

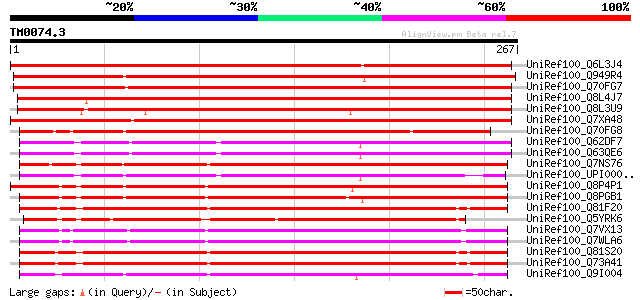
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0074.3
(267 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6L3J4 Putative dioxygenase [Solanum demissum] 387 e-106
UniRef100_Q949R4 4,5-DOPA dioxygenase extradiol-like protein [Ar... 376 e-103
UniRef100_Q70FG7 4,5-DOPA dioxygenase extradiol [Beta vulgaris] 367 e-100
UniRef100_Q8L4J7 P0018C10.9 protein [Oryza sativa] 353 2e-96
UniRef100_Q8L3U9 P0018C10.10 protein [Oryza sativa] 335 9e-91
UniRef100_Q7XA48 4,5-DOPA dioxygenase extradiol [Portulaca grand... 327 3e-88
UniRef100_Q70FG8 4,5 dioxygenase extradiol [Physcomitrella patens] 287 2e-76
UniRef100_Q62DF7 Class III extradiol-type catecholic dioxygenase... 217 2e-55
UniRef100_Q63QE6 Hypothetical protein [Burkholderia pseudomallei] 217 3e-55
UniRef100_Q7NS76 Hypothetical protein [Chromobacterium violaceum] 212 7e-54
UniRef100_UPI00002E1BC7 UPI00002E1BC7 UniRef100 entry 210 3e-53
UniRef100_Q8P4P1 Hypothetical protein XCC3666 [Xanthomonas campe... 203 4e-51
UniRef100_Q8PGB1 Hypothetical protein XAC3706 [Xanthomonas axono... 202 9e-51
UniRef100_Q81F20 Hypothetical protein [Bacillus cereus] 200 4e-50
UniRef100_Q5YRK6 Hypothetical protein [Nocardia farcinica] 198 1e-49
UniRef100_Q7VX13 Hypothetical protein [Bordetella pertussis] 197 3e-49
UniRef100_Q7WLA6 Hypothetical protein [Bordetella bronchiseptica] 197 3e-49
UniRef100_Q81S20 Oxidoreductase [Bacillus anthracis] 194 2e-48
UniRef100_Q73A41 Oxidoreductase [Bacillus cereus] 193 3e-48
UniRef100_Q9I004 Hypothetical protein [Pseudomonas aeruginosa] 193 4e-48
>UniRef100_Q6L3J4 Putative dioxygenase [Solanum demissum]
Length = 306
Score = 387 bits (995), Expect = e-106
Identities = 186/265 (70%), Positives = 219/265 (82%), Gaps = 2/265 (0%)
Query: 1 MALKDTFYISHGSPTLSIDESLVARKFLQSWKKE-VFPPRPTSILVISGHWDTAVPTVNV 59
+ +K+TF+ISHGSPTLSIDESL AR FL+S+K+ + +P SILVIS HW+T+ PTVN
Sbjct: 38 LPVKETFFISHGSPTLSIDESLPARNFLKSFKERFMINQKPNSILVISAHWETSEPTVNS 97
Query: 60 VDSTNDTIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAW 119
+ DTI+DFYGFPK MYQLKYPAPG+P LAKRVK++L GF V EDK RGLDHGAW
Sbjct: 98 IRGRQDTIHDFYGFPKSMYQLKYPAPGSPELAKRVKDVLMASGFPIVHEDKNRGLDHGAW 157
Query: 120 VPLLLMYPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALER 179
VPL+LMYPEADIPVCQLSVQ N DGT+HYN+GKALA LKDEGVLI+GSGSA HNLRAL
Sbjct: 158 VPLMLMYPEADIPVCQLSVQPNRDGTYHYNLGKALASLKDEGVLIIGSGSATHNLRALGP 217
Query: 180 HATVAAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGE 239
V++ WA+EFDNWLK+ALL GR++DVN+Y+ KAPHAK AHPWP+H YPLHVA+GAAGE
Sbjct: 218 SKNVSS-WALEFDNWLKDALLSGRHQDVNNYDMKAPHAKVAHPWPEHIYPLHVALGAAGE 276
Query: 240 NSKAKLIHSSIDLGSLSYASYQFTS 264
+LIH S DLG+LSYASY+F S
Sbjct: 277 GVNGELIHHSWDLGALSYASYRFPS 301
>UniRef100_Q949R4 4,5-DOPA dioxygenase extradiol-like protein [Arabidopsis thaliana]
Length = 269
Score = 376 bits (965), Expect = e-103
Identities = 180/267 (67%), Positives = 221/267 (82%), Gaps = 4/267 (1%)
Query: 3 LKDTFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDS 62
