A+RMLAY + AD+
Sbjct: 89 LFRRRFRMSRELFLRIVASVEAHDDYFRQRPNAVGLLGATALQKVYGAIRMLAYDIPADS 148
Query: 61 VDEYIKIGGTTALECLRRFCNGII*LYEHEYLRAPTQEDLQRILQVSEQRGFPGMIGSID 120
+DE ++I +T +E + F ++ ++ +YLRAPT ED R++ ++ RGFPGM+G ID
Sbjct: 149 LDEVVRISESTMIEAFKHFVKAVVDVFADQYLRAPTAEDTARLMAINTPRGFPGMLGCID 208
Query: 121 CMHWEWKNCPKAWEGQFTRGDKGTTTVILEAVASHDLWIWHAFFGCPGTLNDINVLDRSP 180
CMHW WKNCP W+GQ++ G T+ILEAVAS DLWIWH+FFG PG+LNDINVL RSP
Sbjct: 209 CMHWRWKNCPTGWKGQYS-GHVDGPTMILEAVASKDLWIWHSFFGLPGSLNDINVLQRSP 267
Query: 181 VFDDVEQGKAPTVSFIVNQRPYNMAYYLADGIYPSYPTFVKSIRLPQSEPDKLFAQVQER 240
+F + G AP + F+VN Y M YYLADGIY S+ TFVK+I Q +A+VQE
Sbjct: 268 LFQRLTSGTAPELEFMVNGNKYTMGYYLADGIYLSWATFVKTISNSQGNKRIHYAKVQEG 327
Query: 241 CRKDIERAFGVLQARFKIIREPARLWDIADLGIIMRSCIILHNMIVEDERDTFAQRWTDF 300
RKD+ERAFGVLQARF ++R PAR WD L IM +C+I+HNMI+++ERD DF
Sbjct: 328 VRKDVERAFGVLQARFAMVRGPARFWDTETLWYIMTACVIMHNMIIDNERD----ENVDF 383
Query: 301 EQSGEGGS-STPQPYSTEVLPAFANHVRARSELRDSNVHHELQADLVKHIW 350
+ E + Y P ++ E+ D VH +L+ DL++H+W
Sbjct: 384 DYDQEDSEVLRKEEYQRRNKPVLEKFLKIHKEIEDRRVHEQLRDDLMEHLW 434
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.139 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 598,021,542
Number of Sequences: 2790947
Number of extensions: 24502573
Number of successful extensions: 59880
Number of sequences better than 10.0: 159
Number of HSP's better than 10.0 without gapping: 63
Number of HSP's successfully gapped in prelim test: 96
Number of HSP's that attempted gapping in prelim test: 59700
Number of HSP's gapped (non-prelim): 173
length of query: 356
length of database: 848,049,833
effective HSP length: 128
effective length of query: 228
effective length of database: 490,808,617
effective search space: 111904364676
effective search space used: 111904364676
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0075.14


