

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
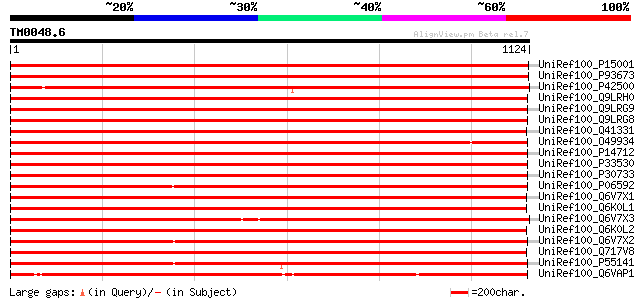
Query= TM0048.6
(1124 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P15001 Phytochrome A [Pisum sativum] 2004 0.0
UniRef100_P93673 Phytochrome type A [Lathyrus sativus] 1998 0.0
UniRef100_P42500 Phytochrome A [Glycine max] 1882 0.0
UniRef100_Q9LRH0 Phytochrome A [Armoracia rusticana] 1816 0.0
UniRef100_Q9LRG9 Phytochrome A [Armoracia rusticana] 1815 0.0
UniRef100_Q9LRG8 Phytochrome A [Armoracia rusticana] 1814 0.0
UniRef100_Q41331 PhyA protein [Lycopersicon esculentum] 1810 0.0
UniRef100_O49934 Phytochrome A [Populus tremuloides] 1808 0.0
UniRef100_P14712 Phytochrome A [Arabidopsis thaliana] 1808 0.0
UniRef100_P33530 Phytochrome A1 [Nicotiana tabacum] 1806 0.0
UniRef100_P30733 Phytochrome A [Solanum tuberosum] 1798 0.0
UniRef100_P06592 Phytochrome A [Cucurbita pepo] 1786 0.0
UniRef100_Q6V7X1 Phytochrome A [Monotropastrum globosum] 1776 0.0
UniRef100_Q6K0L1 PHYA4 photoreceptor [Stellaria longipes] 1699 0.0
UniRef100_Q6V7X3 Phytochrome A [Cuscuta pentagona] 1696 0.0
UniRef100_Q6K0L2 PHYA3 photoreceptor [Stellaria longipes] 1692 0.0
UniRef100_Q6V7X2 Phytochrome A [Orobanche minor] 1678 0.0
UniRef100_Q717V8 Phytochrome A1 [Stellaria longipes] 1667 0.0
UniRef100_P55141 Phytochrome A [Petroselinum crispum] 1657 0.0
UniRef100_Q6VAP1 Phytochrome A [Cyrtosia septentrionalis] 1522 0.0
>UniRef100_P15001 Phytochrome A [Pisum sativum]
Length = 1124
Score = 2004 bits (5191), Expect = 0.0
Identities = 1001/1123 (89%), Positives = 1063/1123 (94%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MS++RPSQSSNNSGRSR+SAR+IAQTTVDAK+HA FEESGSSFDYSSSVR SG+ D D Q
Sbjct: 1 MSTTRPSQSSNNSGRSRNSARIIAQTTVDAKLHATFEESGSSFDYSSSVRVSGSVDGDQQ 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
P+SNKVTTAYL+HIQRGK IQPFGCLLALDEKTCKV+AYSENAPEMLTMVSHAVPSVG+H
Sbjct: 61 PRSNKVTTAYLNHIQRGKQIQPFGCLLALDEKTCKVVAYSENAPEMLTMVSHAVPSVGDH 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
PALGI TDIRT+FTAPSASALQKALGFAEV+LLNPILVHCKTSGKPFYAIIHRVTGSLII
Sbjct: 121 PALGIGTDIRTVFTAPSASALQKALGFAEVSLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSL SGSMERLCDTMVQEVFELTGYDRVM
Sbjct: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLASGSMERLCDTMVQEVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFHEDDHGEVIAEI KPGLEPYLGLHYPATDIPQA+RFLFMKNKVRMIVDC+AK VKV
Sbjct: 241 AYKFHEDDHGEVIAEIAKPGLEPYLGLHYPATDIPQAARFLFMKNKVRMIVDCNAKHVKV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQK 360
L DEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVND+DEDGD +D+V PQK
Sbjct: 301 LQDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDSDEDGDSADAVLPQK 360
Query: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLC 420
+KRLWGLVVCHNT+PRFVPFPLRYACEFLAQVFAIHVNKEIELE QILEKNILRTQTLLC
Sbjct: 361 KKRLWGLVVCHNTTPRFVPFPLRYACEFLAQVFAIHVNKEIELEYQILEKNILRTQTLLC 420
Query: 421 DMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHTD 480
DMLMRDAPLGI++QSPN+MDLVKCDGAAL Y+NK+W+LG TP+E +R+IA W+S+YHTD
Sbjct: 421 DMLMRDAPLGIVSQSPNIMDLVKCDGAALFYRNKLWLLGATPTESQLREIALWMSEYHTD 480
Query: 481 STGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGE 540
STGLSTDSLSDAGFPGALSL D VCGMAAVRIT KD+VFWFRSHTAAEIRWGGAKHEPG+
Sbjct: 481 STGLSTDSLSDAGFPGALSLSDTVCGMAAVRITSKDIVFWFRSHTAAEIRWGGAKHEPGD 540
Query: 541 QDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAID 600
QDDG+KMHPRSSFKAFLEVV+ARS PWKD+EMDAIHSLQLILRNA KDTD +D+NT AI+
Sbjct: 541 QDDGRKMHPRSSFKAFLEVVKARSVPWKDFEMDAIHSLQLILRNASKDTDIIDLNTKAIN 600
Query: 601 TRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAI 660
TRL+DLKIEGMQELEAVTSEMVRLIETATVPILAVD+DG VNGWNIKIAELTGLPVGEAI
Sbjct: 601 TRLNDLKIEGMQELEAVTSEMVRLIETATVPILAVDVDGTVNGWNIKIAELTGLPVGEAI 660
Query: 661 GKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDL 720
GKHLLTLVED STD VKKML+LAL GEEEKNVQFEIKTHG ++ESGPISL+VNACAS+DL
Sbjct: 661 GKHLLTLVEDSSTDIVKKMLNLALQGEEEKNVQFEIKTHGDQVESGPISLIVNACASKDL 720
Query: 721 RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNPA 780
RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPN LIPPIFGTDEFGWCCEWN A
Sbjct: 721 RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNQLIPPIFGTDEFGWCCEWNAA 780
Query: 781 MTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGF 840
M KLTGWKREEVMDKMLLGEVFGT M+ CRLKNQEAFVNFGIVLNKAMTG ETEKV FGF
Sbjct: 781 MIKLTGWKREEVMDKMLLGEVFGTQMSCCRLKNQEAFVNFGIVLNKAMTGLETEKVPFGF 840
Query: 841 FARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALT 900
F+R GKYVECLLSVSKK+D EGLVTGVFCFLQLASPELQQALHIQ LSEQTALKRLK LT
Sbjct: 841 FSRKGKYVECLLSVSKKIDAEGLVTGVFCFLQLASPELQQALHIQRLSEQTALKRLKVLT 900
Query: 901 YMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYL 960
YMKRQIRNPL+GIVFS K LEGTDL EQKR+V+TS+QCQRQLSKILDDSDLD I+DGYL
Sbjct: 901 YMKRQIRNPLAGIVFSSKMLEGTDLETEQKRIVNTSSQCQRQLSKILDDSDLDGIIDGYL 960
Query: 961 DLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLL 1020
DLEMAEFTL +VL+TSLSQ+M RS+ +GIRI NDVAE I E LYGDSLRLQQVLADFLL
Sbjct: 961 DLEMAEFTLHEVLVTSLSQVMNRSNTKGIRIANDVAEHIARETLYGDSLRLQQVLADFLL 1020
Query: 1021 ISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLESE 1080
ISIN TPNGGQVV+AASLTKEQLGKSVHL NLELSITHGGSGVPEA LNQMFGN+ LESE
Sbjct: 1021 ISINSTPNGGQVVIAASLTKEQLGKSVHLVNLELSITHGGSGVPEAALNQMFGNNVLESE 1080