+ TF++SHGSPTLSID+SL AR+F +SW ++V P +P SILVIS HWDT P+VN V
Sbjct: 4 VNQTFFLSHGSPTLSIDDSLEARQFFKSWTQKVLPQKPKSILVISAHWDTKFPSVNTV-L 62
Query: 63 TNDTIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELL-KEGGFSRVDEDKKRGLDHGAWVP 121
N+TI+DF GFP PMY+LKY APGA L KRVKELL KEGG RVDED KRGLDHGAWVP
Sbjct: 63 RNNTIHDFSGFPDPMYKLKYEAPGAIELGKRVKELLMKEGGMKRVDEDTKRGLDHGAWVP 122
Query: 122 LLLMYPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHA 181
L+LMYPEADIP+CQLSVQSN +G++HYN+GKALA LKDEGVLI+GSGSA HNLR L+ +
Sbjct: 123 LMLMYPEADIPICQLSVQSNQNGSYHYNMGKALASLKDEGVLIIGSGSATHNLRKLDFNI 182
Query: 182 TVAA--PWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGE 239
T + PWA+EFD+WL+++LL+GRY DVN +E+KAP+AK AHPWP+H YPLHV +GAAG
Sbjct: 183 TDGSPVPWALEFDHWLRDSLLQGRYGDVNEWEEKAPNAKMAHPWPEHLYPLHVVMGAAGG 242
Query: 240 NSKAKLIHSSIDLGSLSYASYQFTSDV 266
++KA+ IH+S LG+LSY+SY FTS +
Sbjct: 243 DAKAEQIHTSWQLGTLSYSSYSFTSSL 269
>UniRef100_Q70FG7 4,5-DOPA dioxygenase extradiol [Beta vulgaris]
Length = 268
Score = 367 bits (942), Expect = e-100
Identities = 171/262 (65%), Positives = 211/262 (80%), Gaps = 1/262 (0%)
Query: 3 LKDTFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDS 62
+K+TF+ISHG+P ++ID+S ++KFL+SW++++F +P +ILVIS HW+T P+VNVVD
Sbjct: 7 IKETFFISHGTPMMAIDDSKPSKKFLESWREKIFSKKPKAILVISAHWETDQPSVNVVD- 65
Query: 63 TNDTIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPL 122
NDTIYDF GFP +YQ KY APG+P LA R+++LL GF V+ DKKRGLDHGAWVPL
Sbjct: 66 INDTIYDFRGFPARLYQFKYSAPGSPELANRIQDLLAGSGFKSVNTDKKRGLDHGAWVPL 125
Query: 123 LLMYPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHAT 182
+LMYPEADIPVCQLSVQS+LDGTHHY +G+ALAPLKDEGVLI+GSGSA H +
Sbjct: 126 MLMYPEADIPVCQLSVQSHLDGTHHYKLGQALAPLKDEGVLIIGSGSATHPSNGTPPCSD 185
Query: 183 VAAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSK 242
APWA FD+WL+ AL G YE+VN YE KAP+ K AHPWP+HFYPLHVA+GAAGENSK
Sbjct: 186 GVAPWAAAFDSWLETALTNGSYEEVNKYETKAPNWKLAHPWPEHFYPLHVAMGAAGENSK 245
Query: 243 AKLIHSSIDLGSLSYASYQFTS 264
A+LIH+S D G +SY SY+FTS
Sbjct: 246 AELIHNSWDGGIMSYGSYKFTS 267
>UniRef100_Q8L4J7 P0018C10.9 protein [Oryza sativa]
Length = 266
Score = 353 bits (907), Expect = 2e-96
Identities = 163/262 (62%), Positives = 203/262 (77%), Gaps = 2/262 (0%)
Query: 5 DTFYISHGSPTLSIDESLVARKFLQSWKKEVFPPR--PTSILVISGHWDTAVPTVNVVDS 62
DTF++SHG+PTLSID+++ A+ F +SW P +ILV+SGHW+ A PTVNV+
Sbjct: 2 DTFFLSHGAPTLSIDDTIAAQGFFKSWLPAAVAGAELPRAILVVSGHWEAAAPTVNVIRG 61
Query: 63 TNDTIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPL 122
NDTI+DFYGFPK MY+LKYPAPGAP LA + KELL++ GF V E+ RGLDHGAWVPL
Sbjct: 62 NNDTIHDFYGFPKAMYKLKYPAPGAPDLAMKTKELLEQAGFGPVKENHSRGLDHGAWVPL 121
Query: 123 LLMYPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHAT 182
+ MYPEA++PVCQLS+QS DG +HY +G+ALAPL+D+GVL++GSGSA HNLR + T
Sbjct: 122 MFMYPEANVPVCQLSLQSGRDGAYHYELGRALAPLRDDGVLVLGSGSATHNLRRMGPEGT 181
Query: 183 VAAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSK 242
WA EFD WL+EALL GR++DV YE+KAPH + AHP PDHF PLHVA+GAAGE +K
Sbjct: 182 PVPQWAAEFDGWLQEALLGGRHDDVKRYEEKAPHGRVAHPSPDHFLPLHVALGAAGEGAK 241
