

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
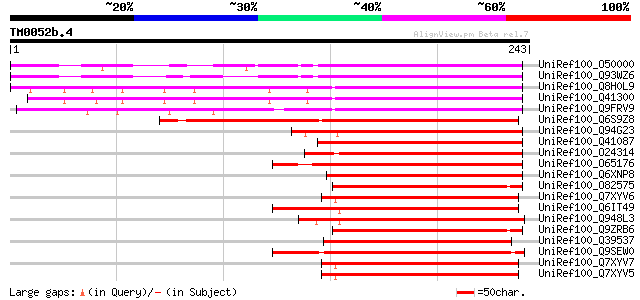
Query= TM0052b.4
(243 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O50000 Abscisic stress ripening protein homolog [Prunu... 234 1e-60
UniRef100_Q93WZ6 Abscisic stress ripening-like protein [Prunus p... 227 2e-58
UniRef100_Q8H0L9 DS2 protein [Solanum tuberosum] 211 2e-53
UniRef100_Q41300 Abscisic stress ripening protein [Solanum chaco... 209 5e-53
UniRef100_Q9FRV9 Putative ripening protein [Calystegia soldanella] 190 2e-47
UniRef100_Q6S9Z8 ASR protein [Ginkgo biloba] 183 4e-45
UniRef100_Q94G23 Putative transcription factor [Vitis vinifera] 174 1e-42
UniRef100_Q41087 LP3-1 [Pinus taeda] 157 3e-37
UniRef100_O24314 Water deficit inducible protein LP3 [Pinus taeda] 155 9e-37
UniRef100_O65176 ABA stress ripening protein [Mesembryanthemum c... 155 1e-36
UniRef100_Q6XNP8 ASR-like protein 1 [Hevea brasiliensis] 147 2e-34
UniRef100_O82575 Fruit-ripening protein [Lycopersicon esculentum] 146 4e-34
UniRef100_Q7XYV6 ASR2 [Lycopersicon hirsutum] 144 2e-33
UniRef100_Q6IT49 Abscisic stress ripening protein-like protein [... 144 2e-33
UniRef100_Q948L3 Drought inducible 22 kD protein [Saccharum offi... 144 2e-33
UniRef100_Q9ZRB6 Ci21A protein [Solanum tuberosum] 144 3e-33
UniRef100_Q39537 ASR [Citrus maxima] 144 3e-33
UniRef100_Q9SEW0 Anth [Lilium longiflorum] 142 6e-33
UniRef100_Q7XYV7 ASR2 [Lycopersicon chilense] 142 1e-32
UniRef100_Q7XYV5 ASR2 [Lycopersicon peruvianum var. humifusum] 141 2e-32
>UniRef100_O50000 Abscisic stress ripening protein homolog [Prunus armeniaca]
Length = 200
Score = 234 bits (597), Expect = 1e-60
Identities = 130/242 (53%), Positives = 146/242 (59%), Gaps = 44/242 (18%)
Query: 1 MAEENRDRGLFHHHKNEDRPTDTETGYNKTSYSTDEPSGGDY-DSGYKKSSYETSGGGYE 59
M+EE GLFHHHK+EDRP +T +D P G Y D G S Y GGGY
Sbjct: 1 MSEEKHHHGLFHHHKDEDRPIET----------SDYPQSGGYSDEGRTGSGYGGGGGGY- 49
Query: 60 TGYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYS-TGGGYGTGG 118
GGGGY G GGGYG A G G GYG GG
Sbjct: 50 ---------------GGGGGYGDG------------GGGYGDNTAYSGEGRPGSGYG-GG 81
Query: 119 GGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGELGTTAAGAY 178
GG GE + GG Y YG +G E+DYK+EEKHHK LEHL E G AAG +
Sbjct: 82 GGYGESADYSDGGRYKE-TAAYGT--TGTNESEIDYKKEEKHHKHLEHLSEPGAAAAGVF 138
Query: 179 ALHEKHKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHH 238
ALHEKH+S+KDPE+AH+HK+EEE+AAAAAVG+GGFAFHEHHEKKEAKEEEEESHGKKHHH
Sbjct: 139 ALHEKHESKKDPEHAHKHKIEEEIAAAAAVGSGGFAFHEHHEKKEAKEEEEESHGKKHHH 198
