

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0065.18
(799 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
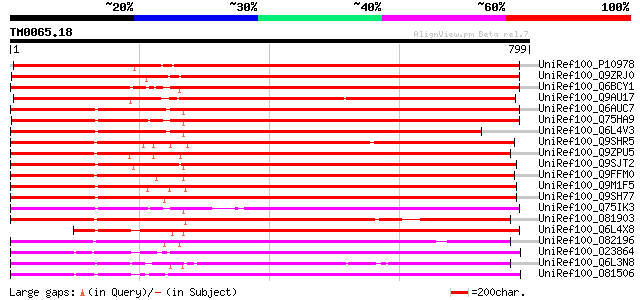
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P10978 Retrovirus-related Pol polyprotein from transpo... 835 0.0
UniRef100_Q9ZRJ0 ORF [Nicotiana tabacum] 828 0.0
UniRef100_Q6BCY1 Gag-Pol [Ipomoea batatas] 775 0.0
UniRef100_Q9AU17 Polyprotein-like [Lycopersicon chilense] 744 0.0
UniRef100_Q6AUC7 Putative polyprotein [Oryza sativa] 742 0.0
UniRef100_Q75HA9 Putative polyprotein [Oryza sativa] 717 0.0
UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa] 687 0.0
UniRef100_Q9SHR5 F28L22.3 protein [Arabidopsis thaliana] 685 0.0
UniRef100_Q9ZPU5 Putative retroelement pol polyprotein [Arabidop... 684 0.0
UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidop... 679 0.0
UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidops... 679 0.0
UniRef100_Q9M1F5 Copia-like polyprotein [Arabidopsis thaliana] 676 0.0
UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidop... 666 0.0
UniRef100_Q75IK3 Putative polyprotein [Oryza sativa] 638 0.0
UniRef100_O81903 Putative transposable element [Arabidopsis thal... 632 e-179
UniRef100_Q6L4X8 Putative polyprotein [Oryza sativa] 622 e-176
UniRef100_O82196 Copia-like retroelement pol polyprotein [Arabid... 591 e-167
UniRef100_O23864 Polyprotein [Oryza australiensis] 580 e-164
UniRef100_Q6L3N8 Putative gag-pol polyprotein [Solanum demissum] 560 e-158
UniRef100_O81506 T7M24.7 protein [Arabidopsis thaliana] 554 e-156
>UniRef100_P10978 Retrovirus-related Pol polyprotein from transposon TNT 1-94
[Contains: Protease (EC 3.4.23.-); Reverse transcriptase
(EC 2.7.7.49); Endonuclease] [Nicotiana tabacum]
Length = 1328
Score = 835 bits (2157), Expect = 0.0
Identities = 430/793 (54%), Positives = 559/793 (70%), Gaps = 16/793 (2%)
Query: 6 VENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNER 65
VE +T K+K L+SDNGGEY S+EF+++CS +GIR KT+ GTP+ NGVAERMNRT+ E+
Sbjct: 536 VERETGRKLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTVPGTPQHNGVAERMNRTIVEK 595
Query: 66 ARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVFGCVS 125
R M + LPK FW +A+ TA YLINR PS+PL +++PE VW+ KEVS SHLKVFGC +
Sbjct: 596 VRSMLRMAKLPKSFWGEAVQTACYLINRSPSVPLAFEIPERVWTNKEVSYSHLKVFGCRA 655
Query: 126 YVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYKDRSS 185
+ + ++R KLD K+I C FIGYG + +GYR WD KK+IRSR+V F ES +
Sbjct: 656 FAHVPKEQRTKLDDKSIPCIFIGYGDEEFGYRLWDPVKKKVIRSRDVVFRESEVRTAADM 715
Query: 186 AESMS-----------SSKQLKLSERVALEEISESDVVKRNQINPENEDDEVEVELEQEP 234
+E + S+ S +E+SE I + DE E+E
Sbjct: 716 SEKVKNGIIPNFVTIPSTSNNPTSAESTTDEVSEQGEQPGEVIEQGEQLDEGVEEVEHP- 774
Query: 235 TTVIETPITQVRRSTRTPKAPQRYSPSLNYLLLTDAGEPEYFGEAMQGNDSIKWELAMKD 294
T E +RRS R P+ R PS Y+L++D EPE E + + + AM++
Sbjct: 775 -TQGEEQHQPLRRSER-PRVESRRYPSTEYVLISDDREPESLKEVLSHPEKNQLMKAMQE 832
Query: 295 EMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR-RYKARLVVKGFQQKQGIDFT 353
EM SLQKNGT+ L +LP+GK+ L+ +WV++LK++ D RYKARLVVKGF+QK+GIDF
Sbjct: 833 EMESLQKNGTYKLVELPKGKRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFEQKKGIDFD 892
Query: 354 EIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNL 413
EIFSPVVKMT+IR ILS+ A+ +L +EQLDVK AFLHGDLEEEIYM QPEGFEV G K++
Sbjct: 893 EIFSPVVKMTSIRTILSLAASLDLEVEQLDVKTAFLHGDLEEEIYMEQPEGFEVAGKKHM 952
Query: 414 VCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADS-YIILALYVDDM 472
VCKL+KSLYGLKQAPRQWY KF+ FM + + + D C + K+F+++ +IIL LYVDDM
Sbjct: 953 VCKLNKSLYGLKQAPRQWYMKFDSFMKSQTYLKTYSDPCVYFKRFSENNFIILLLYVDDM 1012
Query: 473 LIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKLLDR 532
LI G + I +LK +S++F+MKDLGPA+QILGM+I R R+ L LSQEKY+E++L+R
Sbjct: 1013 LIVGKDKGLIAKLKGDLSKSFDMKDLGPAQQILGMKIVRERTSRKLWLSQEKYIERVLER 1072
Query: 533 FNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHA 592
FN+ +A STPL HLK SKK P T EE+ M+ VPY+SAVGSLMYAMVCTRPDIAHA
Sbjct: 1073 FNMKNAKPVSTPLAGHLKLSKKMCPTTVEEKGNMAKVPYSSAVGSLMYAMVCTRPDIAHA 1132
Query: 593 VGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNNLTLQEFSDADLGGDSDGGKS 652
VGVVSRF+ NPGKEHWE VKWILRYL+G++ CLCF ++ L+ ++DAD+ GD D KS
Sbjct: 1133 VGVVSRFLENPGKEHWEAVKWILRYLRGTTGDCLCFGGSDPILKGYTDADMAGDIDNRKS 1192
Query: 653 TTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVPP 712
+TGY+FT G A+SW+SKLQ VALSTTE+EY+A +E KEMIWLK FL+ELG Q
Sbjct: 1193 STGYLFTFSGGAISWQSKLQKCVALSTTEAEYIAATETGKEMIWLKRFLQELGLHQKEYV 1252
Query: 713 LFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTVT 772
++ DSQS I L+KN ++H+R KHI ++YH+IRE++ DE L +LKI +ENP DMLTK V
Sbjct: 1253 VYCDSQSAIDLSKNSMYHARTKHIDVRYHWIREMVDDESLKVLKISTNENPADMLTKVVP 1312
Query: 773 ADNLRLCIASAGL 785
+ LC G+
Sbjct: 1313 RNKFELCKELVGM 1325
>UniRef100_Q9ZRJ0 ORF [Nicotiana tabacum]
Length = 1338
Score = 828 bits (2139), Expect = 0.0
Identities = 425/799 (53%), Positives = 547/799 (68%), Gaps = 21/799 (2%)
Query: 4 TEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLN 63
T VE +T K+K +++DNGGEY Q F +C E+GIR T TP+ NG+AERMNRTL
Sbjct: 532 TLVERETGKKLKCIRTDNGGEYQGQ-FDAYCKEHGIRHQFTPPKTPQLNGLAERMNRTLI 590
Query: 64 ERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVFGC 123
ER RC+ S LPK FW +A+ TAAY++N P +PL Y+ PE++W G+++S L+VFGC
Sbjct: 591 ERTRCLLSHSKLPKAFWGEALVTAAYVLNHSPCVPLQYKAPEKIWLGRDISYDQLRVFGC 650
Query: 124 VSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYKDR 183
+YV + D R KLD K +C FIGYG DM GY+F+D KK++RSR+V F E +D
Sbjct: 651 KAYVHVPKDERSKLDVKTRECVFIGYGQDMLGYKFYDPVEKKLVRSRDVVFVEDQTIEDI 710
Query: 184 SSAE-SMSSSKQLKLSERVALEEISES--------------DVVKRNQINPENEDDEVEV 228
E S S + +L V ++ + D + + N +N DD+ +
Sbjct: 711 DKVEKSTDDSAEFELPPTVVPRQVGDDVQDNQPEAPGLPNEDELADTEGNEDNGDDDADE 770
Query: 229 ELEQEPTTVIETPITQVRRSTRTPKAPQRYSPSLNYLLLTDAGEPEYFGEAMQGNDSIKW 288
E + +P + P RS R + RYSP Y+LLTD GEP+ F EA+ KW
Sbjct: 771 EDQPQPPILNNPPYHT--RSGRVVQQSTRYSPH-EYVLLTDGGEPDSFEEAIDDEHKEKW 827
Query: 289 ELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR-RYKARLVVKGFQQK 347
AM+DE+ SL +N T+ L KLP+GK+AL+N+WV+++K + S R+KARLVVKGF Q+
Sbjct: 828 IEAMQDEIKSLHENKTFELVKLPKGKRALKNKWVFKMKHDEHNSLPRFKARLVVKGFNQR 887
Query: 348 QGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEV 407
+GIDF EIFSPVVKMT+IR +L + A+ NL +EQ+DVK AFLHGDLEEEIYM QP+GF+
Sbjct: 888 KGIDFDEIFSPVVKMTSIRTVLGLAASLNLEVEQMDVKTAFLHGDLEEEIYMEQPDGFQQ 947
Query: 408 LGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADS-YIILA 466
G ++ VC+L KSLYGLKQAPRQWYKKF M G+ + DHC F +KF+D +IIL
Sbjct: 948 KGKEDYVCRLRKSLYGLKQAPRQWYKKFESVMGQHGYKKTTSDHCVFAQKFSDDDFIILL 1007
Query: 467 LYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYV 526
LYVDDMLI G N++ IN LK+Q+S+ F MKDLGPAKQILGMRI R+R L LSQEKY+
Sbjct: 1008 LYVDDMLIVGRNVSRINSLKEQLSKFFAMKDLGPAKQILGMRIMRDREAKKLWLSQEKYI 1067
Query: 527 EKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTR 586
EK+L RFN+ S PL NH + S KQSP TD+E M +PYASAVGSLMYAMVCTR
Sbjct: 1068 EKVLQRFNMEKTKAVSCPLANHFRLSTKQSPSTDDERRKMERIPYASAVGSLMYAMVCTR 1127
Query: 587 PDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNNLTLQEFSDADLGGD 646
PDIAHAVGVVSRF+SNPGKEHW+ VKWILRYL+G+S++CLCF +N L ++DAD+ GD
Sbjct: 1128 PDIAHAVGVVSRFLSNPGKEHWDAVKWILRYLRGTSKLCLCFGEDNPVLVGYTDADMAGD 1187
Query: 647 SDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGK 706
D KST+GY+ G AVSW+SKLQ VALSTTE+E++A +EA KE+IW+K FL ELG
Sbjct: 1188 VDSRKSTSGYLINFSGGAVSWQSKLQKCVALSTTEAEFIAATEACKELIWMKKFLTELGF 1247
Query: 707 EQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDM 766
QD LF DSQS I LAKN FHSR KHI ++Y++IR+++ + L L KI EN +DM
Sbjct: 1248 SQDGYQLFCDSQSAIHLAKNASFHSRSKHIDVRYNWIRDVLEKKMLRLEKIHTDENGSDM 1307