Query: 243 AKLIHSSIDLGSLSYASYQFTS 264
A+LIH S SLSYASY+FT+
Sbjct: 242 AELIHRSWSNASLSYASYRFTT 263
>UniRef100_Q8L3U9 P0018C10.10 protein [Oryza sativa]
Length = 280
Score = 335 bits (858), Expect = 9e-91
Identities = 162/269 (60%), Positives = 208/269 (77%), Gaps = 11/269 (4%)
Query: 5 DTFYISHGSPTLSIDESLVARKFLQSWKKEVF----PPRPTSILVISGHWDTAVPTVNVV 60
DTF++SHG+PTLSID+++ A+ F +SW PPR +IL++SGHW+TA PTVNVV
Sbjct: 2 DTFFLSHGTPTLSIDDTMPAQHFFRSWLPAAVAGAQPPR--AILIVSGHWETATPTVNVV 59
Query: 61 DSTNDTIYDF--YGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGA 118
NDTI+DF YGFPK M+QL+YPAPGAP +AK+ KELL++ GF V ED RGLDHGA
Sbjct: 60 RGNNDTIHDFEGYGFPKSMFQLEYPAPGAPDVAKKAKELLEQAGFGPVKEDHGRGLDHGA 119
Query: 119 WVPLLLMYPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALE 178
WVPL+ MYPEA++PVCQLS+Q+ DG +HY++G+ALAPL+D+GVLI+GSG+A HNL +
Sbjct: 120 WVPLMFMYPEANVPVCQLSLQTGRDGAYHYDLGRALAPLRDDGVLILGSGNATHNLSCMA 179
Query: 179 --RHATVAAPWAVEFDNWLKEALLE-GRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIG 235
T WA EFD WL+EALL GR++DV YE+KAP+ K AHP PDHF PLHVA+G
Sbjct: 180 PVAEGTPVPQWAAEFDGWLQEALLAGGRHDDVKQYEEKAPNGKMAHPSPDHFLPLHVALG 239
Query: 236 AAGENSKAKLIHSSIDLGSLSYASYQFTS 264
AAGE++KA+LIH S +LS+ASY+FT+
Sbjct: 240 AAGEDAKAELIHHSWYNATLSHASYRFTT 268
>UniRef100_Q7XA48 4,5-DOPA dioxygenase extradiol [Portulaca grandiflora]
Length = 271
Score = 327 bits (837), Expect = 3e-88
Identities = 158/265 (59%), Positives = 193/265 (72%), Gaps = 2/265 (0%)
Query: 1 MALKDTFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVV 60
++ K++F++SHG+P + DES +AR FL WKK VFP +P SILV+S HW+T VP V+
Sbjct: 7 VSFKESFFLSHGNPAMLADESFIARNFLLGWKKNVFPVKPKSILVVSAHWETDVPCVSAG 66
Query: 61 DSTNDTIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWV 120
N IYDF P M+Q+KYPAPG P LAKRV+ELL GGF D++RG DH +WV
Sbjct: 67 QYPN-VIYDFTEVPASMFQMKYPAPGCPKLAKRVQELLIAGGFKSAKLDEERGFDHSSWV 125
Query: 121 PLLLMYPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERH 180
PL +M PEADIPVCQLSVQ LD THH+N+G+ALAPLK EGVL +GSG AVH
Sbjct: 126 PLSMMCPEADIPVCQLSVQPGLDATHHFNVGRALAPLKGEGVLFIGSGGAVHPSDDTPHW 185
Query: 181 ATVAAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPHA-KKAHPWPDHFYPLHVAIGAAGE 239
APWA EFD WL++ALLEGRYEDVN+Y+ KAP K AHP P+HF PLHVA+GA GE
Sbjct: 186 FDGVAPWAAEFDQWLEDALLEGRYEDVNNYQTKAPEGWKLAHPIPEHFLPLHVAMGAGGE 245
Query: 240 NSKAKLIHSSIDLGSLSYASYQFTS 264
SKA+LI+ + D G+L YASY+FTS
Sbjct: 246 KSKAELIYRTWDHGTLGYASYKFTS 270
>UniRef100_Q70FG8 4,5 dioxygenase extradiol [Physcomitrella patens]
Length = 264
Score = 287 bits (734), Expect = 2e-76
Identities = 139/248 (56%), Positives = 176/248 (70%), Gaps = 4/248 (1%)
Query: 6 TFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDSTND 65
TFY+SHGSP + ++++ + R+F +W E +P RP +IL IS HWDT P VN V S N
Sbjct: 9 TFYVSHGSPMMPLEDTPI-REFFSTWT-ERYPTRPKAILAISAHWDTREPAVNAV-SQNS 65
Query: 66 TIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPLLLM 125
TI+DFYGFP+ +YQL+Y PGAP +AKRV LLK+ GF V ED KRGLDHGAW PL+LM
Sbjct: 66 TIHDFYGFPRELYQLQYTPPGAPDVAKRVTSLLKDAGFKTVLEDNKRGLDHGAWTPLMLM 125