Query: 239 LF 240
LF
Sbjct: 199 LF 200
>UniRef100_Q93WZ6 Abscisic stress ripening-like protein [Prunus persica]
Length = 193
Score = 227 bits (578), Expect = 2e-58
Identities = 124/240 (51%), Positives = 142/240 (58%), Gaps = 47/240 (19%)
Query: 1 MAEENRDRGLFHHHKNEDRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYET 60
M+EE GLFHHHK+EDRP +T +D P G Y + S GGGY
Sbjct: 1 MSEEKHHHGLFHHHKDEDRPIET----------SDYPQSGGYSDEGRTGSGYGGGGGY-- 48
Query: 61 GYNKTSSYSTDEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGG 120
GGGGY + TAYS + G GYG GGGG
Sbjct: 49 -------------GDGGGGYG---DNTAYSGEGRPGSGYG----------------GGGG 76
Query: 121 GGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGELGTTAAGAYAL 180
GE + GG Y YG +G E+DYK+EEKHHK LEHL E G AAG +AL
Sbjct: 77 YGESADYSDGGRYKE-TAAYGT--TGTHESEIDYKKEEKHHKHLEHLSEAGAAAAGVFAL 133
Query: 181 HEKHKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHLF 240
HEKH+S+KDPE+AHRHK+EEE+AAAAAVG+GGFAFHEHHEKKEAKEEEEESHGKKHHHLF
Sbjct: 134 HEKHESKKDPEHAHRHKIEEEIAAAAAVGSGGFAFHEHHEKKEAKEEEEESHGKKHHHLF 193
>UniRef100_Q8H0L9 DS2 protein [Solanum tuberosum]
Length = 268
Score = 211 bits (536), Expect = 2e-53
Identities = 128/268 (47%), Positives = 156/268 (57%), Gaps = 29/268 (10%)
Query: 1 MAEENRDR--GLFHHHKNEDRPTDTE-TGYNKTSYSTDEP--------SGGDYDSGYKKS 49
MAE+ + GLFHHHKNE+ T E T Y +T+Y E S GD G K +
Sbjct: 1 MAEQKKHHFGGLFHHHKNEEEDTPVEKTTYEETTYGESEKTRTYGEKTSYGDDTYGEKTT 60
Query: 50 SY-------ETSGGGYETGYNKTSSYSTD----EPNSGGGGYDSGYN-KTAYSTDEPSGG 97
++ E + G +T K +SY D E S G G D+ Y KT+Y + +
Sbjct: 61 TFGDDNKYGEKTSYGDDTYGEKPTSYGGDNTYGEKTSYGEGDDNKYGEKTSYGEGDDNQY 120
Query: 98 GYGTGGAGGGYSTGGGYGTGGGG--GGEYSTGGGGGGYSTGGG---GYGNDDSGNRRDEV 152
G T GY YG G G + S G GGY G G Y ++S + E
Sbjct: 121 GEKTSYGDSGYGEKPSYGGGDDNKYGEKTSYGNEEGGYGGGVGETTNYEENESETKTSE- 179
Query: 153 DYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAAAAAVGAGG 212
DYKEE+KHHK LE +G LG AAGA+ALHEKHK+EKDPENAH+HK+EE +AAAAA+GAGG
Sbjct: 180 DYKEEKKHHKHLEEIGGLGAVAAGAFALHEKHKAEKDPENAHKHKIEEGIAAAAAIGAGG 239
Query: 213 FAFHEHHEKKEAKEEEEESHGKKHHHLF 240
FAFHEHHEKKEAKEEEEE+ GKK HH F
Sbjct: 240 FAFHEHHEKKEAKEEEEEAEGKKKHHFF 267
>UniRef100_Q41300 Abscisic stress ripening protein [Solanum chacoense]
Length = 263
Score = 209 bits (532), Expect = 5e-53
Identities = 124/258 (48%), Positives = 151/258 (58%), Gaps = 27/258 (10%)
Query: 9 GLFHHHKNEDRPTDTE-TGYNKTSYSTDEPSG--------GDYDSGYKKSSY-------E 52
GLFHHHKNE+ T E T Y +T+Y E + GD G K +++ E
Sbjct: 6 GLFHHHKNEEEDTPVEKTTYEETTYGESEKTSTYGEKTSYGDDTYGEKTTTFGDDNKYGE 65