Query: 767 LTKTVTADNLRLCIASAGL 785
LTKT+ C +AG+
Sbjct: 1308 LTKTLPKGKFEFCREAAGI 1326
>UniRef100_Q6BCY1 Gag-Pol [Ipomoea batatas]
Length = 1298
Score = 775 bits (2001), Expect = 0.0
Identities = 399/791 (50%), Positives = 533/791 (66%), Gaps = 24/791 (3%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
+K VE + KIK ++DNGGEY S+EF FC + GI+ T+ TP+QNGVAERMNRT
Sbjct: 522 FKARVELDSGKKIKCFRTDNGGEYTSEEFDDFCKKEGIKRQFTVAYTPQQNGVAERMNRT 581
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
L ER R M +GL K FW +A+NTA YL+NR PS ++ + P E+W+GK V S+L +F
Sbjct: 582 LLERTRAMLRAAGLEKSFWAEAVNTACYLVNRAPSTAIELKTPMEMWTGKPVDYSNLHIF 641
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
G + Y + ++ KLDPK+ KC F+GY + GYR WD K++ SR+V F E L +
Sbjct: 642 GSIVYAMYNAQEITKLDPKSRKCRFLGYADGVKGYRLWDPTAHKVVISRDVIFVEDRLQR 701
Query: 182 DRSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVEVELEQEPTTVIETP 241
S+ K+ + ++ +EE E D ++ P +E E E E PTT
Sbjct: 702 GEVDD---STEKEKPETTQIQVEEEFEQD---SSEAEPAHE--EPEPESSGAPTT----- 748
Query: 242 ITQVRRSTRTPKAPQRYSP-----SLNYLLLTDAGEPEYFGEAMQGNDSIKWELAMKDEM 296
R+S R + P +S ++ Y LLT+ GEP F EA+ +D +W AM++E+
Sbjct: 749 ----RQSDREKRRPTWHSDYVMEGNVAYCLLTEDGEPSTFQEAINSSDVSQWTAAMQEEI 804
Query: 297 TSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGS-RRYKARLVVKGFQQKQGIDFTEI 355
+L KN TW L LP+G+K + N+WV+++K D RY+ARLVVKG+ QK+GIDF EI
Sbjct: 805 EALHKNNTWDLVPLPQGRKPIGNKWVFKIKRNGDDQVERYRARLVVKGYAQKEGIDFNEI 864
Query: 356 FSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNLVC 415
FSPVV++TT+RV+L++ A NLHLEQLDVK AFLHGDLEEEIYM QPEGFE +NLVC
Sbjct: 865 FSPVVRLTTVRVVLAMCATFNLHLEQLDVKTAFLHGDLEEEIYMLQPEGFEDKENQNLVC 924
Query: 416 KLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFA-DSYIILALYVDDMLI 474
+L+KSLYGLKQAPR WYK+F+ F+ G+NR + D C + K+F D+++IL LYVDDML+
Sbjct: 925 RLNKSLYGLKQAPRCWYKRFDSFIMCLGYNRLNADPCAYFKRFGKDNFVILLLYVDDMLV 984
Query: 475 AGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKLLDRFN 534
AG N I+ LK Q++ FEMKDLGPA +ILGM+I R+R + LSQ+ Y++K+L RF+
Sbjct: 985 AGPNKDHIDELKAQLAREFEMKDLGPANKILGMQIHRDRGNRKIWLSQKNYLKKILSRFS 1044
Query: 535 VGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVG 594
+ D + STPL +LK S SP +E MS VPYASAVGSLM+AM+CTRPDIA AVG
Sbjct: 1045 MQDCKSISTPLPINLKVSSSMSPSNEEGRMEMSRVPYASAVGSLMFAMICTRPDIAQAVG 1104
Query: 595 VVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNNLTLQEFSDADLGGDSDGGKSTT 654
VVSR+M+NPG+EHW CVK ILRY+KG+S + LC+ ++ + + D+D GD D KSTT
Sbjct: 1105 VVSRYMANPGREHWNCVKRILRYIKGTSDVALCYGGSDFIINGYVDSDYAGDLDKSKSTT 1164
Query: 655 GYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVPPLF 714
GY+F + G AVSW SKLQ VA STTE+EYVA ++A+KE IWLK L+ELG +Q+ LF
Sbjct: 1165 GYVFKVAGGAVSWVSKLQAVVATSTTEAEYVAATQASKEAIWLKMLLEELGHKQEFVSLF 1224
Query: 715 SDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTVTAD 774
DSQS + LA+NP FHSR KHI+++YHFIRE + + + L KI ++N D LTK + D
Sbjct: 1225 CDSQSALHLARNPAFHSRTKHIRVQYHFIREKVKEGTVDLQKIHTADNVADFLTKIINVD 1284
Query: 775 NLRLCIASAGL 785
C +S GL
Sbjct: 1285 KFTWCRSSCGL 1295
>UniRef100_Q9AU17 Polyprotein-like [Lycopersicon chilense]
Length = 1328
Score = 744 bits (1921), Expect = 0.0
Identities = 397/795 (49%), Positives = 536/795 (66%), Gaps = 35/795 (4%)
Query: 6 VENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTLNER 65
VE +T K K L++DNGGEY S+EF+++CS +GIR KT+ GTP+ NGVAERMNRT+ E+
Sbjct: 537 VERETGRKRKRLRTDNGGEYTSREFEEYCSNHGIRHEKTVPGTPQHNGVAERMNRTIVEK 596
Query: 66 ARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVFGCVS 125
R M + LPK FW +A+ TA YLINR PS+PL++ +PE VW+ KE+S SHLKVFGC +
Sbjct: 597 VRSMLRMAKLPKTFWGEAVRTACYLINRSPSVPLEFDIPERVWTNKEMSYSHLKVFGCKA 656
Query: 126 YVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVL--YKDR 183
+ + ++R KLD K++ C FIGYG + +GYR WD KK+IRSR+V F ES + D
Sbjct: 657 FAHVPKEQRTKLDDKSVPCIFIGYGDEEFGYRLWDLVKKKVIRSRDVIFRESEVGTAADL 716
Query: 184 S-----------------SAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDDEV 226
S S+ + +S + + E V EE + V + Q+ E E
Sbjct: 717 SEKAKKKNGIIPNLVTIPSSSNHPTSAESTIDEVVEQEEQPDEIVEQGEQLGDNTEQMEY 776
Query: 227 EVELEQEPTTVIETPITQVRRSTRTPKAPQRYSPSLNYLLLTDAGEPEYFGEAMQGNDSI 286
E + +P +RRS R +Y PS Y+L+ GEPE E + +
Sbjct: 777 PEEEQSQP----------LRRSERQRVESTKY-PSSEYVLIKYEGEPENLKEVLSHPEKS 825
Query: 287 KWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR-RYKARLVVKGFQ 345
+W AM +EM SLQKNGT+ L +LP+GK+ L+ +WV++LK++ +G RYKARLVVKGF+
Sbjct: 826 QWMKAMHEEMGSLQKNGTYQLVELPKGKRPLKCKWVFKLKKDGNGKLVRYKARLVVKGFE 885
Query: 346 QKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGF 405
QK+GIDF EIFSPVVKMT+IR ILSI A+ +L +EQLDVK AFLHGDLEEEIYM Q EGF
Sbjct: 886 QKKGIDFDEIFSPVVKMTSIRTILSIAASLDLEVEQLDVKTAFLHGDLEEEIYMEQGEGF 945
Query: 406 EVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFAD-SYII 464
EV G K++VCKL+KSLYGLKQAPRQWYKKF+ FM + + C + K+F+D ++II
Sbjct: 946 EVSGKKHMVCKLNKSLYGLKQAPRQWYKKFDSFMKSQTYRNTYSHPCVYFKRFSDKNFII 1005
Query: 465 LALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEK 524
L LY D MLI G + I +L++ S++F+MKDLGPAKQILGM+I+R + L LS EK
Sbjct: 1006 LLLYTDYMLIVGKDKELIAKLRKDFSKSFDMKDLGPAKQILGMKIAREEQK-KLGLSHEK 1064
Query: 525 YVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDE-EESYMSTVPYASAVGSLMYAMV 583
Y+E++L+RFN+ A STPL ++LK +K+ P + E+ M+ VPY+SAVGS MYAMV
Sbjct: 1065 YIERVLERFNMKSAKPISTPLVSYLKLTKQMFPTKKKGEKGDMAKVPYSSAVGSFMYAMV 1124
Query: 584 CTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNNLTLQEFSDADL 643
CTRP+I AV VVSRF+ PGKEH E VKWILRYL+ ++R CF ++ + +++ D+
Sbjct: 1125 CTRPNIV-AVCVVSRFLEIPGKEHLEAVKWILRYLRRTTRDYFCFEGSDPISKGYTNVDM 1183
Query: 644 GGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKE 703
GD D KSTT Y+FT G +SW+SKLQ VALSTTE++Y+A +E KEM+WLK FL+E
Sbjct: 1184 EGDLDNRKSTTCYLFTFSGGDISWQSKLQKYVALSTTEAKYIAGTEVCKEMLWLKRFLQE 1243
Query: 704 LGKEQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENP 763
G Q ++ +SQS + L+K ++H+ KHI M+YH+IRE++ D L ++KI SENP
Sbjct: 1244 HGLHQKEYVVYCESQSAMDLSKKAMYHATTKHIDMRYHWIREMVDDGSLQVVKIPTSENP 1303
Query: 764 TDMLTKTVTADNLRL 778
DM+TK V + L
Sbjct: 1304 ADMVTKVVQNEKFEL 1318
>UniRef100_Q6AUC7 Putative polyprotein [Oryza sativa]
Length = 1241
Score = 742 bits (1916), Expect = 0.0
Identities = 388/797 (48%), Positives = 519/797 (64%), Gaps = 22/797 (2%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
WKT VE QT K+K L++DNG E S+ FK +C GI T+ TP+QNGVAERMNRT
Sbjct: 452 WKTMVERQTERKVKILRTDNGMELCSKIFKSYCKSEGIVRHYTVPHTPQQNGVAERMNRT 511
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
+ +ARCM + LPK FW +A++TA YLINR PS +D + P EVWSG + S L+VF
Sbjct: 512 IISKARCMLSNASLPKQFWAEAVSTACYLINRSPSYAIDKKTPIEVWSGSPANYSDLRVF 571
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
GC +Y +D+ KL+P+ IKC F+GY + GY+ W + KK++ SRNV F+ES++
Sbjct: 572 GCTAYAHVDNG---KLEPRVIKCIFLGYLSGVKGYKLWCPETKKVVISRNVVFHESIMLH 628
Query: 182 DRSSAESMSSSKQ---LKLSERVALEEISESDVVKRNQINPENEDDEVEVELEQEPTTVI 238
D+ S S++ +++ ++ E + V NQ P ED + + ++Q P I
Sbjct: 629 DKPSTNVPVESQEKVSVQVEHLISSGHAPEKEDVAINQDAPVIEDSDSSI-VQQSPKRSI 687
Query: 239 ETPITQVRRSTRTPKAPQRYSPSLNYLL--------LTDAGEPEYFGEAMQGNDSIKWEL 290
R R K P+RY N + + EP + +A+ +D +W
Sbjct: 688 AKD-----RPKRNTKPPRRYIEEANIVAYALSVAEEIEGNAEPSTYSDAIVSDDCNRWIT 742
Query: 291 AMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGS--RRYKARLVVKGFQQKQ 348
AM DEM SL+KN +W L KLP+ KK ++ +W+++ KE S RYKARLV KG+ Q
Sbjct: 743 AMHDEMESLEKNHSWELEKLPKEKKPIRCKWIFKRKEGMSPSDEARYKARLVAKGYSQIP 802
Query: 349 GIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVL 408
GIDF ++FSPVVK ++IR +LSIVA + LEQ+DVK AFLHG+LEE+IYM QPEGF V
Sbjct: 803 GIDFNDVFSPVVKHSSIRTLLSIVAMHDYELEQMDVKTAFLHGELEEDIYMEQPEGFVVP 862