Query: 126 YPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHATVAA 185
YP ADIPV Q+S+QSN DG HHY +G+ALAPLKDEGVLI SG+ VHNLR ++ A
Sbjct: 126 YPNADIPVLQVSIQSNKDGLHHYQLGRALAPLKDEGVLIFASGTTVHNLREIDFSAKKPT 185
Query: 186 PWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSKAKL 245
WA FD WL + LL ++++ +E KAP+A KAHP PDHF P+ V +GAAGE +A+
Sbjct: 186 VWAKAFDGWLTDVLLNSKHKEAMEWE-KAPYASKAHPHPDHFLPVLVGLGAAGEQCQAEK 244
Query: 246 IHSSIDLG 253
I+ G
Sbjct: 245 IYEEFAYG 252
>UniRef100_Q62DF7 Class III extradiol-type catecholic dioxygenase family protein
[Burkholderia mallei]
Length = 262
Score = 217 bits (553), Expect = 2e-55
Identities = 108/261 (41%), Positives = 156/261 (59%), Gaps = 8/261 (3%)
Query: 6 TFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDSTND 65
+ Y+SHG+PTL ID +L + F + + PRP ++L++S HW T P V+ + D
Sbjct: 6 SLYLSHGAPTLPIDPTLPSGAFTELGAEL---PRPRAVLMLSAHWLTQQPVVSTAERP-D 61
Query: 66 TIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPLLLM 125
TI+DFYGFP+ +Y+++YPAPGAP +A LL+ G GLDHGAWVP+LLM
Sbjct: 62 TIHDFYGFPRALYEIRYPAPGAPDVAAHAAALLRSAGIETAATP--HGLDHGAWVPMLLM 119
Query: 126 YPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHATV-- 183
+P AD+PV QLS+Q D HH+ +G+AL PL+DEGV+++GSG HNLRA + A
Sbjct: 120 FPHADVPVAQLSIQPRADAAHHFRVGRALRPLRDEGVMVIGSGQITHNLRAADFSAAPDD 179
Query: 184 AAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSKA 243
P EF W +EAL + + Y ++APHA HP +H P+ A+GAA ++ +
Sbjct: 180 VDPRVAEFTGWFEEALAARDVDALVDYRRRAPHAALMHPTDEHLLPVFAALGAANDDYRL 239
Query: 244 KLIHSSIDLGSLSYASYQFTS 264
++ L+ +Y F S
Sbjct: 240 RIQSLGTYQRMLAMTNYVFDS 260
>UniRef100_Q63QE6 Hypothetical protein [Burkholderia pseudomallei]
Length = 262
Score = 217 bits (552), Expect = 3e-55
Identities = 108/261 (41%), Positives = 156/261 (59%), Gaps = 8/261 (3%)
Query: 6 TFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDSTND 65
+ Y+SHG+PTL ID +L + F + + PRP ++L++S HW T P V+ + D
Sbjct: 6 SLYLSHGAPTLPIDPTLPSGAFTELGAEL---PRPRAVLMLSAHWLTQQPVVSTAERP-D 61
Query: 66 TIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPLLLM 125
TI+DFYGFP+ +Y+++YPAPGAP +A LL+ G GLDHGAWVP+LLM
Sbjct: 62 TIHDFYGFPRALYEIRYPAPGAPDVAAHAAALLRSAGIETAATP--HGLDHGAWVPMLLM 119
Query: 126 YPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHATV-- 183
+P AD+PV QLS+Q D HH+ +G+AL PL+DEGV+++GSG HNLRA + A
Sbjct: 120 FPHADVPVAQLSIQPRADAAHHFRVGRALRPLRDEGVMVIGSGQITHNLRAADFSAAPDD 179
Query: 184 AAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSKA 243
P EF W +EAL + + Y ++APHA HP +H P+ A+GAA ++ +
Sbjct: 180 VDPRVAEFTGWFEEALAARDVDALVDYRRRAPHAALMHPTDEHLLPVFAALGAADDDYRL 239
Query: 244 KLIHSSIDLGSLSYASYQFTS 264
++ L+ +Y F S
Sbjct: 240 RIQSLGTYQRMLAMTNYVFDS 260
>UniRef100_Q7NS76 Hypothetical protein [Chromobacterium violaceum]
Length = 259
Score = 212 bits (540), Expect = 7e-54
Identities = 107/257 (41%), Positives = 164/257 (63%), Gaps = 5/257 (1%)
Query: 6 TFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDSTND 65
+ +ISHG+PTL I++S R FL+ ++ PRP +I+++S HW+ TVN+ +
