

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0057b.10
(306 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
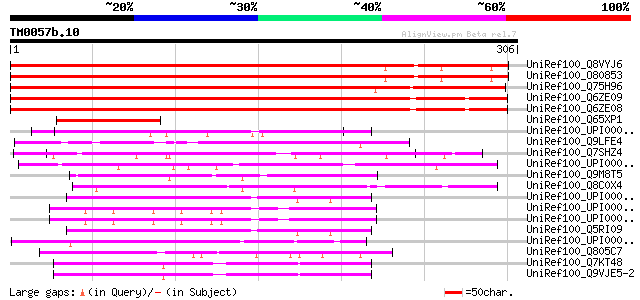
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8VYJ6 At2g30880/F7F1.9 [Arabidopsis thaliana] 340 4e-92
UniRef100_O80853 Hypothetical protein At2g30880 [Arabidopsis tha... 340 4e-92
UniRef100_Q75H96 Putative PH domain containing protein [Oryza sa... 314 2e-84
UniRef100_Q6ZE09 Hypothetical protein P0495H05.40-2 [Oryza sativa] 280 4e-74
UniRef100_Q6ZE08 Hypothetical protein P0495H05.40-1 [Oryza sativa] 280 4e-74
UniRef100_Q65XP1 Hypothetical protein OSJNBa0072C16.20 [Oryza sa... 81 3e-14
UniRef100_UPI000034DD7C UPI000034DD7C UniRef100 entry 62 2e-08
UniRef100_Q9LFE4 Hypothetical protein F5E19_70 [Arabidopsis thal... 62 2e-08
UniRef100_Q7SHZ4 Hypothetical protein [Neurospora crassa] 60 6e-08
UniRef100_UPI0000436284 UPI0000436284 UniRef100 entry 59 2e-07
UniRef100_Q9M8T5 F13E7.12 protein [Arabidopsis thaliana] 59 2e-07
UniRef100_Q8C0X4 Mus musculus adult male testis cDNA, RIKEN full... 58 3e-07
UniRef100_UPI000024976E UPI000024976E UniRef100 entry 57 5e-07
UniRef100_UPI00002B91B8 UPI00002B91B8 UniRef100 entry 57 5e-07
UniRef100_UPI0000299C6A UPI0000299C6A UniRef100 entry 57 5e-07
UniRef100_Q5RI09 Novel protein similar to translocated promoter ... 57 5e-07
UniRef100_UPI000042F053 UPI000042F053 UniRef100 entry 57 6e-07
UniRef100_Q805C7 Tropomyosin2 [Fugu rubripes] 57 8e-07
UniRef100_Q7KT48 CG5020-PD, isoform D [Drosophila melanogaster] 57 8e-07
UniRef100_Q9VJE5-2 Splice isoform B of Q9VJE5 [Drosophila melano... 57 8e-07
>UniRef100_Q8VYJ6 At2g30880/F7F1.9 [Arabidopsis thaliana]
Length = 504
Score = 340 bits (871), Expect = 4e-92
Identities = 193/310 (62%), Positives = 238/310 (76%), Gaps = 11/310 (3%)
Query: 1 MQISLRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLS 60
MQISLRN L + NK DGP+DDLTIMKETL VKDEEL N AR+LR+RDS I++IA+KLS
Sbjct: 180 MQISLRNALKITPNKPIDGPLDDLTIMKETLRVKDEELHNLARELRSRDSMIKEIADKLS 239
Query: 61 ETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKE 120
ETAEAA AAASAAHT+DEQ + C E E L DS++Q E + KLKE E K +LSKEK+
Sbjct: 240 ETAEAAVAAASAAHTMDEQRKIVCVEFERLTTDSQRQQEATKLKLKELEEKTFTLSKEKD 299
Query: 121 QLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGES 180
QL K+R+AA QEA+MWRSELGKARE VILEGA+ RAEEKVRVAEA+ EAK KEA Q E+
Sbjct: 300 QLVKERDAALQEAHMWRSELGKARERVVILEGAVVRAEEKVRVAEASGEAKSKEASQREA 359
Query: 181 AAVSEKQELLAYVDKLKAQLQRQHIDSTEVF-EKTESCSDTKHVDL---TEENVDKACLS 236
A +EKQELLAYV+ L+ QLQRQ +++ +V EKTES + + + TE+NVDKACLS
Sbjct: 360 TAWTEKQELLAYVNMLQTQLQRQQLETKQVCEEKTESTNGEASLPMTKETEKNVDKACLS 419
Query: 237 VSRSVPDPAAAECVVQMPTDQVI--IQPVGDNEWSDIQATEARVADVREVASESE---VS 291
+SR+ P E VV M +QV+ PVG+NEW+DIQATEARV+DVRE+++E+E +
Sbjct: 420 ISRTASIP--GENVVHMSEEQVVNAQPPVGENEWNDIQATEARVSDVREISAETERDRRN 477
Query: 292 SLDIPVISQQ 301
SLDIPV+S +
Sbjct: 478 SLDIPVVSPE 487
>UniRef100_O80853 Hypothetical protein At2g30880 [Arabidopsis thaliana]
Length = 534
Score = 340 bits (871), Expect = 4e-92
Identities = 193/310 (62%), Positives = 238/310 (76%), Gaps = 11/310 (3%)
Query: 1 MQISLRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLS 60
MQISLRN L + NK DGP+DDLTIMKETL VKDEEL N AR+LR+RDS I++IA+KLS
Sbjct: 210 MQISLRNALKITPNKPIDGPLDDLTIMKETLRVKDEELHNLARELRSRDSMIKEIADKLS 269
Query: 61 ETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKE 120
ETAEAA AAASAAHT+DEQ + C E E L DS++Q E + KLKE E K +LSKEK+
Sbjct: 270 ETAEAAVAAASAAHTMDEQRKIVCVEFERLTTDSQRQQEATKLKLKELEEKTFTLSKEKD 329
Query: 121 QLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGES 180
QL K+R+AA QEA+MWRSELGKARE VILEGA+ RAEEKVRVAEA+ EAK KEA Q E+
Sbjct: 330 QLVKERDAALQEAHMWRSELGKARERVVILEGAVVRAEEKVRVAEASGEAKSKEASQREA 389
Query: 181 AAVSEKQELLAYVDKLKAQLQRQHIDSTEVF-EKTESCSDTKHVDL---TEENVDKACLS 236
A +EKQELLAYV+ L+ QLQRQ +++ +V EKTES + + + TE+NVDKACLS
Sbjct: 390 TAWTEKQELLAYVNMLQTQLQRQQLETKQVCEEKTESTNGEASLPMTKETEKNVDKACLS 449
Query: 237 VSRSVPDPAAAECVVQMPTDQVI--IQPVGDNEWSDIQATEARVADVREVASESE---VS 291
+SR+ P E VV M +QV+ PVG+NEW+DIQATEARV+DVRE+++E+E +
Sbjct: 450 ISRTASIP--GENVVHMSEEQVVNAQPPVGENEWNDIQATEARVSDVREISAETERDRRN 507
Query: 292 SLDIPVISQQ 301
SLDIPV+S +
Sbjct: 508 SLDIPVVSPE 517
>UniRef100_Q75H96 Putative PH domain containing protein [Oryza sativa]
Length = 500
Score = 314 bits (804), Expect = 2e-84
Identities = 172/307 (56%), Positives = 227/307 (73%), Gaps = 9/307 (2%)
Query: 1 MQISLRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLS 60
M++S+R LG+ N +G +DDLTIMKETL VKDEELQN A+D+R RD+TI++IA KL+
Sbjct: 180 MKVSMRAALGLGANNPKEGQLDDLTIMKETLRVKDEELQNLAKDIRARDATIKEIANKLT 239
Query: 61 ETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKE 120
ETAEAAEAAASAAHT+DEQ R C EIE L++ E+Q+E S KL++ E K+ISLSKEK+
Sbjct: 240 ETAEAAEAAASAAHTMDEQRRLLCSEIERLRQAMERQMEQSMLKLRQSEEKVISLSKEKD 299
Query: 121 QLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGES 180
QL K+R+AA QEA+MWR+ELGKARE VI E +ARA+EKVR +EA+A A+IKEA +
Sbjct: 300 QLLKERDAALQEAHMWRTELGKAREQAVIQEATIARADEKVRASEADAAARIKEAAEKLH 359
Query: 181 AAVSEKQELLAYVDKLKAQLQRQHIDSTEVF-EKTESCSD-------TKHVDLTEENVDK 232
A EK+ELL+ V L++Q+QR+ + +V E++ESCS TKHVD ++++VDK
Sbjct: 360 AVEKEKEELLSLVGILQSQVQREQSSTKQVCEERSESCSGTDNSPPLTKHVDASDDDVDK 419
Query: 233 ACLSVSRSVPDPAAAECVVQMPTDQVIIQPVGDNEWSDIQATEARVADVREVASESEVSS 292
AC+S SRSV + VQ+ D V I+P+GD EW Q +EA +ADVREV+ ESE S
Sbjct: 420 ACVSDSRSVL-VSNDNTEVQLAVDGVDIRPIGDAEWGSFQQSEALIADVREVSPESEGGS 478
Query: 293 LDIPVIS 299
LDIPV++
Sbjct: 479 LDIPVVN 485
>UniRef100_Q6ZE09 Hypothetical protein P0495H05.40-2 [Oryza sativa]
Length = 484
Score = 280 bits (715), Expect = 4e-74
Identities = 157/302 (51%), Positives = 211/302 (68%), Gaps = 7/302 (2%)
Query: 1 MQISLRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLS 60
M++SLR LG TNK + G +DDLTIM ETL VKD+EL +D+R RD+TIR+I +KL
Sbjct: 179 MKVSLRAALGSTTNKLSKGQLDDLTIMMETLRVKDDELHQLLQDIRARDATIREITDKLQ 238
Query: 61 ETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKE 120
ETAEAAE AASAAH+IDEQ R E+E LK+D EKQ+E S +L+E E K LS+E+E
Sbjct: 239 ETAEAAETAASAAHSIDEQRRFLSSELERLKQDQEKQIEFSLLRLRESEEKAKLLSEERE 298
Query: 121 QLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGES 180
L K+R++A QEA MWRSELGKAR + VILE A+ RAEEK RV+ A+A+ +I +A
Sbjct: 299 HLLKERDSALQEAQMWRSELGKARGNAVILEAAVVRAEEKARVSAADADMRINDAASRLD 358
Query: 181 AAVSEKQELLAYVDKLKAQLQRQHIDSTEVF-EKTESCS-DTKHVDLTEENVDKACLSVS 238
+A EK+EL+A VD L+ Q++ Q + +V E++E CS +KHVD+ ++NVDKACLS +
Sbjct: 359 SATKEKEELVALVDALQLQIRSQDTSTKQVCEERSELCSTSSKHVDMEDDNVDKACLSDT 418
Query: 239 RSVPDPAAAECVVQMPTDQVIIQPVGDNEWSDIQATEARVADVREVASESEVSSLDIPVI 298
+P E +V + D V I +G EW++ ++E V+DVREV +E E +SLDIPV
Sbjct: 419 DPIP---ITENIVDLDDDGVDIPTIGVTEWNNPHSSE--VSDVREVTTEPEDNSLDIPVD 473
Query: 299 SQ 300
SQ
Sbjct: 474 SQ 475
>UniRef100_Q6ZE08 Hypothetical protein P0495H05.40-1 [Oryza sativa]
Length = 387
Score = 280 bits (715), Expect = 4e-74
Identities = 157/302 (51%), Positives = 211/302 (68%), Gaps = 7/302 (2%)
Query: 1 MQISLRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLS 60
M++SLR LG TNK + G +DDLTIM ETL VKD+EL +D+R RD+TIR+I +KL