Query: 409 GTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADSYIILALY 468
G +NLVC+L KSLYGLKQ+PRQWYK+F+ FM + F R + D C ++K S I L LY
Sbjct: 863 GKENLVCRLKKSLYGLKQSPRQWYKRFDSFMLSQKFRRSNYDSCVYLKVVDGSAIYLLLY 922
Query: 469 VDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEK 528
VDDMLIA + +EI +LK Q+S FEMKDLG AK+ILGM I+R R G L LSQ+ Y+EK
Sbjct: 923 VDDMLIAAKDKSEIAKLKAQLSSEFEMKDLGAAKKILGMEITRERHSGKLYLSQKCYIEK 982
Query: 529 LLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPD 588
+L RFN+ DA ST L H + S PQ+ + YMS VPY+SAV SLMYAMVC+RPD
Sbjct: 983 VLHRFNMHDAKLVSTLLAAHFRLSSDLCPQSAYDIEYMSRVPYSSAVSSLMYAMVCSRPD 1042
Query: 589 IAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNNLTLQEFSDADLGGDSD 648
++HA+ VVSR+M+NPGKEHW+ V+WI RYL+G+S CL F R++ L + D+D GD D
Sbjct: 1043 LSHALSVVSRYMANPGKEHWKAVQWIFRYLRGTSSACLQFGRSSDGLVGYVDSDFAGDLD 1102
Query: 649 GGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQ 708
+S TGY+FT+GG AVSWK+ LQ VALSTTE+EY+AISEA KE+IWL+ EL
Sbjct: 1103 RRRSLTGYVFTVGGCAVSWKASLQATVALSTTEAEYMAISEACKEVIWLRGLYTELCGVT 1162
Query: 709 DVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLT 768
+F DSQS I L K+ +FH R KHI ++YHFIR +I++ ++ + KI +NP DM+T
Sbjct: 1163 SCINIFCDSQSAICLTKDQMFHERTKHIDLRYHFIRGVIAEGDVKVCKISTHDNPVDMMT 1222
Query: 769 KTVTADNLRLCIASAGL 785
K V A LC + G+
Sbjct: 1223 KPVPATKFELCSSLVGV 1239
>UniRef100_Q75HA9 Putative polyprotein [Oryza sativa]
Length = 1322
Score = 717 bits (1852), Expect = 0.0
Identities = 373/796 (46%), Positives = 511/796 (63%), Gaps = 25/796 (3%)
Query: 3 KTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRTL 62
K +E QT ++K L +DNGGE+ S F +C + GI TI TP+QNGVAERMNRT+
Sbjct: 537 KVMIERQTEKEVKVLCTDNGGEFCSDAFDDYCRKEGIVRHHTIPYTPQQNGVAERMNRTI 596
Query: 63 NERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVFG 122
+ARCM + + K FW +A NTA YLINR PSIPL+ + P E+WSG S L+VFG
Sbjct: 597 ISKARCMLSNARMNKRFWAEAANTACYLINRSPSIPLNKKTPIEIWSGMPADYSQLRVFG 656
Query: 123 CVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYKD 182
C +Y +D+ KL+P+AIKC F+GYG + GY+ W+ + K SRNV FNE V++ D
Sbjct: 657 CTAYAHVDNG---KLEPRAIKCLFLGYGSGVKGYKLWNPETNKTFMSRNVIFNEFVMFND 713
Query: 183 RSSAESM---SSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVEVELEQEPTTVIE 239
+ + S +Q +S +V + E+++V N +N + ++ + EP
Sbjct: 714 SLPTDVIPGGSDEEQQYVSVQVEHVDDQETEIVG-NDVNDTVQHSPSVLQPQDEPIAH-- 770
Query: 240 TPITQVRRSTRTPKAPQRYSPSLNYLL--------LTDAGEPEYFGEAMQGNDSIKWELA 291
RR+ R+ AP R + + + + EP + EA+ D KW A
Sbjct: 771 ------RRTKRSCGAPVRLIEECDMVYYAFSYAEQVENTLEPATYTEAVVSGDREKWISA 824
Query: 292 MKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR--RYKARLVVKGFQQKQG 349
+++EM SL+KNGTW L LP+ KK ++ +W+++ KE S R+K RLV KGF Q G
Sbjct: 825 IQEEMQSLEKNGTWELVHLPKQKKPVRCKWIFKRKEGLSPSEPPRFKVRLVAKGFSQIAG 884
Query: 350 IDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLG 409
+D+ ++FSPVVK ++IR SIV +L LEQLDVK FLHG+LEEEIYM QPEGF V G
Sbjct: 885 VDYNDVFSPVVKHSSIRTFFSIVTMHDLELEQLDVKTTFLHGELEEEIYMDQPEGFIVPG 944
Query: 410 TKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADSYIILALYV 469
++ VCKL +SLYGLKQ+PRQWYK+F+ FM + GF R + D C ++K S I L LYV
Sbjct: 945 KEDYVCKLKRSLYGLKQSPRQWYKRFDSFMLSHGFKRSEFDSCVYIKFVNGSPIYLLLYV 1004
Query: 470 DDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKL 529
DDMLIA + +I LK+Q+S F+MKDLG AK+ILGM I+R+R+ G+L LSQ+ Y++K+
Sbjct: 1005 DDMLIAAKSKEQITTLKKQLSSEFDMKDLGAAKKILGMEITRDRNSGLLFLSQQSYIKKV 1064
Query: 530 LDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPDI 589
L RFN+ DA STP+ H K S Q TDE+ YMS VPY+SAVGSLMYAMVC+ PD+
Sbjct: 1065 LQRFNMHDAKPVSTPIAPHFKLSALQCASTDEDVEYMSRVPYSSAVGSLMYAMVCSWPDL 1124
Query: 590 AHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNNLTLQEFSDADLGGDSDG 649
+HA+ +VSR+M+NPGKEHW+ V+WI RYL+G++ CL F R + L + D+D D D
Sbjct: 1125 SHAMSLVSRYMANPGKEHWKAVQWIFRYLRGTADACLKFGRIDKGLVGYVDSDFAADLDK 1184
Query: 650 GKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQD 709
+S TGY+FT+G AVSWK+ LQ VA STTE+EY+AI+EA KE +WLK EL
Sbjct: 1185 RRSLTGYVFTIGSCAVSWKATLQPVVAQSTTEAEYMAIAEACKESVWLKGLFAELCGVDS 1244
Query: 710 VPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTK 769
LF DSQS I L K+ +FH R KHI +KYH++R++++ +L + KI +NP DM+TK
Sbjct: 1245 CINLFCDSQSAICLTKDQMFHERTKHIDIKYHYVRDIVAQGKLKVCKISIHDNPADMMTK 1304
Query: 770 TVTADNLRLCIASAGL 785
+ LC + G+
Sbjct: 1305 PIPVAKFELCSSLVGI 1320
>UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa]
Length = 1243
Score = 687 bits (1773), Expect = 0.0
Identities = 363/738 (49%), Positives = 479/738 (64%), Gaps = 22/738 (2%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
WKT VE QT K+K L++DNG E+ S+ FK +C GI T TP+QN VAERMNRT
Sbjct: 515 WKTMVERQTERKVKILRTDNGMEFCSKIFKSYCKSEGIVCHYTAPHTPQQNDVAERMNRT 574
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
+ +ARCM +GLPK FW +A++TA YLINR P +D + P EVWSG + S L+VF
Sbjct: 575 IISKARCMLSNAGLPKQFWAEAVSTACYLINRSPGYAIDKKTPIEVWSGSPTNYSDLRVF 634
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
GC +Y +D+ KL+P+AIKC F+GY + GY+ W + KK++ SRNV F+ESV+
Sbjct: 635 GCTAYAHVDNG---KLEPRAIKCIFLGYASGVKGYKLWCPETKKVVISRNVVFHESVILH 691
Query: 182 DRSSAESMSSSKQ---LKLSERVALEEISESDVVKRNQINPENEDDEVEVELEQEPTTVI 238
D+ S S++ +++ ++ E + V NQ P ED + + + Q P I
Sbjct: 692 DKPSTNVPVESQEKASVQVEHLISSGHAPEKEDVAINQDAPVIEDSDSSI-VHQSPKRSI 750
Query: 239 ETPITQVRRSTRTPKAPQRY---SPSLNYLL-----LTDAGEPEYFGEAMQGNDSIKWEL 290
+ R K P+RY + + Y L + EP + EA+ +D +W
Sbjct: 751 AKD-----KPKRNIKPPRRYIEEAKIVAYALSVAEKIEGNAEPSTYSEAIVSDDCNRWIT 805
Query: 291 AMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGS--RRYKARLVVKGFQQKQ 348
AM DEM SL+KN TW L KLP+ KK ++ +W+++ KE S RYKARLV KG+ Q
Sbjct: 806 AMHDEMESLEKNHTWELVKLPKEKKPIRCKWIFKRKEGMSPSDEARYKARLVAKGYSQIP 865
Query: 349 GIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVL 408
GIDF ++FSPVVK ++IR +L IVA + LEQ++VK AFLHG+LEE+IYM QPEGF V
Sbjct: 866 GIDFNDVFSPVVKHSSIRTLLGIVAMHDYELEQMNVKTAFLHGELEEDIYMEQPEGFVVP 925
Query: 409 GTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADSYIILALY 468
G +NLVC+L KSLYGLKQ+PRQWYK+F+ FM + F + D C ++K S I L LY
Sbjct: 926 GKENLVCRLKKSLYGLKQSPRQWYKRFDSFMLSQKFRISNYDSCVYLKVVDGSVIYLLLY 985
Query: 469 VDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEK 528
VDDMLIA + +EI +LK Q+S FEMKDLG AK+ILGM I+R R G L LSQ+ Y+EK
Sbjct: 986 VDDMLIAAKDKSEIEKLKAQLSSEFEMKDLGAAKKILGMEITRERHSGKLYLSQKGYIEK 1045
Query: 529 LLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPD 588
+L RFN+ DA STPL H + S P +D + YMS VPY+SAVGSLMYAMVC RPD
Sbjct: 1046 VLRRFNMHDAKPVSTPLAAHFRLSSDLCPLSDYDIEYMSRVPYSSAVGSLMYAMVCCRPD 1105
Query: 589 IAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNNLTLQEFSDADLGGDSD 648
++HA+ VV+R+M+NPGKEHW+ V+WI RYL+G+S CL F R+ L + D+D GD D
Sbjct: 1106 LSHALSVVNRYMANPGKEHWKAVQWIFRYLRGTSSACLQFERSRDGLVGYVDSDFAGDLD 1165
Query: 649 GGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQ 708
+S TGY+FT+GG AVSWK+ LQ VALSTTE+EY+AI EA KE IWL+ EL
Sbjct: 1166 RRRSITGYVFTIGGCAVSWKASLQATVALSTTEAEYMAIFEACKEAIWLRGLYTELCGVT 1225
Query: 709 DVPPLFSDSQSVIFLAKN 726
+F DSQS I+L K+
Sbjct: 1226 SCINIFCDSQSAIYLTKD 1243
>UniRef100_Q9SHR5 F28L22.3 protein [Arabidopsis thaliana]
Length = 1356
Score = 685 bits (1767), Expect = 0.0
Identities = 361/805 (44%), Positives = 525/805 (64%), Gaps = 37/805 (4%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
WK+ VENQ K+K L++DNG E+ + F +C E+GI +T TP+QNGVAERMNRT
Sbjct: 548 WKSLVENQVNKKVKCLRTDNGLEFCNSRFDSYCKEHGIERHRTCTYTPQQNGVAERMNRT 607
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
+ E+ RC+ +SG+ ++FW +A TAAYLINR P+ +++ +PEE+W ++ HL+ F
