

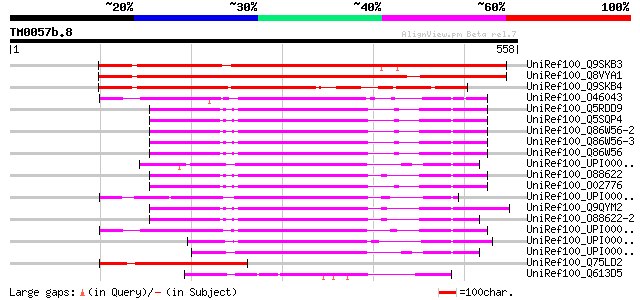
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0057b.8
(558 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SKB3 Poly(ADP-ribose) glycohydrolase [Arabidopsis th... 529 e-149
UniRef100_Q8VYA1 Putative poly(ADP-ribose) glycohydrolase [Arabi... 462 e-128
UniRef100_Q9SKB4 Putative poly(ADP-ribose) glycohydrolase [Arabi... 309 1e-82
UniRef100_O46043 Poly(ADP-ribose) glycohydrolase [Drosophila mel... 216 2e-54
UniRef100_Q5RDD9 Hypothetical protein DKFZp469F0213 [Pongo pygma... 202 2e-50
UniRef100_Q5SQP4 Poly (ADP-ribose) glycohydrolase [Homo sapiens] 202 2e-50
UniRef100_Q86W56-2 Splice isoform 2 of Q86W56 [Homo sapiens] 202 2e-50
UniRef100_Q86W56-3 Splice isoform 3 of Q86W56 [Homo sapiens] 202 2e-50
UniRef100_Q86W56 Poly(ADP-ribose) glycohydrolase [Homo sapiens] 202 2e-50
UniRef100_UPI000027C995 UPI000027C995 UniRef100 entry 197 5e-49
UniRef100_O88622 Poly(ADP-ribose) glycohydrolase [Mus musculus] 196 1e-48
UniRef100_O02776 Poly(ADP-ribose) glycohydrolase [Bos taurus] 196 1e-48
UniRef100_UPI00002F9CB0 UPI00002F9CB0 UniRef100 entry 196 1e-48
UniRef100_Q9QYM2 Poly(ADP-ribose) glycohydrolase [Rattus norvegi... 192 2e-47
UniRef100_O88622-2 Splice isoform 2 of O88622 [Mus musculus] 192 3e-47
UniRef100_UPI00003AE137 UPI00003AE137 UniRef100 entry 188 3e-46
UniRef100_UPI0000318FC5 UPI0000318FC5 UniRef100 entry 174 5e-42
UniRef100_UPI00002B206C UPI00002B206C UniRef100 entry 174 6e-42
UniRef100_Q75LD2 Putative poly(ADP-ribose) glycohydrolase [Oryza... 161 5e-38
UniRef100_Q613D5 Hypothetical protein CBG16441 [Caenorhabditis b... 123 2e-26
>UniRef100_Q9SKB3 Poly(ADP-ribose) glycohydrolase [Arabidopsis thaliana]
Length = 548
Score = 529 bits (1363), Expect = e-149
Identities = 274/466 (58%), Positives = 332/466 (70%), Gaps = 30/466 (6%)
Query: 98 EESRKWFQEVVPALGILLLRLPSLLEAHYQNADRVLDEDGGLVRTGLRLLDSQEPGIVFL 157
+ES++WF E++PAL LLL+ PSLLE H+QNAD ++ ++TGLRLL+SQ+ GIVFL
Sbjct: 86 KESKRWFDEIIPALASLLLQFPSLLEVHFQNADNIVSG----IKTGLRLLNSQQAGIVFL 141
Query: 158 SQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHYFQR 217
SQELI ALL CSFFCLFP + R KHL INFD LF SLY YSQ QESKI+CI+HYF+R
Sbjct: 142 SQELIGALLACSFFCLFPDDNRGAKHLPVINFDHLFASLYISYSQSQESKIRCIMHYFER 201
Query: 218 ITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLSEAV 277
S +P G+VSFERK+ + P+AD WS S + LC F+VHS GLIEDQ A+
Sbjct: 202 FCSCVPIGIVSFERKIT---------AAPDADFWSKSDVSLCAFKVHSFGLIEDQPDNAL 252
Query: 278 EVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSSYTG 337
EVDFAN+YLGGG+L RGCVQEEIRFMI+PELI MLFLP M DNEAIEIVG ERFS YTG
Sbjct: 253 EVDFANKYLGGGSLSRGCVQEEIRFMINPELIAGMLFLPRMDDNEAIEIVGAERFSCYTG 312
Query: 338 YASSFRFSGNYVDERDVDTLGRRKTRIVAIDALCSPGMRQYRPKFLLREINKAFCGFLYG 397
YASSFRF+G Y+D++ +D RR+TRIVAIDALC+P MR ++ LLREINKA CGFL
Sbjct: 313 YASSFRFAGEYIDKKAMDPFKRRRTRIVAIDALCTPKMRHFKDICLLREINKALCGFLNC 372
Query: 398 SKHQLYQKIL--------------QEKGCPSTLFDAATSTPM---ETSEGKCSNHEIRDS 440
SK +Q I ++ G T A+ TP+ E + K +N+ IRD
Sbjct: 373 SKAWEHQNIFMDEGDNEIQLVRNGRDSGLLRTETTASHRTPLNDVEMNREKPANNLIRDF 432
Query: 441 QNDYHRMEQCNNIGVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTFRLEALHN 500
+ E + GVATGNWGCG FGGDPE+K IQWLAASQ RPFI+YYTF +EAL N
Sbjct: 433 YVEGVDNEDHEDDGVATGNWGCGVFGGDPELKATIQWLAASQTRRPFISYYTFGVEALRN 492
Query: 501 IDKVAHWILSHRWTVGDLWNMLTEYSMNRSKGETNVGFLQWVLPSM 546
+D+V WILSH+WTVGDLWNM+ EYS R +T+VGF W+LPS+
Sbjct: 493 LDQVTKWILSHKWTVGDLWNMMLEYSAQRLYKQTSVGFFSWLLPSL 538
>UniRef100_Q8VYA1 Putative poly(ADP-ribose) glycohydrolase [Arabidopsis thaliana]
Length = 522
Score = 462 bits (1188), Expect = e-128
Identities = 244/450 (54%), Positives = 304/450 (67%), Gaps = 18/450 (4%)
Query: 98 EESRKWFQEVVPALGILLLRLPSLLEAHYQNADRVLDEDGGLVRTGLRLLDSQEPGIVFL 157
EES +F EVVPAL LLL+LPS+LE HYQ AD VLD V++GLRLL QE GIV L
Sbjct: 88 EESANFFGEVVPALCRLLLQLPSMLEKHYQKADHVLDG----VKSGLRLLGPQEAGIVLL 143
Query: 158 SQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHYFQR 217
