

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0065.1
(378 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
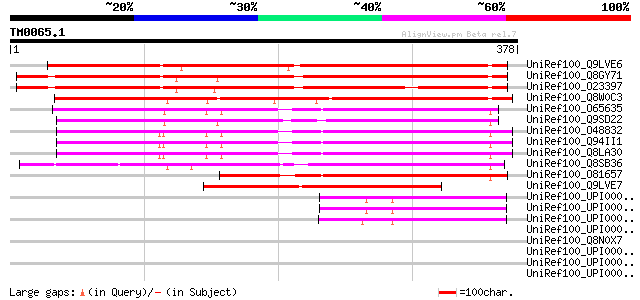
UniRef100_Q9LVE6 Similarity to senescence-associated protein [Ar... 377 e-103
UniRef100_Q8GY71 Hypothetical protein At4g15450/dl3768w [Arabido... 373 e-102
UniRef100_O23397 Hypothetical protein AT4g15450 [Arabidopsis tha... 342 1e-92
UniRef100_Q8W0C3 P0690B02.29 protein [Oryza sativa] 298 2e-79
UniRef100_O65635 Hypothetical protein T19K4.110 [Arabidopsis tha... 208 2e-52
UniRef100_Q9SD22 Hypothetical protein F24M12.290 [Arabidopsis th... 204 3e-51
UniRef100_O48832 Putative senescence-related protein [Arabidopsi... 196 7e-49
UniRef100_Q94II1 ERD7 protein [Arabidopsis thaliana] 195 2e-48
UniRef100_Q8LA30 Putative senescence-associated protein 12 [Arab... 193 6e-48
UniRef100_Q8SB36 Putative senescence-associated protein [Oryza s... 187 3e-46
UniRef100_O81657 Senescence-associated protein 12 [Hemerocallis ... 153 7e-36
UniRef100_Q9LVE7 Emb|CAB45990.1 [Arabidopsis thaliana] 147 4e-34
UniRef100_UPI00003645F4 UPI00003645F4 UniRef100 entry 47 9e-04
UniRef100_UPI00003645F3 UPI00003645F3 UniRef100 entry 47 9e-04
UniRef100_UPI0000369A2B UPI0000369A2B UniRef100 entry 47 9e-04
UniRef100_UPI0000072FD6 UPI0000072FD6 UniRef100 entry 47 0.001
UniRef100_Q8N0X7 Spartin [Homo sapiens] 47 0.001
UniRef100_UPI000023CC30 UPI000023CC30 UniRef100 entry 46 0.001
UniRef100_UPI00002BB14B UPI00002BB14B UniRef100 entry 44 0.006
UniRef100_UPI0000249FF2 UPI0000249FF2 UniRef100 entry 44 0.010
>UniRef100_Q9LVE6 Similarity to senescence-associated protein [Arabidopsis thaliana]
Length = 374
Score = 377 bits (968), Expect = e-103
Identities = 192/352 (54%), Positives = 262/352 (73%), Gaps = 17/352 (4%)
Query: 29 KPKNLIRQEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWP 88
+P+ + +EVL+QIP CRV+L+ E +AVELA G F ++KV D+ V+LA I+++G+ +QWP
Sbjct: 28 QPQTMRAEEVLLQIPRCRVHLIGESEAVELASGDFKLVKVSDNGVTLAMIVRIGHDLQWP 87
Query: 89 LTKDEPVVKVDTLHYLFTLPVKDGGSEPLSYGVTFPEQ-----CYGSMGLLESFLKDHSC 143
+ +DEPVVK+D YLFTLPVKDG +PLSYGVTF S+ LL+ FL ++SC
Sbjct: 88 VIRDEPVVKLDARDYLFTLPVKDG--DPLSYGVTFSGDDRDVALVNSLKLLDQFLSENSC 145
Query: 144 FSDLKTGK-KGGLDWEQFAPRVEDYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQ 202
FS + K G+DW++FAPR+EDYN+ +AKAIAGGTG I++GIF SNAY+NQV GG
Sbjct: 146 FSSTASSKVNNGIDWQEFAPRIEDYNNVVAKAIAGGTGHIIRGIFSLSNAYSNQVHKGGD 205
Query: 203 TILT---DNKNNGLVRQSVSNNKTADATKKNAMNENLKRVRQLTTMTEKLSKSLLDGVGI 259
++T +++ NG S +N ++ KKN +N +L+RVR+L+ TE LSK++L+G G+
Sbjct: 206 IMITKAEESQRNG----SYNNGNSSGNEKKNGINTHLQRVRKLSKATENLSKTMLNGAGV 261
Query: 260 MSGSVMAPVLKSQPGQAFLKMLPGEVLLASLDAVNKVFEATEAAQKQSLSATSQAATRMV 319
+SGSVM P++KS+PG AF M+PGEVLLASLDA+NK+ +ATEAA++Q+LSATS+AATRMV
Sbjct: 262 VSGSVMVPMMKSKPGMAFFSMVPGEVLLASLDALNKILDATEAAERQTLSATSRAATRMV 321
Query: 320 SNRYGEEAGEATEHVFATAGHAANTAWNVSKIRKAINPVTPASTAGGALRNS 371
S R+G+ AGEAT V ATAGHAA TAWNV KIRK P +S G ++N+
Sbjct: 322 SERFGDNAGEATGDVLATAGHAAGTAWNVLKIRKTFYP--SSSLTSGIVKNA 371
>UniRef100_Q8GY71 Hypothetical protein At4g15450/dl3768w [Arabidopsis thaliana]