Sbjct: 608 IMEKVRCLLNKSGVEEVFWAEAAATAAYLINRSPASAINHNVPEEMWLNRKPGYKHLRKF 667
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
G ++YV D + KL P+A+K FF+GY GY+ W + +K + SRNV F ESV+Y+
Sbjct: 668 GSIAYVHQD---QGKLKPRALKGFFLGYPAGTKGYKVWLLEEEKCVISRNVVFQESVVYR 724
Query: 182 DRSSAESMSSSKQLKLSERVALE-----EISESDVVKRNQINPE-------NEDDEVEVE 229
D E + + K + +E E S S V + Q + E + D E EVE
Sbjct: 725 DLKVKEDDTDNLNQKETTSSEVEQNKFAEASGSGGVIQLQSDSEPITEGEQSSDSEEEVE 784
Query: 230 L-EQEPTTVIETPITQVR----RSTRTPKAPQRYSP--SLNYLLLTDAG----EPEYFGE 278
E+ T T +T + R R P R++ S+ + L+ EP+ + E
Sbjct: 785 YSEKTQETPKRTGLTTYKLARDRVRRNINPPTRFTEESSVTFALVVVENCIVQEPQSYQE 844
Query: 279 AMQGNDSIKWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR--RYK 336
AM+ D KW++A DEM SL KNGTW L P+ +K + RW+++LK G R+K
Sbjct: 845 AMESQDCEKWDMATHDEMDSLMKNGTWDLVDKPKDRKIIGCRWLFKLKSGIPGVEPTRFK 904
Query: 337 ARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEE 396
ARLV KG+ Q++G+D+ EIF+PVVK +IR+++S+V ++L LEQ+DVK FLHGDLEEE
Sbjct: 905 ARLVAKGYTQREGVDYQEIFAPVVKHVSIRILMSLVVDKDLELEQMDVKTTFLHGDLEEE 964
Query: 397 IYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVK 456
+YM QPEGF ++N VC+L KSLYGLKQ+PRQW K+F+ FMS+ F R + D C +VK
Sbjct: 965 LYMEQPEGFVSDSSENKVCRLKKSLYGLKQSPRQWNKRFDRFMSSQQFIRSEHDACVYVK 1024
Query: 457 KFAD-SYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSE 515
++ +I L LYVDDMLIAG++ EINR+K+Q+S FEMKD+G A +ILG+ I R+R
Sbjct: 1025 HVSEHDFIYLLLYVDDMLIAGASKAEINRVKEQLSTEFEMKDMGGASRILGIDIYRDRKG 1084
Query: 516 GVLKLSQEKYVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMST--VPYAS 573
GVLKLSQE Y+ K+LDRFN+ A + P+G H K + + EE+ + T VPY+S
Sbjct: 1085 GVLKLSQEIYIRKVLDRFNMSGAKMTNAPVGAHFKLAAVR-----EEDECVDTDVVPYSS 1139
Query: 574 AVGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCF-RRNN 632
AVGS+MYAM+ TRPD+A+A+ ++SR+MS PG HWE VKW++RYLKG+ + L F + +
Sbjct: 1140 AVGSIMYAMLGTRPDLAYAICLISRYMSKPGSMHWEAVKWVMRYLKGAQDLNLVFTKEKD 1199
Query: 633 LTLQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAK 692
T+ + D++ D D +S +GY+FT+GG VSWK+ LQ VA+STTE+EY+A++EAAK
Sbjct: 1200 FTVTGYCDSNYAADLDRRRSISGYVFTIGGNTVSWKASLQPVVAMSTTEAEYIALAEAAK 1259
Query: 693 EMIWLKSFLKELGKEQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEEL 752
E +W+K L+++G +QD ++ DSQS I L+KN V+H R KHI +++++IR+++ ++
Sbjct: 1260 EAMWIKGLLQDMGMQQDKVKIWCDSQSAICLSKNSVYHERTKHIDVRFNYIRDVVESGDV 1319
Query: 753 SLLKILGSENPTDMLTKTVTADNLR 777
+LKI S NP D LTK + + +
Sbjct: 1320 DVLKIHTSRNPVDALTKCIPVNKFK 1344
>UniRef100_Q9ZPU5 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1335
Score = 684 bits (1765), Expect = 0.0
Identities = 370/797 (46%), Positives = 507/797 (63%), Gaps = 30/797 (3%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
WK VENQ+ K+K L++DNG EY + F+KFC E GI KT TP+QNG+AER+NRT
Sbjct: 525 WKKMVENQSDRKVKKLRTDNGLEYCNHYFEKFCKEEGIVRHKTCAYTPQQNGIAERLNRT 584
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
+ ++ R M +SG+ K FW +A +TA YLINR PS +++ LPEE W+G LS L+ F
Sbjct: 585 IMDKVRSMLSRSGMEKKFWAEAASTAVYLINRSPSTAINFDLPEEKWTGALPDLSSLRKF 644
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
GC++Y+ D + KL+P++ K F Y + GY+ W ++KK + SRNV F E V++K
Sbjct: 645 GCLAYIHAD---QGKLNPRSKKGIFTSYPEGVKGYKVWVLEDKKCVISRNVIFREQVMFK 701
Query: 182 D----RSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPE-----------NEDDEV 226
D + S S + L+++ + +E ++ ++ NP +
Sbjct: 702 DLKGDSQNTISESDLEDLRVNPDMNDQEFTDQGGATQDNSNPSEATTSHNPVLNSPTHSQ 761
Query: 227 EVELEQEPTTVIETPITQ--VR-RSTRTPKAPQRYSPS----LNYLLLTDAG-EPEYFGE 278
+ E E+E + +E T VR R RT KA +Y+ S Y D EP+ + E
Sbjct: 762 DEESEEEDSDAVEDLSTYQLVRDRVRRTIKANPKYNESNMVGFAYYSEDDGKPEPKSYQE 821
Query: 279 AMQGNDSIKWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDG--SRRYK 336
A+ D KW AMK+EM S+ KN TW L PE K + RWV+ K G + R+
Sbjct: 822 ALLDPDWEKWNAAMKEEMVSMSKNHTWDLVTKPEKVKLIGCRWVFTRKAGIPGVEAPRFI 881
Query: 337 ARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEE 396
ARLV KGF QK+G+D+ EIFSPVVK +IR +LS+V N+ L+Q+DVK AFLHG LEEE
Sbjct: 882 ARLVAKGFTQKEGVDYNEIFSPVVKHVSIRYLLSMVVHYNMELQQMDVKTAFLHGFLEEE 941
Query: 397 IYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVK 456
IYM QPEGFE+ N VC L +SLYGLKQ+PRQW +F+EFM + R D C + K
Sbjct: 942 IYMAQPEGFEIKRGSNKVCLLKRSLYGLKQSPRQWNLRFDEFMRGIKYTRSAYDSCVYFK 1001
Query: 457 KF-ADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSE 515
K D+YI L LYVDDMLIA +N +E+N LKQ +S FEMKDLG AK+ILGM ISR+R
Sbjct: 1002 KCNGDTYIYLLLYVDDMLIASANKSEVNELKQLLSREFEMKDLGDAKKILGMEISRDRDA 1061
Query: 516 GVLKLSQEKYVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAV 575
G+L LSQE YV+K+L F + +A STPLG H K + +E+ M VPYA+ +
Sbjct: 1062 GLLTLSQEGYVKKVLRSFQMDNAKPVSTPLGIHFKLKAATDKEYEEQFERMKIVPYANTI 1121
Query: 576 GSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRR-NNLT 634
GS+MY+M+ TRPD+A+++GV+SRFMS P K+HW+ VKW+LRY++G+ + LCFR+ +
Sbjct: 1122 GSIMYSMIGTRPDLAYSLGVISRFMSKPLKDHWQAVKWVLRYMRGTEKKKLCFRKQEDFL 1181
Query: 635 LQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEM 694
L+ + D+D G + D +S TGY+FT+GG +SWKSKLQ VA+S+TE+EY+A++EA KE
Sbjct: 1182 LRGYCDSDYGSNFDTRRSITGYVFTVGGNTISWKSKLQKVVAISSTEAEYMALTEAVKEA 1241
Query: 695 IWLKSFLKELGKEQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSL 754
+WLK F ELG QD + SDSQS I LAKN V H R KHI ++ HFIR++I + +
Sbjct: 1242 LWLKGFAAELGHSQDYVEVHSDSQSAITLAKNSVHHERTKHIDIRLHFIRDIICAGLIKV 1301
Query: 755 LKILGSENPTDMLTKTV 771
+KI NP ++ TKTV
Sbjct: 1302 VKIATECNPANIFTKTV 1318
>UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1333
Score = 679 bits (1752), Expect = 0.0
Identities = 361/803 (44%), Positives = 520/803 (63%), Gaps = 28/803 (3%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
W VENQT +IK+L++DNG E+ ++ F +FCS+ GI +T TP+QNGVAERMNRT
Sbjct: 527 WANLVENQTDKRIKTLRTDNGLEFCNRSFDEFCSQKGILWHRTCAYTPQQNGVAERMNRT 586
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
L E+ R M SGLPK FW +A +T A LIN+ PS L+Y++P++ WSGK S+L+ F
Sbjct: 587 LMEKVRSMLSDSGLPKKFWAEATHTTAILINKTPSSALNYEVPDKRWSGKSPIYSYLRRF 646
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
GC+++V D KL+P+A K +GY + GY+ W + KK + SRNV F E+ YK
Sbjct: 647 GCIAFVHTDDG---KLNPRAKKGILVGYPIGVKGYKIWLLEEKKCVVSRNVIFQENASYK 703
Query: 182 DRSSAESM---------SSSKQLKLSERVALEEISESDVVK-RNQINPENEDDEVEVELE 231
D ++ SS L L + + +V+ ++ NP + E
Sbjct: 704 DMMQSKDAEKDENEAPPSSYLDLDLDHEEVITSGGDDPIVEAQSPFNPSPATTQTYSEGV 763
Query: 232 QEPTTVIETPITQ--VR-RSTRTPKAPQRYSPSLNYLL-----LTDAGE--PEYFGEAMQ 281
T +I++P++ VR R RT +AP R+ +YL D+GE P + EA +
Sbjct: 764 NSETDIIQSPLSYQLVRDRDRRTIRAPVRFDDE-DYLAEALYTTEDSGEIEPADYSEAKR 822
Query: 282 GNDSIKWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR--RYKARL 339
+ KW+LAM +EM S KN TW++ K P+ +K + +RW+Y+ K G R+KARL
Sbjct: 823 SMNWNKWKLAMNEEMESQIKNHTWTVVKRPQHQKVIGSRWIYKFKLGIPGVEEGRFKARL 882
Query: 340 VVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYM 399
V KG+ Q++GID+ EIF+PVVK +IR+++SIVA E+L LEQLDVK AFLHG+L+E+IYM
Sbjct: 883 VAKGYAQRKGIDYHEIFAPVVKHVSIRILMSIVAQEDLELEQLDVKTAFLHGELKEKIYM 942
Query: 400 TQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFA 459
PEG+E + ++ VC L+KSLYGLKQAP+QW +KFN +MS GF R D C ++K+ +
Sbjct: 943 VPPEGYEEMFKEDEVCLLNKSLYGLKQAPKQWNEKFNAYMSEIGFIRSLYDSCAYIKELS 1002
Query: 460 D-SYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVL 518
D S + L LYVDDML+A N +I++LK+++S+ F+MKDLG AK+ILGM I RNR E L
Sbjct: 1003 DGSRVYLLLYVDDMLVAAKNKEDISQLKEELSQRFDMKDLGAAKRILGMEIIRNREENTL 1062
Query: 519 KLSQEKYVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSL 578
LSQ Y+ K+L+ +N+ ++ TPLG HLK + +++E YM ++PY+SAVGS+
Sbjct: 1063 WLSQNGYLNKILETYNMAESKHVVTPLGAHLKMRAATVEKQEQDEDYMKSIPYSSAVGSI 1122
Query: 579 MYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRR-NNLTLQE 637