Sbjct: 82 MKVSLRAALGSTTNKLSKGQLDDLTIMMETLRVKDDELHQLLQDIRARDATIREITDKLQ 141
Query: 61 ETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKE 120
ETAEAAE AASAAH+IDEQ R E+E LK+D EKQ+E S +L+E E K LS+E+E
Sbjct: 142 ETAEAAETAASAAHSIDEQRRFLSSELERLKQDQEKQIEFSLLRLRESEEKAKLLSEERE 201
Query: 121 QLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGES 180
L K+R++A QEA MWRSELGKAR + VILE A+ RAEEK RV+ A+A+ +I +A
Sbjct: 202 HLLKERDSALQEAQMWRSELGKARGNAVILEAAVVRAEEKARVSAADADMRINDAASRLD 261
Query: 181 AAVSEKQELLAYVDKLKAQLQRQHIDSTEVF-EKTESCS-DTKHVDLTEENVDKACLSVS 238
+A EK+EL+A VD L+ Q++ Q + +V E++E CS +KHVD+ ++NVDKACLS +
Sbjct: 262 SATKEKEELVALVDALQLQIRSQDTSTKQVCEERSELCSTSSKHVDMEDDNVDKACLSDT 321
Query: 239 RSVPDPAAAECVVQMPTDQVIIQPVGDNEWSDIQATEARVADVREVASESEVSSLDIPVI 298
+P E +V + D V I +G EW++ ++E V+DVREV +E E +SLDIPV
Sbjct: 322 DPIP---ITENIVDLDDDGVDIPTIGVTEWNNPHSSE--VSDVREVTTEPEDNSLDIPVD 376
Query: 299 SQ 300
SQ
Sbjct: 377 SQ 378
>UniRef100_Q65XP1 Hypothetical protein OSJNBa0072C16.20 [Oryza sativa]
Length = 313
Score = 81.3 bits (199), Expect = 3e-14
Identities = 40/63 (63%), Positives = 51/63 (80%)
Query: 29 ETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIE 88
ETL VKD ELQN A ++R RD+TI++IA+KL++TA+AAEAAASA HT+DE R C EIE
Sbjct: 236 ETLQVKDGELQNLANNIRARDATIKEIADKLTQTAQAAEAAASATHTMDEHRRLLCSEIE 295
Query: 89 SLK 91
L+
Sbjct: 296 RLR 298
>UniRef100_UPI000034DD7C UPI000034DD7C UniRef100 entry
Length = 4089
Score = 62.0 bits (149), Expect = 2e-08
Identities = 51/202 (25%), Positives = 92/202 (45%), Gaps = 15/202 (7%)
Query: 28 KETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEI 87
+ETL E Q +L R+ TI+ + E++ AA S A ++ +I
Sbjct: 1281 EETLRNVKRECQQHKEELNVRNETIKSLTEQMGLLRGAAGELESGAELRQKELIQLHSQI 1340
Query: 88 ESLKKD----------SEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWR 137
++L +D +EK+L L +Q+L + + ++ ++ LS + + TQ+ R
Sbjct: 1341 QALTEDKQQLQAARRTTEKELALQSQRLCDLQGQLKEALEQNSSLSAELGSLTQKNRALR 1400
Query: 138 SELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLK 197
+L + ES LA ++ + E +I E Q V +K+EL + VD+LK
Sbjct: 1401 EDLAQKLES----VSELAADRSALQQQLSGLEEQIAEDRQATDRLVKQKEELGSTVDELK 1456
Query: 198 AQLQRQH-IDSTEVFEKTESCS 218
L+ H ++ + EKT C+
Sbjct: 1457 KVLEESHQSNAAGLLEKTNECA 1478
Score = 47.0 bits (110), Expect = 6e-04
Identities = 48/212 (22%), Positives = 92/212 (42%), Gaps = 24/212 (11%)
Query: 14 NKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAA 73
N+ G D ++ +L+ K+ L + L S ++ K + EA++ A
Sbjct: 646 NQQLKGLTDTQESLESSLVEKETALARTCQQLELVSSLQEALSLKELQLREASDKLLQAE 705
Query: 74 HTIDEQWRNA------CEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKE----KEQLS 123
H++D W+ C E+++ D ++L + +K ++ EV I SL +E E+L
Sbjct: 706 HSLDSIWQKCSGSEKQCSELKAEVADLTQKLGVLKEKTQKQEVTIESLQREVDQTNEELD 765
Query: 124 KQREAATQE-ANMWRSELGKARE----SGVILE---------GALARAEEKVRVAEANAE 169
K A +E A + G RE V+LE G ++ E++ + + +
Sbjct: 766 KLNTACLEERAQLIHDLQGCEREIDSLKDVLLEKDREISALSGHVSECTEQLSLLKHELK 825
Query: 170 AKIKEAVQGESAAVSEKQELLAYVDKLKAQLQ 201
K + +Q E+A ++ELL + ++ Q
Sbjct: 826 LKEQNLIQVENALSKAERELLLLRESRSSEQQ 857
Score = 44.3 bits (103), Expect = 0.004
Identities = 56/257 (21%), Positives = 113/257 (43%), Gaps = 16/257 (6%)
Query: 1 MQISLRNTLGMMTNKTTD-GPMDDLT-IMKETLLVKDEELQNFARDLRTRDSTIRDIAEK 58
+Q+ L LG M +TT+ G +L ++++ + ++ +L +++ I+++ E
Sbjct: 3150 LQLQLEEALGRMETQTTELGAQVELNNLLQKEKQNLSQHMEAMQTELGKKEALIQELQEV 3209
Query: 59 LSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKE 118
+S ++ + +++ EE+E++++ S+K + KEY ++ + E
Sbjct: 3210 VSRHSQETVSLNEKVRILEDDKSLLQEELENVQETSDK-----VKNEKEYLETVLLQNSE 3264
Query: 119 KEQLSKQREAATQEANM-WRSELGKARESGVILEGALARAEEKVRVAEANAEA--KIKEA 175
K + A Q N+ S+L + + + + EE++R+ E ++
Sbjct: 3265 KVDELTESVAVLQSQNLELNSQLAASSHTNHRVRQE--KEEEQLRLVRELEEKLRAVQRG 3322
Query: 176 VQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTKH--VDLTEENVDKA 233
QG E QELL + QLQ+ I EV + ES S + V+ + ++K+
Sbjct: 3323 SQGSKTINKELQELLKEKHQEINQLQQNCIRYQEVILQLESSSKSSQAAVEQLQRELEKS 3382
Query: 234 C--LSVSRSVPDPAAAE 248
LS R A AE
Sbjct: 3383 SEQLSAVRQKCSRAEAE 3399
Score = 43.1 bits (100), Expect = 0.009
Identities = 55/271 (20%), Positives = 110/271 (40%), Gaps = 52/271 (19%)
Query: 9 LGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEA 68
L +K + +L KE + + + ++ +T + + T +A
Sbjct: 950 LQQQLSKKEESLEKELKASKEERNRLHSQAEEYRKEAQTVSQQLEEQKRSQGITRGEMKA 1009
Query: 69 AASAAHTIDEQWRNACEEIESLK-----KDSEKQ-----LELSAQKLKEYEVKIISLSKE 118
A A ++ Q R A +E + L+ +DSEK+ L+ A+ + + + SL E
Sbjct: 1010 TAETAAALEAQLREAEKERQRLEAELKTRDSEKEKLSSDLQSKAENISNLQNLLNSLKSE 1069
Query: 119 KEQLSKQREAATQEANMWRSELGKARESGVI--------------LEGALARAEEKV--- 161
K+QL ++ EA T+E ++ + ++ + + L AR ++++
Sbjct: 1070 KQQLQEELEALTEELDLQKEKVRQLSQEAASALDSRTSYQNQAQQLSAEAARLQQELDHL 1129
Query: 162 --RVAEANAEAK-------IKEAVQGESAAV-----SEKQELLAYVDKLKA--------- 198
++E EA+ + EA E+AAV EK+EL +L +
Sbjct: 1130 QRTLSELGCEAESRRDRVSVLEAQVSENAAVIKALREEKEELTLQKQELSSEHVQGLAST 1189
Query: 199 --QLQRQHIDSTEVFEKTESCSDTKHVDLTE 227
QL+R + E ++ +D + +LT+
Sbjct: 1190 AEQLRRSLAERDEALADLQARADAQQKELTQ 1220
Score = 42.7 bits (99), Expect = 0.012
Identities = 44/220 (20%), Positives = 92/220 (41%), Gaps = 24/220 (10%)
Query: 28 KETLLVKDE---ELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNAC 84
+E LL+++ E Q+ L +++D +L + E ++ A+ + EQ
Sbjct: 843 RELLLLRESRSSEQQSLEDRLIQLGDSLKDAQTELVKVREHRDSLAAQVGALQEQAHQDE 902
Query: 85 EEIESLKKDSEKQLE---------------------LSAQKLKEYEVKIISLSKEKEQLS 123
E I L+ + +KQ+ +AQKL+E LSK++E L
Sbjct: 903 ESILELRGEVQKQMRSHGQRLSEGEAHITSLKDQLVAAAQKLQESSQLQQQLSKKEESLE 962
Query: 124 KQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAV 183
K+ +A+ +E N S+ + R+ + L + + +A + A E+
Sbjct: 963 KELKASKEERNRLHSQAEEYRKEAQTVSQQLEEQKRSQGITRGEMKATAETAAALEAQLR 1022
Query: 184 SEKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTKHV 223
++E +LK + + S+++ K E+ S+ +++
Sbjct: 1023 EAEKERQRLEAELKTRDSEKEKLSSDLQSKAENISNLQNL 1062
Score = 42.0 bits (97), Expect = 0.020
Identities = 59/289 (20%), Positives = 116/289 (39%), Gaps = 24/289 (8%)
Query: 27 MKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEE 86
+KE EELQ+ L R + + ++L ++A + AA A T+ + +
Sbjct: 1903 LKEKYATSLEELQDARGQLSQRMEEVSSLQKQLEDSASQHQRAAGAVETLRSEISAVGRK 1962
Query: 87 IESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARES 146
+E + + + L ++ K+ L+ E E+L Q + N+ L + RE
Sbjct: 1963 LERAEDLNSSLSREKEEALASHQAKVSLLTVEIEKLKSQHVQVAAQVNVLTENL-EQREM 2021
Query: 147 GV----ILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEK-QELLAYVDKLKAQLQ 201
+ + AR ++ ++E ++ + +Q E A+ E+ Q+LLA + A+L+
Sbjct: 2022 ALHAINSQHSSQARRTSQL-LSEMQTLQEVNQRLQEEMASAKEEHQKLLAASSQENARLK 2080
Query: 202 RQHIDSTEVFEKTESCSDTKH----------VDLTEENVDKACLSVSRSVPDPAAAECVV 251
D+ + E D H + ++ L SV D ++
Sbjct: 2081 E---DAGRFLAEKEELEDRCHRLERQSSSLGETMARTTSERDDLQTKVSVQDKELSQLKD 2137
Query: 252 QMPTDQVIIQPVGDNEW---SDIQATEARVADVREVASESEVSSLDIPV 297
+ + I+Q + EW + + E R + E+E+SS D+ V
Sbjct: 2138 NVRKVEQILQD-SEREWLLVLEREKEEKNQLVERLTSVENEMSSKDVKV 2185
Score = 40.0 bits (92), Expect = 0.078
Identities = 40/200 (20%), Positives = 82/200 (41%), Gaps = 28/200 (14%)