Length = 381
Score = 373 bits (957), Expect = e-102
Identities = 196/384 (51%), Positives = 268/384 (69%), Gaps = 32/384 (8%)
Query: 6 SKPEEEEPTMMRTSTLVPIAEYSKPKNLIRQEVLIQIPGCRVYLMDEGQAVELAQGQFMI 65
SK E ++P+ + Y + EVL+QI GCR +L++ +AVELA G F +
Sbjct: 9 SKTETQQPSSYGQQQDAMYSSYGT----VSDEVLLQIHGCRAHLINGSEAVELAAGDFEL 64
Query: 66 IKVMDDNVSLATIIKVGNGVQWPLTKDEPVVKVDTLHYLFTLPVKDGGSEPLSYGVTF-- 123
++V+D NV+LA ++++GN +QWP+ KDEPVVK+D+ YLFTLPVKDG EPLSYG+TF
Sbjct: 65 VQVLDSNVALAMVVRIGNDLQWPVIKDEPVVKLDSRDYLFTLPVKDG--EPLSYGITFFP 122
Query: 124 ----PEQCYGSMGLLESFLKDHSCFSDLKTGKKG-----------GLDWEQFAPRVEDYN 168
S+ LL+ FL+++SCFS + G+DW++FAP++EDYN
Sbjct: 123 IDENDIVFVNSIELLDDFLRENSCFSSSPSSSSSSSSSSSSSVNNGIDWKEFAPKIEDYN 182
Query: 169 HFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQTILTD-NKNNGLVRQSVSNNKTADAT 227
+ +AKAIAGGTG I++G+F CSNAYTNQV GG+ ++T K NG +++K T
Sbjct: 183 NVVAKAIAGGTGHIIRGMFKCSNAYTNQVHKGGEIMITKAEKKNG------ASSKRNATT 236
Query: 228 KKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEVLL 287
KN +N+NL+RVR+L+ TEKLSK++L+GVG++SGSVM PV+KS+PG+AF M+PGEVLL
Sbjct: 237 NKNQINKNLQRVRKLSRATEKLSKTMLNGVGVVSGSVMGPVVKSKPGKAFFSMVPGEVLL 296
Query: 288 ASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTAWN 347
ASLDA+NK+ +A EAA++Q+LSATS+A TRMVS R GE AGEAT+ V T GHAA TAWN
Sbjct: 297 ASLDALNKLLDAAEAAERQTLSATSKATTRMVSERLGESAGEATKDVLGTVGHAAGTAWN 356
Query: 348 VSKIRKAINPVTPASTAGGALRNS 371
V IRKA +P +S G L+N+
Sbjct: 357 VFNIRKAFSP--SSSVTSGILKNA 378
>UniRef100_O23397 Hypothetical protein AT4g15450 [Arabidopsis thaliana]
Length = 372
Score = 342 bits (876), Expect = 1e-92
Identities = 185/384 (48%), Positives = 256/384 (66%), Gaps = 41/384 (10%)
Query: 6 SKPEEEEPTMMRTSTLVPIAEYSKPKNLIRQEVLIQIPGCRVYLMDEGQAVELAQGQFMI 65
SK E ++P+ + Y + EVL+QI GCR +L++ +AVELA G F +
Sbjct: 9 SKTETQQPSSYGQQQDAMYSSYGT----VSDEVLLQIHGCRAHLINGSEAVELAAGDFEL 64
Query: 66 IKVMDDNVSLATIIKVGNGVQWPLTKDEPVVKVDTLHYLFTLPVKDGGSEPLSYGVTF-- 123
++V+D NV+LA ++++GN +QWP+ KDEPVVK+D+ YLFTLPVKDG EPLSYG+TF
Sbjct: 65 VQVLDSNVALAMVVRIGNDLQWPVIKDEPVVKLDSRDYLFTLPVKDG--EPLSYGITFFP 122
Query: 124 ----PEQCYGSMGLLESFLKDHSCFSDLKTGK-----------KGGLDWEQFAPRVEDYN 168
S+ LL+ FL+++SCFS + G+DW++FAP++EDYN
Sbjct: 123 IDENDIVFVNSIELLDDFLRENSCFSSSPSSSSSSSSSSSSSVNNGIDWKEFAPKIEDYN 182
Query: 169 HFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQTILTD-NKNNGLVRQSVSNNKTADAT 227
+ +AKAIAGGTG I++G+F CSNAYTNQV GG+ ++T K NG +++K T
Sbjct: 183 NVVAKAIAGGTGHIIRGMFKCSNAYTNQVHKGGEIMITKAEKKNG------ASSKRNATT 236
Query: 228 KKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEVLL 287
KN +N+NL+RVR+L+ TEKLSK++L+GVG++SGSVM PV+KS+PG+AF M+PGEVLL
Sbjct: 237 NKNQINKNLQRVRKLSRATEKLSKTMLNGVGVVSGSVMGPVVKSKPGKAFFSMVPGEVLL 296
Query: 288 ASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTAWN 347
ASLDA++K + + Q++TRMVS R GE AGEAT+ V T GHAA TAWN
Sbjct: 297 ASLDALSK---------ETNSFCYVQSSTRMVSERLGESAGEATKDVLGTVGHAAGTAWN 347
Query: 348 VSKIRKAINPVTPASTAGGALRNS 371
V IRKA +P +S G L+N+
Sbjct: 348 VFNIRKAFSP--SSSVTSGILKNA 369
>UniRef100_Q8W0C3 P0690B02.29 protein [Oryza sativa]