MYAM+ TRPD+A+ VG++SR+MS P +EHW VKW+LRY+KGS L ++R ++ +
Sbjct: 1123 MYAMIGTRPDLAYPVGIISRYMSQPAREHWLGVKWVLRYIKGSLGTKLQYKRSSDFKVVG 1182
Query: 638 FSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWL 697
+ DAD D +S TG +FTLGG+ +SWKS Q VALSTTE+EY++++EA KE +W+
Sbjct: 1183 YCDADHAACKDRRRSITGLVFTLGGSTISWKSGQQRVVALSTTEAEYMSLTEAVKEAVWM 1242
Query: 698 KSFLKELGKEQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKI 757
K LKE G EQ +F DSQS I L+KN V H R KHI ++Y +IR++I++ + ++KI
Sbjct: 1243 KGLLKEFGYEQKSVEIFCDSQSAIALSKNNVHHERTKHIDVRYQYIRDIIANGDGDVVKI 1302
Query: 758 LGSENPTDMLTKTVTADNLRLCI 780
+NP D+ TK V + + +
Sbjct: 1303 DTEKNPADIFTKIVPVNKFQAAL 1325
>UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidopsis thaliana]
Length = 1342
Score = 679 bits (1751), Expect = 0.0
Identities = 355/799 (44%), Positives = 505/799 (62%), Gaps = 26/799 (3%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
WKT++ENQ K+K L +DNG E+ +QEF FC + G+ +T TP+QNGVAERMNRT
Sbjct: 536 WKTQIENQQDKKLKILITDNGLEFCNQEFDSFCRKEGVIRHRTCAYTPQQNGVAERMNRT 595
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
+ + RCM +SGL K FW +A +TA +LIN+ PS +++ +PEE W+G LK F
Sbjct: 596 IMNKVRCMLSESGLGKQFWAEAASTAVFLINKSPSSSIEFDIPEEKWTGHPPDYKILKKF 655
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
G V+Y+ D + KL+P+A K F+GY + ++ W +++K + SR++ F E+ +YK
Sbjct: 656 GSVAYIHSD---QGKLNPRAKKGIFLGYPDGVKRFKVWLLEDRKCVVSRDIVFQENQMYK 712
Query: 182 DRSSAESMSSSKQLKLSER--VALEEISESDVVKRNQINPENEDD---------EVEVEL 230
+ + KQL ER + L+ +S D + + N++ + +VE
Sbjct: 713 ELQKNDMSEEDKQLTEVERTLIELKNLSADDENQSEGGDNSNQEQASTTRSASKDKQVEE 772
Query: 231 EQEPTTVIETPITQVRRSTRTPKAPQRYSPSLNYLL-----LTDAGE---PEYFGEAMQG 282
+E + R R +APQR+ + L+ +T+ GE PE + EAM+
Sbjct: 773 TDSDDDCLENYLLARDRIRRQIRAPQRFVEEDDSLVGFALTMTEDGEVYEPETYEEAMRS 832
Query: 283 NDSIKWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR--RYKARLV 340
+ KW+ A +EM S++KN TW + PEGK+ + +W+++ K G RYKARLV
Sbjct: 833 PECEKWKQATIEEMDSMKKNDTWDVIDKPEGKRVIGCKWIFKRKAGIPGVEPPRYKARLV 892
Query: 341 VKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMT 400
KGF Q++GID+ EIFSPVVK +IR +LSIV ++ LEQLDVK AFLHG+L+E I M+
Sbjct: 893 AKGFSQREGIDYQEIFSPVVKHVSIRYLLSIVVQFDMELEQLDVKTAFLHGNLDEYILMS 952
Query: 401 QPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFAD 460
QPEG+E + VC L KSLYGLKQ+PRQW ++F+ FM NSG+ R + C + ++ D
Sbjct: 953 QPEGYEDEDSTEKVCLLKKSLYGLKQSPRQWNQRFDSFMINSGYQRSKYNPCVYTQQLND 1012
Query: 461 -SYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLK 519
SYI L LYVDDMLIA N +I +LK+ ++ FEMKDLGPA++ILGM I+RNR +G+L
Sbjct: 1013 GSYIYLLLYVDDMLIASQNKDQIQKLKESLNREFEMKDLGPARKILGMEITRNREQGILD 1072
Query: 520 LSQEKYVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLM 579
LSQ +YV +L F + + TPLG H K + YM VPY +A+GS+M
Sbjct: 1073 LSQSEYVAGVLRAFGMDQSKVSQTPLGAHFKLRAANEKTLARDAEYMKLVPYPNAIGSIM 1132
Query: 580 YAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNN-LTLQEF 638
Y+M+ +RPD+A+ VGVVSRFMS P KEHW+ VKW++RY+KG+ CL F++++ ++ +
Sbjct: 1133 YSMIGSRPDLAYPVGVVSRFMSKPSKEHWQAVKWVMRYMKGTQDTCLRFKKDDKFEIRGY 1192
Query: 639 SDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLK 698
D+D D D +S TG++FT GG +SWKS LQ VALSTTE+EY+A++EA KE IWL+
Sbjct: 1193 CDSDYATDLDRRRSITGFVFTAGGNTISWKSGLQRVVALSTTEAEYMALAEAVKEAIWLR 1252
Query: 699 SFLKELGKEQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKIL 758
E+G EQD + DSQS I L+KN V H R KHI ++YHFIRE I+D E+ ++KI
Sbjct: 1253 GLAAEMGFEQDAVEVMCDSQSAIALSKNSVHHERTKHIDVRYHFIREKIADGEIQVVKIS 1312
Query: 759 GSENPTDMLTKTVTADNLR 777
+ NP D+ TKTV L+
Sbjct: 1313 TTWNPADIFTKTVPVSKLQ 1331
>UniRef100_Q9M1F5 Copia-like polyprotein [Arabidopsis thaliana]
Length = 1363
Score = 676 bits (1744), Expect = 0.0
Identities = 367/803 (45%), Positives = 508/803 (62%), Gaps = 27/803 (3%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
W + VENQ+ ++K+L++DNG E+ ++ F FC E G + +T TP+QNGV ERMNRT
Sbjct: 556 WISLVENQSGERVKTLRTDNGLEFCNRMFDGFCEEKGFQRHRTCAYTPQQNGVVERMNRT 615
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
+ E+ R M SGLPK FW +A +TA LIN+ P ++++ P++ WSGK S+L+ +
Sbjct: 616 IMEKVRSMLCDSGLPKRFWAEATHTAVLLINKTPCSAINFEFPDKRWSGKAPIYSYLRRY 675
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
GCV++V D KL+ +A K IGY + GY+ W + KK + SRNV+F E+ +YK
Sbjct: 676 GCVTFVHTDGG---KLNLRAKKGVLIGYPSGVKGYKVWLIEEKKCVVSRNVSFQENAVYK 732
Query: 182 D-RSSAESMSSSKQLKLSERVALEEISESD-----VVKRNQINPENEDDEVEVELEQEPT 235
D E +S + + L+ ++ D + Q+ P E
Sbjct: 733 DLMQRKEQVSCEEDDHAGSYIDLDLEADKDNSSGGEQSQAQVTPATRGAVTSTPPRYETD 792
Query: 236 TVIETPITQ-------VR-RSTRTPKAPQRYSPSLNYL--LLT----DAGEPEYFGEAMQ 281
+ ET + Q VR R R +AP+R+ Y L T DA EP + EA++
Sbjct: 793 DIEETDVHQSPLSYHLVRDRERREIRAPRRFDDEDYYAEALYTTEDGDAVEPADYKEAVR 852
Query: 282 GNDSIKWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR--RYKARL 339
+ KW LAM +E+ S KN TW+ PE ++ + +RW+Y+ K+ G R+KARL
Sbjct: 853 DENWDKWRLAMNEEIESQLKNDTWTTVTRPEKQRIIGSRWIYKYKQGIPGVEEPRFKARL 912
Query: 340 VVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYM 399
V KG+ Q++G+D+ EIF+PVVK +IR++LSIVA ENL LEQLDVK AFLHG+L+E+IYM
Sbjct: 913 VAKGYAQREGVDYHEIFAPVVKHVSIRILLSIVAQENLELEQLDVKTAFLHGELKEKIYM 972
Query: 400 TQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFA 459
PEG E L +N VC L+KSLYGLKQAPRQW +KFN +M+ GF R D D C + KK +
Sbjct: 973 MPPEGCESLFKENEVCLLNKSLYGLKQAPRQWNEKFNHYMTEIGFKRSDYDSCAYTKKLS 1032
Query: 460 D-SYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVL 518
D S + L YVDDML+A +NM I+ LK+++S FEMKDLG AK+ILG+ I +R GVL
Sbjct: 1033 DDSTMYLLFYVDDMLVAANNMQAIDALKKELSIKFEMKDLGAAKKILGIEIIIDREAGVL 1092
Query: 519 KLSQEKYVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSL 578
LSQE Y+ K+L FN+ ++ TPLG HLK + EE YM++VPY+SAVGS+
Sbjct: 1093 WLSQESYLNKVLKTFNMLESKPALTPLGAHLKMKSATEEKLSTEEEYMNSVPYSSAVGSI 1152
Query: 579 MYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRN-NLTLQE 637
MYAM+ TRPD+A+ VGVVSRFMS P KEHW VKW+LRY+KG+ LC++RN + ++
Sbjct: 1153 MYAMIGTRPDLAYPVGVVSRFMSQPAKEHWLGVKWVLRYIKGTVDTRLCYKRNSDFSICG 1212
Query: 638 FSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWL 697
+ DAD D D +S TG +FTLGG +SWKS LQ VA S+TE EY++++EA KE IWL
Sbjct: 1213 YCDADYAADLDKRRSITGLVFTLGGNTISWKSGLQRVVAQSSTECEYMSLTEAVKEAIWL 1272
Query: 698 KSFLKELGKEQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKI 757
K LK+ G EQ +F DSQS I L+KN V H R KHI +K+HFIRE+I+D ++ + KI
Sbjct: 1273 KGLLKDFGYEQKNVEIFCDSQSAIALSKNNVHHERTKHIDVKFHFIREIIADGKVEVSKI 1332
Query: 758 LGSENPTDMLTKTVTADNLRLCI 780
+NP D+ TK + + + +
Sbjct: 1333 STEKNPADIFTKVLPVNKFQTAL 1355
>UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1356
Score = 666 bits (1719), Expect = 0.0
Identities = 361/803 (44%), Positives = 506/803 (62%), Gaps = 27/803 (3%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
W VENQT ++K+L++DNG E+ ++ F FC GI +T TP+QNGVAERMNRT
Sbjct: 549 WVNLVENQTDRRVKTLRTDNGLEFCNKLFDGFCESIGIHRHRTCAYTPQQNGVAERMNRT 608
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
+ E+ R M SGLPK FW +A +T LIN+ PS L++++P++ WSG S+L+ +
Sbjct: 609 IMEKVRSMLSDSGLPKRFWAEATHTTVLLINKTPSSALNFEIPDKKWSGNPPVYSYLRRY 668
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
GCV++V D KL+P+A K IGY + GY+ W +K + SRN+ F E+ +YK
Sbjct: 669 GCVAFVHTDDG---KLEPRAKKGVLIGYPVGVKGYKVWILDERKCVVSRNIIFQENAVYK 725
Query: 182 D-RSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVEVELEQEPTT---- 236
D E++S+ + + + + +E DV+ N E + PTT
Sbjct: 726 DLMQRQENVSTEEDDQTGSYLEFDLEAERDVISGGDQEMVNTIPAPESPVVSTPTTQDTN 785
Query: 237 ------VIETPITQ--VR-RSTRTPKAPQRYSPSLNY---LLLTDAGE---PEYFGEAMQ 281
V ++P++ VR R R +AP+R+ Y L T+ GE PE + +A