SQELIAALL CSFFCLFP +R K+LQ INF LF Y + KQE+KI+C+IHYF R
Sbjct: 144 SQELIAALLACSFFCLFPEVDRSLKNLQGINFSGLFSFPYMRHCTKQENKIKCLIHYFGR 203
Query: 218 ITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLSEAV 277
I MP G VSFERK+LPLE +SYP AD W+ S+ PLC E+H+SG IEDQ EA+
Sbjct: 204 ICRWMPTGFVSFERKILPLEYHPHFVSYPKADSWANSVTPLCSIEIHTSGAIEDQPCEAL 263
Query: 278 EVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSSYTG 337
EVDFA+EY GG L +QEEIRF+I+PELI M+FLP M NEAIEIVGVERFS YTG
Sbjct: 264 EVDFADEYFGGLTLSYDTLQEEIRFVINPELIAGMIFLPRMDANEAIEIVGVERFSGYTG 323
Query: 338 YASSFRFSGNYVDERDVDTLGRRKTRIVAIDALCSPGMRQYRPKFLLREINKAFCGFLYG 397
Y SF+++G+Y D +D+D RRKTR++AIDA+ PGM QY+ L+RE+NKAF G+++
Sbjct: 324 YGPSFQYAGDYTDNKDLDIFRRRKTRVIAIDAMPDPGMGQYKLDALIREVNKAFSGYMHQ 383
Query: 398 SKHQLYQKILQEKGCPST-LFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNIGVA 456
K+ + K E L + S +E+S C +HE + IGVA
Sbjct: 384 CKYNIDVKHDPEASSSHVPLTSDSASQVIESSHRWCIDHEEK-------------KIGVA 430
Query: 457 TGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTFRLEALHNIDKVAHWILSHRWTVG 516
TGNWGCG FGGDPE+K ++QWLA SQ+ RPF++YYTF L+AL N+++V + TVG
Sbjct: 431 TGNWGCGVFGGDPELKIMLQWLAISQSGRPFMSYYTFGLQALQNLNQVIEMVALQEMTVG 490
Query: 517 DLWNMLTEYSMNRSKGETNVGFLQWVLPSM 546
DLW L EYS R T +GF W++ S+
Sbjct: 491 DLWKKLVEYSSERLSRRTWLGFFSWLMTSL 520
>UniRef100_Q9SKB4 Putative poly(ADP-ribose) glycohydrolase [Arabidopsis thaliana]
Length = 364
Score = 309 bits (792), Expect = 1e-82
Identities = 194/409 (47%), Positives = 250/409 (60%), Gaps = 50/409 (12%)
Query: 98 EESRKWFQEVVPALGILLLRLPSLLEAHYQNADRVLDEDGGLVRTGLRLLDSQEPGIVFL 157
EES +WF E +PA+ LLLR PSLLE+HY N+D +++ +TGLR+L + GIVFL
Sbjct: 4 EESSRWFNEFLPAMACLLLRFPSLLESHYLNSDNLING----TKTGLRVLVPNKAGIVFL 59
Query: 158 SQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDY-YSQKQESKIQCIIHYFQ 216
SQELI ALL CSFFCLFPV++R HL INFD+LFGSL + ++ QE+KI+CIIHYFQ
Sbjct: 60 SQELIGALLSCSFFCLFPVDDRGSNHLPIINFDKLFGSLINTGRNEHQENKIKCIIHYFQ 119
Query: 217 RITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLSEA 276
R++S++ G VSFERK+L LE D S + W S + LC EV +SGLIEDQ EA
Sbjct: 120 RLSSSISPGFVSFERKILSLEQDS---STLDEGFWGKSTVNLCPVEVRTSGLIEDQSVEA 176
Query: 277 VEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSSYT 336
+EVDFAN+ LGGGALR+GCVQEEIRFMI+PELIV MLFLP+M EAIE+VG ERFS YT
Sbjct: 177 LEVDFANKNLGGGALRKGCVQEEIRFMINPELIVGMLFLPTMEVTEAIEVVGAERFSLYT 236
Query: 337 GYASSFRFSGNYVDERDVDTLGRRKTRIVAIDALCSPGMRQYRPKFLLREINKAFCGFLY 396
G FR + KTRIVAIDAL PG+ QY+ + LL +
Sbjct: 237 G---CFR---------------KAKTRIVAIDALRHPGVSQYKLESLLSVL--------- 269
Query: 397 GSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRME-QCNNIGV 455
IL G P L+ + S ++ EI S ++ + +N+ +
Sbjct: 270 ---------ILSSSGRPIRLYMGSVS--LQGIGDVVLMVEILSSSLFFNGLRFHRSNLYL 318
Query: 456 ATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTFRLEALHNIDKV 504
+ C P V + L + QA RPF++YYTF EAL N+++V
Sbjct: 319 FSPLIACNY---QPMVVVWLTTLHSFQARRPFMSYYTFGFEALQNLNQV 364
>UniRef100_O46043 Poly(ADP-ribose) glycohydrolase [Drosophila melanogaster]
Length = 768
Score = 216 bits (549), Expect = 2e-54
Identities = 156/440 (35%), Positives = 219/440 (49%), Gaps = 91/440 (20%)
Query: 99 ESRKWFQEVVPALGILLLRLPSLLEAHYQNADRVLDEDGGLVRTGLRLLDSQEPGIVFLS 158
E+R +F++++P + L LRLP L+++ + LL + + LS
Sbjct: 204 ETRVFFEDLLPRIIRLALRLPDLIQSP------------------VPLLKHHKNASLSLS 245
Query: 159 QELIAALLVCSFFCLFPVNERYGKHLQSINFDEL-FGSLYDYYSQKQESKIQCIIHYFQR 217
Q+ I+ LL +F C FP + + F ++ F LY K++CI+HYF+R
Sbjct: 246 QQQISCLLANAFLCTFPRRNTLKRKSEYSTFPDINFNRLYQSTGPAVLEKLKCIMHYFRR 305
Query: 218 I------TSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVH--SSGLI 269
+ SN+P GVV+F R+ L + I WS S PL +H + G I
Sbjct: 306 VCPTERDASNVPTGVVTFVRRS-GLPEHLID--------WSQSAAPLGDVPLHVDAEGTI 356
Query: 270 EDQLSEAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGV 329
ED+ ++VDFAN+YLGGG L GCVQEEIRF+I PEL+V LF + EA+ ++G
Sbjct: 357 EDEGIGLLQVDFANKYLGGGVLGHGCVQEEIRFVICPELLVGKLFTECLRPFEALVMLGA 416