Length = 395
Score = 298 bits (763), Expect = 2e-79
Identities = 170/383 (44%), Positives = 237/383 (61%), Gaps = 46/383 (12%)
Query: 34 IRQEVLIQIPGCRVYLMDEGQA-VELAQGQFMIIKVMDDNVSLATIIKVGNGVQWPLTKD 92
IR+E L+++PG V+L+D + VELA+G ++++ D V++AT+ +VG G+ WP+T+D
Sbjct: 17 IREETLLRVPGASVHLLDGAEGPVELARGDLAVVRIAKDGVAVATVARVGRGLGWPITRD 76
Query: 93 EPVVKVDTLHYLFTLPVKDGGSEP-------LSYGVTFPEQCYGSMGLLESFLKDHSCFS 145
EPVV++D LHYLFTLP GG L+YGV+F + L++FLK ++CFS
Sbjct: 77 EPVVRLDRLHYLFTLPDSTGGGGGGGGGALFLNYGVSFAAPDDALLASLDAFLKANACFS 136
Query: 146 D-------------------LKTGKKGGLDWEQFAPRVEDYNHFLAKAIAGGTGQIVKGI 186
T G W FAPR++ YN+ LAKAIA GTGQ+V+GI
Sbjct: 137 TPSSPAPSRSSATTTTRPAPTTTATADGY-WNDFAPRMDSYNNVLAKAIAAGTGQLVRGI 195
Query: 187 FMCSNAYTNQ--------VQSGGQTILTDNKNNGLVRQSVSNNKTADAT------KKNAM 232
FMCS AY Q VQ G I + R + A T K+ +
Sbjct: 196 FMCSEAYATQILIQPRVQVQRGADLIRPQATGSVTKRSGGAGGGGASRTTGQPDAKRGGV 255
Query: 233 NENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEVLLASLDA 292
N++LKR QL+ MTEK+S+SLLD V ++GS+ AP+L+S+ G+AFL +PGEV+LASLDA
Sbjct: 256 NKSLKR--QLSEMTEKMSQSLLDTVIAVTGSMAAPLLRSKQGKAFLATVPGEVILASLDA 313
Query: 293 VNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTAWNVSKIR 352
+NKV +A EAA+++SL+ATS + VS RYGE AGEATE FATAGHA TAWN+ KIR
Sbjct: 314 INKVMDAVEAAERRSLAATSNVVSGAVSRRYGESAGEATEDAFATAGHAVGTAWNLFKIR 373
Query: 353 KAINPVTPASTAGGALRNSIKNR 375
KA+ P +S G ++++++NR
Sbjct: 374 KAVTP--SSSLPGNMVKSAVRNR 394
>UniRef100_O65635 Hypothetical protein T19K4.110 [Arabidopsis thaliana]
Length = 512
Score = 208 bits (530), Expect = 2e-52
Identities = 130/375 (34%), Positives = 202/375 (53%), Gaps = 54/375 (14%)
Query: 33 LIRQEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWPLTKD 92
L +EV++ I G V+L+D+ +VELA G I++++ ++++A +VG+ +QWPLTKD
Sbjct: 66 LATEEVILTIHGAMVHLIDKSYSVELACGDLEILRLVQGDITVAVFARVGDEIQWPLTKD 125
Query: 93 EPVVKVDTLHYLFTL-PVKDGGS----------EPLSYGVTFPEQCYGSM-GLLESFLKD 140
EP VKVD HY F+L PVK+ S E L+YG+T + M L+ L D
Sbjct: 126 EPAVKVDESHYFFSLRPVKESESSDHSVNETENEMLNYGLTMASKGQEPMLEKLDKILAD 185
Query: 141 HSCFS----------------------DLKTGKKGGLD------WEQFAPRVEDYNHFLA 172
+S F+ +LK +K ++ W AP VEDY+ A
Sbjct: 186 YSSFTAEEKQKEENVLDLTAAKETSPEELKGKRKKMVEKQCTAYWTTLAPNVEDYSGVAA 245
Query: 173 KAIAGGTGQIVKGIFMCSNAYTNQVQSGGQTILTDNKNNGLVRQSVSNNKTADATKKNAM 232
K IA G+GQ++KGI C + +++ G N +++ +S + +
Sbjct: 246 KLIAAGSGQLIKGILWCGDLTMDRLMWG----------NDFMKKKLSKAEKERQVSPGTL 295
Query: 233 NENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEVLLASLDA 292
+ LKRV+++T MTEK++ +L GV +SG + V+ S+ GQ +LPGE++LA+LD
Sbjct: 296 -KRLKRVKKMTKMTEKVANGVLSGVVKVSGFFSSSVINSKAGQKLFGLLPGEMVLATLDG 354
Query: 293 VNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTAWNVSKIR 352
NKV +A E A + + TS T +V ++YG + +AT + AGHA TAW V KIR
Sbjct: 355 FNKVCDAVEVAGRHVMKTTSDVTTEIVDHKYGAKTAQATNEGLSAAGHAFGTAWTVFKIR 414
Query: 353 KAINP---VTPASTA 364
+A+NP + P+S A
Sbjct: 415 QALNPKSAMKPSSLA 429
>UniRef100_Q9SD22 Hypothetical protein F24M12.290 [Arabidopsis thaliana]
Length = 463
Score = 204 bits (520), Expect = 3e-51
Identities = 126/387 (32%), Positives = 198/387 (50%), Gaps = 69/387 (17%)