Sbjct: 7 SLFISHGAPTLPIEDS-PTRHFLEQLGRDW--PRPRAIVIVSAHWEARALTVNL-QPRPE 62
Query: 66 TIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPLLLM 125
T+YDF GFP + L+YPA LA+RV LL G V ++R LDHG W PL+LM
Sbjct: 63 TMYDFGGFPPVLRTLRYPAATDAALAERVIGLLDAAGLP-VAATEERPLDHGMWTPLMLM 121
Query: 126 YPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHATVAA 185
+P+ADIP+ +S++ HY +G+AL PL+D+ VL++GSGS HNL L + A
Sbjct: 122 FPQADIPLVMISLRHQAGVAEHYALGQALKPLRDDNVLVIGSGSYTHNLWELSPEGSEPA 181
Query: 186 PWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSKAKL 245
PWA +F W+ +ALL+ R +D+ H++Q+AP+ ++ HP +HF PL VA+GA+ +++
Sbjct: 182 PWAADFAGWMDQALLDNRRDDIVHWQQRAPYPRENHPSDEHFQPLLVALGASDDDAVPVK 241
Query: 246 IHSSIDLGSLSYASYQF 262
+H +GSLS A ++F
Sbjct: 242 LHDDWRMGSLSMACWRF 258
>UniRef100_UPI00002E1BC7 UPI00002E1BC7 UniRef100 entry
Length = 301
Score = 210 bits (535), Expect = 3e-53
Identities = 108/258 (41%), Positives = 154/258 (58%), Gaps = 17/258 (6%)
Query: 6 TFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDSTND 65
+ Y+SHG+PTL ID +L + F + PRP ++L++S HW T P ++ + +
Sbjct: 44 SLYLSHGAPTLPIDPALPSGAFTHLAAEL---PRPRAVLMLSAHWGTQRPVASIA-AHPE 99
Query: 66 TIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPLLLM 125
TI+DFYGFP+ +Y ++YPAPGAP +A+R LL G + GLDHGAWVP+LLM
Sbjct: 100 TIHDFYGFPQALYDIRYPAPGAPDVAERAAGLLNAAGIETATTE--HGLDHGAWVPMLLM 157
Query: 126 YPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHATV-- 183
+PEAD+PV QLS+Q D HH+ +G+AL PL+DEGV+++GSG HNLRA + A
Sbjct: 158 FPEADVPVAQLSIQPRADAAHHFALGRALRPLRDEGVMVIGSGQITHNLRAADFGAAPED 217
Query: 184 AAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSKA 243
A P EF +W + L + + Y ++APHA HP +H P+ A+GAA ++
Sbjct: 218 ADPRVTEFTDWFEAKLAARDVDALLDYRRQAPHAALMHPTDEHLLPVFTALGAADDD--- 274
Query: 244 KLIHSSIDLGSLSYASYQ 261
LG S +YQ
Sbjct: 275 ------YQLGIQSLGTYQ 286
>UniRef100_Q8P4P1 Hypothetical protein XCC3666 [Xanthomonas campestris]
Length = 272
Score = 203 bits (516), Expect = 4e-51
Identities = 113/265 (42%), Positives = 162/265 (60%), Gaps = 8/265 (3%)
Query: 1 MALKDTFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVV 60
M++ + YISHGSP ++ + + L + + + PRP +I+V S HW P V
Sbjct: 1 MSVLPSLYISHGSPMTALRPGELGTQ-LAALAQGL--PRPRAIVVASAHWLARRPLVGAA 57
Query: 61 DSTNDTIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWV 120
S TI+DF GFP P+Y+++YPAPGAP LA++V LL+ G +D +RGLDHG WV
Sbjct: 58 -SKPVTIHDFGGFPPPLYEIEYPAPGAPALAQQVTRLLELAGL-HPHQDPQRGLDHGIWV 115
Query: 121 PLLLMYPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALER- 179
PL ++YP+ADIPV LS+Q L H + +G+ALAPL+ EGVL++GSGS HNL L +
Sbjct: 116 PLRMLYPQADIPVVPLSIQPELGPAHQFAVGRALAPLRAEGVLVIGSGSITHNLHDLHQG 175
Query: 180 -HATVAAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAA- 237
A AP+ F +W + L + + Y ++APHA +AHP +H PL+VA+GAA
Sbjct: 176 YSAEREAPYVRPFIDWTERRLADDDVPALLDYRRQAPHAARAHPTDEHLLPLYVAMGAAG 235
Query: 238 GENSKAKLIHSSIDLGSLSYASYQF 262
G+ A+ I + ID G + Y+F
Sbjct: 236 GDRLGAQRIDAGIDQGLIGMDIYRF 260
>UniRef100_Q8PGB1 Hypothetical protein XAC3706 [Xanthomonas axonopodis]
Length = 272