Query: 1081 EGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHKLK 1123
EGISL ISRKLLKLM+GDVRYL+EAGKSSFILSVELAAAHKLK
Sbjct: 1081 EGISLHISRKLLKLMNGDVRYLKEAGKSSFILSVELAAAHKLK 1123
>UniRef100_P93673 Phytochrome type A [Lathyrus sativus]
Length = 1124
Score = 1998 bits (5176), Expect = 0.0
Identities = 999/1123 (88%), Positives = 1060/1123 (93%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MS++RPSQSSNNSGRSR+SAR+IAQTTVDAK+HA FEESGSSFDYSS VR SG+ D D Q
Sbjct: 1 MSTTRPSQSSNNSGRSRNSARIIAQTTVDAKLHATFEESGSSFDYSSWVRVSGSVDGDQQ 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
P+SNKVTTAYL+HIQRGK IQPFGCLLALDEKTCKV+AYSENAPEMLTMVSHAVPSVG+H
Sbjct: 61 PRSNKVTTAYLNHIQRGKQIQPFGCLLALDEKTCKVVAYSENAPEMLTMVSHAVPSVGDH 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
PALGI TDIRT+FTAPSASALQKALGFAEV+LLNPILVHCKTSGKPFYAIIHRVTGSLII
Sbjct: 121 PALGIGTDIRTVFTAPSASALQKALGFAEVSLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSL SGSMERLCDTMVQEVFELTGYDRVM
Sbjct: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLASGSMERLCDTMVQEVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFHEDDHGEVIAEI KPGLEPYLGLHYPATDIPQA+RFLFMKNKVRMIVDC+AK VKV
Sbjct: 241 AYKFHEDDHGEVIAEIAKPGLEPYLGLHYPATDIPQAARFLFMKNKVRMIVDCNAKHVKV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQK 360
L DEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVND+DEDGD +D+V PQK
Sbjct: 301 LQDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDSDEDGDSADAVLPQK 360
Query: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLC 420
+KRLWGLVVCHNT+PRFVPFPLRYACEFLAQVFAIHVNKEIELE QILEKNILRTQTLLC
Sbjct: 361 KKRLWGLVVCHNTTPRFVPFPLRYACEFLAQVFAIHVNKEIELEYQILEKNILRTQTLLC 420
Query: 421 DMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHTD 480
DMLMRDAPLGI++QSPN+MDLVKCDGAAL Y+NK+W+LG TP+E IR+IA W+S+YHTD
Sbjct: 421 DMLMRDAPLGIVSQSPNIMDLVKCDGAALFYRNKLWLLGATPTEYQIREIALWMSEYHTD 480
Query: 481 STGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGE 540
STGLSTDSL DAGFPGALSL D VCGMAAVRIT KD+VFWFRSHTAAEIRWGGAKHEPGE
Sbjct: 481 STGLSTDSLLDAGFPGALSLSDTVCGMAAVRITSKDIVFWFRSHTAAEIRWGGAKHEPGE 540
Query: 541 QDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAID 600
QDDG+KMHPRSSFKAFLEVV+ARS PWKD+EMDAIHSLQLILRNA KDTD +D+NT AI+
Sbjct: 541 QDDGRKMHPRSSFKAFLEVVKARSVPWKDFEMDAIHSLQLILRNASKDTDIIDLNTKAIN 600
Query: 601 TRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAI 660
TRL+DLKIEGMQELEAVTSEMVRLIETATVPILAVD+DG VNGWNIKIAELTGLPVGEAI
Sbjct: 601 TRLNDLKIEGMQELEAVTSEMVRLIETATVPILAVDVDGTVNGWNIKIAELTGLPVGEAI 660
Query: 661 GKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDL 720
GKHLLTLVED STD VKKML+LAL GEEEKNVQFEIKTHG ++E GPISL+VNACASRDL
Sbjct: 661 GKHLLTLVEDSSTDIVKKMLNLALQGEEEKNVQFEIKTHGDQVEFGPISLIVNACASRDL 720
Query: 721 RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNPA 780
RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPN LIPPIFGTDEFGWCCEWN A
Sbjct: 721 RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNQLIPPIFGTDEFGWCCEWNAA 780
Query: 781 MTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGF 840
M KLTGWKREEVMDKMLLGEVFGT M+ CRLKNQEAFVNFGIVLNKAMTG ETEKV FGF
Sbjct: 781 MIKLTGWKREEVMDKMLLGEVFGTQMSCCRLKNQEAFVNFGIVLNKAMTGLETEKVAFGF 840
Query: 841 FARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALT 900
F+R GKYVECLLSVSKK+D EGLVTGVFCFLQLASPELQQALHIQ LSEQTALKRLK LT
Sbjct: 841 FSRKGKYVECLLSVSKKIDAEGLVTGVFCFLQLASPELQQALHIQRLSEQTALKRLKVLT 900
Query: 901 YMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYL 960
YMKRQIRNPL+GIVFS K LEGTDL EQK++V+TS+QCQRQLSKILDDSDLD I+DGYL
Sbjct: 901 YMKRQIRNPLAGIVFSSKMLEGTDLETEQKQIVNTSSQCQRQLSKILDDSDLDGIIDGYL 960
Query: 961 DLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLL 1020
DLEMAEFTL +VL+TSLSQ+M RS+ +GIRI NDVAE I E LYGDSLRLQQVLADFLL
Sbjct: 961 DLEMAEFTLHEVLVTSLSQVMNRSNTKGIRIANDVAEHIAKESLYGDSLRLQQVLADFLL 1020
Query: 1021 ISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLESE 1080
ISIN TPNGGQVV+A+SLTKEQLGKSVHL NLELSITHGGSGVPEA LNQMFGN+ LESE
Sbjct: 1021 ISINSTPNGGQVVIASSLTKEQLGKSVHLVNLELSITHGGSGVPEAALNQMFGNNVLESE 1080
Query: 1081 EGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHKLK 1123
EGISL ISRKLLKLM+GDVRYL+EAGKSSFILSVELAAAHKLK
Sbjct: 1081 EGISLHISRKLLKLMNGDVRYLKEAGKSSFILSVELAAAHKLK 1123
>UniRef100_P42500 Phytochrome A [Glycine max]
Length = 1131
Score = 1882 bits (4875), Expect = 0.0
Identities = 961/1137 (84%), Positives = 1032/1137 (90%), Gaps = 19/1137 (1%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MS+SRPSQSS+NS RSRHSAR+ AQ TVDAKIHA FEESGSSFDYSSSVR SGTAD +Q
Sbjct: 1 MSTSRPSQSSSNSRRSRHSARM-AQATVDAKIHATFEESGSSFDYSSSVRVSGTADGVNQ 59
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEK----TCKVIAYSENAPEMLTMVSHAVPS 116
P+S+KVTTAYL RGK+IQPFGCLLA+DEK TCKVIAYSEN PEMLTMVSHAVPS
Sbjct: 60 PRSDKVTTAYL----RGKMIQPFGCLLAIDEKNHMQTCKVIAYSENEPEMLTMVSHAVPS 115
Query: 117 VGEHPALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTG 176
VG+HPALGI TDI+T+FTAPS S LQKALG A+V+LLNPILVHCKTSGKPFYAI+HRVTG
Sbjct: 116 VGDHPALGIGTDIKTLFTAPSVSGLQKALGCADVSLLNPILVHCKTSGKPFYAIVHRVTG 175
Query: 177 SLIIDFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGY 236
SLI+DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSG+MERLCDTMVQEVFELTGY