Query: 42 ARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELS 101
AR+L R + I D+++ + ++ S + EE+ LK + +Q L
Sbjct: 1635 ARELEQRKAEITDLSDNVQALSQQRATFKSELRETGTALARSQEEVAQLKAECSRQEGLQ 1694
Query: 102 AQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKV 161
+EKEQL +Q+EA Q+ ++EL + L R +
Sbjct: 1695 VAL------------QEKEQLLRQKEALIQQMTASKAELDQ-----------LLRQKTDE 1731
Query: 162 RVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTK 221
V+ + + ++E+++ + + V L+ LQ++ S E ++ + +T
Sbjct: 1732 AVSLSTQTSDLQESIR---RLRGQLERSALEVSTLQRSLQQKEESSLEGLSRSAAALETL 1788
Query: 222 HVDLTEENVDKACLSVSRSV 241
DL ++ + CLS+ +
Sbjct: 1789 RTDLQDKQAE--CLSLKEQL 1806
Score = 39.3 bits (90), Expect = 0.13
Identities = 50/204 (24%), Positives = 81/204 (39%), Gaps = 44/204 (21%)
Query: 23 DLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRN 82
DL ++ T +EL + L+ RD ++RD+ L + A+ A N
Sbjct: 2574 DLEELRSTCSSDQDELAALRQLLQERDESLRDLKLSLDQHQSASLA-------------N 2620
Query: 83 ACEEIESLKKDS----------EKQLELSAQKLKEYEVKIIS-----------LSKEKEQ 121
EE+E LK + E+ L + Q+ + + K+++ L + EQ
Sbjct: 2621 LKEELEDLKSQNGHLSEELASKEEALMVGEQRAQALDSKLLTVVEHLETAQAELRDKSEQ 2680
Query: 122 LSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESA 181
+ K +EA + E G +L AL EE+ R AE + + E Q +A
Sbjct: 2681 VEKHQEALRAQELTAEQEKGALESQLDLLTSAL--EEERRRCAEQQSRLDLSEREQAGAA 2738
Query: 182 AVSE--KQEL------LAYVDKLK 197
+ K EL LA DK+K
Sbjct: 2739 TLIRQLKDELGAGGTKLAQFDKVK 2762
Score = 37.0 bits (84), Expect = 0.66
Identities = 30/127 (23%), Positives = 61/127 (47%), Gaps = 8/127 (6%)
Query: 49 DSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSE-KQLELSAQKLKE 107
++T+ +++E+L +T + A I E+ C + E LK + E + +LS +L+
Sbjct: 27 ETTVEEMSERLGQTEQLV---AQLKELIREKDAALCSKDEQLKAEKEASEAKLSKLRLQN 83
Query: 108 YEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEAN 167
+ K+ SL+ + E+L KQR A + + G + + G + ++KV E
Sbjct: 84 -KAKVTSLTTQLEELRKQRGALDTPTH---GKKGSSEGAEQASRGKIVLLKKKVEDLEQQ 139
Query: 168 AEAKIKE 174
+++E
Sbjct: 140 LAQRVEE 146
Score = 36.6 bits (83), Expect = 0.86
Identities = 47/219 (21%), Positives = 95/219 (42%), Gaps = 26/219 (11%)
Query: 30 TLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIES 89
T++ + EE Q D T S + L T E E A + +++ E+E
Sbjct: 2285 TMITQLEEGQQGQVD--TLRSEVNKSVADLERTQEKLEEAERRSEQKEQEAAGLQTEVEL 2342
Query: 90 LKKDSEKQLELSAQ----------KLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSE 139
L+ Q++++ Q +L+E +I +S + +Q +Q++ ++A + ++
Sbjct: 2343 LQSQLHAQVDITNQAAAKLERLSSQLQEKGDQISRMSVQLQQQQQQQQLVDKDAAVAQAM 2402
Query: 140 LGKARESGVI--LEGALARAEEKVRVAEANAEAKIK-EAVQGE----SAAVSEKQELLAY 192
+A + V+ LE + V+ E E K K E ++ E +A+SEK+E L
Sbjct: 2403 ESQANQESVLAQLESLQQEHQRSVKRREQILEQKAKSEQLRSEKQLLESALSEKEERL-- 2460
Query: 193 VDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVD 231
+Q + ++ + V E+ ++ + K +E D
Sbjct: 2461 -----SQAVQTLMEKSSVLEQLQASAAQKDAAFEQERKD 2494
Score = 35.4 bits (80), Expect = 1.9
Identities = 36/175 (20%), Positives = 74/175 (41%), Gaps = 16/175 (9%)
Query: 52 IRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQ---KLKEY 108
+R++ EKL ++ + + + E + +EI L+++ + E+ Q K
Sbjct: 3309 VRELEEKLRAVQRGSQGSKTINKELQELLKEKHQEINQLQQNCIRYQEVILQLESSSKSS 3368
Query: 109 EVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEK-------- 160
+ + L +E E+ S+Q A Q+ + +EL + R + AE +
Sbjct: 3369 QAAVEQLQRELEKSSEQLSAVRQKCSRAEAELSEQRNLLQQAQQKRPGAESQRDPTALDL 3428
Query: 161 ---VRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDK--LKAQLQRQHIDSTEV 210
R +E + E++ E E+ + +L+ D+ K Q +Q +DS +V
Sbjct: 3429 SHPSRSSEGHRESQPAEKADKEAVLQQQMAQLVLSKDQESKKVQELKQQLDSGDV 3483
Score = 34.7 bits (78), Expect = 3.3
Identities = 33/175 (18%), Positives = 77/175 (43%), Gaps = 12/175 (6%)
Query: 37 ELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEK 96
E++ L + A KL + + + Q + ++ + + KD+
Sbjct: 2339 EVELLQSQLHAQVDITNQAAAKLERLSSQLQEKGDQISRMSVQLQQQQQQQQLVDKDAAV 2398
Query: 97 QLELSAQKLKEYEV-KIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALA 155
+ +Q +E + ++ SL +E ++ K+RE ++ +++ + R +LE AL+
Sbjct: 2399 AQAMESQANQESVLAQLESLQQEHQRSVKRREQILEQ----KAKSEQLRSEKQLLESALS 2454
Query: 156 RAEEKVRVAEANAEAKIK-------EAVQGESAAVSEKQELLAYVDKLKAQLQRQ 203
EE++ A K A Q ++A E+++ + +D+L+ +LQ++
Sbjct: 2455 EKEERLSQAVQTLMEKSSVLEQLQASAAQKDAAFEQERKDWMQKLDQLQKELQKE 2509
Score = 33.1 bits (74), Expect = 9.5
Identities = 27/116 (23%), Positives = 50/116 (42%), Gaps = 9/116 (7%)
Query: 55 IAEKLSETAEAA-------EAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKE 107
+ +KLSE+ EA A + + E+ ++L+ +++L S
Sbjct: 3946 LPQKLSESESQGRSARLQNEALRKAMAALQDDRDRLIEDFKTLRNGYDQELRESRAAFSR 4005
Query: 108 YEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVI--LEGALARAEEKV 161
E + S + L+KQR+ + + +S A SG++ L GALA E ++
Sbjct: 4006 VERSLQEASSDLAVLAKQRDVLLLQMSALKSSQTHAELSGLVDQLSGALAEKEREL 4061
>UniRef100_Q9LFE4 Hypothetical protein F5E19_70 [Arabidopsis thaliana]
Length = 853
Score = 62.0 bits (149), Expect = 2e-08
Identities = 59/242 (24%), Positives = 111/242 (45%), Gaps = 26/242 (10%)
Query: 4 SLRNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETA 63
S R + N+ D++ ++K L E + F +++ ++ I EKL+
Sbjct: 254 STREKTAISDNEMVAKLEDEIVVLKRDL----ESARGFEAEVKEKEM----IVEKLNVDL 305
Query: 64 EAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLS 123
EAA+ A S AH++ +W++ +E+ E+QLE A KL+ S S E +
Sbjct: 306 EAAKMAESNAHSLSNEWQSKAKEL-------EEQLE-EANKLER------SASVSLESVM 351
Query: 124 KQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAV 183
KQ E + + + +E+ +E V LE +A+ +E + V+E + +E + E
Sbjct: 352 KQLEGSNDKLHDTETEITDLKERIVTLETTVAKQKEDLEVSEQRLGSVEEEVSKNEKEVE 411
Query: 184 SEKQELLAYVDKLKAQLQRQHIDSTEV----FEKTESCSDTKHVDLTEENVDKACLSVSR 239
K EL ++ L+++ ++ V EK++ SD + EE KA S++
Sbjct: 412 KLKSELETVKEEKNRALKKEQDATSRVQRLSEEKSKLLSDLESSKEEEEKSKKAMESLAS 471
Query: 240 SV 241
++
Sbjct: 472 AL 473
>UniRef100_Q7SHZ4 Hypothetical protein [Neurospora crassa]
Length = 4007
Score = 60.5 bits (145), Expect = 6e-08
Identities = 57/239 (23%), Positives = 99/239 (40%), Gaps = 30/239 (12%)
Query: 23 DLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRN 82
D+T +K+ L KD L A ++ +RD+ + + E+L A A ++++ +
Sbjct: 2519 DVTKLKQELSTKDANLTQKAGEIGSRDAGLAKLREELRAKEAALAKKTEEASSLEKNVKK 2578
Query: 83 ACEEIESLKKD---SEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSE 139
+E LKKD + QL + + E I L++E T E +E
Sbjct: 2579 LTDEATGLKKDVTSRDTQLAQDKDAISKLEKDIAKLNQELSTKDASLTQKTGEVGSKNAE 2638
Query: 140 LGKARESGVILEGALARAEEKVRVAEANAEAK-------------IKEAVQGESAAVSEK 186
L K RE + E ALA+ E+++ + +AK +++ V+G +A + +
Sbjct: 2639 LAKLREEIRVKETALAKKTEELKGLNQSVDAKDTQLAQDKIKIERLEKEVKGLTADIVKL 2698
Query: 187 QELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVDKACLSVSRSVPDPA 245
+E +A+ DK F K D D+TE N + A L + D A
Sbjct: 2699 REDVAFKDK--------------SFAKKAEAVDHLKADITELNSEVAKLKKEGTNKDAA 2743
Score = 50.4 bits (119), Expect = 6e-05
Identities = 68/313 (21%), Positives = 131/313 (41%), Gaps = 41/313 (13%)
Query: 3 ISLRNTLGMMTNKTTDGPMDDLT----IMKETLLVKDEELQNFARDLRTRDSTIRDIAEK 58
+SLR + +TN+ D + L+K++E + F +L+ ++D AE+