Query: 330 ERFSSYTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREIN 388
ER+S+YTGYA SF +SGN+ D D+ GRR+T IVAIDAL + QYR + RE+N
Sbjct: 417 ERYSNYTGYAGSFEWSGNFEDSTPRDSSGRRQTAIVAIDALHFAQSHHQYREDLMERELN 476
Query: 389 KAFCGFLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRME 448
KA+ G H + T P
Sbjct: 477 KAYI----GFVHWM-----------------VTPPP------------------------ 491
Query: 449 QCNNIGVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTFRLEALHNIDKVAHWI 508
GVATGNWGCGAFGGD +K ++Q + +Q RP +AYYTF + D W+
Sbjct: 492 -----GVATGNWGCGAFGGDSYLKALLQLMVCAQLGRP-LAYYTFGNVEFRD-DFHEMWL 544
Query: 509 L--SHRWTVGDLWNMLTEYS 526
L + TV LW++L YS
Sbjct: 545 LFRNDGTTVQQLWSILRSYS 564
>UniRef100_Q5RDD9 Hypothetical protein DKFZp469F0213 [Pongo pygmaeus]
Length = 978
Score = 202 bits (514), Expect = 2e-50
Identities = 136/373 (36%), Positives = 193/373 (51%), Gaps = 62/373 (16%)
Query: 155 VFLSQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHY 214
+ +SQE IA+LL +FFC FP K S D F L++ S ++ K++ + Y
Sbjct: 625 ITMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCY 684
Query: 215 FQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
F+R+T P G+V+F R+ LED +P W PL R V G IE+
Sbjct: 685 FRRVTEKKPTGLVTFTRQ--SLED------FPE---WERCEKPLTRLHVTYEGTIEENGR 733
Query: 275 EAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSS 334
++VDFAN ++GGG G VQEEIRF+I+PELI+ LF + NE + I G E++S
Sbjct: 734 GMLQVDFANRFVGGGVTSAGLVQEEIRFLINPELIISRLFTEVLDHNECLIITGTEQYSE 793
Query: 335 YTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCG 393
YTGYA ++R+S ++ D + D RR T IVAIDAL + Q+ P+ + RE+NKA+CG
Sbjct: 794 YTGYAETYRWSRSHEDGSERDGWQRRCTEIVAIDALHFRRYLDQFVPEKMRRELNKAYCG 853
Query: 394 FLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNI 453
FL P +SE N
Sbjct: 854 FL---------------------------RPGVSSE---------------------NLS 865
Query: 454 GVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHR 512
VATGNWGCGAFGGD +K +IQ LAA+ A R + Y+TF E + +I + ++ +
Sbjct: 866 AVATGNWGCGAFGGDARLKALIQILAAAAAERD-VVYFTFGDSELMRDIYSMHIFLTERK 924
Query: 513 WTVGDLWNMLTEY 525
TVGD++ +L +Y
Sbjct: 925 LTVGDVYKLLLQY 937
>UniRef100_Q5SQP4 Poly (ADP-ribose) glycohydrolase [Homo sapiens]
Length = 491
Score = 202 bits (514), Expect = 2e-50
Identities = 136/373 (36%), Positives = 192/373 (51%), Gaps = 62/373 (16%)
Query: 155 VFLSQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHY 214
+ +SQE IA+LL +FFC FP K S D F L++ S ++ K++ + Y
Sbjct: 138 ITMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCY 197
Query: 215 FQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
F+R+T P G+V+F R+ LED +P W PL R V G IE+
Sbjct: 198 FRRVTEKKPTGLVTFTRQ--SLED------FPE---WERCEKPLTRLHVTYEGTIEENGQ 246
Query: 275 EAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSS 334
++VDFAN ++GGG G VQEEIRF+I+PELI+ LF + NE + I G E++S
Sbjct: 247 GMLQVDFANRFVGGGVTSAGLVQEEIRFLINPELIISRLFTEVLDHNECLIITGTEQYSE 306
Query: 335 YTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCG 393
YTGYA ++R+S ++ D + D RR T IVAIDAL + Q+ P+ + RE+NKA+CG
Sbjct: 307 YTGYAETYRWSRSHEDGSERDDWQRRCTEIVAIDALHFRRYLDQFVPEKMRRELNKAYCG 366
Query: 394 FLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNI 453
FL P +SE N
Sbjct: 367 FL---------------------------RPGVSSE---------------------NLS 378
Query: 454 GVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHR 512
VATGNWGCGAFGGD +K +IQ LAA+ A R + Y+TF E + +I + ++ +
Sbjct: 379 AVATGNWGCGAFGGDARLKALIQILAAAAAERD-VVYFTFGDSELMRDIYSMHIFLTERK 437
Query: 513 WTVGDLWNMLTEY 525
TVGD++ +L Y
Sbjct: 438 LTVGDVYKLLLRY 450
>UniRef100_Q86W56-2 Splice isoform 2 of Q86W56 [Homo sapiens]
Length = 894
Score = 202 bits (514), Expect = 2e-50
Identities = 136/373 (36%), Positives = 192/373 (51%), Gaps = 62/373 (16%)
Query: 155 VFLSQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHY 214
+ +SQE IA+LL +FFC FP K S D F L++ S ++ K++ + Y
Sbjct: 541 ITMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCY 600