Sbjct: 786 DDEDSDVNQSPLSYHLVRDRDKREIRAPRRFDDEDYYAEALYTTEDGEAVEPENYRKAKL 845
Query: 282 GNDSIKWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR--RYKARL 339
+ KW+LAM +E+ S +KN TW++ PE ++ + RW+++ K G R+KARL
Sbjct: 846 DANFDKWKLAMDEEIDSQEKNNTWTIVTRPENQRIIGCRWIFKYKLGILGVEEPRFKARL 905
Query: 340 VVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYM 399
V KG+ QK+GID+ EIF+PVVK +IRV+LSIVA E+L LEQLDVK AFLHG+L+E+IYM
Sbjct: 906 VAKGYAQKEGIDYHEIFAPVVKHVSIRVLLSIVAQEDLELEQLDVKTAFLHGELKEKIYM 965
Query: 400 TQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFA 459
+ PEG+E + N VC L+K+LYGLKQAP+QW +KF+ FM F + D C + K
Sbjct: 966 SPPEGYESMFKANEVCLLNKALYGLKQAPKQWNEKFDNFMKEICFVKSAYDSCAYTKVLP 1025
Query: 460 D-SYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVL 518
D S + L +YVDD+L+A N I LK + FEMKDLG AK+ILGM I R+R+ GVL
Sbjct: 1026 DGSVMYLLIYVDDILVASKNKEAITALKANLGMRFEMKDLGAAKKILGMEIIRDRTLGVL 1085
Query: 519 KLSQEKYVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSL 578
LSQE Y+ K+L+ +N+ +A TPLG H KF + +E +M +VPY+SAVGS+
Sbjct: 1086 WLSQEGYLNKILETYNMAEAKPAMTPLGAHFKFQAATEQKLIRDEDFMKSVPYSSAVGSI 1145
Query: 579 MYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRR-NNLTLQE 637
MYAM+ TRPD+A+ VG++SRFMS P KEHW VKW+LRY+KG+ + LC+++ ++ ++
Sbjct: 1146 MYAMLGTRPDLAYPVGIISRFMSQPIKEHWLGVKWVLRYIKGTLKTRLCYKKSSSFSIVG 1205
Query: 638 FSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWL 697
+ DAD D D +S TG +FTLGG +SWKS LQ VA STTESEY++++EA KE IWL
Sbjct: 1206 YCDADYAADLDKRRSITGLVFTLGGNTISWKSGLQRVVAQSTTESEYMSLTEAVKEAIWL 1265
Query: 698 KSFLKELGKEQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKI 757
K LK+ G EQ +F DSQS I L+KN V H R KHI +KYHFIRE+ISD + +LKI
Sbjct: 1266 KGLLKDFGYEQKSVEIFCDSQSAIALSKNNVHHERTKHIDVKYHFIREIISDGTVEVLKI 1325
Query: 758 LGSENPTDMLTKTVTADNLRLCI 780
+NP D+ TK + + +
Sbjct: 1326 STEKNPADIFTKVLAVSKFQAAL 1348
>UniRef100_Q75IK3 Putative polyprotein [Oryza sativa]
Length = 1175
Score = 638 bits (1645), Expect = 0.0
Identities = 346/795 (43%), Positives = 479/795 (59%), Gaps = 63/795 (7%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
WK ++ QT ++K L++DNGG + S F +C + GI M TI TP+QNGVAERMNRT
Sbjct: 431 WKVMIKRQTEKEVKVLRTDNGGGFCSDAFDDYCRKEGIVMHHTIPYTPQQNGVAERMNRT 490
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
+ +ARCM + + K FW +A TA YLINR PSI L+ + P EVWSG + S L+VF
Sbjct: 491 IISKARCMLSNARMNKRFWAEAAKTACYLINRSPSISLNKKTPIEVWSGMPANYSQLRVF 550
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
GC +Y +++ KL+P+AIKC F+GYG + GY+ W+ + K SR+V FNESV++
Sbjct: 551 GCTAYAHVNNG---KLEPRAIKCLFLGYGSGVKGYKLWNPETNKTFMSRSVVFNESVMFN 607
Query: 182 DRSSAESMSSS---KQLKLSERVALEEISESDVVKRNQINPENEDDEVEVELEQEPTTVI 238
D + + +Q +S +V + ++++V N +N + ++ + EP
Sbjct: 608 DSLPTDVIPGGFDEEQQYVSVQVEHVDDQKTEIVG-NDVNDTVQHSPPVLQPQDEPIAH- 665
Query: 239 ETPITQVRRSTRTPKAPQRYSPSLNYLL--------LTDAGEPEYFGEAMQGNDSIKWEL 290
RR+ R+ AP R + + + + E + E + D KW
Sbjct: 666 -------RRTKRSCGAPVRLIEECDMVYYAFICAEQVENTLELATYTEVVVSGDREKWIS 718
Query: 291 AMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSRRYKARLVVKGFQQKQGI 350
AM++EM SL+KN TW L LP+ QK+ +
Sbjct: 719 AMQEEMQSLEKNDTWELVHLPK---------------------------------QKKPV 745
Query: 351 DFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGT 410
F VK ++IR SIVA +L LEQLDVK AFLHG+LEEEIYM QPEGF V G
Sbjct: 746 HF-------VKHSSIRTFFSIVAMHDLELEQLDVKTAFLHGELEEEIYMDQPEGFIVPGK 798
Query: 411 KNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADSYIILALYVD 470
++ VCKL +SLYGLKQ+PRQWYK+F+ FM + GF R + D C ++K S I L LYVD
Sbjct: 799 EDYVCKLKRSLYGLKQSPRQWYKRFDSFMLSHGFKRSEFDSCVYIKFVNVSPIYLLLYVD 858
Query: 471 DMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKLL 530
DMLIA + +I LK+Q+S F+MKDL AK+ILGM I+R+R+ G L LSQ+ Y++K+L
Sbjct: 859 DMLIAAKSKEQITTLKKQLSSEFDMKDLDAAKKILGMEITRDRNSGWLFLSQQSYIKKVL 918
Query: 531 DRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPDIA 590
RFN+ D ST + H K S Q TDE+ YMS VPY+S VGSLMYAMVC+R D++
Sbjct: 919 QRFNMHDTKPVSTHIAPHFKLSALQCASTDEDVEYMSRVPYSSVVGSLMYAMVCSRLDLS 978
Query: 591 HAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNNLTLQEFSDADLGGDSDGG 650
HA+ +VSR+M+NPGKEHW+ ++WI RYL+ ++ CL F R N L + D+D D D
Sbjct: 979 HAMSLVSRYMANPGKEHWKAIQWIFRYLRDTANACLKFGRTNKGLIGYVDSDFAADLDKR 1038
Query: 651 KSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDV 710
+S TGY+FT+G AVSWK+ L++ VA STTE+EY+AI+EA KE +WLK EL
Sbjct: 1039 RSLTGYVFTIGSCAVSWKATLRHVVAQSTTEAEYMAIAEACKESVWLKGLFAELCGVYSC 1098
Query: 711 PPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKT 770
LF DSQS I L K+ +FH R KHI +KYH++R++++ +L + KI +NP DM+TK
Sbjct: 1099 INLFCDSQSAICLTKDQMFHERTKHIDIKYHYVRDVVAQGKLKVCKISTHDNPVDMMTKH 1158
Query: 771 VTADNLRLCIASAGL 785
V LC + G+
Sbjct: 1159 VPVAKFELCSSLVGI 1173
>UniRef100_O81903 Putative transposable element [Arabidopsis thaliana]
Length = 1308
Score = 632 bits (1631), Expect = e-179
Identities = 337/788 (42%), Positives = 493/788 (61%), Gaps = 52/788 (6%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
WK VENQ K+K L++DNG E+ + +F +FC +NGI +T TP+QNGVA+RMNRT
Sbjct: 539 WKELVENQVNKKVKILRTDNGLEFCNLKFDEFCKQNGIERHRTCTYTPQQNGVAKRMNRT 598
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
L E+ RC+ +SGL ++FW +A TAAYL+NR P+ +D+ +PEE+W K+ HL+ F
Sbjct: 599 LMEKVRCLLNESGLEEVFWAEAAATAAYLVNRSPASAVDHNVPEELWLDKKPGYKHLRRF 658
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
GC++YV +D + KL P+A+K F+GY GY+ W +K + SRN+ FNE+ +YK
Sbjct: 659 GCIAYVHLD---QGKLKPRALKGVFLGYPQGTKGYKVWLLDEEKCVISRNIVFNENQVYK 715
Query: 182 D--RSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVEVE--LEQEPTTV 237
D SS +S+ L+ + + K + E D E + E + QEP
Sbjct: 716 DIRESSEQSVKDISDLEGYNEFQVSVKEHGECSKTGGVTIEEIDQESDSENSVTQEPLIA 775
Query: 238 ---IETPITQVRRSTRTPKAPQRYSPSLNYLLLT------DAGEPEYFGEAMQGNDSIKW 288
+ + R R P PQ+ + ++ L ++ EP+ + +A + IKW
Sbjct: 776 SIDLSNYQSARDRERRAPNPPQKLADYTHFALALVMAEEIESEEPQCYHDAKKDKHWIKW 835
Query: 289 ELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDG--SRRYKARLVVKGFQQ 346
MK+E+ SL KNGTW + + P+ +K + RW+++LK G ++RYKARLV +GF Q
Sbjct: 836 NGGMKEEIDSLLKNGTWDIVEWPKEQKVISCRWLFKLKPGIPGVEAQRYKARLVARGFTQ 895
Query: 347 KQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFE 406
++GID+ E+F+PVVK +IR+++S V +++ LEQ+DVK FLHG+L++ +YM QPEGFE
Sbjct: 896 QKGIDYEEVFAPVVKHISIRILMSAVVKDDMELEQMDVKTTFLHGELDQVLYMEQPEGFE 955
Query: 407 VLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKF-ADSYIIL 465
V K+ VC L KSLYGLKQAPRQW KKF+ FM + F R + D C +VK+ ++ L
Sbjct: 956 VNPEKDQVCLLKKSLYGLKQAPRQWNKKFHAFMLSLQFARSEHDSCVYVKEVNPGEFVYL 1015
Query: 466 ALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKY 525
LYVDDML+A + +EI++LK+ +S FEMKD+G A +ILG+ I RNR EG L+LSQ +Y
Sbjct: 1016 LLYVDDMLLAAKSKSEISKLKEALSLKFEMKDMGAASRILGIDIIRNRKEGTLRLSQTRY 1075
Query: 526 VEKLLDRFNVGDANTRSTPLGNHLKFSK--KQSPQTDEEESYMSTVPYASAVGSLMYAMV 583
V+K++ RF + DA STP+G H K + + D E VPY+SAVGS+MYAM+
Sbjct: 1076 VDKVIQRFRMADAKVVSTPMGAHFKLTSLIDEIGSVDPE-----VVPYSSAVGSVMYAMI 1130
Query: 584 CTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFRRNNLTLQEFSDADL 643
T PD+A+A+G+VSRFMS PG NL +Q + D+D
Sbjct: 1131 GTIPDVAYAMGLVSRFMSRPGA--------------------------NLEVQGYCDSDH 1164
Query: 644 GGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKE 703
D D +S +GY+FT+GG VSWKS LQ+ VALS+T++E++A++EA KE IW++ L++
Sbjct: 1165 AADLDKRRSISGYVFTVGGNTVSWKSSLQHVVALSSTQAEFIALTEAVKEAIWIRGLLED 1224
Query: 704 LGKEQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENP 763
+G + ++ DSQS I L+KN FH R KH+++K++FIR++I E+ + KI S NP
Sbjct: 1225 MGLQPKPATVWCDSQSAICLSKNNAFHDRTKHVEVKFYFIRDIIEAGEVKVRKIHTSVNP 1284
Query: 764 TDMLTKTV 771
DMLTK +
Sbjct: 1285 ADMLTKCI 1292
