

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0057b.6
(396 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
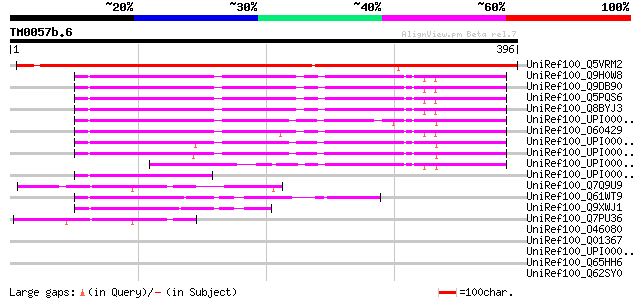
UniRef100_Q5VRM2 Hypothetical protein OSJNBa0019F11.6 [Oryza sat... 402 e-110
UniRef100_Q9H0W8 Hypothetical protein DKFZp564H1322 [Homo sapiens] 137 7e-31
UniRef100_Q9DB90 Mus musculus adult male cerebellum cDNA, RIKEN ... 136 1e-30
UniRef100_Q5PQS6 Hypothetical protein [Rattus norvegicus] 135 2e-30
UniRef100_Q8BYJ3 Mus musculus adult male spinal cord cDNA, RIKEN... 133 8e-30
UniRef100_UPI000024AA87 UPI000024AA87 UniRef100 entry 131 4e-29
UniRef100_O60429 F17127_1 [Homo sapiens] 127 7e-28
UniRef100_UPI00002F9DE4 UPI00002F9DE4 UniRef100 entry 115 3e-24
UniRef100_UPI000001B3D4 UPI000001B3D4 UniRef100 entry 110 7e-23
UniRef100_UPI00001CE803 UPI00001CE803 UniRef100 entry 71 6e-11
UniRef100_UPI0000436C1F UPI0000436C1F UniRef100 entry 69 2e-10
UniRef100_Q7Q9U9 ENSANGP00000021895 [Anopheles gambiae str. PEST] 57 9e-07
UniRef100_Q61WT9 Hypothetical protein CBG04259 [Caenorhabditis b... 57 1e-06
UniRef100_Q9XWJ1 Hypothetical protein Y54E2A.2 [Caenorhabditis e... 54 8e-06
UniRef100_Q7PU36 ENSANGP00000015543 [Anopheles gambiae str. PEST] 50 1e-04
UniRef100_O46080 EG:39E1.3 protein [Drosophila melanogaster] 43 0.014
UniRef100_Q01367 Stage III sporulation protein AA [Bacillus subt... 42 0.030
UniRef100_UPI000033FD31 UPI000033FD31 UniRef100 entry 39 0.20
UniRef100_Q65HH6 SpoIIIAA [Bacillus licheniformis] 39 0.20
UniRef100_Q62SY0 Hypothetical protein spoIIIAA [Bacillus licheni... 39 0.20
>UniRef100_Q5VRM2 Hypothetical protein OSJNBa0019F11.6 [Oryza sativa]
Length = 412
Score = 402 bits (1032), Expect = e-110
Identities = 212/394 (53%), Positives = 276/394 (69%), Gaps = 9/394 (2%)
Query: 6 PSPSPKILLAKPGLVTGGPVAGKFGRGAAGEDDSAQHRSRLPSV-ASLNLLSDSWDFHID 64
P P PKILLAKP L P G G RSR + SL+L+SD+W+ H D
Sbjct: 25 PPPPPKILLAKPPL----PPPSSSGADDDGGGGGGAGRSRQATQPGSLSLVSDAWEVHTD 80
Query: 65 RFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPFATQSEENRAMARHC 124
+ LP+LTEN DF VIGVIGP GVGKSTIMNELYG+D SSPGMLPPFATQ+EE +AMA+HC
Sbjct: 81 KILPYLTENNDFMVIGVIGPTGVGKSTIMNELYGYDGSSPGMLPPFATQTEEIKAMAKHC 140
Query: 125 STGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGESMSAELAHEIMGIQL 184
+ GI+ RIS ER+ILLDTQPVFS SVL +MM+PDGSS + +LSG+ +SA+LAHE+MGIQL
Sbjct: 141 TAGIDIRISNERVILLDTQPVFSPSVLIDMMKPDGSSAIPILSGDPLSADLAHELMGIQL 200
Query: 185 AVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASSHSQSSSSGHEKDNK 244
V LASVC+ILLVVSEG++D MW L+LTVDLLKH IPDPSLL SS +Q ++ DN+
Sbjct: 201 GVFLASVCNILLVVSEGINDLSMWDLILTVDLLKHNIPDPSLLTSSTTQDKE--NKNDNQ 258
Query: 245 LSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVREHAEHKPEEHISS-- 302
+Y+A FVH +L +Q+F+P + L++ L ++FK SSF + P+ SS
Sbjct: 259 SGIEDYIADLCFVHARLREQDFSPSKLMVLKRVLEKHFKSSSFSIGSSGATPQVSDSSVP 318
Query: 303 SMVHDSPMDSNVPSLFAIPFKKKDENPRAQHGSYISALWKLRDQILSMKSPSFTRPVSER 362