Sbjct: 176 SLIVDFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGNMERLCDTMVQEVFELTGY 235
Query: 237 DRVMAYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAK 296
DRVMAYKFHEDDHGEVI EITKP LEPYLGLHYPATDIPQASRFLF KNKVRMIVDCHAK
Sbjct: 236 DRVMAYKFHEDDHGEVIREITKPCLEPYLGLHYPATDIPQASRFLFRKNKVRMIVDCHAK 295
Query: 297 QVKVLIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSV 356
V+VL DEKL FDL LCGSTLRAPHSCH QYMANMDSIASLV+AVVVNDN+EDGD +D+V
Sbjct: 296 HVRVLQDEKLQFDLILCGSTLRAPHSCHAQYMANMDSIASLVLAVVVNDNEEDGD-TDAV 354
Query: 357 QPQKRKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQ 416
QPQK +RLWGLVVCHNT+PRFVPFPLRYA EFL QVFA HV+KEIELE QI+EKNIL
Sbjct: 355 QPQKTERLWGLVVCHNTTPRFVPFPLRYAREFLPQVFADHVHKEIELEYQIIEKNILHHP 414
Query: 417 TLLCDMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSK 476
L MLMRDAPLGI ++SPN+MDLVKCDGAAL+Y+NKVW LGVTPSE IR+IA WLS+
Sbjct: 415 GHLLCMLMRDAPLGIASESPNIMDLVKCDGAALIYRNKVWRLGVTPSEPQIREIALWLSE 474
Query: 477 YHTDSTGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKH 536
YH DST STDSL DAGFP ALSLGD+VCGMA+VR+T KD+VFWFRSHTAAEIRWGGAKH
Sbjct: 475 YHMDSTSFSTDSLFDAGFPSALSLGDVVCGMASVRVTAKDMVFWFRSHTAAEIRWGGAKH 534
Query: 537 EPGEQDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFK-DTDSMDIN 595
E GE+DD ++MHPRSSFKAFLEVV+ARS PWK+YEMDAIHSLQ+ILRNAFK DT+S+D+N
Sbjct: 535 EAGEKDDSRRMHPRSSFKAFLEVVKARSLPWKEYEMDAIHSLQIILRNAFKEDTESLDLN 594
Query: 596 TTAIDTRLSDLKIEG--------MQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIK 647
AI+TRL DLKIEG MQELEAVTSE+VRL TATVPILAVD+DGLVNGWNIK
Sbjct: 595 AKAINTRLRDLKIEGINDLKIERMQELEAVTSEIVRLDYTATVPILAVDVDGLVNGWNIK 654
Query: 648 IAELTGLPVGEAIGKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGP 707
IAELTGLP+GEA GKHLLTLVED STDRVKKML+LAL GEEEKNVQFEIKT GSKM+SGP
Sbjct: 655 IAELTGLPIGEATGKHLLTLVEDSSTDRVKKMLNLALLGEEEKNVQFEIKTLGSKMDSGP 714
Query: 708 ISLVVNACASRDLRENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFG 767
ISLVVN CASRDLR+NVVGVCFVA DITAQK VMDKF RIEGDYKAIVQN NPLIPPIFG
Sbjct: 715 ISLVVNRCASRDLRDNVVGVCFVAHDITAQKNVMDKFIRIEGDYKAIVQNRNPLIPPIFG 774
Query: 768 TDEFGWCCEWNPAMTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKA 827
TDEFGWCCEWNPAM KLTGWKREEVMDKMLLGE+FGT MAACRLKNQEAFVN G+VLNKA
Sbjct: 775 TDEFGWCCEWNPAMMKLTGWKREEVMDKMLLGEIFGTQMAACRLKNQEAFVNLGVVLNKA 834
Query: 828 MTGSETEKVGFGFFARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHL 887
MTGSETEKV FGFFAR+GKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQ L
Sbjct: 835 MTGSETEKVPFGFFARNGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQRL 894
Query: 888 SEQTALKRLKALTYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKIL 947
SEQTA KRL AL+YMKRQIRNPL GIVFSRK LEGTDLG EQK+L+ TSAQCQ+QLSKIL
Sbjct: 895 SEQTASKRLNALSYMKRQIRNPLCGIVFSRKMLEGTDLGTEQKQLLRTSAQCQQQLSKIL 954
Query: 948 DDSDLDSIMDGYLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGD 1007
DDSDLD+I+DGYLDLEMAEFTL +VL+TSLSQ+M +S+ + IRIVNDVA IM+E LYGD
Sbjct: 955 DDSDLDTIIDGYLDLEMAEFTLHEVLVTSLSQVMEKSNGKSIRIVNDVAGHIMMETLYGD 1014
Query: 1008 SLRLQQVLADFLLISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEAL 1067
SLRLQQVLADFLLISIN TPNGGQVVVA SLTKEQLGKSVHL LELSITHGGSGVPE L
Sbjct: 1015 SLRLQQVLADFLLISINFTPNGGQVVVAGSLTKEQLGKSVHLVKLELSITHGGSGVPEVL 1074
Query: 1068 LNQMFGNDGLESEEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHKLKA 1124
LNQMFGN+GLESEEGISLLI KLLKLM+GDVRYLREAGKS+FILS ELAAAH LKA
Sbjct: 1075 LNQMFGNNGLESEEGISLLIRAKLLKLMNGDVRYLREAGKSAFILSAELAAAHNLKA 1131
>UniRef100_Q9LRH0 Phytochrome A [Armoracia rusticana]
Length = 1122
Score = 1816 bits (4705), Expect = 0.0
Identities = 890/1122 (79%), Positives = 1010/1122 (89%), Gaps = 1/1122 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MS SRPSQSS S RSRHSAR+IAQTTVDAK+HA+FEESGSSFDYS+SVR +G +
Sbjct: 1 MSGSRPSQSSEGSRRSRHSARIIAQTTVDAKLHADFEESGSSFDYSTSVRVTGPVVENQP 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
P+S+KVTT YLHHIQ+GKLIQPFGCLLALDEKT KVIAYSENAPE+LTM SHAVPSVGEH
Sbjct: 61 PRSDKVTTTYLHHIQKGKLIQPFGCLLALDEKTFKVIAYSENAPELLTMASHAVPSVGEH 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P LGI TDIR++FTAPSASALQKALGF +V+LLNPILVHCKTS KPFYAI+HRVTGS+I+
Sbjct: 121 PVLGIGTDIRSLFTAPSASALQKALGFGDVSLLNPILVHCKTSAKPFYAIVHRVTGSIIV 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM
Sbjct: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFHEDDHGEV++E+TKPG+EPYLGLHYPATDIPQA+RFLFMKNKVRMIVDC+AK +V
Sbjct: 241 AYKFHEDDHGEVVSEVTKPGMEPYLGLHYPATDIPQAARFLFMKNKVRMIVDCNAKHARV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSV-QPQ 359
L DEKL FDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVN+ D +GD DS QPQ
Sbjct: 301 LQDEKLSFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNEEDGEGDAPDSTTQPQ 360
Query: 360 KRKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLL 419
KRKRLWGLVVCHNT+PRFVPFPLRYACEFLAQVFAIHVNKE+ELE QI+EKNILRTQTLL
Sbjct: 361 KRKRLWGLVVCHNTTPRFVPFPLRYACEFLAQVFAIHVNKEVELENQIVEKNILRTQTLL 420
Query: 420 CDMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHT 479
CDMLMRDAPLGI++QSPN+MDLVKCDGAALLYK+K+W LG TPSE H+++IASWL +YHT
Sbjct: 421 CDMLMRDAPLGIVSQSPNIMDLVKCDGAALLYKDKIWKLGTTPSEFHLQEIASWLYEYHT 480
Query: 480 DSTGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPG 539
DSTGLSTDSL DAGFP ALSLGD VCGMAAVRI+ KD++FWFRSHTA E+RWGGAKH+P