Sbjct: 2752 VSLRKAVRDLTNQAKQSAQDSKKSAEDLANRDALLKEKEKKIF--ELQQEIQKVKDTAEE 2809
Query: 59 LSETAEAAEAAASAAHT----IDEQWRNACEEIESLKKDSE----------KQLELSAQK 104
L++T + ++ S + + EQ + +E SLK D E LE Q+
Sbjct: 2810 LNQTTKTRDSTLSQKNEELRKLREQIKQLEDEANSLKMDKETLGRTINTRDSSLEQKEQE 2869
Query: 105 LKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVA 164
+ E +I LS++ L++++ Q + L +A + L+G++ EEK
Sbjct: 2870 ISGLEKEIKRLSEQAANLTQEKVDLGQIVGARDASLLQANKDIDGLKGSIKILEEKA--- 2926
Query: 165 EANAEAKIKEAVQGESAAVSEK----QELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDT 220
A++ + G+ + EK Q+ +D LK +Q+ + + E+ T
Sbjct: 2927 -----AELSKLNAGQDQTIGEKDASLQKANEDIDNLKGSVQKLENKAATLAEEKAQMGQT 2981
Query: 221 KHVDLT-----EENVDKACLSVSRSVPDPAAAECVVQMPTDQVIIQ--PVGDNEWSDIQA 273
T +E++ K ++ R + + ++ T + IQ + E DIQ
Sbjct: 2982 IGAHETSLLKKDEDIKKLTANIQRLTAEANDLKKGIENLTGDIAIQNRALAQKE-KDIQN 3040
Query: 274 TEARVADVR-EVA 285
E + D+ EVA
Sbjct: 3041 MEKTIQDLNTEVA 3053
Score = 39.7 bits (91), Expect = 0.10
Identities = 40/191 (20%), Positives = 83/191 (42%), Gaps = 23/191 (12%)
Query: 33 VKDEELQN--FARDLRTRDSTIRDIAEK---LSETAEAAEAAASAAHTIDEQWRNACEEI 87
+KD E QN D+ + + I I E+ L+ A+ + A ++ + E
Sbjct: 2027 IKDFEAQNAKLQIDIENKKAEIERIKEERRTLNTEADKSIARIEGLERKIKELTGSSAEK 2086
Query: 88 ESLKKDSEKQLELSAQ---KLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKAR 144
E+ K + L A+ ++K+ E + + S + K+ + A +AN +RS L
Sbjct: 2087 EAQMKQYQADLAAKAETEARIKQLERDLATKSNSLAEFEKKYKRANMDANNYRSSLAHT- 2145
Query: 145 ESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQH 204
+G +A+ EE+++ + + + + + + E Q++ VD+LK ++
Sbjct: 2146 ------QGEVAKLEEEIKTTKGDVQYWEDQMIMNQ----EETQKIQDQVDRLKMDVK--- 2192
Query: 205 IDSTEVFEKTE 215
D ++ E E
Sbjct: 2193 -DKNKILEDHE 2202
Score = 37.7 bits (86), Expect = 0.39
Identities = 27/128 (21%), Positives = 60/128 (46%), Gaps = 3/128 (2%)
Query: 22 DDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWR 81
D+ T +K+ + E+ +++ ++ R + I + + + E + + T +
Sbjct: 3162 DETTKLKKDTVKLKEDSKSWEETVKQRQTEINKLNDNIKNLQEEIKRKEALLATRQGEIN 3221
Query: 82 NACEEIESLKKD-SEKQLELSAQ--KLKEYEVKIISLSKEKEQLSKQREAATQEANMWRS 138
+EI LKKD +EK +L ++ +L ++ I + E+L K++ A ++
Sbjct: 3222 ALKDEIVGLKKDLAEKDAQLKSRDGELGKFRKSIAAKETALERLEKEKTALREKVEHLEG 3281
Query: 139 ELGKARES 146
E+G+ R S
Sbjct: 3282 EVGRRRRS 3289
Score = 36.2 bits (82), Expect = 1.1
Identities = 39/189 (20%), Positives = 87/189 (45%), Gaps = 23/189 (12%)
Query: 23 DLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRN 82
D++ TL K E + D++T +STI + + + + + A A T D R+
Sbjct: 2393 DISTKNATLDDKREIIDQLKDDIKTANSTIDTLRKDVKD-----KDAILAHKTKDVVARD 2447
Query: 83 ACEEIESLKKDSEKQLELSAQKLKE---YEVKIISLSKEKEQLSKQREAATQEANMWRSE 139
A E+ LK + + A+K +E +E + +L+ + + L++ T + R+
Sbjct: 2448 A--ELAKLKAEIASKNAALAKKTEEAKAFEKNVQTLTDQAKGLNQDVATKTTQLAQDRAT 2505
Query: 140 LGKARESGVILEGALARAEEKVRVAEANAE-------------AKIKEAVQGESAAVSEK 186
+ K + L+ + + ++++ +AN AK++E ++ + AA+++K
Sbjct: 2506 ISKLNKDIFDLKTDVTKLKQELSTKDANLTQKAGEIGSRDAGLAKLREELRAKEAALAKK 2565
Query: 187 QELLAYVDK 195
E + ++K
Sbjct: 2566 TEEASSLEK 2574
Score = 34.7 bits (78), Expect = 3.3
Identities = 39/198 (19%), Positives = 81/198 (40%), Gaps = 22/198 (11%)
Query: 34 KDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKD 93
+D L +D+ +I+ + EK +E ++ D + A E+I++LK
Sbjct: 2901 RDASLLQANKDIDGLKGSIKILEEKAAELSKLNAGQDQTIGEKDASLQKANEDIDNLKGS 2960
Query: 94 SEKQLELSAQKLKE-----------YEVKIISLSKEKEQLSKQREAATQEANMWRSELGK 142
+K LE A L E +E ++ ++ ++L+ + T EAN + +
Sbjct: 2961 VQK-LENKAATLAEEKAQMGQTIGAHETSLLKKDEDIKKLTANIQRLTAEANDLKKGIEN 3019
Query: 143 ARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLA----------Y 192
I ALA+ E+ ++ E + E + ++ A Q+ +A
Sbjct: 3020 LTGDIAIQNRALAQKEKDIQNMEKTIQDLNTEVARLKTNAAEHNQKTIAKDATLTAKNDQ 3079
Query: 193 VDKLKAQLQRQHIDSTEV 210
+ KL Q+++ + T++
Sbjct: 3080 ISKLNDQIKQLRAEVTKL 3097
Score = 34.7 bits (78), Expect = 3.3
Identities = 48/210 (22%), Positives = 87/210 (40%), Gaps = 19/210 (9%)
Query: 23 DLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRN 82
DL+ +K + +L+N + I+ +KL+ + EA +A+ + ++
Sbjct: 1959 DLSSLKADYQKETTKLKNEISQKEKELAEIQKTNKKLNADIKEKEATLTASQA---KVKD 2015
Query: 83 ACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGK 142
E++ KKD K E KL ++ I + E E++ ++R EA+ + +
Sbjct: 2016 LNREVQQ-KKDQIKDFEAQNAKL---QIDIENKKAEIERIKEERRTLNTEADKSIARIEG 2071
Query: 143 ARESGVILEGALARAEEKVR------VAEANAEAKIKE-----AVQGESAAVSEKQELLA 191
L G+ A E +++ A+A EA+IK+ A + S A EK+ A
Sbjct: 2072 LERKIKELTGSSAEKEAQMKQYQADLAAKAETEARIKQLERDLATKSNSLAEFEKKYKRA 2131
Query: 192 YVDKLKAQLQRQHIDSTEVFEKTESCSDTK 221
+D + H EV + E TK
Sbjct: 2132 NMDANNYRSSLAHTQG-EVAKLEEEIKTTK 2160
Score = 33.5 bits (75), Expect = 7.3
Identities = 56/295 (18%), Positives = 125/295 (41%), Gaps = 28/295 (9%)
Query: 22 DDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIA---EKLSE-----TAEAA------E 67
D L E + +E++NF +D+ ++T+ + EKLS TAE + +
Sbjct: 2280 DQLMKRGEDIKKLRDEIKNFKKDISDHETTLEETMAEIEKLSADNKQLTAEISSYKDKLK 2339
Query: 68 AAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLS---- 123
+ + A ++ ++ E L +D++ + + A+K+KE + S+++ + +S
Sbjct: 2340 QSQTEADALNNDIKDMKSTKEKLGQDAKAKETVLAEKMKEIQGLKDSINRLNQDISTKNA 2399
Query: 124 ---KQREAATQ---EANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQ 177
+RE Q + S + R+ + LA + V +A AK+K +
Sbjct: 2400 TLDDKREIIDQLKDDIKTANSTIDTLRKDVKDKDAILAHKTKDVVARDAEL-AKLKAEIA 2458
Query: 178 GESAAVSEKQELLAYVDKLKAQL--QRQHIDSTEVFEKTESCSDTKHVDLTEENVDKACL 235
++AA+++K E +K L Q + ++ + T+ D + +++
Sbjct: 2459 SKNAALAKKTEEAKAFEKNVQTLTDQAKGLNQDVATKTTQLAQDRATISKLNKDIFDLKT 2518
Query: 236 SVSRSVPDPAAAECVVQMPTDQVIIQPVGDNEW-SDIQATEARVADVREVASESE 289
V++ + + + + ++ + G + +++A EA +A E AS E
Sbjct: 2519 DVTKLKQELSTKDANLTQKAGEIGSRDAGLAKLREELRAKEAALAKKTEEASSLE 2573
Score = 33.1 bits (74), Expect = 9.5
Identities = 42/186 (22%), Positives = 77/186 (40%), Gaps = 11/186 (5%)
Query: 37 ELQNFARDLR----TRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKK 92
+L++ A DL ++D + E+++ A T D + +EI LK
Sbjct: 3096 KLKSDAADLNQATTSKDIVLAQRMEEINGLRNEMVELNKALDTRDTTFLQNTDEINRLK- 3154
Query: 93 DSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEG 152
++ ++L KLK+ VK+ SK E+ KQR+ E N + +E E
Sbjct: 3155 ENVRRLGDETTKLKKDTVKLKEDSKSWEETVKQRQT---EINKLNDNIKNLQEEIKRKEA 3211
Query: 153 ALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQ---LQRQHIDSTE 209
LA + ++ + K+ + ++ S EL + + A+ L+R + T
Sbjct: 3212 LLATRQGEINALKDEIVGLKKDLAEKDAQLKSRDGELGKFRKSIAAKETALERLEKEKTA 3271
Query: 210 VFEKTE 215
+ EK E
Sbjct: 3272 LREKVE 3277
>UniRef100_UPI0000436284 UPI0000436284 UniRef100 entry
Length = 841
Score = 58.9 bits (141), Expect = 2e-07
Identities = 69/323 (21%), Positives = 141/323 (43%), Gaps = 46/323 (14%)
Query: 6 RNTLGMMTNKTTDGPMDDLTIMKETLLVKDEELQN-FARDLRTRDSTIRDIAEKLSETAE 64
R +G +TN+ ++ + T K+ L+K EEL RD+R + D+ E+++ +
Sbjct: 90 RERIGGLTNRMSELETELATARKD--LLKSEELSTKHQRDIREAMAQKEDMEERITTLEK 147
Query: 65 ---AAEAAASAAHTIDEQWRNACEEIESLKKDSEKQ-------LELSAQKLK-------- 106
AA+ ++ H ++++ N +SL + SE++ LEL+ Q+L+
Sbjct: 148 RYLAAQRETTSIHDLNDKLENELATKDSLHRQSEEKVRHLQELLELAEQRLQQTMRKAET 207
Query: 107 ------EYEVKIISLSKEKEQLS-------KQREAATQEANMWRSELGKARESGVILEGA 153