S + + SN ++ +P + D + ++ + S L LRDQILS S SF++ ++ER
Sbjct: 319 SSMKIEDLSSNQQDIYLLPLRTPDNSTNFEYRTCPSMLGMLRDQILSRPSRSFSKNLTER 378
Query: 363 EWLKNSAKIWEQVKNSPTMLEYCRTLQQSGMFRR 396
+WL++SAKIW+ VK SP +LEYC+TLQ SG FR+
Sbjct: 379 DWLRSSAKIWDMVKKSPVILEYCKTLQDSGFFRK 412
>UniRef100_Q9H0W8 Hypothetical protein DKFZp564H1322 [Homo sapiens]
Length = 520
Score = 137 bits (344), Expect = 7e-31
Identities = 104/358 (29%), Positives = 171/358 (47%), Gaps = 38/358 (10%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GV+G G GKS +M+ L F
Sbjct: 180 SIKLVDDQMNW-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRTYVF 238
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
QS E + + ++GI+ I+ ERI+ LDTQP+ S S+L ++ D L E
Sbjct: 239 RAQSAEMKERGGNQTSGIDFFITQERIVFLDTQPILSPSILDHLINNDRK-----LPPEY 293
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASS 230
E+ +Q+A L +VCH+++VV + D ++ + T +++K P P S
Sbjct: 294 NLPHTYVEMQSLQIAAFLFTVCHVVIVVQDWFTDLSLYRFLQTAEMVKPSTPSP-----S 348
Query: 231 HSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVRE 290
H SSSSG ++ G EY VF+ K ++F P+ Q+ + Q S +
Sbjct: 349 HESSSSSGSDE-----GTEYYPHLVFLQNKARREDFCPRKLRQMHLMIDQLMAHSHLRYK 403
Query: 291 HAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF---KKKDEN-PRA--------------- 331
+ ++ + D +DS V +LF +PF + + EN PRA
Sbjct: 404 GTLSMLQCNVFPGLPPDF-LDSEV-NLFLVPFMDSEAESENPPRAGPGSSPLFSLLPGYR 461
Query: 332 QHGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 388
H S+ S + KLR Q++SM P + + +E+ W +A+IW+ V+ S + EY R L
Sbjct: 462 GHPSFQSLVSKLRSQVMSMARPQLSHTILTEKNWFHYAARIWDGVRKSSALAEYSRLL 519
>UniRef100_Q9DB90 Mus musculus adult male cerebellum cDNA, RIKEN full-length enriched
library, clone:1500002O20 product:CDNA FLJ12886 FIS,
CLONE NT2RP2004041, WEAKLY SIMILAR TO SYNAPSINS IA AND
IB homolog [Mus musculus]
Length = 520
Score = 136 bits (342), Expect = 1e-30
Identities = 103/358 (28%), Positives = 172/358 (47%), Gaps = 38/358 (10%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GV+G G GKS +M+ L F
Sbjct: 180 SIKLVDDQMNW-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRAYVF 238
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
QS E + + ++GI+ I+ ERI+ LDTQP+ S S+L ++ D L E
Sbjct: 239 RAQSAEMKERGGNQTSGIDFFITQERIVFLDTQPILSPSILDHLINNDRK-----LPPEY 293
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASS 230
E+ +Q+A L +VCH+++VV + D ++ + T +++K P P S
Sbjct: 294 NLPHTYVEMQSLQIAAFLFTVCHVVIVVQDWFTDLSLYRFLQTAEMVKPSTPSP-----S 348
Query: 231 HSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVRE 290
H SSS+G ++ G EY VF+ K ++F P+ Q+ + Q S +
Sbjct: 349 HESSSSAGSDE-----GTEYYPHLVFLQNKARREDFCPRKLRQMHLMIDQLMAHSHLRYK 403
Query: 291 HAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF---KKKDEN-PRA--------------- 331
+ ++ + D +D+ V +LF +PF + ++EN PRA
Sbjct: 404 GTLSMLQCNVFPGLPPDF-LDAEV-NLFLVPFMDSEAENENPPRAGPGSSPLFSLLPGYR 461
Query: 332 QHGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 388