Query: 53 TSGGGYETGYNKTSSYSTD----EPNSGGGGYDSGYN-KTAYSTDEPSGGGYGTGGAGGG 107
+ G +T K +SY D E S G G D+ Y KT+Y + + G T G
Sbjct: 66 KTSYGDDTYGEKPTSYGGDNTYGEKTSYGKGDDNKYGEKTSYGEGDDNKYGEKTSYGDSG 125
Query: 108 YSTGGGYGTGGGG--GGEYSTGGGGGGYSTGGG---GYGNDDSGNRRDEVDYKEEEKHHK 162
Y YG G G + S G GGY G G Y ++S + E DYKEE+KHHK
Sbjct: 126 YGEKPSYGGGDDNKYGEKTSYGNEEGGYGGGVGETTNYEENESETKTSE-DYKEEKKHHK 184
Query: 163 KLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKK 222
LE +G LG AAGA+ALHEKHK+EKDPENAH+HK+EE +AAAAA+GAGGFAFHEHHEKK
Sbjct: 185 HLEEIGGLGAVAAGAFALHEKHKAEKDPENAHKHKIEEGIAAAAAIGAGGFAFHEHHEKK 244
Query: 223 EAKEEEEESHGKKHHHLF 240
EAKEEEEE+ GKK HH F
Sbjct: 245 EAKEEEEEAEGKKKHHFF 262
>UniRef100_Q9FRV9 Putative ripening protein [Calystegia soldanella]
Length = 248
Score = 190 bits (483), Expect = 2e-47
Identities = 114/251 (45%), Positives = 136/251 (53%), Gaps = 19/251 (7%)
Query: 4 ENRDRGLFHHHKNEDRPTDTETGYNKTSYSTD--EPSGGDYDSGYKKS-------SYETS 54
E + LF HHK+++ G N S D E D Y+K SYE
Sbjct: 3 EEKHHHLFGHHKDKEEERQPSYGENTYGSSDDSYERKNTYGDDSYEKKNTYGGDDSYERK 62
Query: 55 GGGYETGYNKTSSYSTDEP---NSGGGGYDSGYNKTAYSTDEP--SGGGYGTGGAGGGYS 109
E Y K ++Y DE + GG +S K Y DE YG +
Sbjct: 63 NTYGEDSYEKKNTYGGDESYERKNTYGGDESYEKKNTYGGDESYERKNNYGDNNESSYEN 122
Query: 110 TGGGYGTGGGGGGEYSTGGGGGGYSTGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGE 169
GYG G Y GGG G G ++ G R E DY++E+KHHK LE LG
Sbjct: 123 KPTGYGKTSYGEDSY----GGGQTDKYGSATGIEEEGGRTHE-DYEKEKKHHKHLEELGG 177
Query: 170 LGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEE 229
LGT AAGAYAL+EKH+++KDPENAHRHK+EEEVAAA AVG+GGFAFHEHHEKKE KEEEE
Sbjct: 178 LGTVAAGAYALYEKHEAKKDPENAHRHKIEEEVAAAVAVGSGGFAFHEHHEKKETKEEEE 237
Query: 230 ESHGKKHHHLF 240
E+ GKK HH F
Sbjct: 238 EAEGKKKHHFF 248
Score = 38.1 bits (87), Expect = 0.20
Identities = 23/77 (29%), Positives = 30/77 (38%)
Query: 3 EENRDRGLFHHHKNEDRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYETGY 62
+E+ +R + NE + TGY KTSY D GG D + E GG Y
Sbjct: 103 DESYERKNNYGDNNESSYENKPTGYGKTSYGEDSYGGGQTDKYGSATGIEEEGGRTHEDY 162
Query: 63 NKTSSYSTDEPNSGGGG 79
K + GG G
Sbjct: 163 EKEKKHHKHLEELGGLG 179
>UniRef100_Q6S9Z8 ASR protein [Ginkgo biloba]
Length = 181
Score = 183 bits (464), Expect = 4e-45
Identities = 93/168 (55%), Positives = 115/168 (68%), Gaps = 4/168 (2%)
Query: 71 DEPNSGGGGYDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGGGGGEYSTGGGG 130
DE N G GYN + ++PS GYG Y+TG GY G G + +G G
Sbjct: 15 DEDNVDSEG---GYNTGGDAYNKPSDDGYGGAYGSSDYNTGSGYNDTGSGYNDTGSGYGY 71