Query: 215 FQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
F+R+T P G+V+F R+ LED +P W PL R V G IE+
Sbjct: 601 FRRVTEKKPTGLVTFTRQ--SLED------FPE---WERCEKPLTRLHVTYEGTIEENGQ 649
Query: 275 EAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSS 334
++VDFAN ++GGG G VQEEIRF+I+PELI+ LF + NE + I G E++S
Sbjct: 650 GMLQVDFANRFVGGGVTSAGLVQEEIRFLINPELIISRLFTEVLDHNECLIITGTEQYSE 709
Query: 335 YTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCG 393
YTGYA ++R+S ++ D + D RR T IVAIDAL + Q+ P+ + RE+NKA+CG
Sbjct: 710 YTGYAETYRWSRSHEDGSERDDWQRRCTEIVAIDALHFRRYLDQFVPEKMRRELNKAYCG 769
Query: 394 FLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNI 453
FL P +SE N
Sbjct: 770 FL---------------------------RPGVSSE---------------------NLS 781
Query: 454 GVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHR 512
VATGNWGCGAFGGD +K +IQ LAA+ A R + Y+TF E + +I + ++ +
Sbjct: 782 AVATGNWGCGAFGGDARLKALIQILAAAAAERD-VVYFTFGDSELMRDIYSMHIFLTERK 840
Query: 513 WTVGDLWNMLTEY 525
TVGD++ +L Y
Sbjct: 841 LTVGDVYKLLLRY 853
>UniRef100_Q86W56-3 Splice isoform 3 of Q86W56 [Homo sapiens]
Length = 868
Score = 202 bits (514), Expect = 2e-50
Identities = 136/373 (36%), Positives = 192/373 (51%), Gaps = 62/373 (16%)
Query: 155 VFLSQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHY 214
+ +SQE IA+LL +FFC FP K S D F L++ S ++ K++ + Y
Sbjct: 515 ITMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCY 574
Query: 215 FQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
F+R+T P G+V+F R+ LED +P W PL R V G IE+
Sbjct: 575 FRRVTEKKPTGLVTFTRQ--SLED------FPE---WERCEKPLTRLHVTYEGTIEENGQ 623
Query: 275 EAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSS 334
++VDFAN ++GGG G VQEEIRF+I+PELI+ LF + NE + I G E++S
Sbjct: 624 GMLQVDFANRFVGGGVTSAGLVQEEIRFLINPELIISRLFTEVLDHNECLIITGTEQYSE 683
Query: 335 YTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCG 393
YTGYA ++R+S ++ D + D RR T IVAIDAL + Q+ P+ + RE+NKA+CG
Sbjct: 684 YTGYAETYRWSRSHEDGSERDDWQRRCTEIVAIDALHFRRYLDQFVPEKMRRELNKAYCG 743
Query: 394 FLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNI 453
FL P +SE N
Sbjct: 744 FL---------------------------RPGVSSE---------------------NLS 755
Query: 454 GVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHR 512
VATGNWGCGAFGGD +K +IQ LAA+ A R + Y+TF E + +I + ++ +
Sbjct: 756 AVATGNWGCGAFGGDARLKALIQILAAAAAERD-VVYFTFGDSELMRDIYSMHIFLTERK 814
Query: 513 WTVGDLWNMLTEY 525
TVGD++ +L Y
Sbjct: 815 LTVGDVYKLLLRY 827
>UniRef100_Q86W56 Poly(ADP-ribose) glycohydrolase [Homo sapiens]
Length = 976
Score = 202 bits (514), Expect = 2e-50
Identities = 136/373 (36%), Positives = 192/373 (51%), Gaps = 62/373 (16%)
Query: 155 VFLSQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHY 214
+ +SQE IA+LL +FFC FP K S D F L++ S ++ K++ + Y
Sbjct: 623 ITMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCY 682
Query: 215 FQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
F+R+T P G+V+F R+ LED +P W PL R V G IE+
Sbjct: 683 FRRVTEKKPTGLVTFTRQ--SLED------FPE---WERCEKPLTRLHVTYEGTIEENGQ 731
Query: 275 EAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSS 334
++VDFAN ++GGG G VQEEIRF+I+PELI+ LF + NE + I G E++S
Sbjct: 732 GMLQVDFANRFVGGGVTSAGLVQEEIRFLINPELIISRLFTEVLDHNECLIITGTEQYSE 791
Query: 335 YTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCG 393
YTGYA ++R+S ++ D + D RR T IVAIDAL + Q+ P+ + RE+NKA+CG
Sbjct: 792 YTGYAETYRWSRSHEDGSERDDWQRRCTEIVAIDALHFRRYLDQFVPEKMRRELNKAYCG 851
Query: 394 FLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNI 453
FL P +SE N
Sbjct: 852 FL---------------------------RPGVSSE---------------------NLS 863
Query: 454 GVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHR 512
VATGNWGCGAFGGD +K +IQ LAA+ A R + Y+TF E + +I + ++ +
Sbjct: 864 AVATGNWGCGAFGGDARLKALIQILAAAAAERD-VVYFTFGDSELMRDIYSMHIFLTERK 922
Query: 513 WTVGDLWNMLTEY 525
TVGD++ +L Y
Sbjct: 923 LTVGDVYKLLLRY 935
>UniRef100_UPI000027C995 UPI000027C995 UniRef100 entry
Length = 327