H S+ S + KLR Q++SM P + + +E+ W +A+IW+ VK S + EY R L
Sbjct: 462 GHPSFQSLVSKLRSQVMSMARPQLSHTILTEKNWFHYAARIWDGVKKSSALAEYSRLL 519
>UniRef100_Q5PQS6 Hypothetical protein [Rattus norvegicus]
Length = 520
Score = 135 bits (340), Expect = 2e-30
Identities = 103/358 (28%), Positives = 172/358 (47%), Gaps = 38/358 (10%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GV+G G GKS +M+ L F
Sbjct: 180 SIKLVDDQMNW-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRAYVF 238
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
QS E + + ++GI+ I+ ERI+ LDTQP+ S S+L ++ D L E
Sbjct: 239 RAQSAEMKERGGNQTSGIDFFITQERIVFLDTQPILSPSILDHLINNDRK-----LPPEY 293
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASS 230
E+ +Q+A L +VCH+++VV + D ++ + T +++K P P S
Sbjct: 294 NLPHTYVEMQSLQIAAFLFTVCHVVIVVQDWFTDLSLYRFLQTAEMVKPSTPSP-----S 348
Query: 231 HSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVRE 290
H SS++G ++ G EY VF+ K ++F P+ Q+ + Q S +
Sbjct: 349 HESSSAAGSDE-----GTEYYPHLVFLQNKARREDFCPRKLRQMHLMIDQLMAHSHLRYK 403
Query: 291 HAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF---KKKDEN-PRA--------------- 331
+ +I + D +D+ V +LF +PF + ++EN PRA
Sbjct: 404 GTLSMLQCNIFPGLPPDF-LDAEV-NLFLVPFMDSEAENENPPRAGPGSSPLFSLLPGYR 461
Query: 332 QHGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 388
H S+ S + KLR Q++SM P + + +E+ W +A+IW+ VK S + EY R L
Sbjct: 462 GHPSFQSLVSKLRSQVMSMARPQLSHTILTEKNWFHYAARIWDGVKKSSALAEYSRLL 519
>UniRef100_Q8BYJ3 Mus musculus adult male spinal cord cDNA, RIKEN full-length
enriched library, clone:A330033E23 product:CDNA FLJ12886
FIS, CLONE NT2RP2004041, WEAKLY SIMILAR TO SYNAPSINS IA
AND IB homolog [Mus musculus]
Length = 520
Score = 133 bits (335), Expect = 8e-30
Identities = 102/358 (28%), Positives = 171/358 (47%), Gaps = 38/358 (10%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GV+G G GKS +M+ L F
Sbjct: 180 SIKLVDDQMNW-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRAYVF 238
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
QS E + + ++GI+ I+ ERI+ LDTQP+ S S+L ++ D L E
Sbjct: 239 RAQSAEMKERGGNQTSGIDFFITQERIVFLDTQPILSPSILDHLINNDRK-----LPPEY 293
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASS 230
E+ +Q+A L +VCH+++VV + D ++ + T +++K P P S
Sbjct: 294 NLPHTYVEMQSLQIAAFLFTVCHVVIVVQDWFTDLSLYRFLQTAEMVKPSTPSP-----S 348
Query: 231 HSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVRE 290
H SSS+G ++ G EY VF+ K ++F P+ Q+ + Q S +
Sbjct: 349 HESSSSAGSDE-----GTEYYPHLVFLQNKARREDFCPRKLRQMHLMIDQLMAHSHLRYK 403
Query: 291 HAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF---KKKDEN-PRA--------------- 331
+ ++ + D +D+ V +LF +PF + ++EN PRA
Sbjct: 404 GTLSMLQCNVFPGLPPDF-LDAEV-NLFLVPFMDSEAENENPPRAGPGSSPLFSLLPGYR 461
Query: 332 QHGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 388
H S+ S + KLR Q++SM P + + + + W +A+IW+ VK S + EY R L
Sbjct: 462 GHPSFQSLVSKLRSQVMSMARPQLSHTILTGKNWFHYAARIWDGVKKSSALAEYSRLL 519