Sbjct: 481 DSTGLSTDSLYDAGFPKALSLGDAVCGMAAVRISSKDMIFWFRSHTAGEVRWGGAKHDPD 540
Query: 540 EQDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAI 599
++DD ++MHPRSSFKAFLEVV+ RS PWKDYEMDAIHSLQLILRNAFKD +S D+NT I
Sbjct: 541 DRDDARRMHPRSSFKAFLEVVKTRSLPWKDYEMDAIHSLQLILRNAFKDGESTDVNTKFI 600
Query: 600 DTRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEA 659
++L+DLKI+G+QELEAVTSEMVRLIETATVPILAVD DGLVNGWN KIAELTGLPV EA
Sbjct: 601 HSKLNDLKIDGIQELEAVTSEMVRLIETATVPILAVDSDGLVNGWNTKIAELTGLPVDEA 660
Query: 660 IGKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRD 719
IGKHLLTLVED S + VK+ML+ AL G EE+NVQFEIKTH S+ ++GPISLVVNACASRD
Sbjct: 661 IGKHLLTLVEDSSVEIVKRMLENALEGTEEQNVQFEIKTHLSRADAGPISLVVNACASRD 720
Query: 720 LRENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNP 779
L ENVVGVCFVA D+T QKTVMDKFTRIEGDYKAI+QNPNPLIPPIFGTDEFGWC EWNP
Sbjct: 721 LHENVVGVCFVAHDLTGQKTVMDKFTRIEGDYKAIIQNPNPLIPPIFGTDEFGWCTEWNP 780
Query: 780 AMTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFG 839
AM+KLTG KREEV+DKMLLGEVFGT A CRLKNQEAFVN GIVLN A+T E+EKV F
Sbjct: 781 AMSKLTGLKREEVIDKMLLGEVFGTQTACCRLKNQEAFVNLGIVLNSAVTSQESEKVSFA 840
Query: 840 FFARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKAL 899
FF R GKY+ECLL VSKKLD EG+VTGVFCFLQLAS ELQQALH+Q L+E+TALKRLK L
Sbjct: 841 FFTRGGKYIECLLCVSKKLDREGVVTGVFCFLQLASHELQQALHVQRLAERTALKRLKTL 900
Query: 900 TYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGY 959
Y+KRQIRNPLSGI+F+RK +EGT+LG EQ++++ TS+ CQ+QLSK+LDDSDL+ I++G
Sbjct: 901 AYIKRQIRNPLSGIMFTRKMMEGTELGPEQRQILQTSSLCQKQLSKVLDDSDLERIIEGC 960
Query: 960 LDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFL 1019
LDLEM EF+L +VL S SQ+M +S+ + +RI N+ EE+M + LYGDS+RLQQVLADF+
Sbjct: 961 LDLEMKEFSLNEVLTASTSQVMMKSNGKSVRITNETGEEVMSDTLYGDSIRLQQVLADFM 1020
Query: 1020 LISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLES 1079
L+S+N TP+GGQ+ V ASL K+QLG+SVHLA LE+ +TH G+G+PE LLNQMFG + S
Sbjct: 1021 LMSVNFTPSGGQLTVTASLRKDQLGRSVHLAYLEIRLTHTGAGIPELLLNQMFGTEKDVS 1080
Query: 1080 EEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
EEG+SL++SRKL+KLM+GDV+YLR+AGKSSFI+S ELAAA+K
Sbjct: 1081 EEGLSLMVSRKLVKLMNGDVQYLRQAGKSSFIISAELAAANK 1122
>UniRef100_Q9LRG9 Phytochrome A [Armoracia rusticana]
Length = 1122
Score = 1815 bits (4701), Expect = 0.0
Identities = 889/1122 (79%), Positives = 1010/1122 (89%), Gaps = 1/1122 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MS SRPSQSS S RSRHSAR+IAQTTVDAK+HA+FEESGSSFDYS+SVR +G +
Sbjct: 1 MSGSRPSQSSEGSRRSRHSARIIAQTTVDAKLHADFEESGSSFDYSTSVRVTGPVVENQP 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
P+S+KVTT YLHHIQ+GKLIQPFGCLLALDEKT KVIAYSEN+PE+LTM SHAVPSVGEH
Sbjct: 61 PRSDKVTTTYLHHIQKGKLIQPFGCLLALDEKTFKVIAYSENSPELLTMASHAVPSVGEH 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P LGI TDIR++FTAPSASALQKALGF +V+LLNPILVHCKTS KPFYAI+HRVTGS+I+
Sbjct: 121 PVLGIGTDIRSLFTAPSASALQKALGFGDVSLLNPILVHCKTSAKPFYAIVHRVTGSIIV 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM
Sbjct: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFHEDDHGEV++E+TKPGLEPYLGLHYPATDIPQA+RFLFMKNKVRMIVDC+AK +V
Sbjct: 241 AYKFHEDDHGEVVSEVTKPGLEPYLGLHYPATDIPQAARFLFMKNKVRMIVDCNAKHARV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSV-QPQ 359
L DEKL FDLTLCGSTLRAPHSCHLQYMANMDSIAS VMAVVVN+ D +GD +DS QPQ
Sbjct: 301 LQDEKLSFDLTLCGSTLRAPHSCHLQYMANMDSIASHVMAVVVNEEDGEGDATDSTTQPQ 360
Query: 360 KRKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLL 419
KRKRLWGLVVCHNT+PRFVPFPLRYACEFLAQVFAIHVNKE+ELE QI+EKNILRTQTLL
Sbjct: 361 KRKRLWGLVVCHNTTPRFVPFPLRYACEFLAQVFAIHVNKEVELENQIVEKNILRTQTLL 420
Query: 420 CDMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHT 479
CDMLMRDAPLGI++QSPN+MDLVKCDGAALLYK+K+W LG TPSELH+++IASWL +YHT
Sbjct: 421 CDMLMRDAPLGIVSQSPNIMDLVKCDGAALLYKDKIWKLGTTPSELHLQEIASWLYEYHT 480
Query: 480 DSTGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPG 539
DSTGLSTDSL DAGFP ALSLGD VCGMAAVRI+ KD++FWFRSHTA E+RWGGAKH P
Sbjct: 481 DSTGLSTDSLHDAGFPKALSLGDAVCGMAAVRISLKDMIFWFRSHTAGEVRWGGAKHNPD 540
Query: 540 EQDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAI 599
++DD ++MHPRSSFKAFLEVV+ RS PWKDYEMDAIHSLQLILRNAFKD +S D+NT I
Sbjct: 541 DRDDARRMHPRSSFKAFLEVVKTRSLPWKDYEMDAIHSLQLILRNAFKDGESTDVNTNII 600
Query: 600 DTRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEA 659
++L+DLKI+G+QELEAVTSEMVRLIETATVPILAVD DGLVNGWN KIAELTGLPV EA
Sbjct: 601 HSKLNDLKIDGIQELEAVTSEMVRLIETATVPILAVDSDGLVNGWNTKIAELTGLPVDEA 660
Query: 660 IGKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRD 719
IGKHLLTLVED S + VK+ML+ AL G EE+NVQFEIKTH S+ ++G ISL VNACASRD
Sbjct: 661 IGKHLLTLVEDSSVEIVKRMLENALEGTEEQNVQFEIKTHQSRADAGTISLAVNACASRD 720
Query: 720 LRENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNP 779
L ENVVGVCFVA D+T QKTVMDKFTRIEGDYKAI+QNPNPLIPPIFGTDEFGWC EWNP
Sbjct: 721 LHENVVGVCFVAHDLTGQKTVMDKFTRIEGDYKAIIQNPNPLIPPIFGTDEFGWCTEWNP 780
Query: 780 AMTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFG 839
AM+KLTG KREEV+DKMLLGEVFGT + CRLKNQEAFVN GIVLN A+T E+EKV F
Sbjct: 781 AMSKLTGLKREEVIDKMLLGEVFGTQKSCCRLKNQEAFVNLGIVLNGAVTSQESEKVSFA 840
Query: 840 FFARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKAL 899
FF R GKY+ECLL VSKKLD EG+VTGVFCFLQLAS ELQQALH+Q L+E+TALKRLKA+
Sbjct: 841 FFTRGGKYIECLLCVSKKLDREGVVTGVFCFLQLASHELQQALHVQRLAERTALKRLKAI 900