>UniRef100_Q6L4X8 Putative polyprotein [Oryza sativa]
Length = 1211
Score = 622 bits (1604), Expect = e-176
Identities = 328/707 (46%), Positives = 448/707 (62%), Gaps = 36/707 (5%)
Query: 99 LDYQLPEEVWSGKEVSLSHLKVFGCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRF 158
+D + P EVWSG + S L+VFGC +Y +D+ KL+P+AIKC F+GY + Y+
Sbjct: 519 IDKKTPIEVWSGSPANYSDLRVFGCTAYAHVDNG---KLEPRAIKCIFLGYPSGVKDYKL 575
Query: 159 WDEQNKKIIRSRNVTFNESVLYKDRSSAESMSSSKQLKLSERVALEEISESDVVKRNQIN 218
W + KK++ SRNV F+ESV+ D+ S V +E ++ V + I+
Sbjct: 576 WCPKTKKVVISRNVVFHESVMLHDKPSTN-------------VPVESQEKASVQVEHLIS 622
Query: 219 PENEDDEVEVELEQEPTTVIETPITQVRRST----------RTPKAPQRYSPSLNYLL-- 266
+ ++ +V + Q+ + + ++ + V++S R K P+RY N +
Sbjct: 623 SGHAPEKEDVAINQDASVIEDSDSSAVQQSPKRSIAKDKPKRNIKPPRRYIEEANIVAYA 682
Query: 267 ------LTDAGEPEYFGEAMQGNDSIKWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNR 320
+ EP + EA+ + +W AM DEM SL+KN TW L KLP+ KK ++ +
Sbjct: 683 LSVAEEIEGNAEPSTYSEAIVSDGCNRWITAMHDEMESLEKNHTWELVKLPKEKKPIRCK 742
Query: 321 WVYRLKEESDGS--RRYKARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLH 378
W+++ K+ S RYKARLV KG+ Q GIDF ++FSPV+K ++IR +LSIVA +
Sbjct: 743 WIFKRKKGMSPSDEARYKARLVAKGYSQIPGIDFNDVFSPVLKHSSIRTLLSIVAVHDYE 802
Query: 379 LEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEF 438
LEQ+DVK AFLHG+LEE+IYM QPEGF V G +NLV +L KSLYGLKQ+PRQWYK+F+ F
Sbjct: 803 LEQMDVKTAFLHGELEEDIYMEQPEGFVVPGKENLVYRLKKSLYGLKQSPRQWYKRFDSF 862
Query: 439 MSNSGFNRCDMDHCCFVKKFADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKDL 498
M + F R + D C ++K S I L LYVDDMLIA + +EI +LK Q+S FEMKDL
Sbjct: 863 MFSQKFRRSNYDSCVYLKVVDGSSIYLLLYVDDMLIAAKDKSEIEKLKAQLSSEFEMKDL 922
Query: 499 GPAKQILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQ 558
G AK+ILGM I+R R G L LSQ+ Y+EK+L RFN+ DA STPL H + S PQ
Sbjct: 923 GAAKKILGMEITRERHSGKLYLSQKGYIEKVLRRFNMHDAKPVSTPLAAHFRLSSDLCPQ 982
Query: 559 TDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYL 618
+D + YMS VPY S VGSLMYAMVC+R D++HA+ VVSR+M+NPGKEHW+ V+WI +YL
Sbjct: 983 SDYDIEYMSRVPYLSVVGSLMYAMVCSRLDLSHALSVVSRYMANPGKEHWKVVQWIFKYL 1042
Query: 619 KGSSRMCLCFRRNNLTLQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALS 678
+G+S CL F R+ L + D+D GD D +S TGY+FT+GG AVSWK+ LQ VALS
Sbjct: 1043 RGTSSACLQFGRSRDGLVGYVDSDFAGDLDRRRSLTGYVFTIGGCAVSWKASLQATVALS 1102
Query: 679 TTESEYVAISEAAKEMIWLKSFLKELGKEQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQM 738
TTE EY+AISEA KE IWL+ EL +F DSQS I K+ +FH R KHI +
Sbjct: 1103 TTEVEYMAISEACKEAIWLRGLYTELCGVTSCINIFCDSQSAICFTKDQMFHERTKHIDV 1162
Query: 739 KYHFIRELISDEELSLLKILGSENPTDMLTKTVTADNLRLCIASAGL 785
+YHFIR +I++ ++ + KI +NP DM+TK V LC + G+
Sbjct: 1163 RYHFIRGVIAEGDVKVCKISTHDNPADMMTKPVPTTKFELCSSLVGV 1209
>UniRef100_O82196 Copia-like retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1137
Score = 591 bits (1524), Expect = e-167
Identities = 330/797 (41%), Positives = 467/797 (58%), Gaps = 46/797 (5%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
WK VE Q+ K+K L++DNG E+ + +F + C + GI +T TP+QNGVAER+NRT
Sbjct: 343 WKKMVETQSERKLKHLRTDNGLEFCNHKFDEVCKKEGIVRHRTCTYTPQQNGVAERLNRT 402
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
+ + R M +SGL K FW A +TA YLINR PS ++ ++PEE+W+ + S LK F
Sbjct: 403 IMNKVRSMLSESGLDKKFWAKAASTAVYLINRSPSSSIENKIPEELWTSAVPNFSGLKRF 462
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
GCV YV + KLDP+A K F+GY + G+R W + ++ SRNV F E V+YK
Sbjct: 463 GCVVYVY---SQEGKLDPRAKKGVFVGYPNGVKGFRVWMIEEERCSISRNVVFREDVMYK 519
Query: 182 DRSSAESMSSSKQLKLS-ERVALEEISESDVVKRNQINPENEDDEVEVELEQEPTTV--- 237
D + + S L+ R+ E + + + DD+ + E+ P +
Sbjct: 520 DILNQSTSGMSFDFPLATNRIPSFECAGNRKEDEISVQGGVSDDDTKQSSEESPISTGSS 579
Query: 238 --------IETPITQVRRSTRTPKAPQRYS---------PSLNYLLLTDAGEPEY--FGE 278
+ + +R T+ P + Y Y++ D G PE + +
Sbjct: 580 GQNSGQRTYQIARDKPKRQTKIPDKLRDYELNEEVLDEIAGYAYMITEDGGNPEPNDYQK 639
Query: 279 AMQGNDSIKWELAMKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSR--RYK 336
A+Q +D W A+ +E+ SL KN TW L + +K + +WV++ K G R+K
Sbjct: 640 ALQDSDYKMWLKAVDEEIESLLKNNTWVLVNRDQFQKPIGCKWVFKRKSGIVGVEKPRFK 699
Query: 337 ARLVVKGFQQKQGIDFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEE 396
ARLVVKG+ QK+GID+ EIFSPVVK +IR++LS+V ++ L+Q+DVK AFLHG L+E
Sbjct: 700 ARLVVKGYSQKEGIDYQEIFSPVVKHVSIRLLLSMVTHCDMELQQMDVKTAFLHGYLDET 759
Query: 397 IYMTQPEGFEVLGTKNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVK 456
IY+ QPEG+ + VC L +SLYGL+Q+PRQW +FNEFM G+ R D C + K
Sbjct: 760 IYIEQPEGYVHKRYPDKVCLLKRSLYGLRQSPRQWNNRFNEFMQKIGYERSKYDSCVYFK 819
Query: 457 KF-ADSYIILALYVDDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSE 515
+ + YI L LYVDD+LIA + + LK ++ FEMKDLG AK+ILGM I R+R
Sbjct: 820 ELQSGEYIYLLLYVDDILIASRDKRTVCDLKALLNSEFEMKDLGDAKKILGMEIVRDRKA 879
Query: 516 GVLKLSQEKYVEKLLDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAV 575
G + +SQE Y+ K+L F + A TP+G H K + + M VPY SAV
Sbjct: 880 GTMSISQEGYLLKVLGNFGMDQAKPVFTPMGAHFKLKPATDEEVMRQSEVMRAVPYQSAV 939
Query: 576 GSLMYAMVCTRPDIAHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCFR-RNNLT 634
GSLMY+M+ TRPD+AH+VG+V RFMS P KEHW+ VKWILRY++GS LC++ L
Sbjct: 940 GSLMYSMIGTRPDLAHSVGLVCRFMSKPLKEHWQAVKWILRYIRGSIDRKLCYKNEGELI 999
Query: 635 LQEFSDADLGGDSDGGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEM 694
L+ + D+D D +G +ST+G VALS+TE+EY+A+++ AKE
Sbjct: 1000 LEGYCDSDYAADKEGRRSTSGV----------------KVVALSSTEAEYMALTDGAKEA 1043
Query: 695 IWLKSFLKELGKEQDVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSL 754
IWLK + ELG Q + DSQS I LAKN V+H R KHI +KYHFIR+L+++ E+ +
Sbjct: 1044 IWLKGHVSELGFVQKTVNIHCDSQSAIALAKNAVYHERTKHIDVKYHFIRDLVNNGEVQV 1103
Query: 755 LKILGSENPTDMLTKTV 771
LKI +NP D+ TK +
Sbjct: 1104 LKIDTEDNPADIFTKVL 1120
>UniRef100_O23864 Polyprotein [Oryza australiensis]
Length = 1317
Score = 580 bits (1496), Expect = e-164
Identities = 324/791 (40%), Positives = 470/791 (58%), Gaps = 30/791 (3%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
++ EV+N IK L+SD GGEY SQEF + GI T GTP+ NGV+ER NRT
Sbjct: 547 FQNEVQNHLGKTIKFLRSDRGGEYVSQEFGNHLKDCGIVPQLTPPGTPQWNGVSERRNRT 606
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
L + R M QS LP FW A+ TAA +NR PS ++ + P E+W+G+ SLS LK++
Sbjct: 607 LLDMVRSMMSQSDLPLSFWGYALETAALTLNRVPSKSVE-KTPYEIWTGQPPSLSFLKIW 665
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
GC +YV + DKL PK+ KCF +GY + GY F++ + K+ +R+ F E
Sbjct: 666 GCEAYV--KRLQSDKLTPKSDKCFVVGYPKETKGYYFYNREQAKVFVARHGVFLEKEFLS 723
Query: 182 DRSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVEVELEQEPTTVIETP 241
R S RV LEE+ E+ P+ ED V V++TP
Sbjct: 724 RRVSGI------------RVHLEEVQETPETVSATTEPQQEDQSVA-------PPVVDTP 764
Query: 242 ITQVRRSTRTPKAPQRYSPSLNY-LLLTDAGEPEYFGEAMQGNDSIKWELAMKDEMTSLQ 300
RRS R+ +AP RY+ + +LL D EP+ + EAM G+DS KW AMK E+ S+
Sbjct: 765 AP--RRSERSRRAPDRYTGAEQRDILLLDNDEPKTYEEAMVGHDSNKWLGAMKSEIESMY 822
Query: 301 KNGTWSLTKLPEGKKALQNRWVYRLKEESDGSRR-YKARLVVKGFQQKQGIDFTEIFSPV 359
N W+L P+G K ++ +W+++ K + DG+ YKARLV KGF+Q QG+D+ E FSPV
Sbjct: 823 DNQVWNLVDPPDGVKTIECKWLFKKKADMDGNVHIYKARLVAKGFKQIQGVDYDETFSPV 882
Query: 360 VKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNLVCKLHK 419
+ +IR+IL+I A + + Q+DVK AFL+G+L E++YM QP+GF + +CKL K
Sbjct: 883 AMLKSIRIILAIAAYFDYEIWQMDVKTAFLNGNLSEDVYMIQPQGFVDPESPGKICKLQK 942
Query: 420 SLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADSYIILALYVDDMLIAGSNM 479
S+YGLKQA R W +F+E + GF + + + C + K + + L LYVDD+L+ G+++
Sbjct: 943 SIYGLKQASRSWNIRFDEVIKGFGFIKNEEEACVYKKVSGSAIVFLILYVDDILLIGNDI 1002
Query: 480 TEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKLLDRFNVGDAN 539
+ +K + +F MKDLG A ILG+RI R+RS+ ++ LSQ Y++K+L RFN+ D+