Query: 36 QEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWPLTKDEPV 95
+EVLI++PG + L+D+ +VELA G F I++++ +A + VGN +QWPLTK+E
Sbjct: 75 EEVLIRVPGAILNLIDKSYSVELACGDFTIVRIIQGQNIVAVLANVGNEIQWPLTKNEVA 134
Query: 96 VKVDTLHYLFTL-PVKDGGS--------------------EPLSYGVTFPEQCYGSMGL- 133
KVD HY F++ P K+ G E L+YG+T + ++ L
Sbjct: 135 AKVDGSHYFFSIHPPKEKGQGSGSDSDDEQGQKSKSKSDDEILNYGLTIASKGQENVLLV 194
Query: 134 LESFLKDHSCFSDLKTGKKG---------------------------------GLDWEQF 160
L+ L+D+SCF++ + +K W
Sbjct: 195 LDQVLRDYSCFTEQRMSEKAKETGEEVLGNSVVADTSPEELKGERKDVVEGQCAAYWTTL 254
Query: 161 APRVEDYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQTILTDNKNNGLVRQSVSN 220
AP VEDY H AK IA G+G++++GI C + +++ G + + N L R
Sbjct: 255 APNVEDYTHSTAKMIASGSGKLIRGILWCGDVTVERLKKGNEVM-----KNRLSRAEKEK 309
Query: 221 NKTADATKKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKM 280
+ + + ++ +KRV+++T MTEK++ +L GV +SG + + S+ G+ +
Sbjct: 310 DVSPETLRR------IKRVKRVTQMTEKVATGVLSGVVKVSGFITGSMANSKAGKKLFGL 363
Query: 281 LPGEVLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGH 340
LPGE++LASLD +K+ +A E A K +S +S T +V++RYG +A EAT AGH
Sbjct: 364 LPGEIVLASLDGFSKICDAVEVAGKNVMSTSSTVTTELVNHRYGTKAAEATNEGLDAAGH 423
Query: 341 AANTAWNVSKIRKAINP---VTPASTA 364
A TAW KIRKA NP + P+S A
Sbjct: 424 AFGTAWVAFKIRKAFNPKNVIKPSSLA 450
>UniRef100_O48832 Putative senescence-related protein [Arabidopsis thaliana]
Length = 452
Score = 196 bits (499), Expect = 7e-49
Identities = 132/393 (33%), Positives = 205/393 (51%), Gaps = 64/393 (16%)
Query: 36 QEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWPLTKDEPV 95
+EV+++I G ++L+D+ +VELA G II+++ +A + V + +QWPLTKDE
Sbjct: 69 EEVILKISGAILHLIDKSYSVELACGDLEIIRIVQGENVVAVLASVSDEIQWPLTKDENS 128
Query: 96 VKVDTLHYLFTL-PVK------------DGG---SEPLSYGVTFPEQCYGSMGL-LESFL 138
VKVD HY FTL P K DGG +E L+YG+T + + + LE L
Sbjct: 129 VKVDESHYFFTLRPTKEISHDSSDEEDGDGGKNTNEMLNYGLTIASKGQEHLLVELEKIL 188
Query: 139 KDHSCFS------DLKTGKKGGLD---------------------------WEQFAPRVE 165
+D+S FS + K + LD W AP VE
Sbjct: 189 EDYSSFSVQEVSEEAKEAGEKVLDVTVARETSPVELTGERKEIVERQCSAYWTTLAPNVE 248
Query: 166 DYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQTILTDNKNNGLVRQSVSNNKTAD 225
DY+ AK IA G+G ++KGI C + +++ G NG +++ +S +
Sbjct: 249 DYSGKAAKLIATGSGHLIKGILWCGDVTMDRLIWG----------NGFMKRRLSKAEKES 298
Query: 226 ATKKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEV 285
+ + + ++RV+++T MTE ++ S+L GV +SG + V ++ G+ F +LPGEV
Sbjct: 299 EVHPDTL-KRIRRVKRMTKMTESVANSILSGVLKVSGFFTSSVANTKVGKKFFSLLPGEV 357
Query: 286 LLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTA 345
+LASLD NKV +A E A + +S +S T +V ++YG +A EAT AG+A TA
Sbjct: 358 ILASLDGFNKVCDAVEVAGRNVMSTSSTVTTELVDHKYGGKAAEATNEGLDAAGYALGTA 417
Query: 346 WNVSKIRKAINP---VTPASTAGGALRNSIKNR 375
W KIRKAINP + P++ A A+R++ +
Sbjct: 418 WVAFKIRKAINPKSVLKPSTLAKTAIRSAASQK 450
>UniRef100_Q94II1 ERD7 protein [Arabidopsis thaliana]
Length = 441
Score = 195 bits (495), Expect = 2e-48
Identities = 131/393 (33%), Positives = 204/393 (51%), Gaps = 64/393 (16%)