Query: 900 TYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGY 959
Y+KRQIRNPLSG++F+RK +EGT+LG EQ++++HTSA CQ QLSK+LDDSDL+SI++G
Sbjct: 901 AYIKRQIRNPLSGVMFTRKMMEGTELGPEQRQILHTSALCQEQLSKVLDDSDLESIIEGC 960
Query: 960 LDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFL 1019
LDLEM EF+L +VL S SQ+M +S+ + +RI N+ EE+M + LYGDS+RLQQVLADF+
Sbjct: 961 LDLEMKEFSLNEVLTASTSQVMMKSNGKSVRITNETGEEVMSDTLYGDSIRLQQVLADFM 1020
Query: 1020 LISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLES 1079
L+S+N TP+GGQ+ V ASL K+QLG+SVHLA LE+ +TH G+G+PE LLNQMFG + S
Sbjct: 1021 LMSVNFTPSGGQLTVTASLRKDQLGRSVHLAYLEIRLTHTGAGIPEFLLNQMFGTEEDVS 1080
Query: 1080 EEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
EEG+SL++SRKL+KLM+GDV+YLR+AGKSSFI+S ELAAA+K
Sbjct: 1081 EEGLSLMVSRKLVKLMNGDVQYLRQAGKSSFIISAELAAANK 1122
>UniRef100_Q9LRG8 Phytochrome A [Armoracia rusticana]
Length = 1122
Score = 1814 bits (4698), Expect = 0.0
Identities = 889/1122 (79%), Positives = 1009/1122 (89%), Gaps = 1/1122 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MS SRPSQSS S RSRHSAR+IAQTTVDAK+HA+FEESGSSFDYS+SVR +G +
Sbjct: 1 MSGSRPSQSSEGSRRSRHSARIIAQTTVDAKLHADFEESGSSFDYSTSVRVTGPVVENQP 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
P+S+KVTT YLHHIQ+GKLIQPFGCLLALDEKT KVIAYSENAPE+LTM SHAVPSVGEH
Sbjct: 61 PRSDKVTTTYLHHIQKGKLIQPFGCLLALDEKTFKVIAYSENAPELLTMASHAVPSVGEH 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P LGI TDIR++FTAPSASALQKALGF +V+LLNPILVHCKTS KPFYAI+HRVTGS+I+
Sbjct: 121 PVLGIGTDIRSLFTAPSASALQKALGFGDVSLLNPILVHCKTSAKPFYAIVHRVTGSIIV 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM
Sbjct: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFHEDDHGEV++E+TKPGLEPYLGLHYPATDIPQA+RFLFMKNKVRMIVDC+AK +V
Sbjct: 241 AYKFHEDDHGEVVSEVTKPGLEPYLGLHYPATDIPQAARFLFMKNKVRMIVDCNAKHARV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSV-QPQ 359
L DEKL FDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVN+ D +GD DS QPQ
Sbjct: 301 LQDEKLSFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNEEDGEGDAPDSTTQPQ 360
Query: 360 KRKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLL 419
KRKRLWGLVVCHNT+PRFVPFPLRYACEFLAQVFAIHVNKE+ELE QI+EKNILRTQTLL
Sbjct: 361 KRKRLWGLVVCHNTTPRFVPFPLRYACEFLAQVFAIHVNKEVELENQIVEKNILRTQTLL 420
Query: 420 CDMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHT 479
CDMLMRDAPLGI++QSPN+MDLVKCDGAALLYK+K+W LG TPSELH+++IASWL +YHT
Sbjct: 421 CDMLMRDAPLGIVSQSPNIMDLVKCDGAALLYKDKIWKLGTTPSELHLQEIASWLYEYHT 480
Query: 480 DSTGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPG 539
DSTGLSTDSL DAGFP ALSLGD VCGMAAVRI+ KD++FWFRSHTA E+RWGGAKH P
Sbjct: 481 DSTGLSTDSLHDAGFPKALSLGDAVCGMAAVRISLKDMIFWFRSHTAGEVRWGGAKHNPD 540
Query: 540 EQDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAI 599
++DD ++MHPRSSFKAFLEVV+ RS PWKDYEMDAIHSLQLILRNAFKD +S D+NT I
Sbjct: 541 DRDDARRMHPRSSFKAFLEVVKTRSLPWKDYEMDAIHSLQLILRNAFKDGESTDVNTNII 600
Query: 600 DTRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEA 659
++L+DLKI+G+QELEAVTSEMVRLIETATVPILAVD DGLVNGWN KIAELTGLPV EA
Sbjct: 601 HSKLNDLKIDGIQELEAVTSEMVRLIETATVPILAVDSDGLVNGWNTKIAELTGLPVDEA 660
Query: 660 IGKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRD 719
IGKHLLTLVED S + VK+ML+ AL G EE+NVQFEIKTH S+ ++G ISL VNACASRD
Sbjct: 661 IGKHLLTLVEDSSVEIVKRMLENALEGTEEQNVQFEIKTHQSRADAGTISLAVNACASRD 720
Query: 720 LRENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNP 779
L ENVVGVCFVA D+T QKTVMDKFTRIEGDYKAI+QNPNPLIPPIFGTDEFGWC EWNP
Sbjct: 721 LHENVVGVCFVAHDLTGQKTVMDKFTRIEGDYKAIIQNPNPLIPPIFGTDEFGWCTEWNP 780
Query: 780 AMTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFG 839
AM+KLTG KREEV+DKMLLGEVFGT + CRLKNQEAFVN GIVLN A+T E+EKV F
Sbjct: 781 AMSKLTGLKREEVIDKMLLGEVFGTQKSCCRLKNQEAFVNLGIVLNGAVTSQESEKVSFA 840
Query: 840 FFARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKAL 899
FF R GKY+ECLL VSKKLD EG+VTGVFCFLQLAS ELQQALH+Q L+E+TALKRLKA+
Sbjct: 841 FFTRGGKYIECLLCVSKKLDREGVVTGVFCFLQLASHELQQALHVQRLAERTALKRLKAI 900
Query: 900 TYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGY 959
Y+KRQIRNPLSG++F+R+ +EGT+LG EQ++++HTSA CQ QLSK+LDDSDL+SI++G
Sbjct: 901 AYIKRQIRNPLSGVMFTREMIEGTELGPEQRQILHTSALCQEQLSKVLDDSDLESIIEGC 960
Query: 960 LDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFL 1019
LDLEM EF+L +VL S SQ+M +S+ + +RI N+ EE+M + LYGDS+RLQQVLADF+
Sbjct: 961 LDLEMKEFSLNEVLTASTSQVMMKSNGKSVRITNETGEEVMSDTLYGDSIRLQQVLADFM 1020
Query: 1020 LISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLES 1079
L+S+N TP+GGQ+ V ASL K+Q G+SVHLA LE+ +TH G+G+PE LLNQMFG + S
Sbjct: 1021 LMSVNFTPSGGQLTVTASLRKDQPGRSVHLAYLEIRLTHTGAGIPEFLLNQMFGTEEDVS 1080
Query: 1080 EEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
EEG+SL++SRKL+KLM+GDV+YLR+AGKSSFI+S ELAAA+K
Sbjct: 1081 EEGLSLMVSRKLVKLMNGDVQYLRQAGKSSFIISAELAAANK 1122
>UniRef100_Q41331 PhyA protein [Lycopersicon esculentum]
Length = 1123
Score = 1810 bits (4689), Expect = 0.0
Identities = 884/1119 (78%), Positives = 1003/1119 (88%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MSSSRPSQSS S RS+HSAR++AQT++DAK+HA+FEESG SFDYSSSVR + A + +
Sbjct: 1 MSSSRPSQSSTTSSRSKHSARIVAQTSIDAKLHADFEESGDSFDYSSSVRVTSVAGDEEK 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
PKS+KVTTAYLH IQ+GK IQPFGCLLALDEKT KVIA+SENAPEMLTMVSHAVPSVGEH