>UniRef100_UPI000024AA87 UPI000024AA87 UniRef100 entry
Length = 506
Score = 131 bits (329), Expect = 4e-29
Identities = 102/362 (28%), Positives = 169/362 (46%), Gaps = 47/362 (12%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GVIG G GKSTIM+ L F
Sbjct: 167 SIKLVDDQMNW-CDSAMEYLRDQTDMLVVGVIGLQGTGKSTIMSLLSANSPEEDQRAYVF 225
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
Q++E + A + S+GI+ I+ ER+I LDTQPV S S+L ++ D L E
Sbjct: 226 RAQTQEIKERAGNQSSGIDFYITQERVIFLDTQPVLSPSILDHLINNDRK-----LPPEY 280
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASS 230
E+ +Q+ L +VCH+++V+ + D ++ + T ++LK PS ++S
Sbjct: 281 NLPHTYVEMQSLQITAFLFTVCHVVIVIQDWFTDINLYRFLQTAEMLK-----PSTPSAS 335
Query: 231 HSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVRE 290
H + SSG + G EY VF+ K +EF P+N ++ + + S
Sbjct: 336 HDSTGSSGPD-----DGSEYYPHIVFLQNKARREEFCPRNLKKMHMAVDKLMAHS----- 385
Query: 291 HAEHKPEE------HISSSMVHDSPMDSNVPSLFAIPFKKKDENPRAQ------------ 332
H ++K + +I + D M++ V +LF +P + D
Sbjct: 386 HLKYKGQTLSMLDCNIFPGLSRDY-METEV-NLFLLPLMENDGEDALTRAGSGPPLFSLL 443
Query: 333 -----HGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCR 386
H ++ S + K R QIL+M + + +E+ W +A+IW+ VK S + EY R
Sbjct: 444 PGYRGHPNFSSLVSKFRSQILAMSRSQLSHTILTEKNWFHYAARIWDGVKKSSALSEYSR 503
Query: 387 TL 388
L
Sbjct: 504 LL 505
>UniRef100_O60429 F17127_1 [Homo sapiens]
Length = 528
Score = 127 bits (318), Expect = 7e-28
Identities = 105/377 (27%), Positives = 172/377 (44%), Gaps = 57/377 (15%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GV+G G GKS +M+ L F
Sbjct: 169 SIKLVDDQMNW-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLLSANTPEEDQRTYVF 227
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
QS E + + ++GI+ I+ ERI+ LDTQP+ S S+L ++ D L E
Sbjct: 228 RAQSAEMKERGGNQTSGIDFFITQERIVFLDTQPILSPSILDHLINNDRK-----LPPEY 282
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHL-------------------M 211
E+ +Q+A L +VCH+++VV + D ++ L +
Sbjct: 283 NLPHTYVEMQSLQIAAFLFTVCHVVIVVQDWFTDLSLYRLWDLGCKCKSNSHSPQTPRFL 342
Query: 212 LTVDLLKHGIPDPSLLASSHSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNF 271
T +++K P P SH SSSSG ++ G EY VF+ K ++F P+
Sbjct: 343 QTAEMVKPSTPSP-----SHESSSSSGSDE-----GTEYYPHLVFLQNKARREDFCPRKL 392
Query: 272 VQLRKTLIQYFKPSSFVREHAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF---KKKDEN 328
Q+ + Q S + + ++ + D +DS V +LF +PF + + EN
Sbjct: 393 RQMHLMIDQLMAHSHLRYKGTLSMLQCNVFPGLPPDF-LDSEV-NLFLVPFMDSEAESEN 450
Query: 329 -PRA---------------QHGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKI 371
PRA H S+ S + KLR Q++SM P + + +E+ W +A+I
Sbjct: 451 PPRAGPGSSPLFSLLPGYRGHPSFQSLVSKLRSQVMSMARPQLSHTILTEKNWFHYAARI 510
Query: 372 WEQVKNSPTMLEYCRTL 388
W+ V+ S + EY R L
Sbjct: 511 WDGVRKSSALAEYSRLL 527
>UniRef100_UPI00002F9DE4 UPI00002F9DE4 UniRef100 entry
Length = 511
Score = 115 bits (287), Expect = 3e-24