Query: 131 GGYSTGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDP 190
G T Y +DS ++ D +EEKHHK++EH+GELGT AAGAYA++EKH+++KDP
Sbjct: 72 GEKVTETTVYATEDS-SQDDYEKAMKEEKHHKRMEHVGELGTMAAGAYAMYEKHEAKKDP 130
Query: 191 ENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHH 238
E+AHRHK+EEEVAAAAAVGAGG+AFHEHHEKKE KEE EE+ GKKHHH
Sbjct: 131 EHAHRHKIEEEVAAAAAVGAGGYAFHEHHEKKEDKEEAEEASGKKHHH 178
>UniRef100_Q94G23 Putative transcription factor [Vitis vinifera]
Length = 149
Score = 174 bits (442), Expect = 1e-42
Identities = 83/111 (74%), Positives = 95/111 (84%), Gaps = 3/111 (2%)
Query: 133 YSTGG-GGYGNDDSGNRRDEV--DYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKD 189
Y+T G GY + + D+ DY++EEKHHK LEHLGELG AAGAYALHEKHKSEKD
Sbjct: 39 YATDGVSGYAAETTEVLADDPAPDYRKEEKHHKHLEHLGELGVAAAGAYALHEKHKSEKD 98
Query: 190 PENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHLF 240
PE+AH+HK+EEE+AAAAAVGAGGFAFHEHHEKKEAKEE+EE+HGKKHHHLF
Sbjct: 99 PEHAHKHKIEEEIAAAAAVGAGGFAFHEHHEKKEAKEEDEEAHGKKHHHLF 149
Score = 32.7 bits (73), Expect = 8.4
Identities = 19/49 (38%), Positives = 26/49 (52%), Gaps = 13/49 (26%)
Query: 1 MAEENRDRGLFHHHKNEDRPTD----------TETGYNKTSYSTDEPSG 39
M+EE LFHH +D+P D ++T Y+ TSY+TD SG
Sbjct: 1 MSEEKHHHHLFHH---KDKPVDDAVPYSDNAYSDTTYSDTSYATDGVSG 46
>UniRef100_Q41087 LP3-1 [Pinus taeda]
Length = 126
Score = 157 bits (396), Expect = 3e-37
Identities = 70/96 (72%), Positives = 84/96 (86%)
Query: 145 SGNRRDEVDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAA 204
+GN + K++EKHHK +EHLGE+GT AAGA+ALHEKH +KDPE+AHRHK+EEEVAA
Sbjct: 31 AGNVDEYEKAKKDEKHHKHMEHLGEMGTVAAGAFALHEKHADKKDPEHAHRHKIEEEVAA 90
Query: 205 AAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHLF 240
AAAVGAGG+ FHEHHEKKE+KEEE+E+ GKKHHHLF
Sbjct: 91 AAAVGAGGYVFHEHHEKKESKEEEKEAEGKKHHHLF 126
>UniRef100_O24314 Water deficit inducible protein LP3 [Pinus taeda]
Length = 153
Score = 155 bits (392), Expect = 9e-37
Identities = 73/102 (71%), Positives = 85/102 (82%), Gaps = 2/102 (1%)
Query: 139 GYGNDDSGNRRDEVDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKL 198
G NDD ++ ++EEKHHK LE LG LGT AAGA+ALHEKH S+KDPENAHRHK+
Sbjct: 49 GEVNDDKFAEYEKA--RKEEKHHKHLEELGGLGTVAAGAFALHEKHASKKDPENAHRHKI 106
Query: 199 EEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHLF 240
EEE+AAAAAVGAGG+ FHEHHEKKE+KEEE+E+ GKKHHHLF
Sbjct: 107 EEEIAAAAAVGAGGYVFHEHHEKKESKEEEKEAEGKKHHHLF 148
>UniRef100_O65176 ABA stress ripening protein [Mesembryanthemum crystallinum]
Length = 143
Score = 155 bits (391), Expect = 1e-36