E ++ +L+K E+ +Q EA +E N ELG+AR+ + E
Sbjct: 208 LPEVEAELAQRVAALTKASEERHGNIEERLRQLEAQLEEKNQ---ELGRARQREKMNEEH 264
Query: 154 LARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFEK 213
R + V + +++ ++ AA+ +K+ L+ ++KL+A+L + +
Sbjct: 265 NKRLSDTVDRLLTESNERLQLHLKERMAALEDKERLIGEIEKLRAELD-------HLKRR 317
Query: 214 TESCSDTKHVDLTEENVDKACLSVSRSVPDPAAAECVVQMPTD--QVIIQPVGDNEWSDI 271
+ + D H + SV D ++ V++ V ++ +W
Sbjct: 318 SGAFGDGTHPRSHLGSATDLRFSVVEGQGDHYSSTGVIRRAQKGRMVALRDEPTKDWDRS 377
Query: 272 QATEARVADVREVASESEVSSLD 294
Q + R + + S++E+S LD
Sbjct: 378 QPSSLRASRTHLMGSDTELSDLD 400
>UniRef100_Q9M8T5 F13E7.12 protein [Arabidopsis thaliana]
Length = 806
Score = 58.5 bits (140), Expect = 2e-07
Identities = 53/202 (26%), Positives = 95/202 (46%), Gaps = 23/202 (11%)
Query: 37 ELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSE- 95
+L+N AR L + + I E+L+ EAA+ A S AH ++W+N +E+E +++
Sbjct: 269 DLEN-ARSLEAKVKELEMIIEQLNVDLEAAKMAESYAHGFADEWQNKAKELEKRLEEANK 327
Query: 96 -------------KQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSE--L 140
KQLE+S +L + E +I L ++ E L + A+Q+ ++ +SE L
Sbjct: 328 LEKCASVSLVSVTKQLEVSNSRLHDMESEITDLKEKIELL--EMTVASQKVDLEKSEQKL 385
Query: 141 GKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQL 200
G A E E + EK++ + +A++ E A S Q LL K+ ++L
Sbjct: 386 GIAEEESSKSE----KEAEKLKNELETVNEEKTQALKKEQDATSSVQRLLEEKKKILSEL 441
Query: 201 QRQHIDSTEVFEKTESCSDTKH 222
+ + + + ES + H
Sbjct: 442 ESSKEEEEKSKKAMESLASALH 463
Score = 38.5 bits (88), Expect = 0.23
Identities = 42/189 (22%), Positives = 85/189 (44%), Gaps = 16/189 (8%)
Query: 23 DLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRN 82
++T +KE + + + + + DL + + E+ S++ + AE + T++E
Sbjct: 356 EITDLKEKIELLEMTVASQKVDLEKSEQKLGIAEEESSKSEKEAEKLKNELETVNE---- 411
Query: 83 ACEEIESLKKDSEKQLELSAQKLKEYEVKIIS-LSKEKEQLSKQREAATQEANMWRSELG 141
E+ ++LKK E+ S Q+L E + KI+S L KE+ K ++A A+
Sbjct: 412 --EKTQALKK--EQDATSSVQRLLEEKKKILSELESSKEEEEKSKKAMESLASALHEVSS 467
Query: 142 KARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQE----LLAYVDKLK 197
++RE + E L+R ++ + + IK + E + L+ V++ K
Sbjct: 468 ESRE---LKEKLLSRGDQNYETQIEDLKLVIKATNNKYENMLDEARHEIDVLVNAVEQTK 524
Query: 198 AQLQRQHID 206
Q + +D
Sbjct: 525 KQFESAMVD 533
Score = 34.7 bits (78), Expect = 3.3
Identities = 46/219 (21%), Positives = 87/219 (39%), Gaps = 21/219 (9%)
Query: 27 MKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEE 86
+KE LL + + QN+ + I+ K + A EQ + ++
Sbjct: 472 LKEKLLSRGD--QNYETQIEDLKLVIKATNNKYENMLDEARHEIDVLVNAVEQTK---KQ 526
Query: 87 IESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARES 146
ES D E + +KE++ ++ S+ KE +L + +EA+ + + R+
Sbjct: 527 FESAMVDWEMREAGLVNHVKEFDEEVSSMGKEMNRLGNLVKRTKEEADASWEKESQMRDC 586
Query: 147 GVILEGALARAEEKVRVAEANA---EAKIKEAVQGESAAVSEKQELLAYVDK-------- 195
+E + +E +R A+A + K+ + + V E EL D
Sbjct: 587 LKEVEDEVIYLQETLREAKAETLKLKGKMLDKETEFQSIVHENDELRVKQDDSLKKIKEL 646
Query: 196 ---LKAQLQRQHIDSTEVFEKTESCSD--TKHVDLTEEN 229
L+ L ++HI+ ++E D K V+ +EEN
Sbjct: 647 SELLEEALAKKHIEENGELSESEKDYDLLPKVVEFSEEN 685
Score = 34.3 bits (77), Expect = 4.3
Identities = 37/178 (20%), Positives = 75/178 (41%), Gaps = 21/178 (11%)
Query: 27 MKETLLVKDEELQNFARD---LRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNA 83
+K +L K+ E Q+ + LR + D +K+ E +E E A + H + +
Sbjct: 611 LKGKMLDKETEFQSIVHENDELRVKQD---DSLKKIKELSELLEEALAKKHIEENGELSE 667
Query: 84 CEE--------IESLKKDSEKQLELSAQKLKEYEVKIISLS-----KEKEQLSKQREAAT 130
E+ +E +++ + E + K++ + + L KEK++ S + E
Sbjct: 668 SEKDYDLLPKVVEFSEENGYRSAEEKSSKVETLDGMNMKLEEDTEKKEKKERSPEDETVE 727
Query: 131 QEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQE 188
E MW S + +E V + + EE + V + + + + GE + EK++
Sbjct: 728 VEFKMWESCQIEKKE--VFHKESAKEEEEDLNVVDQSQKTSPVNGLTGEDELLKEKEK 783
>UniRef100_Q8C0X4 Mus musculus adult male testis cDNA, RIKEN full-length enriched
library, clone:4921511I23 product:weakly similar to
CENP-F KINETOCHORE PROTEIN [Mus musculus]
Length = 952
Score = 58.2 bits (139), Expect = 3e-07
Identities = 52/268 (19%), Positives = 124/268 (45%), Gaps = 21/268 (7%)
Query: 39 QNFARDLRTRDST----IRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDS 94
QNFA +++ ++++ +RD+ EKL++ + D + + C ++ K++
Sbjct: 496 QNFAEEMKAKNTSQEIMLRDLQEKLNQQENSLTLEKLKLALADLERQRNCSQVLLKKREH 555
Query: 95 E-KQLELSAQKL-KEYEVKIISLSKEKEQLSKQREAATQEANMWRSE----LGKARESGV 148
QL K+ KE+E + +L +K++ + +E Q + W+ + + +
Sbjct: 556 HIDQLNNKLNKIEKEFETLLSALELKKKECEELKEEKNQ-ISFWKIDSEKLINQIESEKE 614
Query: 149 ILEGALARAEEKVRVAEANAEA--KIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHID 206
IL G + E ++ + + ++ +I+ E + L + +D +++ Q
Sbjct: 615 ILLGKINHLETSLKTQQVSPDSNERIRTLEMERENFTVEIKNLQSMLDSKMVEIKTQKQA 674
Query: 207 STEVFEKTESCSDTKHVDLTEENVDKACLSVSRSVPDPAAAECVVQMPTDQVIIQPVGDN 266
E+ +K+ES SD KH ++ ++ CL ++ + EC +Q+ + +V+ + D
Sbjct: 675 YLELQQKSES-SDQKH----QKEIENMCLKANKLTGQVESLECKLQLLSSEVVTK---DQ 726
Query: 267 EWSDIQATEARVADVREVASESEVSSLD 294
++ D++ + D+ + S V++ D
Sbjct: 727 QYQDLRMEYETLRDLLKSRGSSLVTNED 754
Score = 41.6 bits (96), Expect = 0.027
Identities = 45/212 (21%), Positives = 94/212 (44%), Gaps = 24/212 (11%)
Query: 32 LVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLK 91
L++ E + N +TRD +R A+ + AA+ T++++ + EE+ +
Sbjct: 305 LIEKERILN-----KTRDEVVRSTAQY--------DQAAAKCTTLEQKLKTLTEELSCHR 351
Query: 92 KDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQR---EAATQEANMWRSELGKARESGV 148
+++E Q++KE E ++ +E+LS+Q +A E ++ L + +
Sbjct: 352 QNAESAKRSLEQRIKEKEKEL------QEELSRQHQSFQALDSEYTQMKTRLTQELQQVK 405
Query: 149 ILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDST 208
L L EKV + E ++E S A Q ++L+ + +++
Sbjct: 406 HLHSTLQLELEKVTSVKQQLERNLEEIRLKLSRAEQALQASQVAENELRRSSEEMKKENS 465
Query: 209 EVFEKTESCSDTKHVDLTEENVDKACLSVSRS 240
+ +++S T+ V EE + K +S+S+S
Sbjct: 466 LI--RSQSEQRTREVCHLEEELGKVKVSLSKS 495
Score = 34.7 bits (78), Expect = 3.3
Identities = 47/216 (21%), Positives = 83/216 (37%), Gaps = 24/216 (11%)
Query: 27 MKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAH-------TIDEQ 79
+KE EEL + + DS + +L++ + + S ++ +Q
Sbjct: 365 IKEKEKELQEELSRQHQSFQALDSEYTQMKTRLTQELQQVKHLHSTLQLELEKVTSVKQQ 424
Query: 80 WRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSE 139
EEI +E+ L+ S E + KE + Q E T+E E
Sbjct: 425 LERNLEEIRLKLSRAEQALQASQVAENELRRSSEEMKKENSLIRSQSEQRTREVCHLEEE 484
Query: 140 LGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLK-- 197
LGK + S L++++ E A+ +E + + +QE ++KLK
Sbjct: 485 LGKVKVS-------LSKSQNFAE--EMKAKNTSQEIMLRDLQEKLNQQENSLTLEKLKLA 535
Query: 198 -AQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVDK 232
A L+RQ S + +K E H+D ++K
Sbjct: 536 LADLERQRNCSQVLLKKRE-----HHIDQLNNKLNK 566
>UniRef100_UPI000024976E UPI000024976E UniRef100 entry
Length = 2295
Score = 57.4 bits (137), Expect = 5e-07
Identities = 41/194 (21%), Positives = 90/194 (46%), Gaps = 13/194 (6%)
Query: 35 DEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDS 94
+ +++ +R ++ I ++ E+L T + E + + +++E + E ++
Sbjct: 944 ERDMEVLRAQIRQTENRIEELTERLKNTTASMEQYKAMSLSLEESLDKEKQVTEQVRSSV 1003
Query: 95 EKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGAL 154