Identities = 96/362 (26%), Positives = 164/362 (44%), Gaps = 42/362 (11%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GVIG G GKST+M+ L F
Sbjct: 168 SIKLVDDQMNW-CDSAMEYLRDQTDMLVVGVIGLQGAGKSTVMSLLSANTPEEDQRGYVF 226
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQ-----PVFSASVLAEMMRPDGSSTVSV 165
Q++E + + STGI+ I+ ER+I LDTQ P+ S S+L ++ D
Sbjct: 227 RAQAQEIKERGGNQSTGIDFFITQERVIFLDTQASNTHPILSPSILDHLINNDRK----- 281
Query: 166 LSGESMSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPS 225
L E E +Q+A L +VCH++++V + D ++ + T ++LK PS
Sbjct: 282 LPPEYNLPHTYVETQSLQIAAFLFTVCHVVILVQDWFTDVNLYRFLQTAEMLK-----PS 336
Query: 226 LLASSHSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPS 285
++SH + SSG + G EY VFV K +F P+N + + + S
Sbjct: 337 TPSTSHDSTGSSGTD-----DGAEYYPHIVFVQNKARLDDFCPRNLKNMHLVVDKLMAHS 391
Query: 286 SFVREHAEHKPEEHISSSMVHDSPMDSNVPSLFAIPFKKKDENPRAQ------------- 332
+ + +I + D + + V ++F +P + D +
Sbjct: 392 HLKYKGTLSMLDCNIFPGLGPDY-LTTEV-NMFLLPVQDSDSDDGPTKAAPGTYPLFSLL 449
Query: 333 -----HGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCR 386
H ++ + + KLR +IL+M + + +E+ W +A+IW+ VK S + EY R
Sbjct: 450 PGYKGHPTFSTTVSKLRSRILAMPRCQLSHTILTEKNWFHYAARIWDGVKKSSALSEYSR 509
Query: 387 TL 388
L
Sbjct: 510 LL 511
>UniRef100_UPI000001B3D4 UPI000001B3D4 UniRef100 entry
Length = 529
Score = 110 bits (275), Expect = 7e-23
Identities = 95/379 (25%), Positives = 165/379 (43%), Gaps = 59/379 (15%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GVIG G GKST+M+ L F
Sbjct: 169 SIKLVDDQMNW-CDSAMEYLRDQTDMLVVGVIGLQGAGKSTVMSLLSANTPEEDQRAYVF 227
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDT----------------------QPVFSA 148
Q++E + + STGI+ I+ ER+I LDT QP+ S
Sbjct: 228 RAQTQEIKERGGNQSTGIDFFITQERVIFLDTAKHIAITTGILKKRLINTFYFLQPILSP 287
Query: 149 SVLAEMMRPDGSSTVSVLSGESMSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMW 208
S+L ++ D L E E+ +Q+A L +VCH+++VV + D ++
Sbjct: 288 SILDHLINNDRK-----LPPEYNLPHTYVEMQSLQIAAFLFTVCHVVIVVQDWFTDLNLY 342
Query: 209 HLMLTVDLLKHGIPDPSLLASSHSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTP 268
+ T ++ K PS ++SH + SSG + G EY VF+ K +F P
Sbjct: 343 RFLQTAEMCK-----PSTPSASHDSTGSSGSD-----DGAEYYPHIVFIQNKAGLDDFCP 392
Query: 269 KNFVQLRKTLIQYFKPSSFVREHAEHKPEEHISSSMVHDSPMDSNVPSLFAIPFKKKDEN 328
+N + + + S + + +I + D + + V ++F +P ++ D +
Sbjct: 393 RNLKNMHMVVDKLMAHSHLKYKGTLSMLDCNIFPGLGQDY-LTTEV-NMFLLPVQENDSD 450
Query: 329 PRAQ------------------HGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSA 369
H ++ + + KLR +IL+M + + +E+ W +A
Sbjct: 451 DGLTKAAPGTYPLFSLLPGYKGHPAFSTLVHKLRSRILAMPRCQLSHTILTEKNWFHYAA 510
Query: 370 KIWEQVKNSPTMLEYCRTL 388
+IW+ VK S + EY R L
Sbjct: 511 RIWDGVKKSSALSEYSRLL 529
>UniRef100_UPI00001CE803 UPI00001CE803 UniRef100 entry
Length = 624
Score = 70.9 bits (172), Expect = 6e-11
Identities = 76/300 (25%), Positives = 125/300 (41%), Gaps = 49/300 (16%)
Query: 110 FATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGE 169