Identities = 75/117 (64%), Positives = 85/117 (72%), Gaps = 6/117 (5%)
Query: 124 YSTGGGGGGYSTGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGELGTTAAGAYALHEK 183
YS G G GYS + E DY +EE+HHK EH+GELG AAGA+ALHEK
Sbjct: 33 YSGDGYGTGYSE------TTTAVIAEPEKDYAKEERHHKHKEHMGELGAVAAGAFALHEK 86
Query: 184 HKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHLF 240
HK EKDPE+AHRHK+EEE+AAAA VGAGG+ FHEHHEKKEAKEE +E GK HHHLF
Sbjct: 87 HKIEKDPEHAHRHKIEEEIAAAARVGAGGYVFHEHHEKKEAKEERKEHEGKHHHHLF 143
>UniRef100_Q6XNP8 ASR-like protein 1 [Hevea brasiliensis]
Length = 108
Score = 147 bits (371), Expect = 2e-34
Identities = 66/92 (71%), Positives = 79/92 (85%)
Query: 149 RDEVDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAAAAAV 208
++ VD ++EEKHHK L LG+LG AAG YALHEKH ++KDPE+AH HK++EEVAAAAA+
Sbjct: 17 QEAVDCRKEEKHHKHLCQLGKLGAAAAGTYALHEKHAAKKDPEHAHGHKIKEEVAAAAAI 76
Query: 209 GAGGFAFHEHHEKKEAKEEEEESHGKKHHHLF 240
GAGGFAFHEHHEKKEAK++ EE HGKKHHH F
Sbjct: 77 GAGGFAFHEHHEKKEAKKKNEEGHGKKHHHPF 108
>UniRef100_O82575 Fruit-ripening protein [Lycopersicon esculentum]
Length = 110
Score = 146 bits (369), Expect = 4e-34
Identities = 66/89 (74%), Positives = 82/89 (91%), Gaps = 1/89 (1%)
Query: 152 VDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAAAAAVGAG 211
VDY++E KHHK LE +G+LGT AAGAYALHEKH+++KDPE+AH+HK+EEE+AAAAAVGAG
Sbjct: 23 VDYEKEIKHHKHLEQIGKLGTVAAGAYALHEKHEAKKDPEHAHKHKIEEEIAAAAAVGAG 82
Query: 212 GFAFHEHHEKKEAKEEEEESHGKKHHHLF 240
GFAFHEHHEKK+AK+EE+++ G HHHLF
Sbjct: 83 GFAFHEHHEKKDAKKEEKKAEG-GHHHLF 110
>UniRef100_Q7XYV6 ASR2 [Lycopersicon hirsutum]
Length = 114
Score = 144 bits (364), Expect = 2e-33
Identities = 65/95 (68%), Positives = 80/95 (83%), Gaps = 3/95 (3%)
Query: 147 NRRDE---VDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVA 203
N+ DE VDY++E KHH LE +GELG AAGA+ALHEKHK++KDPENAH+HK+EEE+A
Sbjct: 17 NKEDEGGPVDYEKEVKHHSHLEKIGELGAVAAGAFALHEKHKAKKDPENAHKHKIEEEIA 76
Query: 204 AAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHH 238
A AAVGAGGFAFHEHH+KK+AK+E++E G HHH
Sbjct: 77 AVAAVGAGGFAFHEHHQKKDAKKEKKEVEGGHHHH 111
>UniRef100_Q6IT49 Abscisic stress ripening protein-like protein [Musa acuminata]
Length = 143
Score = 144 bits (363), Expect = 2e-33
Identities = 67/116 (57%), Positives = 87/116 (74%), Gaps = 1/116 (0%)
Query: 124 YSTGGGGGGYSTGGGGYGNDDSGNRRDEVD-YKEEEKHHKKLEHLGELGTTAAGAYALHE 182
YS GG G + + DE + YK+EEKHHK EHLGE+G AAGA+AL+E
Sbjct: 25 YSETAYSGGDDYASGYTETVVAESASDEYEKYKKEEKHHKHKEHLGEMGAVAAGAFALYE 84
Query: 183 KHKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHH 238