Score = 197 bits (502), Expect = 5e-49
Identities = 132/384 (34%), Positives = 194/384 (50%), Gaps = 70/384 (18%)
Query: 143 GLRLLDSQEPGIVFLSQELIAALLVCSFFCLFPVNERYGKHLQ-----SINFDELFGSLY 197
G+ LL + LSQ I+ LL +FFC FP H SINF LF
Sbjct: 5 GIPLLQQGRAAAITLSQAQISCLLANAFFCTFPHRNSTSFHSDYHTYPSINFTRLFS--- 61
Query: 198 DYYSQKQESKIQCIIHYFQRITSNMPQ--GVVSFERKVLPLEDDYIHISYPNADVWSTSI 255
++S+++ K++ I+HYF T + G+V+FER+ L D A WS
Sbjct: 62 -HWSERKMEKLKAIVHYFHVATDEKTKLDGLVTFERRCLANTD---------ARTWSCCK 111
Query: 256 IPLCRFEVHSSGLIEDQLSEAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFL 315
+ + V S G IE + S ++VDFA+ +LGGG L G +QEEI F++SPELIV LF
Sbjct: 112 EEMNKLYVSSCGAIETEGSGLLQVDFASSWLGGGVLDSGLLQEEILFLMSPELIVSRLFT 171
Query: 316 PSMADNEAIEIVGVERFSSYTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPG 374
+ DNE + + G ++FS+Y+GY +FR+ G Y D D RR+ +++A+DAL + G
Sbjct: 172 EKLQDNECVIVTGCQQFSTYSGYGDTFRWKGPYADPTGRDGWARRQRQVLAMDALRFTHG 231
Query: 375 MRQYRPKFLLREINKAFCGFLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSN 434
QY K ++RE+NKA+CGF K
Sbjct: 232 RDQYSMKLVVRELNKAYCGF------------------------------------KRCE 255
Query: 435 HEIRDSQNDYHRMEQCNNIGVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTFR 494
E R + D +ATG WGCGAF GDP++K +IQ +AA++A R +A++TF+
Sbjct: 256 EERRGEEPD-----------IATGKWGCGAFKGDPQLKAVIQLMAAARAGRG-LAFFTFK 303
Query: 495 LEAL-HNIDKVAHWILSHRWTVGD 517
E L H + + + + TVG+
Sbjct: 304 DEKLEHGLRQAYRLLRTKGTTVGE 327
>UniRef100_O88622 Poly(ADP-ribose) glycohydrolase [Mus musculus]
Length = 969
Score = 196 bits (499), Expect = 1e-48
Identities = 135/373 (36%), Positives = 188/373 (50%), Gaps = 62/373 (16%)
Query: 155 VFLSQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHY 214
V +SQE IA+LL +FFC FP K S D F L++ S ++ K++ + Y
Sbjct: 616 VTMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCY 675
Query: 215 FQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
F+R+T P G+V+F R+ LED +P W PL R V G IE
Sbjct: 676 FRRVTEKKPTGLVTFTRQ--SLED------FPE---WERCEKPLTRLHVTYEGTIEGNGR 724
Query: 275 EAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSS 334
++VDFAN ++GGG G VQEEIRF+I+PELIV LF + NE + I G E++S
Sbjct: 725 GMLQVDFANRFVGGGVTGAGLVQEEIRFLINPELIVSRLFTEVLDHNECLIITGTEQYSE 784
Query: 335 YTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCG 393
YTGYA ++R++ ++ D + D RR T IVAIDAL + Q+ P+ + RE+NKA+CG
Sbjct: 785 YTGYAETYRWARSHEDGSEKDDWQRRCTEIVAIDALHFRRYLDQFVPEKVRRELNKAYCG 844
Query: 394 FLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNI 453
FL G PS N
Sbjct: 845 FL-------------RPGVPSE-----------------------------------NLS 856
Query: 454 GVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHR 512
VATGNWGCGAFGGD +K +IQ LAA+ A R + Y+TF E + +I + ++ +
Sbjct: 857 AVATGNWGCGAFGGDARLKALIQILAAAAAERD-VVYFTFGDSELMRDIYSMHTFLTERK 915
Query: 513 WTVGDLWNMLTEY 525
VG ++ +L Y
Sbjct: 916 LDVGKVYKLLLRY 928
>UniRef100_O02776 Poly(ADP-ribose) glycohydrolase [Bos taurus]
Length = 977
Score = 196 bits (499), Expect = 1e-48
Identities = 134/373 (35%), Positives = 190/373 (50%), Gaps = 62/373 (16%)
Query: 155 VFLSQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHY 214
+ +SQE IA+LL +FFC FP K S D F L++ S ++ K++ + Y
Sbjct: 624 ITMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCY 683
Query: 215 FQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
F+R+T P G+V+F R+ LED +P W L R V G IE
Sbjct: 684 FRRVTEKKPTGLVTFTRQ--SLED------FPE---WERCEKLLTRLHVTYEGTIEGNGQ 732
Query: 275 EAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSS 334
++VDFAN ++GGG G VQEEIRF+I+PELIV LF + NE + I G E++S
Sbjct: 733 GMLQVDFANRFVGGGVTSAGLVQEEIRFLINPELIVSRLFTEVLDHNECLIITGTEQYSE 792
Query: 335 YTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCG 393
YTGYA ++R++ ++ D + D RR T IVAIDAL + Q+ P+ + RE+NKA+CG
Sbjct: 793 YTGYAETYRWARSHEDRSERDDWQRRTTEIVAIDALHFRRYLDQFVPEKIRRELNKAYCG 852
Query: 394 FLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNI 453
FL P +SE N