F QS E + + ++GI+ I+ ERI+ LDTQ + V ++ G
Sbjct: 352 FRAQSAEMKERGGNQTSGIDFFITQERIVFLDTQCDWLLRVSPTPCAQHIVGSLLAEQGS 411
Query: 170 SMSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLML-TVDLLKHGIPDPSLLA 228
S L E L+ + +H W L T +++K P PS
Sbjct: 412 DDSGNLPSE--------------EGLMQEFKDLHTS--WARFLQTAEMVKPSTPSPS--- 452
Query: 229 SSHSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFV 288
H SS++G ++ G EY VF+ K ++F P+ Q+ + Q S
Sbjct: 453 --HESSSAAGSDE-----GTEYYPHLVFLQNKARREDFCPRKLRQMHLMIDQLMAHSHLR 505
Query: 289 REHAEHKPEEHISSSMVHDSPMDSNVPSLFAIPF---KKKDENPRAQ------------- 332
+ + +I + D +D+ V +LF +PF + ++ENP
Sbjct: 506 YKGTLSMLQCNIFPGLPPDF-LDAEV-NLFLVPFMDSEAENENPPRAGPGSSPLFSLLPG 563
Query: 333 ---HGSYISALWKLRDQILSMKSPSFTRPV-SEREWLKNSAKIWEQVKNSPTMLEYCRTL 388
H S+ S + KLR Q++SM P + + +E+ W +A+IW+ VK S + EY R L
Sbjct: 564 YRGHPSFQSLVSKLRSQVMSMARPQLSHTILTEKNWFHYAARIWDGVKKSSALAEYSRLL 623
Score = 34.3 bits (77), Expect = 6.3
Identities = 17/46 (36%), Positives = 28/46 (59%), Gaps = 1/46 (2%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNEL 96
S+ L+ D ++ D + +L + TD V+GV+G G GKS +M+ L
Sbjct: 201 SIKLVDDQMNW-CDSAIEYLLDQTDVLVVGVLGLQGTGKSMVMSLL 245
>UniRef100_UPI0000436C1F UPI0000436C1F UniRef100 entry
Length = 241
Score = 69.3 bits (168), Expect = 2e-10
Identities = 41/108 (37%), Positives = 61/108 (55%), Gaps = 1/108 (0%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+ D ++ D + +L + TD V+GVIG G GKSTIM+ L F
Sbjct: 86 SIKLVDDQMNW-CDSAMEYLRDQTDMLVVGVIGLQGTGKSTIMSLLSANSPEEDQRAYVF 144
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPD 158
Q++E + A + S+GI+ I+ ER+I LDTQPV S S+L ++ D
Sbjct: 145 RAQTQEIKERAGNQSSGIDFYITQERVIFLDTQPVLSPSILDHLINND 192
>UniRef100_Q7Q9U9 ENSANGP00000021895 [Anopheles gambiae str. PEST]
Length = 384
Score = 57.0 bits (136), Expect = 9e-07
Identities = 63/218 (28%), Positives = 92/218 (41%), Gaps = 41/218 (18%)
Query: 7 SPSPKILLAKPGLVTGGPVAGKFGRGAAGEDDSAQHRSRLPSVASLNLLSDSWDFHIDRF 66
S +P+I + G + + K A D S P SL LL + + R
Sbjct: 37 STTPQIAVTSKGSESRHAASAKSLPATAAPDPY----SYTPMEGSLVLLKNHNAINY-RT 91
Query: 67 LPFLTE-NTDFTVIGVIGPPGVGKSTIMN----ELYGFDSSSPGMLPPFATQSEENRAMA 121
L FL E N D+ VIG IG PGVGKST++N LY D S F S N A
Sbjct: 92 LDFLHETNQDYVVIGTIGMPGVGKSTVLNLLNTNLYAPDQSDNRKETIFPVHSTLNVAGE 151
Query: 122 RHCSTGIEPRISAERIILLD-TQPVFSASVLAEMMRPDGSSTVSVLSGESMSAELAHEIM 180
+ I+ +R+ILLD ++PVF G + + +E
Sbjct: 152 NE----VRMHITEDRLILLDCSEPVF---------------------GHARKDFIQNEQD 186
Query: 181 GIQLAVLLASVCHILLVVSEGVHD-----DCMWHLMLT 213
++ ++L +CH++LVV E ++ MW M+T
Sbjct: 187 ELKKLMILLRICHVMLVVQEEYYNIRLMRTLMWAEMMT 224
>UniRef100_Q61WT9 Hypothetical protein CBG04259 [Caenorhabditis briggsae]
Length = 314
Score = 56.6 bits (135), Expect = 1e-06
Identities = 62/239 (25%), Positives = 102/239 (41%), Gaps = 30/239 (12%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+D + D LT + +F VI IGP G GKSTI++ L G +S F