E Q+E + ++ K E K++ + KEK+ L +++ A +EL K+ S ++ L
Sbjct: 1004 ENQVEAAQEQYKRLEQKLLEMDKEKQNLLEEKNKAVAAVEQELTELRKSLSS---VQAEL 1060
Query: 155 ARAEEKVRVAEANAEAKI----KEAVQGESAAVSEKQELLAY------VDKLKAQLQRQH 204
A+EK VA A + + ++A A ++E+L + + KAQ Q+
Sbjct: 1061 QSAQEKAAVAAAQEQQALLDGQEQAKMAAEAQDKYEREMLLHAADVEALQAAKAQAQQAS 1120
Query: 205 IDSTEVFEKTESCS 218
+ ++ EK + S
Sbjct: 1121 LVRQQLEEKVQITS 1134
Score = 40.4 bits (93), Expect = 0.060
Identities = 44/178 (24%), Positives = 88/178 (48%), Gaps = 23/178 (12%)
Query: 57 EKLSETAEAAEAAASAAHTIDEQWRNA-CEEIESLKKDSEKQLELSAQKLKEYEVKIISL 115
E+L E AAA++A D++ + A +E+++LK+ L S +++E E ++ +L
Sbjct: 1460 EELKVEYEKLVAAAASAPAQDQEAQQASAQELQNLKES----LNQSETRIRELEGQLENL 1515
Query: 116 SKEKEQLSKQREAATQEANMWRSELGKARES-------GVILEGALARAEEKVRVAEANA 168
++ + + +A ++A+ ++EL + R+ L+ L EE+ + A A
Sbjct: 1516 NRTVGEREMEARSAQEQASRLQTELTRLRQELQEKSSQEERLKQQLTEKEERTKKAILLA 1575
Query: 169 EAKIKE--AVQGESAAVSE-----KQELLAYVDKLKAQ----LQRQHIDSTEVFEKTE 215
+ KI + ++G+ +E K+EL V LK+Q L RQ + ++ E+ E
Sbjct: 1576 KQKISQLTGIKGQFQKENEELKQQKEELEVRVSALKSQYEGRLSRQERELRDLREQQE 1633
Score = 38.1 bits (87), Expect = 0.30
Identities = 41/209 (19%), Positives = 91/209 (42%), Gaps = 23/209 (11%)
Query: 11 MMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAA 70
M +T + M+ +++E ++ELQ+ +R +S I + E SE +E +
Sbjct: 1277 MKKTETMNVLMETNKMLREEKEKLEKELQDTQAKIRKLESDIMPVQESNSELSEKSGMLQ 1336
Query: 71 SAAHTIDE---QWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQRE 127
+ ++E +W+ + + S +KD++ + EY+ L E+E K+ +
Sbjct: 1337 AEKKLLEEDIKRWKARTQHLVSQQKDTDPE---------EYK----RLHSEREAHLKRIQ 1383
Query: 128 AATQEANMWRSELGKARESGVILEGALARAEE---KVRVAEANAEAKIKEAVQGESAAVS 184
+E +++ ++ S L+ + E KV V N +K+ + + +
Sbjct: 1384 QLVEETGRLKADAARSSGSLTTLQSQVQNLRENLGKVMVERDN----LKKDQEAKILDIQ 1439
Query: 185 EKQELLAYVDKLKAQLQRQHIDSTEVFEK 213
EK + + V K+ + + Q+ + +EK
Sbjct: 1440 EKIKTITQVKKIGRRYKTQYEELKVEYEK 1468
Score = 38.1 bits (87), Expect = 0.30
Identities = 47/214 (21%), Positives = 90/214 (41%), Gaps = 37/214 (17%)
Query: 34 KDEELQNFARDLRTRDSTIRDIAEKLSET----AEAAEAAASAAHTIDEQWRNACEEI-- 87
KD + + + R R++ ++ I + + ET A+AA ++ S T+ Q +N E +
Sbjct: 1361 KDTDPEEYKRLHSEREAHLKRIQQLVEETGRLKADAARSSGSLT-TLQSQVQNLRENLGK 1419
Query: 88 -----ESLKKDSEKQLELSAQKLK---------------------EYEVKIISLSKEKEQ 121
++LKKD E ++ +K+K EYE + + + Q
Sbjct: 1420 VMVERDNLKKDQEAKILDIQEKIKTITQVKKIGRRYKTQYEELKVEYEKLVAAAASAPAQ 1479
Query: 122 LSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESA 181
+ ++A+ QE + L ++ LEG L V E A + ++A + ++
Sbjct: 1480 DQEAQQASAQELQNLKESLNQSETRIRELEGQLENLNRTVGEREMEARSAQEQASRLQTE 1539
Query: 182 AVSEKQELLAYVDKLKAQLQRQHIDSTEVFEKTE 215
+QEL + +Q +R TE E+T+
Sbjct: 1540 LTRLRQEL----QEKSSQEERLKQQLTEKEERTK 1569
>UniRef100_UPI00002B91B8 UPI00002B91B8 UniRef100 entry
Length = 659
Score = 57.4 bits (137), Expect = 5e-07
Identities = 67/227 (29%), Positives = 104/227 (45%), Gaps = 40/227 (17%)
Query: 25 TIMKETLLVKDEELQNFARD-------LRTRDSTIRDIAEKLSE-------TAEAAEAAA 70
T+ KE + K+EE + ++ R TIR AEK E + EA E A
Sbjct: 234 TVQKELEIKKNEEFSRYQQEREIAIQKANERTETIRKKAEKEREAEEAEIQSQEAIEVAK 293
Query: 71 SAAHTIDEQWRNACE-----EIESLKKDSEKQLELSA----QKLKEYEVKIISLSKEKE- 120
+ + + E R E EIE +K+ + SA QK E E+KI++L KE E
Sbjct: 294 ISQNQVIEVERKLTETRLIEEIEKRRKEQNALEQDSALDIRQKNLETEIKILNLDKESEY 353
Query: 121 -QLSKQR----EAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEA 175
+L KQR + A ++A + + + K RES E A AE++V+ AKI++
Sbjct: 354 ARLEKQRLVDTKRAQEKAEIVKEQSDKQRES----EEAQIIAEQQVK------NAKIEQQ 403
Query: 176 VQGESAAV-SEKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTK 221
+S + SEKQ ++K K +H E+ EK++ +K
Sbjct: 404 KNIDSHRIESEKQVRTLDIEKAKRIQLEEHQKELEIIEKSKEVLKSK 450
Score = 36.2 bits (82), Expect = 1.1
Identities = 52/236 (22%), Positives = 103/236 (43%), Gaps = 20/236 (8%)
Query: 73 AHTIDEQW-RNACEEIESLKKD-----SEKQLELSAQKLKEYEVKI-ISLSKEKEQLSKQ 125
A+T D Q E+IES KK + ++ + + L+ + ++ I ++E + ++
Sbjct: 194 ANTFDSQGLTQLTEQIESRKKKRNDITQDTKISIENKNLETVQKELEIKKNEEFSRYQQE 253
Query: 126 REAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSE 185
RE A Q+AN + K E E A +++E + V AKI + E
Sbjct: 254 REIAIQKANERTETIRKKAEKEREAEEAEIQSQEAIEV------AKISQNQVIEVERKLT 307
Query: 186 KQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVDKACLSVSRSVPDPA 245
+ L+ ++K + + DS + ++ K ++L +E+ + A L R V
Sbjct: 308 ETRLIEEIEKRRKEQNALEQDSALDIRQKNLETEIKILNLDKES-EYARLEKQRLVDTKR 366
Query: 246 AAEC--VVQMPTDQ----VIIQPVGDNEWSDIQATEARVADVREVASESEVSSLDI 295
A E +V+ +D+ Q + + + + + + + D + SE +V +LDI
Sbjct: 367 AQEKAEIVKEQSDKQRESEEAQIIAEQQVKNAKIEQQKNIDSHRIESEKQVRTLDI 422
>UniRef100_UPI0000299C6A UPI0000299C6A UniRef100 entry
Length = 467
Score = 57.4 bits (137), Expect = 5e-07
Identities = 67/227 (29%), Positives = 104/227 (45%), Gaps = 40/227 (17%)
Query: 25 TIMKETLLVKDEELQNFARD-------LRTRDSTIRDIAEKLSE-------TAEAAEAAA 70
T+ KE + K+EE + ++ R TIR AEK E + EA E A
Sbjct: 234 TVQKELEIKKNEEFSRYQQEREIAIQKANERTETIRKKAEKEREAEEAEIQSQEAIEVAK 293
Query: 71 SAAHTIDEQWRNACE-----EIESLKKDSEKQLELSA----QKLKEYEVKIISLSKEKE- 120
+ + + E R E EIE +K+ + SA QK E E+KI++L KE E
Sbjct: 294 ISQNQVIEVERKLTETRLIEEIEKRRKEQNALEQDSALDIRQKNLETEIKILNLDKESEY 353
Query: 121 -QLSKQR----EAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEA 175
+L KQR + A ++A + + + K RES E A AE++V+ AKI++
Sbjct: 354 ARLEKQRLVDTKRAQEKAEIVKEQSDKQRES----EEAQIIAEQQVK------NAKIEQQ 403
Query: 176 VQGESAAV-SEKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTK 221
+S + SEKQ ++K K +H E+ EK++ +K
Sbjct: 404 KNIDSHRIESEKQVRTLDIEKAKRIQLEEHQKELEIIEKSKEVLKSK 450
Score = 35.0 bits (79), Expect = 2.5
Identities = 51/236 (21%), Positives = 103/236 (43%), Gaps = 20/236 (8%)
Query: 73 AHTIDEQW-RNACEEIESLKKD-----SEKQLELSAQKLKEYEVKI-ISLSKEKEQLSKQ 125
A+T D Q E+IE+ KK + ++ + + L+ + ++ I ++E + ++
Sbjct: 194 ANTFDSQGLTQLTEQIENRKKKRNDITQDTKISIENKNLETVQKELEIKKNEEFSRYQQE 253
Query: 126 REAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSE 185
RE A Q+AN + K E E A +++E + V AKI + E
Sbjct: 254 REIAIQKANERTETIRKKAEKEREAEEAEIQSQEAIEV------AKISQNQVIEVERKLT 307
Query: 186 KQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVDKACLSVSRSVPDPA 245
+ L+ ++K + + DS + ++ K ++L +E+ + A L R V
Sbjct: 308 ETRLIEEIEKRRKEQNALEQDSALDIRQKNLETEIKILNLDKES-EYARLEKQRLVDTKR 366
Query: 246 AAEC--VVQMPTDQ----VIIQPVGDNEWSDIQATEARVADVREVASESEVSSLDI 295
A E +V+ +D+ Q + + + + + + + D + SE +V +LDI
Sbjct: 367 AQEKAEIVKEQSDKQRESEEAQIIAEQQVKNAKIEQQKNIDSHRIESEKQVRTLDI 422
>UniRef100_Q5RI09 Novel protein similar to translocated promoter region [Brachydanio
rerio]
Length = 2352
Score = 57.4 bits (137), Expect = 5e-07
Identities = 41/194 (21%), Positives = 90/194 (46%), Gaps = 13/194 (6%)
Query: 35 DEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKKDS 94
+ +++ +R ++ I ++ E+L T + E + + +++E + E ++
Sbjct: 933 ERDMEVLRAQIRQTENRIEELTERLKNTTASMEQYKAMSLSLEESLDKEKQVTEQVRSSV 992
Query: 95 EKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKARESGVILEGAL 154
E Q+E + ++ K E K++ + KEK+ L +++ A +EL K+ S ++ L
Sbjct: 993 ENQVEAAQEQYKRLEQKLLEMDKEKQNLLEEKNKAVAAVEQELTELRKSLSS---VQAEL 1049