Score = 202 bits (513), Expect = 9e-51
Identities = 113/261 (43%), Positives = 159/261 (60%), Gaps = 10/261 (3%)
Query: 6 TFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDSTND 65
+ YISHGSP ++ V + L + + PRP +I+V S HW P V+
Sbjct: 6 SLYISHGSPMTALQPGQVGER-LADLRHAL--PRPRAIVVASAHWLGRRPLVSGA-LRPP 61
Query: 66 TIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPLLLM 125
TI+DF GFP P+Y L YPAPG P LA+++ L++ G D +RG DHG WVPL L+
Sbjct: 62 TIHDFGGFPAPLYALDYPAPGEPALAQQIVRRLEQAGL-HPQLDPQRGFDHGVWVPLRLL 120
Query: 126 YPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHATVA- 184
YP+ADIPV LS+Q L H + +G+ALAPL+D+GVL++GSGS HNL + RH A
Sbjct: 121 YPDADIPVVSLSIQPELGPAHQFAVGRALAPLRDDGVLVIGSGSITHNLHDV-RHGYSAE 179
Query: 185 --APWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENS- 241
AP+ F W + LL+ + Y ++APHA++AHP +H+ PL+VA+GAAG++
Sbjct: 180 RQAPYVRPFIEWTERKLLDDDIPALLDYRRQAPHAERAHPSDEHWLPLYVAMGAAGDDRL 239
Query: 242 KAKLIHSSIDLGSLSYASYQF 262
A+ I + ID G L+ Y+F
Sbjct: 240 GAQRIDAGIDAGFLAMDLYRF 260
>UniRef100_Q81F20 Hypothetical protein [Bacillus cereus]
Length = 253
Score = 200 bits (508), Expect = 4e-50
Identities = 105/257 (40%), Positives = 164/257 (62%), Gaps = 7/257 (2%)
Query: 6 TFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDSTND 65
+ +++HGSP L+I ++ R FL++ + +P +I++ + HW++ V T++ D+ +
Sbjct: 4 SLFLAHGSPMLAIQDTDYTR-FLKTLGETY---KPKAIVIFTAHWESEVLTISSSDNEYE 59
Query: 66 TIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPLLLM 125
TIYDF GFP +Y++KY A G+ H+A ++ K G S V + RGLDHG+W L M
Sbjct: 60 TIYDFGGFPPELYEIKYRAKGSSHIASILETKFKNKGIS-VHHNMTRGLDHGSWTLLHRM 118
Query: 126 YPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHATVAA 185
YPEA+IPV Q+SV L + IG+AL L E +L++GSG VHNLRAL+ + T
Sbjct: 119 YPEANIPVVQISVNPFLSAKEQFEIGEALKGLGQEDILVIGSGVTVHNLRALKWNQTTPE 178
Query: 186 PWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSKAKL 245
WA+EFD+W+ + + + + ++E+ APHA+ A P +HF PL +A+G +GENS ++
Sbjct: 179 KWAIEFDDWIIKHMQNNDKDALFNWEKNAPHAQLAVPRAEHFVPLFIAMG-SGENS-GEV 236
Query: 246 IHSSIDLGSLSYASYQF 262
IH S +LG+LSY +F
Sbjct: 237 IHRSYELGTLSYLCLRF 253
>UniRef100_Q5YRK6 Hypothetical protein [Nocardia farcinica]
Length = 258
Score = 198 bits (503), Expect = 1e-49
Identities = 111/233 (47%), Positives = 147/233 (62%), Gaps = 11/233 (4%)
Query: 8 YISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDSTNDTI 67
Y+SHG+P L E A L +W E+ PRPT+IL++S HW+ A P +T +
Sbjct: 13 YLSHGAPPLVDHERWPAE--LAAWAAEL--PRPTAILIVSAHWEAA-PIALGATTTVPLV 67
Query: 68 YDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPLLLMYP 127
YDF+GFP+ Y ++YPAPGAP LA++V+ LL G V E RGLDHGA+VPL MYP
Sbjct: 68 YDFWGFPRHYYDVRYPAPGAPDLARKVRGLLGSG----VAEVPDRGLDHGAYVPLTQMYP 123
Query: 128 EADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHATVAAPW 187
+AD+PV QLS+ + LD + IG+ LAPL++EGVL+VGSG HNLRA+ +
Sbjct: 124 DADVPVLQLSMPT-LDPEALFGIGRKLAPLREEGVLVVGSGFFTHNLRAMTEDDVHVHRF 182
Query: 188 AVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGEN 240
EFD+W AL + + + +E KAP A+ AHP DHF PL V +G AGEN