Sbjct: 4 SVRFLNDIGEI-TDSISDLLTNSPNFNVISAIGPQGAGKSTILSMLAGNNSRQMYREYVF 62
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
S E +RH +T I+ ++ + I LD QP+ S S++ + + G + +
Sbjct: 63 RPVSREANEQSRHQTTQIDIYVTNHQ-IFLDCQPMHSFSIMEGLPKLRGG---RLDEATA 118
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHDDCMWHLMLTVDLLKHGIPDPSLLASS 230
MS L +L L + H +LV+SE D + + T + ++ +
Sbjct: 119 MSDTL-------RLTAFLLFISHTVLVISETHFDKTIIDTIRTSEQIRPWL--------- 162
Query: 231 HSQSSSSGHEKDNKLSGREYMATPVFVHTKLHDQEFTPKNFVQLRKTLIQYFKPSSFVR 289
H+ KLS E M VF+ TK + P + + L F+ S +++
Sbjct: 163 --------HQFRPKLS-IERMTNLVFIKTKASSIDTAPSVIRERAQLLRLAFRDSKWLK 212
>UniRef100_Q9XWJ1 Hypothetical protein Y54E2A.2 [Caenorhabditis elegans]
Length = 385
Score = 53.9 bits (128), Expect = 8e-06
Identities = 48/154 (31%), Positives = 70/154 (45%), Gaps = 12/154 (7%)
Query: 51 SLNLLSDSWDFHIDRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPF 110
S+ L+D + D LT + +F VI IGP G GKST+++ L G +S F
Sbjct: 62 SVRFLTDFGEIS-DAISDLLTSSPNFNVISAIGPQGAGKSTLLSMLAGNNSRQMYREYVF 120
Query: 111 ATQSEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGES 170
S E +RH + I+ I I LD QP++S S++ + + G +
Sbjct: 121 RPVSREANEQSRHQTIQIDIYI-VNHQIFLDCQPMYSFSIMEGLPKVRGG---RFDDSTA 176
Query: 171 MSAELAHEIMGIQLAVLLASVCHILLVVSEGVHD 204
MS L +L L V H +LVVSE +D
Sbjct: 177 MSDTL-------RLTAFLLYVSHTVLVVSETHYD 203
>UniRef100_Q7PU36 ENSANGP00000015543 [Anopheles gambiae str. PEST]
Length = 200
Score = 49.7 bits (117), Expect = 1e-04
Identities = 51/153 (33%), Positives = 68/153 (44%), Gaps = 15/153 (9%)
Query: 4 SEPSPSPKILLAKPGLVTGGPVAGKFGRGAAGEDDSAQHR----SRLPSVASLNLLSDSW 59
S P +P IL + + G + A+ + A S P SL LL +
Sbjct: 26 STPRRTPTILASTTPQIAGTSKGSESRHAASAKSLPATAAPDPYSYTPMEGSLVLLKNHN 85
Query: 60 DFHIDRFLPFLTE-NTDFTVIGVIGPPGVGKSTIMN----ELYGFDSSSPGMLPPFATQS 114
+ R L FL E N D+ VIG IG PGVGKST++N LY D S F S
Sbjct: 86 AINY-RTLDFLHETNQDYVVIGTIGMPGVGKSTVLNLLNTNLYAPDQSDNRKETIFPVHS 144
Query: 115 EENRAMARHCSTGIEPRISAERIILLD-TQPVF 146
N A + I+ +R+ILLD ++PVF
Sbjct: 145 TINVAGENE----VRMHITEDRLILLDCSEPVF 173
>UniRef100_O46080 EG:39E1.3 protein [Drosophila melanogaster]
Length = 487
Score = 43.1 bits (100), Expect = 0.014
Identities = 43/149 (28%), Positives = 65/149 (42%), Gaps = 37/149 (24%)
Query: 69 FLTENTDFTVIGVIGPPGVGKSTIMNEL-----YGFDSSSPGMLPP-----FATQ----- 113
F NTDFTVIGV+G GKST++N L +D P FAT+
Sbjct: 187 FHKTNTDFTVIGVLGGQSSGKSTLLNLLAAERSLDYDYYQHLFSPEADECIFATRHKLKP 246
Query: 114 SEENRAMARHCSTGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGESMSA 173
+ +++ R + ++ I+ ER ILLDT P + P G +
Sbjct: 247 NNGQKSILRPRTETLQFFITRERHILLDTPP----------LMPVGKDSDH--------- 287
Query: 174 ELAHEIMGIQLAVLLASVCHILLVVSEGV 202
++ + L SVCHIL++V +G+
Sbjct: 288 ---QDLYSLGTMAQLLSVCHILILVIDGL 313
>UniRef100_Q01367 Stage III sporulation protein AA [Bacillus subtilis]
Length = 307
Score = 42.0 bits (97), Expect = 0.030
Identities = 29/100 (29%), Positives = 47/100 (47%), Gaps = 2/100 (2%)