Sbjct: 1003 PMLESVKSSLKNSFSMKDLGEAAYILGIRIYRDRSKRLIGLSQSTYIDKVLKRFNMHDSK 1062
Query: 540 TRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVGVVSRF 599
P+ + + SK Q PQT +E + M VPYASA+GS+MYAM+CTRPD+++A+ SR+
Sbjct: 1063 KGFLPMSHGINLSKNQCPQTHDERNKMGMVPYASAIGSIMYAMLCTRPDVSYALSATSRY 1122
Query: 600 MSNPGKEHWECVKWILRYLKGSSRMCLCF-RRNNLTLQEFSDADLGGDSDGGKSTTGYIF 658
S+PG+ HW VK IL+YL+ + M L + +L + ++DA D D +S +G++F
Sbjct: 1123 QSDPGEGHWTAVKNILKYLRRTKDMFLVYGGEEDLVVSGYTDASFQTDKDDYRSQSGFVF 1182
Query: 659 TLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVP---PLFS 715
L G AVSWKS Q+ VA STTE+EY+A SEAAKE +W+K F+ ELG L+
Sbjct: 1183 CLNGGAVSWKSSKQDTVADSTTEAEYIAASEAAKEAVWIKKFVSELGVMTSTTGPMSLYC 1242
Query: 716 DSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTVTADN 775
D+ I AK P H + KHI +YH IRE++ ++ + K+ N D LTK +
Sbjct: 1243 DNSGAIAQAKEPRSHQKSKHILRRYHLIREIVDRGDVKICKVHTDLNIADPLTKPLPQPK 1302
Query: 776 LRLCIASAGLR 786
+ G+R
Sbjct: 1303 HEAHTRAMGIR 1313
>UniRef100_Q6L3N8 Putative gag-pol polyprotein [Solanum demissum]
Length = 1333
Score = 560 bits (1444), Expect = e-158
Identities = 318/784 (40%), Positives = 468/784 (59%), Gaps = 42/784 (5%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
+K VENQ+ KIKSL++D GGE+ S +F FC ENGIR T TPEQNGVAER NRT
Sbjct: 554 FKAFVENQSGNKIKSLRTDRGGEFLSNDFNLFCEENGIRRELTAPYTPEQNGVAERKNRT 613
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
+ E AR GLP FW +A+ T Y +N P+ + P E W+GK+ +SHL++F
Sbjct: 614 VVEMARSSLKAKGLPDYFWGEAVATVVYFLNISPTKDVWNTTPLEAWNGKKPRVSHLRIF 673
Query: 122 GCVSYVLIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVLYK 181
GC++Y L++ KLD K+ KC F+GY YR ++ + K+I SRNV FNE V +
Sbjct: 674 GCIAYALVNF--HSKLDEKSTKCIFVGYSLQSKAYRLYNPISGKVIISRNVVFNEDVSWN 731
Query: 182 DRSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVEVELEQEPTTVIETP 241
S +M S+ QL ++ + + S P + V P+T +
Sbjct: 732 FNSG--NMMSNIQLLPTDEESAVDFGNS---------PNSSPVSSSVSSPIAPSTTVAPD 780
Query: 242 ITQV-----RRSTRTPKAPQRYSPSLNY-----LLLTDAGEPEYFGEAMQGNDSIKWELA 291
+ V RRSTR K +YS ++N LL++D P + EA++ ++ W+ A
Sbjct: 781 ESSVEPIPLRRSTREKKPNPKYSNTVNTSCQFALLVSD---PICYEEAVEQSE---WKNA 834
Query: 292 MKDEMTSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGS-RRYKARLVVKGFQQKQGI 350
M +E+ ++++N TW L PEGK + +WV+R K +DGS +++KARLV KG+ Q+QG+
Sbjct: 835 MIEEIQAIERNSTWELVDAPEGKNVIGLKWVFRTKYNADGSIQKHKARLVAKGYSQQQGV 894
Query: 351 DFTEIFSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGT 410
DF E FSPV + T+RV+L++ A +L + Q DVK AFL+GDLEEE+Y++QP+GF + G
Sbjct: 895 DFDETFSPVARFETVRVVLALAAQLHLPVYQFDVKSAFLNGDLEEEVYVSQPQGFMITGN 954
Query: 411 KNLVCKLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKK-FADSYIILALYV 469
+N V KL K+LYGLKQAPR WY K + F SGF R D + ++KK D ++++ LYV
Sbjct: 955 ENKVYKLRKALYGLKQAPRAWYSKIDSFFQGSGFRRSDNEPTLYLKKQGTDEFLLVCLYV 1014
Query: 470 DDMLIAGSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKL 529
DDM+ GS+ + +N K M NFEM DLG K LG+ + +++ +G+ +SQ+KY E L
Sbjct: 1015 DDMIYIGSSKSLVNDFKSNMMRNFEMSDLGLLKYFLGLEVIQDK-DGIF-ISQKKYAEDL 1072
Query: 530 LDRFNVGDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPDI 589
L +F + + +TP+ + K + + + + S VG L Y + TRPDI
Sbjct: 1073 LKKFQMMNCEVATTPMNINEKLQRADGTEKANPKL------FRSLVGGLNY-LTHTRPDI 1125
Query: 590 AHAVGVVSRFMSNPGKEHWECVKWILRYLKGSSRMCLCF-RRNNLTLQEFSDADLGGDSD 648
A +V VVSRF+ +P K+H+ K +LRY+ G++ + + + N L F+D+D G D
Sbjct: 1126 AFSVSVVSRFLQSPTKQHFGAAKRVLRYVAGTTDFGIWYSKAPNFRLVGFTDSDYAGCLD 1185
Query: 649 GGKSTTGYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQ 708
KST+G F+ G V+W SK Q VALST+E+EY A S AA++ +WL+ L++ EQ
Sbjct: 1186 DRKSTSGSCFSFGSGVVTWSSKKQETVALSTSEAEYTAASLAARQALWLRKLLEDFSYEQ 1245
Query: 709 -DVPPLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDML 767
+ +FSDS+S I +AKNP FH R KHI ++YHFIR L++D + L +E D+
Sbjct: 1246 KESTEIFSDSKSAIAMAKNPSFHGRTKHIDVQYHFIRTLVADGRIVLKFCSTNEQAADIF 1305
Query: 768 TKTV 771
TK++
Sbjct: 1306 TKSL 1309
>UniRef100_O81506 T7M24.7 protein [Arabidopsis thaliana]
Length = 964
Score = 554 bits (1427), Expect = e-156
Identities = 311/795 (39%), Positives = 462/795 (57%), Gaps = 35/795 (4%)
Query: 2 WKTEVENQTCLKIKSLKSDNGGEYDSQEFKKFCSENGIRMIKTILGTPEQNGVAERMNRT 61
++ EV+NQ IK+L+SD GGEY SQ F E GI T GTP+ NGV+ER NRT
Sbjct: 191 FQNEVQNQFGKSIKALRSDRGGEYLSQVFSDHLRECGIVSQLTPPGTPQWNGVSERRNRT 250
Query: 62 LNERARCMRIQSGLPKMFWVDAINTAAYLINRGPSIPLDYQLPEEVWSGKEVSLSHLKVF 121
L + R M + LP FW A+ T+A+++NR PS ++ + P E+W+GK +LS LK++
Sbjct: 251 LLDMVRSMMSHTDLPSPFWGYALETSAFMLNRCPSKSVE-KTPYEIWTGKVPNLSFLKIW 309
Query: 122 GCVSYV--LIDSDRRDKLDPKAIKCFFIGYGFDMYGYRFWDEQNKKIIRSRNVTFNESVL 179
GC SY LI DKL PK+ KC+F+GY + GY F+ + K+ RN F E
Sbjct: 310 GCESYAKRLIT----DKLGPKSDKCYFVGYPKETKGYYFYHPTDNKVFVVRNGAFLEREF 365
Query: 180 YKDRSSAESMSSSKQLKLSERVALEEISESDVVKRNQINPENEDDEVEVELEQEPTTVIE 239
+S +V LEE+ E + + E+ ++++ EP V
Sbjct: 366 LSKGTSGS------------KVLLEEVREP----QGDVPTSQEEHQLDLRRVVEPILVEP 409
Query: 240 TPITQVRRSTRTPKAPQRYSPSL---NYLLLTDAGEPEYFGEAMQGNDSIKWELAMKDEM 296
+VRRS R+ P R+ + + L + ++ EP + EA+ G DS KW A K EM
Sbjct: 410 ----EVRRSERSRHEPDRFRDWVMDDHALFMIESDEPTSYEEALMGPDSDKWLEAAKSEM 465
Query: 297 TSLQKNGTWSLTKLPEGKKALQNRWVYRLKEESDGSRR-YKARLVVKGFQQKQGIDFTEI 355
S+ +N W+L LP+G K ++ +W+++ K + DG+ + YKA LV KG++Q GID+ E
Sbjct: 466 ESMSQNKVWTLVDLPDGVKPIECKWIFKKKIDMDGNIQIYKAGLVAKGYKQVHGIDYDET 525
Query: 356 FSPVVKMTTIRVILSIVAAENLHLEQLDVKIAFLHGDLEEEIYMTQPEGFEVLGTKNLVC 415
+SPV + +IR++L+ A + + Q+DVK AFL+G+LEE +YMTQPEGF V VC
Sbjct: 526 YSPVAMLKSIRILLATAAHYDYEIWQMDVKTAFLNGNLEEHVYMTQPEGFTVPEAARKVC 585
Query: 416 KLHKSLYGLKQAPRQWYKKFNEFMSNSGFNRCDMDHCCFVKKFADSYIILALYVDDMLIA 475
KLH+S+YGLKQA R W +FNE + F R + + C + K + L LYVDD+L+
Sbjct: 586 KLHRSIYGLKQASRSWNLRFNEAIKEFDFIRNEEEPCVYKKTSGSAVAFLVLYVDDILLL 645
Query: 476 GSNMTEINRLKQQMSENFEMKDLGPAKQILGMRISRNRSEGVLKLSQEKYVEKLLDRFNV 535
G+++ + +K + F MKD+G A ILG+RI R+R ++ LSQ+ Y++K+L RFN+
Sbjct: 646 GNDIPLLQSVKTWLGSCFSMKDMGEAAYILGIRIYRDRLNKIIGLSQDTYIDKVLHRFNM 705
Query: 536 GDANTRSTPLGNHLKFSKKQSPQTDEEESYMSTVPYASAVGSLMYAMVCTRPDIAHAVGV 595
D+ P+ + + SK Q P T +E MS +PYASA+GS+MYAM+ TRPD+A A+ +
Sbjct: 706 HDSKKGFIPMSHGITLSKTQCPSTHDERERMSKIPYASAIGSIMYAMLYTRPDVACALSM 765
Query: 596 VSRFMSNPGKEHWECVKWILRYLKGSSRMCLCF-RRNNLTLQEFSDADLGGDSDGGKSTT 654
SR+ S+PG+ HW V+ I +YL+ + L + L + ++DA D D +S +
Sbjct: 766 TSRYQSDPGESHWIVVRNIFKYLRRTKDKFLVYGGSEELVVSGYTDASFQTDKDDFRSQS 825
Query: 655 GYIFTLGGTAVSWKSKLQNRVALSTTESEYVAISEAAKEMIWLKSFLKELGKEQDVP--- 711
G+ F L G AVSWKS Q+ VA STTE+EY+A SEAAKE++W++ F+ ELG +
Sbjct: 826 GFFFCLNGGAVSWKSTKQSTVADSTTEAEYIAASEAAKEVVWIRKFITELGVVPSISGPI 885
Query: 712 PLFSDSQSVIFLAKNPVFHSRCKHIQMKYHFIRELISDEELSLLKILGSENPTDMLTKTV 771
L+ D+ I AK P H + KHIQ +YH IRE+I ++ + ++ N D TK +
Sbjct: 886 DLYCDNNGAIAQAKEPKSHQKSKHIQRRYHLIREIIDRGDVKISRVSTDANVADHFTKPL 945
Query: 772 TADNLRLCIASAGLR 786
+ G+R
Sbjct: 946 PQPKHESHTTAIGIR 960
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,318,243,116
Number of Sequences: 2790947
Number of extensions: 56316033
Number of successful extensions: 170021
Number of sequences better than 10.0: 2147
Number of HSP's better than 10.0 without gapping: 1814
Number of HSP's successfully gapped in prelim test: 333
Number of HSP's that attempted gapping in prelim test: 163571
Number of HSP's gapped (non-prelim): 3148
length of query: 799
length of database: 848,049,833
effective HSP length: 136
effective length of query: 663
effective length of database: 468,481,041
effective search space: 310602930183
effective search space used: 310602930183
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 79 (35.0 bits)
Lotus: description of TM0065.18