Query: 36 QEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWPLTKDEPV 95
+EV+++I G ++L+D+ +VELA G II+++ +A + V + +QWPLTKDE
Sbjct: 54 EEVILKISGAILHLIDKSYSVELACGDLEIIRIVQGENVVAVLASVSDEIQWPLTKDENS 113
Query: 96 VKVDTLHYLFTL-PVK------------DGG---SEPLSYGVTFPEQCYGSMGL-LESFL 138
VKVD HY FTL P K DGG +E L+YG+T + + + LE L
Sbjct: 114 VKVDESHYFFTLRPTKEISHDSSDEEDGDGGKNTNEMLNYGLTIASKGQEHLLVELEKIL 173
Query: 139 KDHSCFS------DLKTGKKGGLD---------------------------WEQFAPRVE 165
+D+S FS + K + LD W AP VE
Sbjct: 174 EDYSSFSVQEVSEEAKEAGEKVLDVTVARETSPVELTGERKEIVERQCSAYWTTLAPNVE 233
Query: 166 DYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQTILTDNKNNGLVRQSVSNNKTAD 225
DY+ A IA G+G ++KGI C + +++ G NG +++ +S +
Sbjct: 234 DYSGKAANVIATGSGHLIKGILWCGDVTMDRLIWG----------NGFMKRRLSKAEKES 283
Query: 226 ATKKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEV 285
+ + + ++RV+++T MTE ++ S+L GV +SG + V ++ G+ F +LPGEV
Sbjct: 284 EVHPDTL-KRIRRVKRMTKMTESVANSILSGVLKVSGFFTSSVANTKVGKKFFSLLPGEV 342
Query: 286 LLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTA 345
+LASLD NKV +A E A + +S +S T +V ++YG +A EAT AG+A TA
Sbjct: 343 ILASLDGFNKVCDAVEVAGRNVMSTSSTVTTELVDHKYGGKAAEATNEGLDAAGYALGTA 402
Query: 346 WNVSKIRKAINP---VTPASTAGGALRNSIKNR 375
W KIRKAINP + P++ A A+R++ +
Sbjct: 403 WVAFKIRKAINPKSVLKPSTLAKTAIRSAASQK 435
>UniRef100_Q8LA30 Putative senescence-associated protein 12 [Arabidopsis thaliana]
Length = 452
Score = 193 bits (491), Expect = 6e-48
Identities = 131/393 (33%), Positives = 204/393 (51%), Gaps = 64/393 (16%)
Query: 36 QEVLIQIPGCRVYLMDEGQAVELAQGQFMIIKVMDDNVSLATIIKVGNGVQWPLTKDEPV 95
+EV+++I G ++L+D+ +VELA G II+++ +A + V + +QWPLTKDE
Sbjct: 69 EEVILKISGAILHLIDKSYSVELACGDLEIIRIVQGENVVAVLASVSDEIQWPLTKDENS 128
Query: 96 VKVDTLHYLFTL-PVK------------DGG---SEPLSYGVTFPEQCYGSMGL-LESFL 138
VKVD HY FTL P K DGG +E L+YG+T + + + LE L
Sbjct: 129 VKVDESHYFFTLRPTKEISHDSSDEEDGDGGKNTNEMLNYGLTIASKGQEHLLVELEKIL 188
Query: 139 KDHSCFS------DLKTGKKGGLD---------------------------WEQFAPRVE 165
+D+S FS + K + LD W AP VE
Sbjct: 189 EDYSSFSVQEVSEEAKEAGEKVLDVTVARETSPVELTGERKEIVERQCSAYWTTLAPNVE 248
Query: 166 DYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQTILTDNKNNGLVRQSVSNNKTAD 225
DY+ AK IA G+G ++K I C + +++ G NG +++ +S +
Sbjct: 249 DYSGKAAKLIATGSGHLIKRILWCGDVTMDRLIWG----------NGFMKRRLSKAEKES 298
Query: 226 ATKKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEV 285
+ + + ++RV+++T MTE ++ S+L GV +SG + V ++ G+ F +LPGEV
Sbjct: 299 EVHPDTL-KRIRRVKRITKMTESVANSILSGVLKVSGFFTSSVANTKVGKKFFSLLPGEV 357
Query: 286 LLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTA 345
+LASLD NKV +A E A + +S +S T +V ++YG +A EAT AG+A TA
Sbjct: 358 ILASLDGFNKVCDAVEVAGRNVMSTSSTVTTELVDHKYGGKAAEATNEGLDAAGYALGTA 417
Query: 346 WNVSKIRKAINP---VTPASTAGGALRNSIKNR 375
W KIRKAINP + P++ A A+R++ +
Sbjct: 418 WVAFKIRKAINPKSVLKPSTLAKTAIRSAASQK 450
>UniRef100_Q8SB36 Putative senescence-associated protein [Oryza sativa]
Length = 419
Score = 187 bits (476), Expect = 3e-46
Identities = 121/377 (32%), Positives = 189/377 (50%), Gaps = 28/377 (7%)