Sbjct: 61 PKSDKVTTAYLHQIQKGKFIQPFGCLLALDEKTLKVIAFSENAPEMLTMVSHAVPSVGEH 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P LGI TDIRTIFT PS +ALQKALGF EV+LLNP+LVHCK SGKPFYAI+HRVTGSLI+
Sbjct: 121 PVLGIGTDIRTIFTGPSGAALQKALGFGEVSLLNPVLVHCKNSGKPFYAIVHRVTGSLIL 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM
Sbjct: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
YKFHEDDHGEV++EITKPGLEPYLGLHYPATDIPQA+RFLFMKNKVRMI DC AK VKV
Sbjct: 241 GYKFHEDDHGEVVSEITKPGLEPYLGLHYPATDIPQAARFLFMKNKVRMICDCRAKHVKV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQK 360
+ DEKLPFDLTLCGSTLRAPH CHLQYM NM+SIASLVMAVVVND DE+G+ SDS Q QK
Sbjct: 301 VQDEKLPFDLTLCGSTLRAPHYCHLQYMENMNSIASLVMAVVVNDGDEEGESSDSSQSQK 360
Query: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLC 420
RKRLWGLVVCHNT+PRFVPFPLRYACEFLAQVFAIHVNKE+ELE Q LEKNILRTQTLLC
Sbjct: 361 RKRLWGLVVCHNTTPRFVPFPLRYACEFLAQVFAIHVNKELELENQFLEKNILRTQTLLC 420
Query: 421 DMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHTD 480
DMLMRDAPLGI++QSPN+MDLVKCDGAALLYKNK+ LG+ PS+ ++DI SWL +YHTD
Sbjct: 421 DMLMRDAPLGIVSQSPNIMDLVKCDGAALLYKNKIHRLGMNPSDFQLQDIVSWLCEYHTD 480
Query: 481 STGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGE 540
STGLSTDSL DAGFPGAL+LGD VCGMAAVRI+ KD +FWFRSHTAAE+RWGGAKHEPGE
Sbjct: 481 STGLSTDSLYDAGFPGALALGDAVCGMAAVRISDKDWLFWFRSHTAAEVRWGGAKHEPGE 540
Query: 541 QDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAID 600
+DDG+KMHPRSSFKAFLEVV+ RS PWKDYEMDAIHSLQLILRNAFKD + ++ NT +I
Sbjct: 541 KDDGRKMHPRSSFKAFLEVVKTRSIPWKDYEMDAIHSLQLILRNAFKDAEVVNSNTNSIY 600
Query: 601 TRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAI 660
+L+DLKI+GMQELE+VT+EMVRLIETA VPILAVD+DG VNGWN KIAELTGLPV EAI
Sbjct: 601 KKLNDLKIDGMQELESVTAEMVRLIETALVPILAVDVDGQVNGWNTKIAELTGLPVDEAI 660
Query: 661 GKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDL 720
GKHLLTLVED S D V KML+LAL G+EEKNV+FEIKTHG +S PISL+VNACAS+D+
Sbjct: 661 GKHLLTLVEDSSVDTVNKMLELALQGKEEKNVEFEIKTHGPSRDSSPISLIVNACASKDV 720
Query: 721 RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNPA 780
R+NVVGVCF+A DIT QK++MDKFTRIEGDY+AI+QNP+PLIPPIFGTD+FGWC EWN A
Sbjct: 721 RDNVVGVCFMAHDITGQKSIMDKFTRIEGDYRAIIQNPHPLIPPIFGTDQFGWCSEWNTA 780
Query: 781 MTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGF 840
MTKLTGW+R++VMDKMLLGEVFGT A CRLKNQEAFVNFG+VLN A+TG E+EK+ FGF
Sbjct: 781 MTKLTGWRRDDVMDKMLLGEVFGTQAACCRLKNQEAFVNFGVVLNNAITGQESEKIPFGF 840
Query: 841 FARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALT 900
FAR GKYVECLL VSK+LD EG VTG+FCFLQLAS ELQQAL++Q LSEQTALKRLK L
Sbjct: 841 FARYGKYVECLLCVSKRLDKEGAVTGLFCFLQLASHELQQALYVQRLSEQTALKRLKVLA 900
Query: 901 YMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYL 960
Y++RQIRNPLSGI+FSRK LEGT LG EQK ++HTSAQCQRQL+KILDD+DLDSI+DGYL
Sbjct: 901 YIRRQIRNPLSGIIFSRKMLEGTSLGEEQKNILHTSAQCQRQLNKILDDTDLDSIIDGYL 960
Query: 961 DLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLL 1020
DLEM EF L +VL+ S+SQ+M +S+ + I I ND+ E+++ E LYGDS RLQQVLA+FLL
Sbjct: 961 DLEMLEFKLHEVLVASISQVMMKSNGKNIMISNDMVEDLLNETLYGDSPRLQQVLANFLL 1020
Query: 1021 ISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLESE 1080
+S+N TP+GGQ+ ++ LTK+++G+SV LA LE I H G GVPE LL QMFG++ SE
Sbjct: 1021 VSVNATPSGGQLSISGRLTKDRIGESVQLALLEFRIRHTGGGVPEELLGQMFGSEADASE 1080
Query: 1081 EGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAA 1119
EGISLL+SRKL+KLM+G+V+YLREAG+S+FI+SVELA A
Sbjct: 1081 EGISLLVSRKLVKLMNGEVQYLREAGQSTFIISVELAVA 1119
>UniRef100_O49934 Phytochrome A [Populus tremuloides]
Length = 1125
Score = 1808 bits (4684), Expect = 0.0
Identities = 903/1123 (80%), Positives = 996/1123 (88%), Gaps = 5/1123 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MSSSRPS SS+NS RSRHSAR+IAQTTVDAK+HA+FEESGSSFDYSSSVR + + D
Sbjct: 1 MSSSRPSHSSSNSARSRHSARIIAQTTVDAKLHADFEESGSSFDYSSSVRVTDSVGGDQP 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
P+S+KVTTAYLHHIQ+GKLIQPFGCLLALDEKT +V+AYSENAPE+LTMVSHAVPSVGEH
Sbjct: 61 PRSDKVTTAYLHHIQKGKLIQPFGCLLALDEKTFRVVAYSENAPELLTMVSHAVPSVGEH 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P LGI TDIRTIFTAPSASALQKA+GF +V+LLNPILVHCKTSGKPFYAI+HRVTGSLII
Sbjct: 121 PVLGIGTDIRTIFTAPSASALQKAMGFGDVSLLNPILVHCKTSGKPFYAIVHRVTGSLII 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDR M
Sbjct: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRAM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFH+DDHGEV++E+TKPG+EPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAK VKV
Sbjct: 241 AYKFHDDDHGEVVSEVTKPGMEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKHVKV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQK 360
L DEKLPFDLTLCGSTLRAPHSCHLQYM NM+SIASLVMAVVVND DEDGD DS PQK
Sbjct: 301 LQDEKLPFDLTLCGSTLRAPHSCHLQYMENMNSIASLVMAVVVNDGDEDGDTPDSANPQK 360
Query: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLC 420
RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKE+ELE QI+EKNILRTQTLLC
Sbjct: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKELELENQIVEKNILRTQTLLC 420
Query: 421 DMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHTD 480
DMLMRDAPLGI+TQSPN+MDLVKCDGA L Y+NK+W LG+TPS+L ++DIA WLS+YH D
Sbjct: 421 DMLMRDAPLGIVTQSPNIMDLVKCDGAVLFYRNKIWRLGITPSDLQLQDIAFWLSEYHMD 480
Query: 481 STGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGE 540