Sbjct: 183 TAEFDDWGARALADRDLDALLDFEHKAPAARLAHPRADHFAPLFVTLG-AGEN 234
>UniRef100_Q7VX13 Hypothetical protein [Bordetella pertussis]
Length = 258
Score = 197 bits (500), Expect = 3e-49
Identities = 106/258 (41%), Positives = 152/258 (58%), Gaps = 7/258 (2%)
Query: 6 TFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDSTND 65
T ++SHGSP L+++ L +W + P RP ++LV+S HW V+ D
Sbjct: 5 TLFVSHGSPMLAVEPGRTGPA-LTAWS-DGLPERPRAVLVVSPHWMGQGLAVSTRDR-QV 61
Query: 66 TIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPLLLM 125
+DF GFP +YQL+YPA G+P LA++V ++L G + D +R LDHGAWVPL +
Sbjct: 62 AWHDFGGFPADLYQLQYPAEGSPRLAQQVVDVLAHAGI-QAGNDARRPLDHGAWVPLRYL 120
Query: 126 YPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALER-HATVA 184
YP AD+PV QLS+ D Y +G+ALAPL+D+GVL++GSGS HNLR + H
Sbjct: 121 YPHADLPVVQLSLDMERDARGQYALGQALAPLRDDGVLVIGSGSLTHNLRDVRMPHGAPP 180
Query: 185 APWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSKAK 244
A + F W E L G E + ++ +AP A +AHP DH PL+VA+GA G A+
Sbjct: 181 ADYVAPFQQWYAEHLAAGDVEALLDWQARAPGAARAHPHDDHLMPLYVALGAGG--MPAR 238
Query: 245 LIHSSIDLGSLSYASYQF 262
++ + G+L+ +YQF
Sbjct: 239 RLNDEVAYGALAMDAYQF 256
>UniRef100_Q7WLA6 Hypothetical protein [Bordetella bronchiseptica]
Length = 279
Score = 197 bits (500), Expect = 3e-49
Identities = 106/258 (41%), Positives = 152/258 (58%), Gaps = 7/258 (2%)
Query: 6 TFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDSTND 65
T ++SHGSP L+++ L +W + P RP ++LV+S HW V+ D
Sbjct: 26 TLFVSHGSPMLAVEPGRTGPA-LAAWS-DGLPERPRAVLVVSPHWMGQGLAVSTRDR-QV 82
Query: 66 TIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPLLLM 125
+DF GFP +YQL+YPA G+P LA++V ++L G + D +R LDHGAWVPL +
Sbjct: 83 AWHDFGGFPADLYQLQYPAEGSPRLAQQVVDVLARAGI-QAGNDARRPLDHGAWVPLRYL 141
Query: 126 YPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALER-HATVA 184
YP AD+PV QLS+ D Y +G+ALAPL+D+GVL++GSGS HNLR + H
Sbjct: 142 YPHADLPVVQLSLDMERDARGQYALGQALAPLRDDGVLVIGSGSLTHNLRDVRMPHGAPP 201
Query: 185 APWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSKAK 244
A + F W E L G E + ++ +AP A +AHP DH PL+VA+GA G A+
Sbjct: 202 ADYVAPFQQWYAEHLAAGDVEALLDWQARAPGAARAHPHDDHLMPLYVALGAGG--MPAR 259
Query: 245 LIHSSIDLGSLSYASYQF 262
++ + G+L+ +YQF
Sbjct: 260 RLNDEVAYGALAMDAYQF 277
>UniRef100_Q81S20 Oxidoreductase [Bacillus anthracis]
Length = 253
Score = 194 bits (493), Expect = 2e-48
Identities = 102/257 (39%), Positives = 161/257 (61%), Gaps = 7/257 (2%)
Query: 6 TFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDSTND 65
+ +++HGSP L+I ++ FL++ + +P +I++ + HW++ V T++ D+ +
Sbjct: 4 SLFLAHGSPMLAIQDTDYT-SFLKTLGETY---KPKAIVIFTAHWESEVLTISSTDNEYE 59
Query: 66 TIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPLLLM 125
TIYDF GFP +Y++KY A G+ +A ++ K G V + RGLDHG+W L M
Sbjct: 60 TIYDFGGFPPELYEIKYRAKGSSSIASMLETKFKNKGIP-VHHNMTRGLDHGSWTLLHRM 118
Query: 126 YPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHATVAA 185
YPEA+IPV Q+SV L + IG+AL L E +L++GSG VHNLRAL+ + T
Sbjct: 119 YPEANIPVVQISVNPFLSAKEQFKIGEALKGLGQEDILVIGSGVTVHNLRALKWNQTTPE 178