Query: 8 PEEEEPTMMRTSTLVPIAEYSKPKNLIRQEVLIQIPGCRVYLMDEGQAVELAQGQFMIIK 67
P M S P+ Y+ ++VL++IPG +++L+D ++ LA G +++
Sbjct: 44 PSSPSSPMSPASPATPVDAYANAPPP-SEDVLLRIPGAQLHLIDRHRSYPLAAGDLSLLR 102
Query: 68 VMDDNVSLATIIKVGNGVQWPLTKDEPVVKVDTLHYLFTLPVKDGGSEP----LSYGVTF 123
+ + SLA I + + +QWPL +D VK+D HY F+L V +P L YG+T
Sbjct: 103 IRSGDTSLAAIALL-HPIQWPLARDVASVKLDPCHYSFSLTVPPSADDPNPGPLHYGLTL 161
Query: 124 PEQCYGSMGLL--------ESFLKDHSCFSDLKTGKKGGLDWEQFAPRVEDYNHFLAKAI 175
G+L +S + + S ++ + W AP VE+Y +A AI
Sbjct: 162 SHPDPRLDGILATYTSFSVQSVVGGEALASKVRDEVEAAAYWTAVAPNVEEYGGKVANAI 221
Query: 176 AGGTGQIVKGIFMCSNAYTNQVQSGGQTILTDNKNNGLVRQSVSNNKTADATKKNAMNEN 235
A G G + KGI CS ++++ G + +L G ADA M
Sbjct: 222 ATGAGHLAKGILWCSELTVDRLRWGNE-VLKRRMQPG----------DADAEVSPEMLRR 270
Query: 236 LKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQAFLKMLPGEVLLASLDAVNK 295
+KRV+ +T M+EK++ +L GV ++G + S+ G+ F +LPGE++LASLD K
Sbjct: 271 IKRVKMVTKMSEKVATGILSGVVKVTGYFTNSIANSKAGKKFFNLLPGEIVLASLDGFGK 330
Query: 296 VFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFATAGHAANTAWNVSKIRKAI 355
+ +A E A LS +S T +VS++YGE+A AT AGHA TAW V KIR+A+
Sbjct: 331 ICDAVEVAGTNVLSTSSTVTTGLVSHKYGEKAAAATNEGMDAAGHAIGTAWAVFKIRQAL 390
Query: 356 NP---VTPASTAGGALR 369
NP + P S A ++
Sbjct: 391 NPKSVLKPTSLAKSTIK 407
>UniRef100_O81657 Senescence-associated protein 12 [Hemerocallis sp.]
Length = 222
Score = 153 bits (387), Expect = 7e-36
Identities = 83/218 (38%), Positives = 132/218 (60%), Gaps = 14/218 (6%)
Query: 157 WEQFAPRVEDYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQTILTDNKNNGLVRQ 216
W AP VEDY+ +A+ IA G+G++++GI C + +++ G Q L+++
Sbjct: 6 WTTLAPNVEDYSGSVARVIAMGSGRVIQGILWCGDVTVERLKWGEQ----------LLKR 55
Query: 217 SVSNNKTADATKKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAPVLKSQPGQA 276
+ N + K A+ + +KRV+++T M++K+ +L GV +SG + V+ S+ G++
Sbjct: 56 KMDPNAQSSRVSKEAL-QRMKRVKKVTKMSDKVVTGILSGVVRVSGYFTSSVVNSKAGKS 114
Query: 277 FLKMLPGEVLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFA 336
F MLPGEVLLASLD K+ +A E A K + +S+ T +VS+RYGE+A E T F
Sbjct: 115 FFGMLPGEVLLASLDGFAKICDAAEVAGKNIMETSSEVTTGVVSHRYGEQAAELTHEGFG 174
Query: 337 TAGHAANTAWNVSKIRKAINP---VTPASTAGGALRNS 371
AGHA TAW+V KIRKA+NP + P + A A++++
Sbjct: 175 AAGHAIGTAWSVFKIRKALNPKGALKPTTIAKSAVKSA 212
>UniRef100_Q9LVE7 Emb|CAB45990.1 [Arabidopsis thaliana]
Length = 241
Score = 147 bits (372), Expect = 4e-34
Identities = 77/179 (43%), Positives = 117/179 (65%), Gaps = 2/179 (1%)
Query: 145 SDLKTGKKGG-LDWEQFAPRVEDYNHFLAKAIAGGTGQIVKGIFMCSNAYTNQVQSGGQT 203
S +GK +DW++FAP+ EDY + +AKAIA GTG I+KGIF CSN+Y+ ++ G
Sbjct: 64 SSSTSGKNNNEIDWKKFAPKAEDYKNGVAKAIAVGTGHIIKGIFTCSNSYSKKIHEEGTI 123
Query: 204 ILTDNKNNGLVRQSVSNNKTADATKKNAMNENLKRVRQLTTMTEKLSKSLLDGVGIMSGS 263
D + +G + Q + + KKN +N+NL+R +L ++E + + L+G ++S S
Sbjct: 124 AEEDEERSGDISQ-IDGGGNNETNKKNKLNKNLQRAEKLWKVSEAIGMAALEGGDLVSSS 182
Query: 264 VMAPVLKSQPGQAFLKMLPGEVLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNR 322
++APV+KS+ G+A L PGEV+LASLD+ + + A EAA+ Q+ ATS AATR+VS R
Sbjct: 183 MIAPVVKSKLGKALLSTAPGEVILASLDSFHNIIGAAEAAEIQTHYATSMAATRLVSKR 241