Sbjct: 853 FL---------------------------RPGVSSE---------------------NLS 864
Query: 454 GVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHR 512
VATGNWGCGAFGGD +K +IQ LAA+ A R + Y+TF E + +I + ++ +
Sbjct: 865 AVATGNWGCGAFGGDARLKALIQILAAAVAERD-VVYFTFGDSELMRDIYSMHTFLTERK 923
Query: 513 WTVGDLWNMLTEY 525
TVG+++ +L Y
Sbjct: 924 LTVGEVYKLLLRY 936
>UniRef100_UPI00002F9CB0 UPI00002F9CB0 UniRef100 entry
Length = 463
Score = 196 bits (498), Expect = 1e-48
Identities = 136/399 (34%), Positives = 200/399 (50%), Gaps = 77/399 (19%)
Query: 99 ESRKWFQEVVPALGILLLRLPSLLEAHYQNADRVLDEDGGLVRT-GLRLLDSQEPGIVFL 157
E + +F+ V+PA+ L ++ P L ++R + LL ++ + L
Sbjct: 33 EKQHFFEVVLPAMVELAVQAPFLCTM------------ASILRIFPIPLLKARMNHSLTL 80
Query: 158 SQELIAALLVCSFFCLFPVNERYGKHLQSINFDEL-FGSLYDYYSQKQESKIQCIIHYFQ 216
SQE IA LL +FFC FP R + + N+ E+ F L++ S ++ K++ ++ YF+
Sbjct: 81 SQEQIACLLANAFFCTFP--RRNSRKSEYGNYPEINFYRLFEGSSMRKIEKLKTLLCYFR 138
Query: 217 RITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLSEA 276
R+T N P+G+V+F R+ L W +S L R + G IED
Sbjct: 139 RVTQNRPKGLVTFTRQTLN-----------EPPKWESSQKQLTRLHITCQGTIEDDGYGM 187
Query: 277 VEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSSYT 336
++VDFAN ++GGG G VQEEIRF+I+PELIV LF ++ +E + I G ER+S Y+
Sbjct: 188 LQVDFANRFVGGGVTGHGLVQEEIRFLINPELIVSRLFTEALEHDECLIITGTERYSKYS 247
Query: 337 GYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCGFL 395
GYA S+++ ++DE D RR T IVAIDAL + Q+ P+ + RE+ KA+CGF
Sbjct: 248 GYAESYKWQDCHIDETPSDEWQRRCTEIVAIDALKFRHYLEQFVPQKIRRELYKAYCGFF 307
Query: 396 YGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNIGV 455
C S A V
Sbjct: 308 -------------RNNCESKHLSA-----------------------------------V 319
Query: 456 ATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTFR 494
ATGNWGCGAFGGD +K +IQ +AA++A R +AY+TFR
Sbjct: 320 ATGNWGCGAFGGDTRLKALIQLMAAAEAGRD-VAYFTFR 357
>UniRef100_Q9QYM2 Poly(ADP-ribose) glycohydrolase [Rattus norvegicus]
Length = 972
Score = 192 bits (489), Expect = 2e-47
Identities = 137/398 (34%), Positives = 195/398 (48%), Gaps = 62/398 (15%)
Query: 155 VFLSQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHY 214
V +SQE IA+LL +FFC FP K S D F L++ S ++ K++ + Y
Sbjct: 619 VTMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCY 678
Query: 215 FQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
F+R+T P G+V+F R+ LED +P W PL R V G IE
Sbjct: 679 FRRVTEKKPTGLVTFTRQ--SLED------FPE---WERCDKPLTRLHVTYEGTIEGNGR 727
Query: 275 EAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSS 334
++VDFAN ++GGG G VQEEIRF+I+PELIV LF + NE + I G E++S
Sbjct: 728 GMLQVDFANRFVGGGVTGAGLVQEEIRFLINPELIVSRLFTEVLDHNECLIITGTEQYSE 787
Query: 335 YTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCG 393
YTGYA ++R++ ++ D + D R T IVAIDAL + Q+ P+ + RE+NKA+CG
Sbjct: 788 YTGYAETYRWARSHEDGSEKDDWQRCCTEIVAIDALHFRRYLDQFVPEKVRRELNKAYCG 847
Query: 394 FLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNI 453
FL G P N
Sbjct: 848 FL-------------RPGVPPE-----------------------------------NLS 859
Query: 454 GVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHR 512
VATGNWGCGAFGGD +K +IQ LAA+ A R + Y+TF E + +I + ++ +
Sbjct: 860 AVATGNWGCGAFGGDARLKALIQLLAAAAAERD-VVYFTFGDSELMRDIYSMHTFLTERK 918
Query: 513 WTVGDLWNMLTEYSMNRSKGETNVGFLQWVLPSMYDDA 550
VG ++ +L Y + ++ G + P +Y A
Sbjct: 919 LNVGKVYRLLLRYYREECRDCSSPGPDTKLYPFIYHAA 956
>UniRef100_O88622-2 Splice isoform 2 of O88622 [Mus musculus]
Length = 920
Score = 192 bits (487), Expect = 3e-47
Identities = 133/365 (36%), Positives = 184/365 (49%), Gaps = 62/365 (16%)
Query: 155 VFLSQELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHY 214
V +SQE IA+LL +FFC FP K S D F L++ S ++ K++ + Y
Sbjct: 616 VTMSQEQIASLLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSSRKPEKLKTLFCY 675
Query: 215 FQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLS 274
F+R+T P G+V+F R+ LED +P W PL R V G IE
Sbjct: 676 FRRVTEKKPTGLVTFTRQ--SLED------FPE---WERCEKPLTRLHVTYEGTIEGNGR 724