STGLSTDSL DAG+PGAL+LGD+VCGMAAVRIT KD++FWFRS TAAEIRWGGAKHEPGE
Sbjct: 481 STGLSTDSLYDAGYPGALALGDVVCGMAAVRITSKDMLFWFRSQTAAEIRWGGAKHEPGE 540
Query: 541 QDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAID 600
+DDG++MHPRSSFKAFLEVV+ RS PWKDYEMDAIHSLQLILRN FKD ++MD++T I
Sbjct: 541 KDDGRRMHPRSSFKAFLEVVKTRSLPWKDYEMDAIHSLQLILRNTFKDIETMDVDTKTIH 600
Query: 601 TRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAI 660
RLSDLKIEGMQELEAVTSEMVRLIETATVPILAVD+DGLVNGWN KI+ELTGL V +AI
Sbjct: 601 ARLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDVDGLVNGWNTKISELTGLLVDKAI 660
Query: 661 GKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDL 720
GKHLLTLVED S D VK+ML LAL G+EE+N+QFEIKTHGSK E GPI LVVNACASRDL
Sbjct: 661 GKHLLTLVEDSSVDIVKRMLFLALQGKEEQNIQFEIKTHGSKSECGPICLVVNACASRDL 720
Query: 721 RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNPA 780
ENVVGVCFV QDIT QK VMDKFTRIEGDYKAIVQN NPLIPPIFGTDEFGWC EWNPA
Sbjct: 721 HENVVGVCFVGQDITGQKMVMDKFTRIEGDYKAIVQNRNPLIPPIFGTDEFGWCSEWNPA 780
Query: 781 MTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGF 840
MT LTGWKREEV+DKMLLGEVFG +MA CRLKNQEAFVN G+VLN AMTG E+EKV FGF
Sbjct: 781 MTNLTGWKREEVLDKMLLGEVFGLNMACCRLKNQEAFVNLGVVLNTAMTGQESEKVSFGF 840
Query: 841 FARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALT 900
FAR+GKYVECLL VSKKLD EG VTGVFCFLQLAS ELQQALH+Q LSEQTALKRLKAL
Sbjct: 841 FARTGKYVECLLCVSKKLDREGAVTGVFCFLQLASQELQQALHVQRLSEQTALKRLKALA 900
Query: 901 YMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYL 960
Y+K+QI NPLSGI+FS K +EGT+LG EQK L+HTSAQCQ QLSKILDDSDLDSI++GYL
Sbjct: 901 YLKKQIWNPLSGIIFSGKMMEGTELGAEQKELLHTSAQCQCQLSKILDDSDLDSIIEGYL 960
Query: 961 DLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLL 1020
DLEM EFTL++ S +GI I+ND + M E LYGDS+RLQQVLADF
Sbjct: 961 DLEMVEFTLREYYGCYQSS-HDEKHEKGIPIINDALK--MAETLYGDSIRLQQVLADFCR 1017
Query: 1021 ISINCTPNGGQVVVAASLTKEQLGKSVHLA--NLELSITHGGSGVPEALLNQMFGNDGLE 1078
+ TP+GG + V+AS + +G + + + +L I H G+G+PEAL++QM+G D
Sbjct: 1018 CQLILTPSGGLLTVSASFFQRPVGAILFILVHSGKLRIRHLGAGIPEALVDQMYGEDTGA 1077
Query: 1079 SEEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
S EGISL+ISRKL+KLM+GDVRY+REAGKSSFI+SVELA HK
Sbjct: 1078 SVEGISLVISRKLVKLMNGDVRYMREAGKSSFIISVELAGGHK 1120
>UniRef100_P14712 Phytochrome A [Arabidopsis thaliana]
Length = 1122
Score = 1808 bits (4682), Expect = 0.0
Identities = 886/1122 (78%), Positives = 1010/1122 (89%), Gaps = 1/1122 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MS SRP+QSS S RSRHSAR+IAQTTVDAK+HA+FEESGSSFDYS+SVR +G +
Sbjct: 1 MSGSRPTQSSEGSRRSRHSARIIAQTTVDAKLHADFEESGSSFDYSTSVRVTGPVVENQP 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
P+S+KVTT YLHHIQ+GKLIQPFGCLLALDEKT KVIAYSENA E+LTM SHAVPSVGEH
Sbjct: 61 PRSDKVTTTYLHHIQKGKLIQPFGCLLALDEKTFKVIAYSENASELLTMASHAVPSVGEH 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P LGI TDIR++FTAPSASALQKALGF +V+LLNPILVHC+TS KPFYAIIHRVTGS+II
Sbjct: 121 PVLGIGTDIRSLFTAPSASALQKALGFGDVSLLNPILVHCRTSAKPFYAIIHRVTGSIII 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM
Sbjct: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFHEDDHGEV++E+TKPGLEPYLGLHYPATDIPQA+RFLFMKNKVRMIVDC+AK +V
Sbjct: 241 AYKFHEDDHGEVVSEVTKPGLEPYLGLHYPATDIPQAARFLFMKNKVRMIVDCNAKHARV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSV-QPQ 359
L DEKL FDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVN+ D +GD D+ QPQ
Sbjct: 301 LQDEKLSFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNEEDGEGDAPDATTQPQ 360
Query: 360 KRKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLL 419
KRKRLWGLVVCHNT+PRFVPFPLRYACEFLAQVFAIHVNKE+EL+ Q++EKNILRTQTLL
Sbjct: 361 KRKRLWGLVVCHNTTPRFVPFPLRYACEFLAQVFAIHVNKEVELDNQMVEKNILRTQTLL 420
Query: 420 CDMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHT 479
CDMLMRDAPLGI++QSPN+MDLVKCDGAALLYK+K+W LG TPSE H+++IASWL +YH
Sbjct: 421 CDMLMRDAPLGIVSQSPNIMDLVKCDGAALLYKDKIWKLGTTPSEFHLQEIASWLCEYHM 480
Query: 480 DSTGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPG 539
DSTGLSTDSL DAGFP ALSLGD VCGMAAVRI+ KD++FWFRSHTA E+RWGGAKH+P
Sbjct: 481 DSTGLSTDSLHDAGFPRALSLGDSVCGMAAVRISSKDMIFWFRSHTAGEVRWGGAKHDPD 540
Query: 540 EQDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAI 599
++DD ++MHPRSSFKAFLEVV+ RS PWKDYEMDAIHSLQLILRNAFKD+++ D+NT I
Sbjct: 541 DRDDARRMHPRSSFKAFLEVVKTRSLPWKDYEMDAIHSLQLILRNAFKDSETTDVNTKVI 600
Query: 600 DTRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEA 659
++L+DLKI+G+QELEAVTSEMVRLIETATVPILAVD DGLVNGWN KIAELTGL V EA
Sbjct: 601 YSKLNDLKIDGIQELEAVTSEMVRLIETATVPILAVDSDGLVNGWNTKIAELTGLSVDEA 660
Query: 660 IGKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRD 719
IGKH LTLVED S + VK+ML+ AL G EE+NVQFEIKTH S+ ++GPISLVVNACASRD
Sbjct: 661 IGKHFLTLVEDSSVEIVKRMLENALEGTEEQNVQFEIKTHLSRADAGPISLVVNACASRD 720
Query: 720 LRENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNP 779
L ENVVGVCFVA D+T QKTVMDKFTRIEGDYKAI+QNPNPLIPPIFGTDEFGWC EWNP
Sbjct: 721 LHENVVGVCFVAHDLTGQKTVMDKFTRIEGDYKAIIQNPNPLIPPIFGTDEFGWCTEWNP 780
Query: 780 AMTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFG 839
AM+KLTG KREEV+DKMLLGEVFGT + CRLKNQEAFVN GIVLN A+T + EKV F
Sbjct: 781 AMSKLTGLKREEVIDKMLLGEVFGTQKSCCRLKNQEAFVNLGIVLNNAVTSQDPEKVSFA 840
Query: 840 FFARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKAL 899
FF R GKYVECLL VSKKLD EG+VTGVFCFLQLAS ELQQALH+Q L+E+TA+KRLKAL