>UniRef100_UPI00003645F4 UPI00003645F4 UniRef100 entry
Length = 585
Score = 47.0 bits (110), Expect = 9e-04
Identities = 39/155 (25%), Positives = 71/155 (45%), Gaps = 16/155 (10%)
Query: 232 MNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSV---MAPVLKSQPGQAFLKMLPGE---- 284
+ ++L +Q T K+S+ L+DGV ++G V +AP +K G+ + + +
Sbjct: 421 VTKSLHVAKQATGGAVKVSQFLVDGVCAIAGHVGKELAPHVKKHGGKLIPESMKKDKDGR 480
Query: 285 --------VLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFA 336
V + ++ V+ E A K ++ + V ++YG AG+AT+H
Sbjct: 481 SNMDGAMVVAASGVEGFAAVWTGLEVAAKNITTSVAAETVTTVKHKYGAAAGQATDHAVN 540
Query: 337 TAGHAANTAWNVSKIR-KAINPVTPASTAGGALRN 370
+A + TA+NV + KA+ T TA L +
Sbjct: 541 SAINVGITAFNVDNLGVKAVVKRTGKQTAKALLED 575
>UniRef100_UPI00003645F3 UPI00003645F3 UniRef100 entry
Length = 628
Score = 47.0 bits (110), Expect = 9e-04
Identities = 39/155 (25%), Positives = 71/155 (45%), Gaps = 16/155 (10%)
Query: 232 MNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSV---MAPVLKSQPGQAFLKMLPGE---- 284
+ ++L +Q T K+S+ L+DGV ++G V +AP +K G+ + + +
Sbjct: 464 VTKSLHVAKQATGGAVKVSQFLVDGVCAIAGHVGKELAPHVKKHGGKLIPESMKKDKDGR 523
Query: 285 --------VLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFA 336
V + ++ V+ E A K ++ + V ++YG AG+AT+H
Sbjct: 524 SNMDGAMVVAASGVEGFAAVWTGLEVAAKNITTSVAAETVTTVKHKYGAAAGQATDHAVN 583
Query: 337 TAGHAANTAWNVSKIR-KAINPVTPASTAGGALRN 370
+A + TA+NV + KA+ T TA L +
Sbjct: 584 SAINVGITAFNVDNLGVKAVVKRTGKQTAKALLED 618
>UniRef100_UPI0000369A2B UPI0000369A2B UniRef100 entry
Length = 665
Score = 47.0 bits (110), Expect = 9e-04
Identities = 41/156 (26%), Positives = 70/156 (44%), Gaps = 16/156 (10%)
Query: 231 AMNENLKRVRQLTTMTEKLSKSLLDGVGIMS---GSVMAPVLKSQPGQAFLKMLPGE--- 284
A+ + L +Q T K+S+ L+DGV ++ G +AP +K + + L +
Sbjct: 472 AVTKGLYIAKQATGGAAKVSQFLVDGVCTVANCVGKELAPHVKKHGSKLVPESLKKDKDG 531
Query: 285 ---------VLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVF 335
V +S+ + V++ E A K ++ S + V +YG AGEAT H
Sbjct: 532 KSPLDGAMVVAASSVQGFSTVWQGLECAAKCIVNNVSAETVQTVRYKYGYNAGEATHHAV 591
Query: 336 ATAGHAANTAWNVSKIR-KAINPVTPASTAGGALRN 370
+A + TA+N++ I KA+ T T L +
Sbjct: 592 DSAVNVGVTAYNINNIGIKAMVKKTATQTGHSLLED 627
>UniRef100_UPI0000072FD6 UPI0000072FD6 UniRef100 entry
Length = 686
Score = 46.6 bits (109), Expect = 0.001
Identities = 36/136 (26%), Positives = 63/136 (45%), Gaps = 15/136 (11%)
Query: 231 AMNENLKRVRQLTTMTEKLSKSLLDGVGIMS---GSVMAPVLKSQPGQAFLKMLPGE--- 284
A+ + L +Q T K+S+ L+DGV ++ G +AP +K + + L +
Sbjct: 492 AVTKGLYIAKQATGGAAKVSQFLVDGVCTVANCVGKELAPHVKKHGSKLVPESLKKDKDG 551
Query: 285 ---------VLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVF 335
V +S+ + V++ E A K ++ S + V +YG AGEAT H
Sbjct: 552 KSPLDGAMVVAASSVQGFSTVWQGLECAAKCIVNNVSAETVQTVRYKYGYNAGEATHHAV 611
Query: 336 ATAGHAANTAWNVSKI 351
+A + TA+N++ I
Sbjct: 612 DSAVNVGVTAYNINNI 627
>UniRef100_Q8N0X7 Spartin [Homo sapiens]
Length = 666
Score = 46.6 bits (109), Expect = 0.001
Identities = 36/136 (26%), Positives = 63/136 (45%), Gaps = 15/136 (11%)
Query: 231 AMNENLKRVRQLTTMTEKLSKSLLDGVGIMS---GSVMAPVLKSQPGQAFLKMLPGE--- 284
A+ + L +Q T K+S+ L+DGV ++ G +AP +K + + L +