Query: 275 EAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSS 334
++VDFAN ++GGG G VQEEIRF+I+PELIV LF + NE + I G E++S
Sbjct: 725 GMLQVDFANRFVGGGVTGAGLVQEEIRFLINPELIVSRLFTEVLDHNECLIITGTEQYSE 784
Query: 335 YTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKAFCG 393
YTGYA ++R++ ++ D + D RR T IVAIDAL + Q+ P+ + RE+NKA+CG
Sbjct: 785 YTGYAETYRWARSHEDGSEKDDWQRRCTEIVAIDALHFRRYLDQFVPEKVRRELNKAYCG 844
Query: 394 FLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNI 453
FL G PS N
Sbjct: 845 FL-------------RPGVPSE-----------------------------------NLS 856
Query: 454 GVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHR 512
VATGNWGCGAFGGD +K +IQ LAA+ A R + Y+TF E + +I + ++ +
Sbjct: 857 AVATGNWGCGAFGGDARLKALIQILAAAAAERD-VVYFTFGDSELMRDIYSMHTFLTERK 915
Query: 513 WTVGD 517
VG+
Sbjct: 916 LDVGE 920
>UniRef100_UPI00003AE137 UPI00003AE137 UniRef100 entry
Length = 893
Score = 188 bits (478), Expect = 3e-46
Identities = 137/430 (31%), Positives = 206/430 (47%), Gaps = 81/430 (18%)
Query: 99 ESRKWFQEVVPALGILLLRLPSLLEAHYQNADRVLDEDGGLVRTGLRLLDSQEPGIVFLS 158
E++ FQ ++P + L L LP++ + LL + + +S
Sbjct: 509 EAQHLFQSILPDMVKLALCLPTICTQP------------------IPLLKQKMNHSITMS 550
Query: 159 QELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHYFQRI 218
QE IA+ L +FFC FP K S D F L++ S ++ K++ + YF+R+
Sbjct: 551 QEQIASFLANAFFCTFPRRNAKMKSEYSSYPDINFNRLFEGRSPRKPEKLKTLFCYFRRV 610
Query: 219 TSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRFEVHSSGLIEDQLSEAVE 278
T P G+V+F R+ L +P+ W S L R V G IE ++
Sbjct: 611 TEKKPTGLVTFTRQCLQ--------EFPD---WERSQKKLSRLHVTYEGTIESNGQGMLQ 659
Query: 279 VDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSMADNEAIEIVGVERFSSYTGY 338
VDFAN ++GGG G VQEEIRF+I+PELIV L + NE + I G E++S YTGY
Sbjct: 660 VDFANRFVGGGVTGAGLVQEEIRFLINPELIVSRLITEVLDHNECLIITGTEQYSEYTGY 719
Query: 339 ASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQYRPKFLLREINKA-FCGFLY 396
A ++R++ ++ D+ D RR T IVAIDA + Q+ P+ + RE+NKA +CGF
Sbjct: 720 AETYRWARSHEDKTPRDEWQRRYTEIVAIDAFHFRRFLDQFGPEKIRRELNKASYCGF-- 777
Query: 397 GSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEIRDSQNDYHRMEQCNNIGVA 456
+ + P + N +A
Sbjct: 778 ----------------------SRPNVPPQ------------------------NLSAIA 791
Query: 457 TGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF-RLEALHNIDKVAHWILSHRWTV 515
TGNWGCGAFGGD +K +IQ LAA+++ R I Y+TF +E + +I + ++ TV
Sbjct: 792 TGNWGCGAFGGDSRLKALIQILAAAESGRD-IVYFTFGDVELMRDIYSMHTFLCEKNQTV 850
Query: 516 GDLWNMLTEY 525
GD++ +L Y
Sbjct: 851 GDIYRLLLRY 860
>UniRef100_UPI0000318FC5 UPI0000318FC5 UniRef100 entry
Length = 370
Score = 174 bits (442), Expect = 5e-42
Identities = 110/338 (32%), Positives = 178/338 (52%), Gaps = 62/338 (18%)
Query: 196 LYDYYSQKQESKIQCIIHYFQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSI 255
L++ S ++ K++ ++ YF+ IT MP G+V+F RK L PN W +S
Sbjct: 59 LFEGSSSRKIEKLKTLMCYFKSITEQMPTGLVTFTRKRLDKP--------PN---WRSSQ 107
Query: 256 IPLCRFEVHSSGLIEDQLSEAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFL 315
+ L + + G IED ++VDFAN+++GGG G VQEEIRF+I+PELIV LF
Sbjct: 108 MRLTKLHITCEGTIEDDGYGMLQVDFANQFVGGGVTSAGLVQEEIRFLINPELIVSRLFT 167
Query: 316 PSMADNEAIEIVGVERFSSYTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDALCSPG- 374
++ DN+ + I G +++S YTGYA ++++ G + D D RR T IVA+DAL
Sbjct: 168 EALDDNDCLIITGTQQYSKYTGYAQTYQWRGGHHDTTPRDGWQRRCTEIVAMDALQFRNF 227
Query: 375 MRQYRPKFLLREINKAFCGFLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSN 434
+ Q+ P + RE+NKA+CGF ++ ++ ++ L
Sbjct: 228 LEQFHPLKINRELNKAYCGF---ARPEVEKEAL--------------------------- 257
Query: 435 HEIRDSQNDYHRMEQCNNIGVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTF- 493
VATGNWGCG FGGD +K ++Q LAA++A R +AY+TF
Sbjct: 258 ------------------AAVATGNWGCGVFGGDTRLKALLQMLAAAEAGRD-VAYFTFG 298
Query: 494 RLEALHNIDKVAHWILSHRWTVGDLWNMLTEYSMNRSK 531
+ + ++ + ++ + +VG+++ +L +Y + K
Sbjct: 299 DSQLMTDVHNMHSFLTLNNISVGEVYGLLQQYYRSECK 336