Query: 46 LPSVASLNLLSDSWDFHI-DRFLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSP 104
L +AS N+ I + LP+L +N+ + +IGPP GK+T++ +L S+
Sbjct: 106 LRDIASFNIRIARQKLGIAEPLLPYLYQNSWLNTL-IIGPPQTGKTTLLRDLARLSSTGK 164
Query: 105 GMLPPFATQSEENRAMARHCSTGIEPRISAERIILLDTQP 144
+ P T + R+ C GI +RI +LD P
Sbjct: 165 KNMLPVKTGIVDERSEIAGCLRGIPQHQFGQRIDVLDACP 204
>UniRef100_UPI000033FD31 UPI000033FD31 UniRef100 entry
Length = 282
Score = 39.3 bits (90), Expect = 0.20
Identities = 28/88 (31%), Positives = 45/88 (50%), Gaps = 12/88 (13%)
Query: 36 EDDSAQHRSRLPSVASLNL--LSDSWDFHIDRFLPFLTE---------NTDFTVIGVIGP 84
+DD+ + ++ S N+ +S S + ID FL FL++ DF+ +G+ G
Sbjct: 112 KDDNFKQDKKIDSFGIKNIFFISCSHNIGIDAFLDFLSKYDAKDNKLIKYDFS-LGLFGK 170
Query: 85 PGVGKSTIMNELYGFDSSSPGMLPPFAT 112
VGKST++N+L GF+ S P T
Sbjct: 171 TNVGKSTLLNKLVGFERSIVSNQPKTTT 198
>UniRef100_Q65HH6 SpoIIIAA [Bacillus licheniformis]
Length = 309
Score = 39.3 bits (90), Expect = 0.20
Identities = 39/140 (27%), Positives = 58/140 (40%), Gaps = 18/140 (12%)
Query: 66 FLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPFATQSEENRAMARHCS 125
++P L +N+ + +IGPP GK+T++ +L SS G P + R+ C
Sbjct: 129 YVPHLFQNSWLNTL-IIGPPQTGKTTLLRDLARLISSGSGNAPAKKVGIVDERSEIAGCV 187
Query: 126 TGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGESMSAE--LAHEIMGIQ 183
GI +R +LD P A L M+R SMS E +A EI ++
Sbjct: 188 NGIPQYRLGDRADILDACP--KAEGLMMMIR-------------SMSPEVMIADEIGRME 232
Query: 184 LAVLLASVCHILLVVSEGVH 203
A L H + V H
Sbjct: 233 DAEALLEAVHAGVTVIVSAH 252
>UniRef100_Q62SY0 Hypothetical protein spoIIIAA [Bacillus licheniformis]
Length = 307
Score = 39.3 bits (90), Expect = 0.20
Identities = 39/140 (27%), Positives = 58/140 (40%), Gaps = 18/140 (12%)
Query: 66 FLPFLTENTDFTVIGVIGPPGVGKSTIMNELYGFDSSSPGMLPPFATQSEENRAMARHCS 125
++P L +N+ + +IGPP GK+T++ +L SS G P + R+ C
Sbjct: 127 YVPHLFQNSWLNTL-IIGPPQTGKTTLLRDLARLISSGSGNAPAKKVGIVDERSEIAGCV 185
Query: 126 TGIEPRISAERIILLDTQPVFSASVLAEMMRPDGSSTVSVLSGESMSAE--LAHEIMGIQ 183
GI +R +LD P A L M+R SMS E +A EI ++
Sbjct: 186 NGIPQYRLGDRADILDACP--KAEGLMMMIR-------------SMSPEVMIADEIGRME 230
Query: 184 LAVLLASVCHILLVVSEGVH 203
A L H + V H
Sbjct: 231 DAEALLEAVHAGVTVIVSAH 250
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 686,468,489
Number of Sequences: 2790947
Number of extensions: 29126563
Number of successful extensions: 106527
Number of sequences better than 10.0: 183
Number of HSP's better than 10.0 without gapping: 71
Number of HSP's successfully gapped in prelim test: 113
Number of HSP's that attempted gapping in prelim test: 106381
Number of HSP's gapped (non-prelim): 212
length of query: 396
length of database: 848,049,833
effective HSP length: 129
effective length of query: 267
effective length of database: 488,017,670
effective search space: 130300717890
effective search space used: 130300717890
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0057b.6