Sbjct: 472 AVTKGLYIAKQATGGAAKVSQFLVDGVCTVANCVGKELAPHVKKHGSKLVPESLKKDKDG 531
Query: 285 ---------VLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVF 335
V +S+ + V++ E A K ++ S + V +YG AGEAT H
Sbjct: 532 KSPLDGAMVVAASSVQGFSTVWQGLECAAKCIVNNVSAETVQTVRYKYGYNAGEATHHAV 591
Query: 336 ATAGHAANTAWNVSKI 351
+A + TA+N++ I
Sbjct: 592 DSAVNVGVTAYNINNI 607
>UniRef100_UPI000023CC30 UPI000023CC30 UniRef100 entry
Length = 490
Score = 46.2 bits (108), Expect = 0.001
Identities = 39/136 (28%), Positives = 66/136 (47%), Gaps = 19/136 (13%)
Query: 216 QSVSNNKTADATKKNAM--------NENLKRVRQLTTMTEKLSKSLLDGVGIMSGSVMAP 267
QS ++N T TK NA +E+++R+ + S + + +G ++ ++ A
Sbjct: 279 QSQADNFT-QKTKPNAKPVTFAPTTHEHIRRINTFSNRAATFSATTVGTIGNLAQNLGAT 337
Query: 268 VLK---------SQPGQAFLKMLPGEVLLASLDAVNKVFEATEAAQKQSLSATSQAATRM 318
V + + GQA PG VL SL A N V + E A + L+ TS + T +
Sbjct: 338 VTRRKDGRARGYDKDGQAIDTYKPG-VLNKSLMAFNTVVDGIEQAGRTLLNGTSSSVTTV 396
Query: 319 VSNRYGEEAGEATEHV 334
V +R+G EAGE + ++
Sbjct: 397 VGHRWGAEAGEVSRNL 412
>UniRef100_UPI00002BB14B UPI00002BB14B UniRef100 entry
Length = 596
Score = 44.3 bits (103), Expect = 0.006
Identities = 38/155 (24%), Positives = 69/155 (44%), Gaps = 16/155 (10%)
Query: 232 MNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSV---MAPVLKSQPGQAFLKMLPGE---- 284
+ + L +Q T K+S+ L+DGV ++ V +AP +K G+ + + +
Sbjct: 439 VTKGLHVAKQATGGAVKVSQFLVDGVCAVASRVGRELAPHVKKHGGKLIPESMKKDKDGR 498
Query: 285 --------VLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFA 336
V + ++ V+ E A K ++ + V ++YG AG+AT+H
Sbjct: 499 SNIDGAMVVAASGVEGFAAVWTGLEVAAKNITTSVAAETVTTVKHKYGAAAGQATDHAVN 558
Query: 337 TAGHAANTAWNVSKIR-KAINPVTPASTAGGALRN 370
+A + TA+NV + KA+ T TA L +
Sbjct: 559 SAINVGITAFNVDNLGIKAVVKRTGKQTAKALLED 593
>UniRef100_UPI0000249FF2 UPI0000249FF2 UniRef100 entry
Length = 546
Score = 43.5 bits (101), Expect = 0.010
Identities = 36/153 (23%), Positives = 69/153 (44%), Gaps = 16/153 (10%)
Query: 232 MNENLKRVRQLTTMTEKLSKSLLDGVGIMSGSV---MAPVLKSQPGQAFLKMLPGE---- 284
+ ++L +Q T K+S+ L+DG+ ++G V +AP +K G+ + + +
Sbjct: 389 VTKSLHVAKQATGGAVKVSQFLVDGLCTVAGHVGRELAPHVKKHGGKLIPESMKKDKDGR 448
Query: 285 --------VLLASLDAVNKVFEATEAAQKQSLSATSQAATRMVSNRYGEEAGEATEHVFA 336
V + + ++ E A K + + V ++YG +AG+AT+H
Sbjct: 449 SNIDGAMVVAASGVQGFATMWTGLEVAAKNIAKSVATETVTTVKHKYGPDAGQATDHAVN 508
Query: 337 TAGHAANTAWNVSKIR-KAINPVTPASTAGGAL 368
+A + TA+N+ + KA+ T TA L
Sbjct: 509 SAINVGVTAFNIDNLGIKAMVKKTGKETAHALL 541
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.130 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 603,255,867
Number of Sequences: 2790947
Number of extensions: 24655641
Number of successful extensions: 52475
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 52392
Number of HSP's gapped (non-prelim): 76
length of query: 378
length of database: 848,049,833
effective HSP length: 129
effective length of query: 249
effective length of database: 488,017,670
effective search space: 121516399830
effective search space used: 121516399830
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0065.1