Query: 155 ARAEEKVRVAEANAEAKI----KEAVQGESAAVSEKQELLAY------VDKLKAQLQRQH 204
A+EK VA A + + ++A A ++E+L + + KAQ Q+
Sbjct: 1050 QSAQEKAAVAAAQEQQALLDGQEQAKMAAEAQDKYEREMLLHAADVEALQAAKAQAQQAS 1109
Query: 205 IDSTEVFEKTESCS 218
+ ++ EK + S
Sbjct: 1110 LVRQQLEEKVQITS 1123
Score = 40.4 bits (93), Expect = 0.060
Identities = 44/178 (24%), Positives = 88/178 (48%), Gaps = 23/178 (12%)
Query: 57 EKLSETAEAAEAAASAAHTIDEQWRNA-CEEIESLKKDSEKQLELSAQKLKEYEVKIISL 115
E+L E AAA++A D++ + A +E+++LK+ L S +++E E ++ +L
Sbjct: 1449 EELKVEYEKLVAAAASAPAQDQEAQQASAQELQNLKES----LNQSETRIRELEGQLENL 1504
Query: 116 SKEKEQLSKQREAATQEANMWRSELGKARES-------GVILEGALARAEEKVRVAEANA 168
++ + + +A ++A+ ++EL + R+ L+ L EE+ + A A
Sbjct: 1505 NRTVGEREMEARSAQEQASRLQTELTRLRQELQEKSSQEERLKQQLTEKEERTKKAILLA 1564
Query: 169 EAKIKE--AVQGESAAVSE-----KQELLAYVDKLKAQ----LQRQHIDSTEVFEKTE 215
+ KI + ++G+ +E K+EL V LK+Q L RQ + ++ E+ E
Sbjct: 1565 KQKISQLTGIKGQFQKENEELKQQKEELEVRVSALKSQYEGRLSRQERELRDLREQQE 1622
Score = 38.1 bits (87), Expect = 0.30
Identities = 41/209 (19%), Positives = 91/209 (42%), Gaps = 23/209 (11%)
Query: 11 MMTNKTTDGPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAA 70
M +T + M+ +++E ++ELQ+ +R +S I + E SE +E +
Sbjct: 1266 MKKTETMNVLMETNKMLREEKEKLEKELQDTQAKIRKLESDIMPVQESNSELSEKSGMLQ 1325
Query: 71 SAAHTIDE---QWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQRE 127
+ ++E +W+ + + S +KD++ + EY+ L E+E K+ +
Sbjct: 1326 AEKKLLEEDIKRWKARTQHLVSQQKDTDPE---------EYK----RLHSEREAHLKRIQ 1372
Query: 128 AATQEANMWRSELGKARESGVILEGALARAEE---KVRVAEANAEAKIKEAVQGESAAVS 184
+E +++ ++ S L+ + E KV V N +K+ + + +
Sbjct: 1373 QLVEETGRLKADAARSSGSLTTLQSQVQNLRENLGKVMVERDN----LKKDQEAKILDIQ 1428
Query: 185 EKQELLAYVDKLKAQLQRQHIDSTEVFEK 213
EK + + V K+ + + Q+ + +EK
Sbjct: 1429 EKIKTITQVKKIGRRYKTQYEELKVEYEK 1457
Score = 38.1 bits (87), Expect = 0.30
Identities = 47/214 (21%), Positives = 90/214 (41%), Gaps = 37/214 (17%)
Query: 34 KDEELQNFARDLRTRDSTIRDIAEKLSET----AEAAEAAASAAHTIDEQWRNACEEI-- 87
KD + + + R R++ ++ I + + ET A+AA ++ S T+ Q +N E +
Sbjct: 1350 KDTDPEEYKRLHSEREAHLKRIQQLVEETGRLKADAARSSGSLT-TLQSQVQNLRENLGK 1408
Query: 88 -----ESLKKDSEKQLELSAQKLK---------------------EYEVKIISLSKEKEQ 121
++LKKD E ++ +K+K EYE + + + Q
Sbjct: 1409 VMVERDNLKKDQEAKILDIQEKIKTITQVKKIGRRYKTQYEELKVEYEKLVAAAASAPAQ 1468
Query: 122 LSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESA 181
+ ++A+ QE + L ++ LEG L V E A + ++A + ++
Sbjct: 1469 DQEAQQASAQELQNLKESLNQSETRIRELEGQLENLNRTVGEREMEARSAQEQASRLQTE 1528
Query: 182 AVSEKQELLAYVDKLKAQLQRQHIDSTEVFEKTE 215
+QEL + +Q +R TE E+T+
Sbjct: 1529 LTRLRQEL----QEKSSQEERLKQQLTEKEERTK 1558
>UniRef100_UPI000042F053 UPI000042F053 UniRef100 entry
Length = 1644
Score = 57.0 bits (136), Expect = 6e-07
Identities = 46/217 (21%), Positives = 102/217 (46%), Gaps = 9/217 (4%)
Query: 2 QISLRNTLGMMTNKTTDGPMDDLTIMKETLLVKD--EELQNFARDLRTRDSTIRDIAEKL 59
Q+S+R+ +K +D L+ + ++++ E L++ ++ TR+ I ++ +KL
Sbjct: 669 QLSMRSGQSASPSKNAQDLLDGLSPEERAAVIQEMAERLESLEEEMNTREEVIAELQDKL 728
Query: 60 SETAEAAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIIS-LSKE 118
++ + H E+ EE E ++ D + + + + +I S +
Sbjct: 729 EYAHQSQSPDVQSLHAQIEELARQLEETEQVRVDLQSEFSMKTEDHAAKFTEICSGFESQ 788
Query: 119 KEQLSKQREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQG 178
+ L K +A +EA+ R+E + R G+ + + EE++R E ++ EA++G
Sbjct: 789 VKSLEKDLASAREEADRLRAE--RTRLEGLAEKEGSSEREEELRKQVREMEVEL-EAIKG 845
Query: 179 ESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFEKTE 215
++ + E+ E L +L L ++ ++T+ FE E
Sbjct: 846 QAKDMHEETEELRGKIQL---LNKEKEEATKKFEDAE 879
Score = 36.6 bits (83), Expect = 0.86
Identities = 33/140 (23%), Positives = 60/140 (42%), Gaps = 7/140 (5%)
Query: 33 VKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEEIESLKK 92
V+ E ++ A+D+ +R + L++ E A A E+ + ++ E +
Sbjct: 838 VELEAIKGQAKDMHEETEELRGKIQLLNKEKEEATKKFEDAERRVEEHQKLHQDSEHRAE 897
Query: 93 DSEKQLELSAQKLKEY-------EVKIISLSKEKEQLSKQREAATQEANMWRSELGKARE 145
+E LE + +LKE + K+ KE EQL + E ++ + +SE+ +
Sbjct: 898 RAENDLETLSAELKEASNAQLAADEKLAQYEKELEQLDQLHEEKEKQLDQQQSEIQELNR 957
Query: 146 SGVILEGALARAEEKVRVAE 165
LE A +A E V E
Sbjct: 958 LVQQLEAAQEKAAENEWVKE 977
Score = 35.0 bits (79), Expect = 2.5
Identities = 42/244 (17%), Positives = 104/244 (42%), Gaps = 48/244 (19%)
Query: 86 EIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSK------------QREAATQE- 132
E+ES K + E+ E S +++++ ++ + KE+++L + +RE A +
Sbjct: 1292 EVESFKTEMERMAEESEREVEQAREEVKRVEKERDELKRGIQISKSQVTQLERELADERR 1351
Query: 133 ------------------ANMWRSELGKARESGVILEGALARAE---EKVRVAEANAEAK 171
A ++EL + E+ +LE +L AE +R + A + +
Sbjct: 1352 AYDSLSRRNAQIACDSTTAGQLQAELSQKSEAIRLLESSLRDAESISNHLRESLARRDQE 1411
Query: 172 IKEAVQGESAAVSEKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVD 231
I+E+ + E++ + + + + LQ+ I+S + ++ ++ L E ++
Sbjct: 1412 IRESEAQVNKYRREREIIAQELREFENDLQKHKIESEDFGQQLQA--------LKREQIN 1463
Query: 232 KA------CLSVSRSVPDPAAAECVVQMPTDQVIIQPVGDNEWSDIQATEARVADVREVA 285
K+ L++ R + + E ++ V + +W ++ +AR + + +
Sbjct: 1464 KSSKHAAELLALEREMEEAKERERRLRHEVQDVKTKYEKVEKWRELHECDARPSGLSDTL 1523
Query: 286 SESE 289
+E +
Sbjct: 1524 AEQK 1527
>UniRef100_Q805C7 Tropomyosin2 [Fugu rubripes]
Length = 284
Score = 56.6 bits (135), Expect = 8e-07
Identities = 64/244 (26%), Positives = 113/244 (46%), Gaps = 36/244 (14%)
Query: 19 GPMDDLTIMKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDE 78
G D ++E LL ++L+ +L +++D EKL + + A A + +++
Sbjct: 31 GAEDKCKQLEEELLGLQKKLKGVEDELDKYSESLKDAQEKLEQAEKKAADAEAEVASLNR 90
Query: 79 QWRNACEEIESLKKDSEKQLELSAQKLKEYE---------VKIIS--LSKEKEQLSKQRE 127
+ + EE++ ++++L + QKL+E E +K+I SK++E++ Q E
Sbjct: 91 RIQLVEEELDR----AQERLATALQKLEEAEKAADESERGMKVIENRASKDEEKMEIQ-E 145
Query: 128 AATQEANMWRSELGKARESG----VILEGALARAEEKVRVAEANA---EAKIK------E 174
+EA E + E VILEG L R+EE+ VAEA + E ++K +
Sbjct: 146 MQLKEAKHIAEEADRKYEEVARKLVILEGDLERSEERAEVAEAKSGDLEEELKNVTNNLK 205
Query: 175 AVQGESAAVSEKQ-----ELLAYVDKLKAQLQRQHIDSTEV--FEKTESCSDTKHVDLTE 227
+++ ++ S+K+ E+ DKLK R V EKT + K E
Sbjct: 206 SLEAQAEKYSQKEDKYEEEIRVLTDKLKEAETRAEFAERSVAKLEKTIDDLEEKLAQAKE 265
Query: 228 ENVD 231
EN+D
Sbjct: 266 ENLD 269
>UniRef100_Q7KT48 CG5020-PD, isoform D [Drosophila melanogaster]
Length = 1677
Score = 56.6 bits (135), Expect = 8e-07
Identities = 44/195 (22%), Positives = 89/195 (45%), Gaps = 11/195 (5%)
Query: 27 MKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEE 86
++E L + Q ++ +T + +I + L E ++ + ++E+ R +
Sbjct: 1181 LEEKLKQAQQSEQKLQQESQTSKEKLTEIQQSLQELQDSVKQKEELVQNLEEKVRESSSI 1240
Query: 87 IESLK---KDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKA 143
IE+ +S QLE LKE + +++ K+++QL +EA EL +
Sbjct: 1241 IEAQNTKLNESNVQLENKTSCLKETQDQLLESQKKEKQLQ-------EEAAKLSGELQQV 1293
Query: 144 RESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQ 203
+E+ ++ +L + EE V+V E +A + + + A E QELL + + LQ +
Sbjct: 1294 QEANGDIKDSLVKVEELVKVLEEKLQAATSQ-LDAQQATNKELQELLVKSQENEGNLQGE 1352
Query: 204 HIDSTEVFEKTESCS 218
+ TE ++ E +
Sbjct: 1353 SLAVTEKLQQLEQAN 1367
Score = 44.7 bits (104), Expect = 0.003
Identities = 61/276 (22%), Positives = 123/276 (44%), Gaps = 25/276 (9%)