Query: 186 PWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSKAKL 245
WA+EFD+W+ + + + + ++E+ APHA+ A P +HF PL +A+G +GEN+ ++
Sbjct: 179 QWAIEFDDWIIKHMQAADKDALFNWEKNAPHAQLAVPRAEHFVPLFIAMG-SGENN-GEV 236
Query: 246 IHSSIDLGSLSYASYQF 262
IH S +LG+LSY QF
Sbjct: 237 IHRSYELGTLSYLCLQF 253
>UniRef100_Q73A41 Oxidoreductase [Bacillus cereus]
Length = 253
Score = 193 bits (491), Expect = 3e-48
Identities = 103/257 (40%), Positives = 160/257 (62%), Gaps = 7/257 (2%)
Query: 6 TFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDSTND 65
+ +++HGSP L+I ++ FL+ + +P +I++ + HW++ V T++ D+ +
Sbjct: 4 SLFLAHGSPMLAIQDTDYT-SFLKILGETY---KPKAIVIFTAHWESEVLTISSSDNEYE 59
Query: 66 TIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPLLLM 125
TIYDF GFP +Y++KY A G+ +A ++ K G V + RGLDHG+W L M
Sbjct: 60 TIYDFGGFPPELYEIKYRAKGSSSIASMLETKFKNKGIP-VHHNMTRGLDHGSWTLLHRM 118
Query: 126 YPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHATVAA 185
YPEA+IPV Q+SV L + IG+AL L E +L++GSG VHNLRAL+ + T
Sbjct: 119 YPEANIPVVQISVNPFLSAKEQFKIGEALKGLGQEDILVIGSGVTVHNLRALKWNQTTPE 178
Query: 186 PWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSKAKL 245
WA+EFD+W+ + + + + ++E+ APHA+ A P +HF PL +A+G +GENS ++
Sbjct: 179 QWAIEFDDWIIKHMQAADKDALFNWEKNAPHAQLAVPRAEHFVPLFIAMG-SGENS-GEV 236
Query: 246 IHSSIDLGSLSYASYQF 262
IH S +LG+LSY QF
Sbjct: 237 IHRSYELGTLSYLCLQF 253
>UniRef100_Q9I004 Hypothetical protein [Pseudomonas aeruginosa]
Length = 256
Score = 193 bits (490), Expect = 4e-48
Identities = 111/259 (42%), Positives = 147/259 (55%), Gaps = 9/259 (3%)
Query: 6 TFYISHGSPTLSIDESLVARKFLQSWKKEVFPPRPTSILVISGHWDTAVPTVNVVDSTND 65
T YISHGSP L+++ +Q + PRP +ILV+S HW++ V + D
Sbjct: 4 TLYISHGSPMLALEPGASG---VQLRRLATELPRPKAILVVSAHWESDDLRVTGA-AVPD 59
Query: 66 TIYDFYGFPKPMYQLKYPAPGAPHLAKRVKELLKEGGFSRVDEDKKRGLDHGAWVPLLLM 125
+DFYGFP P+Y ++YPA G P LA+R ELL+ G D +R DHG WVPL LM
Sbjct: 60 IWHDFYGFPPPLYAVRYPAKGEPALAQRTIELLQAAGLP-AQADPQRPFDHGTWVPLSLM 118
Query: 126 YPEADIPVCQLSVQSNLDGTHHYNIGKALAPLKDEGVLIVGSGSAVHNLRALERHA--TV 183
YP+ADIPV QLS+ S L +G+AL L+ EGVL++GSGS HNL L+ A
Sbjct: 119 YPQADIPVLQLSLPSRLGPALQSRVGQALRQLRGEGVLLIGSGSITHNLGELDWRAGPEA 178
Query: 184 AAPWAVEFDNWLKEALLEGRYEDVNHYEQKAPHAKKAHPWPDHFYPLHVAIGAAGENSKA 243
APWA EF +W+ E L ++ Y ++AP A + HP +H PL A GA G +
Sbjct: 179 IAPWAREFRDWMVEKLQADDEAALHDYRRQAPWAARNHPSDEHLLPLFFARGAGGGGMRV 238
Query: 244 KLIHSSIDLGSLSYASYQF 262
+ HS LGSL Y+F
Sbjct: 239 E--HSGFTLGSLGMDIYRF 255
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 492,428,181
Number of Sequences: 2790947
Number of extensions: 21791142
Number of successful extensions: 50064
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 101
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 49647
Number of HSP's gapped (non-prelim): 138
length of query: 267
length of database: 848,049,833
effective HSP length: 125
effective length of query: 142
effective length of database: 499,181,458
effective search space: 70883767036
effective search space used: 70883767036
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0074.3