KH+++KDP++AH+HK+EEE+AAA AVG+GG+AFHEHHEK++AK E EE+ GKKHHH
Sbjct: 85 KHEAKKDPDHAHKHKIEEEIAAAVAVGSGGYAFHEHHEKRDAKNEAEEASGKKHHH 140
Score = 43.9 bits (102), Expect = 0.004
Identities = 28/79 (35%), Positives = 39/79 (48%), Gaps = 6/79 (7%)
Query: 1 MAEENRDRGLFHHHKNEDRPTDTETGYNKTSYSTDEPSGGDYDSGYKKSSYETSGGGYET 60
MAEE LFHHHK E++P + E Y++T+YS G DY SGY ++ S
Sbjct: 1 MAEEKHHHRLFHHHK-EEKPAE-EVIYSETAYS----GGDDYASGYTETVVAESASDEYE 54
Query: 61 GYNKTSSYSTDEPNSGGGG 79
Y K + + + G G
Sbjct: 55 KYKKEEKHHKHKEHLGEMG 73
>UniRef100_Q948L3 Drought inducible 22 kD protein [Saccharum officinarum]
Length = 142
Score = 144 bits (363), Expect = 2e-33
Identities = 71/117 (60%), Positives = 86/117 (72%), Gaps = 11/117 (9%)
Query: 136 GGGGYGN----------DDSGNRRDEVD-YKEEEKHHKKLEHLGELGTTAAGAYALHEKH 184
GGGGYG + DE D YK+EEK HK +HLGE G AAGA+AL+EKH
Sbjct: 26 GGGGYGEAAEYTETTVTEVVSTGEDEYDKYKKEEKEHKHKQHLGEAGAIAAGAFALYEKH 85
Query: 185 KSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHLFG 241
+++KDPE+AHRHK+EEEVAAAAAVG+GGFAFHEHHEKK+ +E EE+ G+K HH FG
Sbjct: 86 EAKKDPEHAHRHKIEEEVAAAAAVGSGGFAFHEHHEKKKDHKEAEEAGGEKKHHFFG 142
>UniRef100_Q9ZRB6 Ci21A protein [Solanum tuberosum]
Length = 109
Score = 144 bits (362), Expect = 3e-33
Identities = 64/89 (71%), Positives = 81/89 (90%), Gaps = 1/89 (1%)
Query: 152 VDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAAAAAVGAG 211
VDY++E KHHK LE +G+LGT AAGAYALHEKH+++KDPE+AH+HK+EEE+AAAAAVGAG
Sbjct: 22 VDYEKETKHHKHLEQIGKLGTVAAGAYALHEKHEAKKDPEHAHKHKIEEEIAAAAAVGAG 81
Query: 212 GFAFHEHHEKKEAKEEEEESHGKKHHHLF 240
GFA HEHHEKK+AK+E++++ G HHHLF
Sbjct: 82 GFALHEHHEKKDAKKEQKKAEG-GHHHLF 109
Score = 43.1 bits (100), Expect = 0.006
Identities = 28/84 (33%), Positives = 36/84 (42%), Gaps = 22/84 (26%)
Query: 156 EEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDP----ENAHRHKLEEEVAAAAAVGAG 211
EEEKHH L H K K E+ P + HK E++ V AG
Sbjct: 2 EEEKHHHHL---------------FHHKDKEEEGPVDYEKETKHHKHLEQIGKLGTVAAG 46
Query: 212 GFAFHEHHEKKEAKEEEEESHGKK 235
+A HE HE AK++ E +H K
Sbjct: 47 AYALHEKHE---AKKDPEHAHKHK 67
>UniRef100_Q39537 ASR [Citrus maxima]
Length = 98
Score = 144 bits (362), Expect = 3e-33
Identities = 63/88 (71%), Positives = 77/88 (86%)
Query: 148 RRDEVDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVAAAAA 207
+ +E DYK+EEKHHK LEHLG LGT AGA+ E HK+EKDP++AHRHK+EEE+AAAA
Sbjct: 4 KEEEFDYKKEEKHHKHLEHLGGLGTAGAGAFPRLESHKAEKDPDHAHRHKIEEEIAAAAG 63
Query: 208 VGAGGFAFHEHHEKKEAKEEEEESHGKK 235
+G+GGFAFHEHHEKKEAKEE++E+HGKK
Sbjct: 64 LGSGGFAFHEHHEKKEAKEEDQEAHGKK 91