Query: 34 KDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWR---NACEEIESL 90
K +E + ++R RD IR++ ++L E + + +D+ R EE +L
Sbjct: 600 KTDECEILQTEVRMRDEQIRELNQQLDEVTTQLNVQKADSSALDDMLRLQKEGTEEKSTL 659
Query: 91 KKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQ----REAATQE---ANMWRSELGKA 143
+ +EK+L S KE K ++ +KEQL KQ ++ A QE M + + +
Sbjct: 660 LEKTEKELVQS----KEQAAKTLN---DKEQLEKQISDLKQLAEQEKLVREMTENAINQI 712
Query: 144 RESGVILEGALARAE---EKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQL 200
+ +E LA + E + ++ +E ++E + E E + KL+ QL
Sbjct: 713 QLEKESIEQQLALKQNELEDFQKKQSESEVHLQEIKAQNTQKDFELVESGESLKKLQQQL 772
Query: 201 QRQHIDSTEVFEKTESCSDTKHVDLTEENVDKACLSVSRSVPDPAAAECVVQMPTDQVII 260
+++ + ++ E K + E+ + L S+S +A + VVQ+ +Q+
Sbjct: 773 EQKTLGHEKLQAALEELKKEKETIIKEKEQELQQLQ-SKSAESESALK-VVQVQLEQLQQ 830
Query: 261 QPVGDNEWSDIQATEARVAD-VREVASESEVSSLDI 295
Q E + T A++ D + ++ S++E + ++
Sbjct: 831 QAAASGE--EGSKTVAKLHDEISQLKSQAEETQSEL 864
Score = 42.4 bits (98), Expect = 0.016
Identities = 44/206 (21%), Positives = 94/206 (45%), Gaps = 18/206 (8%)
Query: 34 KDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWR--------NACE 85
K +L+ ++ ++ I +LS E +A S + I E + E
Sbjct: 1096 KIADLKTLVEAIQVANANISATNAELSTVLEVLQAEKSETNHIFELFEMEADMNSERLIE 1155
Query: 86 EIESLK---KDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGK 142
++ +K K++ QL+ +K +E E K+ + +++L ++ + + ++ + L +
Sbjct: 1156 KVTGIKEELKETHLQLDERQKKFEELEEKLKQAQQSEQKLQQESQTSKEKLTEIQQSLQE 1215
Query: 143 ARESGVILEGALARAEEKVRVAEANAEA---KIKEA---VQGESAAVSEKQELLAYVDKL 196
++S E + EEKVR + + EA K+ E+ ++ +++ + E Q+ L K
Sbjct: 1216 LQDSVKQKEELVQNLEEKVRESSSIIEAQNTKLNESNVQLENKTSCLKETQDQLLESQKK 1275
Query: 197 KAQLQRQHID-STEVFEKTESCSDTK 221
+ QLQ + S E+ + E+ D K
Sbjct: 1276 EKQLQEEAAKLSGELQQVQEANGDIK 1301
Score = 42.4 bits (98), Expect = 0.016
Identities = 41/236 (17%), Positives = 94/236 (39%), Gaps = 29/236 (12%)
Query: 6 RNTLGMMTNKTTDGPMDDLTIMKETLLV-KDEELQNFARDLRTRDSTIRDIAEKLSETAE 64
+ TLG ++ +++L KET++ K++ELQ +S ++ + +L + +
Sbjct: 774 QKTLG---HEKLQAALEELKKEKETIIKEKEQELQQLQSKSAESESALKVVQVQLEQLQQ 830
Query: 65 AAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSK 124
A A+ + + +++S ++++ +L+ + L E SK
Sbjct: 831 QAAASGEEGSKTVAKLHDEISQLKSQAEETQSELKSTQSNL--------------EAKSK 876
Query: 125 QREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVS 184
Q EAA + G E L+ + + + + E+K K+ +A
Sbjct: 877 QLEAANGSLEEEAKKSGHLLEQITKLKSEVGETQAALSSCHTDVESKTKQLEAANAA--- 933
Query: 185 EKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVDKACLSVSRS 240
++K+ + +++++ +K + +DT H +L E + L S
Sbjct: 934 --------LEKVNKEYAESRAEASDLQDKVKEITDTLHAELQAERSSSSALHTKLS 981
>UniRef100_Q9VJE5-2 Splice isoform B of Q9VJE5 [Drosophila melanogaster]
Length = 1689
Score = 56.6 bits (135), Expect = 8e-07
Identities = 44/195 (22%), Positives = 89/195 (45%), Gaps = 11/195 (5%)
Query: 27 MKETLLVKDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWRNACEE 86
++E L + Q ++ +T + +I + L E ++ + ++E+ R +
Sbjct: 1193 LEEKLKQAQQSEQKLQQESQTSKEKLTEIQQSLQELQDSVKQKEELVQNLEEKVRESSSI 1252
Query: 87 IESLK---KDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGKA 143
IE+ +S QLE LKE + +++ K+++QL +EA EL +
Sbjct: 1253 IEAQNTKLNESNVQLENKTSCLKETQDQLLESQKKEKQLQ-------EEAAKLSGELQQV 1305
Query: 144 RESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQLQRQ 203
+E+ ++ +L + EE V+V E +A + + + A E QELL + + LQ +
Sbjct: 1306 QEANGDIKDSLVKVEELVKVLEEKLQAATSQ-LDAQQATNKELQELLVKSQENEGNLQGE 1364
Query: 204 HIDSTEVFEKTESCS 218
+ TE ++ E +
Sbjct: 1365 SLAVTEKLQQLEQAN 1379
Score = 44.7 bits (104), Expect = 0.003
Identities = 61/276 (22%), Positives = 123/276 (44%), Gaps = 25/276 (9%)
Query: 34 KDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWR---NACEEIESL 90
K +E + ++R RD IR++ ++L E + + +D+ R EE +L
Sbjct: 612 KTDECEILQTEVRMRDEQIRELNQQLDEVTTQLNVQKADSSALDDMLRLQKEGTEEKSTL 671
Query: 91 KKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQ----REAATQE---ANMWRSELGKA 143
+ +EK+L S KE K ++ +KEQL KQ ++ A QE M + + +
Sbjct: 672 LEKTEKELVQS----KEQAAKTLN---DKEQLEKQISDLKQLAEQEKLVREMTENAINQI 724
Query: 144 RESGVILEGALARAE---EKVRVAEANAEAKIKEAVQGESAAVSEKQELLAYVDKLKAQL 200
+ +E LA + E + ++ +E ++E + E E + KL+ QL
Sbjct: 725 QLEKESIEQQLALKQNELEDFQKKQSESEVHLQEIKAQNTQKDFELVESGESLKKLQQQL 784
Query: 201 QRQHIDSTEVFEKTESCSDTKHVDLTEENVDKACLSVSRSVPDPAAAECVVQMPTDQVII 260
+++ + ++ E K + E+ + L S+S +A + VVQ+ +Q+
Sbjct: 785 EQKTLGHEKLQAALEELKKEKETIIKEKEQELQQLQ-SKSAESESALK-VVQVQLEQLQQ 842
Query: 261 QPVGDNEWSDIQATEARVAD-VREVASESEVSSLDI 295
Q E + T A++ D + ++ S++E + ++
Sbjct: 843 QAAASGE--EGSKTVAKLHDEISQLKSQAEETQSEL 876
Score = 42.4 bits (98), Expect = 0.016
Identities = 44/206 (21%), Positives = 94/206 (45%), Gaps = 18/206 (8%)
Query: 34 KDEELQNFARDLRTRDSTIRDIAEKLSETAEAAEAAASAAHTIDEQWR--------NACE 85
K +L+ ++ ++ I +LS E +A S + I E + E
Sbjct: 1108 KIADLKTLVEAIQVANANISATNAELSTVLEVLQAEKSETNHIFELFEMEADMNSERLIE 1167
Query: 86 EIESLK---KDSEKQLELSAQKLKEYEVKIISLSKEKEQLSKQREAATQEANMWRSELGK 142
++ +K K++ QL+ +K +E E K+ + +++L ++ + + ++ + L +
Sbjct: 1168 KVTGIKEELKETHLQLDERQKKFEELEEKLKQAQQSEQKLQQESQTSKEKLTEIQQSLQE 1227
Query: 143 ARESGVILEGALARAEEKVRVAEANAEA---KIKEA---VQGESAAVSEKQELLAYVDKL 196
++S E + EEKVR + + EA K+ E+ ++ +++ + E Q+ L K
Sbjct: 1228 LQDSVKQKEELVQNLEEKVRESSSIIEAQNTKLNESNVQLENKTSCLKETQDQLLESQKK 1287
Query: 197 KAQLQRQHID-STEVFEKTESCSDTK 221
+ QLQ + S E+ + E+ D K
Sbjct: 1288 EKQLQEEAAKLSGELQQVQEANGDIK 1313
Score = 42.4 bits (98), Expect = 0.016
Identities = 41/236 (17%), Positives = 94/236 (39%), Gaps = 29/236 (12%)
Query: 6 RNTLGMMTNKTTDGPMDDLTIMKETLLV-KDEELQNFARDLRTRDSTIRDIAEKLSETAE 64
+ TLG ++ +++L KET++ K++ELQ +S ++ + +L + +
Sbjct: 786 QKTLG---HEKLQAALEELKKEKETIIKEKEQELQQLQSKSAESESALKVVQVQLEQLQQ 842
Query: 65 AAEAAASAAHTIDEQWRNACEEIESLKKDSEKQLELSAQKLKEYEVKIISLSKEKEQLSK 124
A A+ + + +++S ++++ +L+ + L E SK
Sbjct: 843 QAAASGEEGSKTVAKLHDEISQLKSQAEETQSELKSTQSNL--------------EAKSK 888
Query: 125 QREAATQEANMWRSELGKARESGVILEGALARAEEKVRVAEANAEAKIKEAVQGESAAVS 184
Q EAA + G E L+ + + + + E+K K+ +A
Sbjct: 889 QLEAANGSLEEEAKKSGHLLEQITKLKSEVGETQAALSSCHTDVESKTKQLEAANAA--- 945
Query: 185 EKQELLAYVDKLKAQLQRQHIDSTEVFEKTESCSDTKHVDLTEENVDKACLSVSRS 240
++K+ + +++++ +K + +DT H +L E + L S
Sbjct: 946 --------LEKVNKEYAESRAEASDLQDKVKEITDTLHAELQAERSSSSALHTKLS 993
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.307 0.122 0.315
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 418,326,353
Number of Sequences: 2790947
Number of extensions: 15295560
Number of successful extensions: 114542
Number of sequences better than 10.0: 7874
Number of HSP's better than 10.0 without gapping: 970
Number of HSP's successfully gapped in prelim test: 7101
Number of HSP's that attempted gapping in prelim test: 91891
Number of HSP's gapped (non-prelim): 21616
length of query: 306
length of database: 848,049,833
effective HSP length: 127
effective length of query: 179
effective length of database: 493,599,564
effective search space: 88354321956
effective search space used: 88354321956
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0057b.10