>UniRef100_UPI00002B206C UPI00002B206C UniRef100 entry
Length = 264
Score = 174 bits (441), Expect = 6e-42
Identities = 112/321 (34%), Positives = 167/321 (51%), Gaps = 61/321 (19%)
Query: 201 SQKQESKIQCIIHYFQRITSNMPQ--GVVSFERKVLPLEDDYIHISYPNADVWSTSIIPL 258
S+++ K++ I+HYF T + G+V+FER+ L D A WS +
Sbjct: 1 SERKMEKLKAIVHYFHVATDEKTKLDGLVTFERRCLANTD---------ARTWSCCKEEM 51
Query: 259 CRFEVHSSGLIEDQLSEAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVDMLFLPSM 318
+ V S G IE + S ++VDFA+ +LGGG L G +QEEI F++SPELIV LF +
Sbjct: 52 NKLYVSSCGAIETEGSGLLQVDFASSWLGGGVLDSGLLQEEILFLMSPELIVSRLFTEKL 111
Query: 319 ADNEAIEIVGVERFSSYTGYASSFRFSGNYVDERDVDTLGRRKTRIVAIDAL-CSPGMRQ 377
DNE + + G ++FS+Y+GY +FR+ G Y D D RR+ +++A+DAL + G Q
Sbjct: 112 QDNECVIVTGCQQFSTYSGYGDTFRWKGPYADPTGRDGWARRQRQVLAMDALRFTHGRDQ 171
Query: 378 YRPKFLLREINKAFCGFLYGSKHQLYQKILQEKGCPSTLFDAATSTPMETSEGKCSNHEI 437
Y K ++RE+NKA+CGF K E
Sbjct: 172 YSMKLVVRELNKAYCGF------------------------------------KRCEEER 195
Query: 438 RDSQNDYHRMEQCNNIGVATGNWGCGAFGGDPEVKTIIQWLAASQALRPFIAYYTFRLEA 497
R + D +ATG WGCGAF GDP++K +IQ +AA++A R +A++TFR E
Sbjct: 196 RGEEPD-----------IATGKWGCGAFKGDPQLKAVIQLMAAARAGRG-LAFFTFRDEK 243
Query: 498 L-HNIDKVAHWILSHRWTVGD 517
L H + + + + TVG+
Sbjct: 244 LEHGLRQAYRLLRTKGTTVGE 264
>UniRef100_Q75LD2 Putative poly(ADP-ribose) glycohydrolase [Oryza sativa]
Length = 242
Score = 161 bits (407), Expect = 5e-38
Identities = 82/163 (50%), Positives = 105/163 (64%), Gaps = 8/163 (4%)
Query: 99 ESRKWFQEVVPALGILLLRLPSLLEAHYQNADRVLDEDGGLVRTGLRLLDSQEPGIVFLS 158
++R WF V P+L LLLRLP+LLE HY+ A G GLR+L SQ+ G+V LS
Sbjct: 87 QARDWFDHVAPSLARLLLRLPTLLEGHYRAA--------GDEARGLRILSSQDAGLVLLS 138
Query: 159 QELIAALLVCSFFCLFPVNERYGKHLQSINFDELFGSLYDYYSQKQESKIQCIIHYFQRI 218
QEL AALL C+ FCLFP +R L +INFD LF +L Q QE K++C++HYF R+
Sbjct: 139 QELAAALLACALFCLFPTADRAEACLPAINFDSLFAALCYNSRQSQEQKVRCLVHYFDRV 198
Query: 219 TSNMPQGVVSFERKVLPLEDDYIHISYPNADVWSTSIIPLCRF 261
T++ P G VSFERKVLP + I+YP+ D W S +PLC F
Sbjct: 199 TASTPTGSVSFERKVLPRRPESDGITYPDMDTWMKSGVPLCTF 241
>UniRef100_Q613D5 Hypothetical protein CBG16441 [Caenorhabditis briggsae]
Length = 331
Score = 123 bits (308), Expect = 2e-26
Identities = 95/304 (31%), Positives = 133/304 (43%), Gaps = 71/304 (23%)
Query: 193 FGSLYDYYSQKQESKIQCIIHYFQRITSNMPQGVVSFERKVLPLEDDYIHISYPNADVW- 251
F L ++ + K++ + YF RI +N P+G VSF K + E+ + W
Sbjct: 59 FHYLLEHDDELSIEKLRFLFAYFDRIATNPPKGCVSFRLKTVRNEEF--------KEEWI 110
Query: 252 STSIIPLCRFEVHSSGLIEDQLSEAVEVDFANEYLGGGALRRGCVQEEIRFMISPELIVD 311
P+ ++ +IE+ + ++DFANEYLGGG L+ G EEIRF++ PE+IV
Sbjct: 111 KNKCNPMPDVRIYDQMVIEET-ALCTQIDFANEYLGGGVLKSGA--EEIRFLMCPEMIVG 167
Query: 312 MLFLPSMADNEAIEIVGVERFSSYTGYASSFRF---SGNYVDERDV---DTLGRRKTRIV 365
ML M D +AI IVG +S YTGYA+ ++ SG Y + D D GR + V
Sbjct: 168 MLLTDKMDDTQAISIVGAYVYSGYTGYANKLKWRPLSGRYARQNDARLRDKYGRLRVETV 227
Query: 366 AIDAL---CSPGMRQYRPKFLLREINKAFCGFLYGSKHQLYQKILQEKGCPSTLFDAATS 422
AIDA+ Q L RE+ KA+ GF
Sbjct: 228 AIDAMYFKTKAIAEQVTSDSLTREMKKAYTGFA--------------------------- 260
Query: 423 TPMETSEGKCSNHEIRDSQNDYHRMEQCNNIGVATGNWGCGAFGGDPEVKTIIQWLAASQ 482
+ E I + TG WGCGAF G +K +IQ +AAS
Sbjct: 261 -----------------------QSEHFGQIPIVTGFWGCGAFNGHKPLKFLIQLMAASI 297
Query: 483 ALRP 486
A RP
Sbjct: 298 AGRP 301
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.138 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 973,305,249
Number of Sequences: 2790947
Number of extensions: 42204258
Number of successful extensions: 102762
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 29
Number of HSP's that attempted gapping in prelim test: 102615
Number of HSP's gapped (non-prelim): 106
length of query: 558
length of database: 848,049,833
effective HSP length: 133
effective length of query: 425
effective length of database: 476,853,882
effective search space: 202662899850
effective search space used: 202662899850
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0057b.8