>UniRef100_Q9SEW0 Anth [Lilium longiflorum]
Length = 142
Score = 142 bits (359), Expect = 6e-33
Identities = 70/118 (59%), Positives = 86/118 (72%), Gaps = 3/118 (2%)
Query: 124 YSTGGGGGGYSTGGGGYGNDDSGNRRDEVDYKEEEKHHKKLEHLGELGTTAAGAYALHEK 183
Y T G YST YG + S + + DY++E+KH K EHLGE+ T AGA+AL+EK
Sbjct: 28 YITDGLNSTYSTSSDVYGGNQS--QANYEDYEKEKKHIKHKEHLGEMATAGAGAFALYEK 85
Query: 184 HKSEKDPENAHRHKLEEEVAAAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHHLFG 241
H+++KDPE+AHRHKLEEE+AAAAA GAGG+ FHEHHEKK K+E EE GKK HH FG
Sbjct: 86 HEAKKDPEHAHRHKLEEEIAAAAAAGAGGYTFHEHHEKKTLKKENEEVEGKK-HHFFG 142
>UniRef100_Q7XYV7 ASR2 [Lycopersicon chilense]
Length = 115
Score = 142 bits (357), Expect = 1e-32
Identities = 65/95 (68%), Positives = 79/95 (82%), Gaps = 3/95 (3%)
Query: 147 NRRDE---VDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVA 203
NR DE VDY++E KHH LE +GELG AAGA+ALHEKHK++KD ENAH+HK+EEE+A
Sbjct: 17 NREDEGGPVDYEKEVKHHSHLEKIGELGAVAAGAFALHEKHKAKKDLENAHKHKIEEEIA 76
Query: 204 AAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHH 238
A AAVGAGGFAFHEHH+KK+AK+E++E G HHH
Sbjct: 77 AVAAVGAGGFAFHEHHQKKDAKKEKKEVEGGHHHH 111
>UniRef100_Q7XYV5 ASR2 [Lycopersicon peruvianum var. humifusum]
Length = 112
Score = 141 bits (355), Expect = 2e-32
Identities = 63/95 (66%), Positives = 79/95 (82%), Gaps = 3/95 (3%)
Query: 147 NRRDE---VDYKEEEKHHKKLEHLGELGTTAAGAYALHEKHKSEKDPENAHRHKLEEEVA 203
N+ DE VDY++E KHH LE +GELG AAGA+ LHEKHK++KDPE+AH+HK+EEE+A
Sbjct: 17 NKEDEGGPVDYEKEVKHHSHLEKIGELGAVAAGAFGLHEKHKAKKDPEHAHKHKIEEEIA 76
Query: 204 AAAAVGAGGFAFHEHHEKKEAKEEEEESHGKKHHH 238
A AAVGAGGFAFHEHH+KK+AK+E +E+ G HHH
Sbjct: 77 AVAAVGAGGFAFHEHHQKKDAKKEGKEAEGGHHHH 111
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.303 0.131 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 577,446,423
Number of Sequences: 2790947
Number of extensions: 36198722
Number of successful extensions: 745417
Number of sequences better than 10.0: 12000
Number of HSP's better than 10.0 without gapping: 7328
Number of HSP's successfully gapped in prelim test: 5073
Number of HSP's that attempted gapping in prelim test: 209798
Number of HSP's gapped (non-prelim): 122917
length of query: 243
length of database: 848,049,833
effective HSP length: 124
effective length of query: 119
effective length of database: 501,972,405
effective search space: 59734716195
effective search space used: 59734716195
T: 11
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.7 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0052b.4