Sbjct: 841 FFTRGGKYVECLLCVSKKLDREGVVTGVFCFLQLASHELQQALHVQRLAERTAVKRLKAL 900
Query: 900 TYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGY 959
Y+KRQIRNPLSGI+F+RK +EGT+LG EQ+R++ TSA CQ+QLSKILDDSDL+SI++G
Sbjct: 901 AYIKRQIRNPLSGIMFTRKMIEGTELGPEQRRILQTSALCQKQLSKILDDSDLESIIEGC 960
Query: 960 LDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFL 1019
LDLEM EFTL +VL S SQ+M +S+ + +RI N+ EE+M + LYGDS+RLQQVLADF+
Sbjct: 961 LDLEMKEFTLNEVLTASTSQVMMKSNGKSVRITNETGEEVMSDTLYGDSIRLQQVLADFM 1020
Query: 1020 LISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLES 1079
L+++N TP+GGQ+ V+ASL K+QLG+SVHLANLE+ +TH G+G+PE LLNQMFG + S
Sbjct: 1021 LMAVNFTPSGGQLTVSASLRKDQLGRSVHLANLEIRLTHTGAGIPEFLLNQMFGTEEDVS 1080
Query: 1080 EEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
EEG+SL++SRKL+KLM+GDV+YLR+AGKSSFI++ ELAAA+K
Sbjct: 1081 EEGLSLMVSRKLVKLMNGDVQYLRQAGKSSFIITAELAAANK 1122
>UniRef100_P33530 Phytochrome A1 [Nicotiana tabacum]
Length = 1124
Score = 1806 bits (4678), Expect = 0.0
Identities = 885/1121 (78%), Positives = 1006/1121 (88%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MSSSRPSQSS S RS+HSAR+IAQTT+DAK+HA+FEESG SFDYSSSVR + A + +
Sbjct: 1 MSSSRPSQSSTTSARSKHSARIIAQTTIDAKLHADFEESGDSFDYSSSVRVTSVAGDERK 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
PKS++VTTAYL+ IQ+GK IQPFGCLLALDEKT KVIA+SENAPEMLTMVSHAVPSVGE
Sbjct: 61 PKSDRVTTAYLNQIQKGKFIQPFGCLLALDEKTFKVIAFSENAPEMLTMVSHAVPSVGEL 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
PALGI TDIRTIFT PSA+ALQKALGF EV+LLNP+LVHCKTSGKP+YAI+HRVTGSLII
Sbjct: 121 PALGIGTDIRTIFTGPSAAALQKALGFGEVSLLNPVLVHCKTSGKPYYAIVHRVTGSLII 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQ+LPSGSMERLCDTMVQEVFELTGYDRVM
Sbjct: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQALPSGSMERLCDTMVQEVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
YKFH+DDHGEV+AEITKPGL+PYLGLHYPATDIPQA+RFLFMKNKVRMI DC AK VKV
Sbjct: 241 TYKFHDDDHGEVVAEITKPGLDPYLGLHYPATDIPQAARFLFMKNKVRMICDCRAKHVKV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQK 360
+ DEKLPFDLTLCGSTLRAPH CHLQYM NM SIASLVMAVVVND DE+G+ SDS Q QK
Sbjct: 301 VQDEKLPFDLTLCGSTLRAPHYCHLQYMENMSSIASLVMAVVVNDGDEEGESSDSTQSQK 360
Query: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLC 420
RKRLWGLVVCHNT+PRFVPFPLRYACEFLAQVFAIHVNKE+ELE QILEKNILRTQTLLC
Sbjct: 361 RKRLWGLVVCHNTTPRFVPFPLRYACEFLAQVFAIHVNKELELESQILEKNILRTQTLLC 420
Query: 421 DMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHTD 480
DMLMR APLGI++QSPN+MDLVKCDGAALLYKNK+ LG+TPS+ + DI SWLS+YHTD
Sbjct: 421 DMLMRVAPLGIVSQSPNIMDLVKCDGAALLYKNKIHRLGMTPSDFQLHDIVSWLSEYHTD 480
Query: 481 STGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGE 540
STGLSTDSL DAGFPGAL+LGD+VCGMAAVRI+ K +FW+RSHTAAE+RWGGAKHEPGE
Sbjct: 481 STGLSTDSLYDAGFPGALALGDVVCGMAAVRISDKGWLFWYRSHTAAEVRWGGAKHEPGE 540
Query: 541 QDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAID 600
+DDG+KMHPRSSFKAFLEVV+ RS PWKDYEMDAIHSLQLILRNA KD D+MD NT I
Sbjct: 541 KDDGRKMHPRSSFKAFLEVVKTRSVPWKDYEMDAIHSLQLILRNASKDADAMDSNTNIIH 600
Query: 601 TRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAI 660
T+L+DLKI+G+QELEAVT+EMVRLIETA+VPI AVD+DG +NGWN KIAELTGLPV EAI
Sbjct: 601 TKLNDLKIDGLQELEAVTAEMVRLIETASVPIFAVDVDGQLNGWNTKIAELTGLPVDEAI 660
Query: 661 GKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDL 720
G HLLTLVED S D V KML+LAL G+EE+NV+FEIKTHG +S PISL+VNACASRD+
Sbjct: 661 GNHLLTLVEDSSVDTVSKMLELALQGKEERNVEFEIKTHGPSGDSSPISLIVNACASRDV 720
Query: 721 RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNPA 780
++VVGVCF+AQDIT QK +MDKFTRIEGDY+AI+QNP+PLIPPIFGTD+FGWC EWN A
Sbjct: 721 GDSVVGVCFIAQDITGQKNIMDKFTRIEGDYRAIIQNPHPLIPPIFGTDQFGWCSEWNSA 780
Query: 781 MTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGF 840
MTKLTGW+R++V+DKMLLGEVFGT A CRLKNQEAFVNFG+VLN AMTG E K+ FGF
Sbjct: 781 MTKLTGWRRDDVIDKMLLGEVFGTQAACCRLKNQEAFVNFGVVLNNAMTGQECAKISFGF 840
Query: 841 FARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALT 900
FAR+GKYVECLL VSK+LD EG VTG+FCFLQLAS ELQQALHIQ LSEQTALKRLK L
Sbjct: 841 FARNGKYVECLLCVSKRLDREGAVTGLFCFLQLASHELQQALHIQRLSEQTALKRLKVLA 900
Query: 901 YMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYL 960
Y++RQIRNPLSGI+FSRK LEGT+LG EQK ++ TS+QCQRQL+KILDD+DLDSI+DGYL
Sbjct: 901 YIRRQIRNPLSGIIFSRKMLEGTNLGEEQKNILRTSSQCQRQLNKILDDTDLDSIIDGYL 960
Query: 961 DLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLL 1020
DLEM EF L +VL+ S+SQIM +S+ + I IVND+ E+++ E LYGDS RLQQVLA+FLL
Sbjct: 961 DLEMLEFKLHEVLVASISQIMMKSNGKNIMIVNDMVEDLLNETLYGDSPRLQQVLANFLL 1020
Query: 1021 ISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLESE 1080
+ +N TP+GGQ+ ++ +LTK+++G+SV LA LE+ I+H G GVPE LL+QMFG + SE
Sbjct: 1021 VCVNSTPSGGQLSISGTLTKDRIGESVQLALLEVRISHTGGGVPEELLSQMFGTEAEASE 1080
Query: 1081 EGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
EGISLLISRKL+KLM+G+V+YLREAG+S+FI+SVELA A K
Sbjct: 1081 EGISLLISRKLVKLMNGEVQYLREAGRSTFIISVELAVATK 1121
>UniRef100_P30733 Phytochrome A [Solanum tuberosum]
Length = 1123
Score = 1798 bits (4658), Expect = 0.0
Identities = 875/1121 (78%), Positives = 1000/1121 (89%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MSSSRPSQSS S RS+HSAR+IAQT++DAK+HA+FEESG SFDYSSSVR + A+ + +
Sbjct: 1 MSSSRPSQSSTTSSRSKHSARIIAQTSIDAKLHADFEESGDSFDYSSSVRVTNVAEGEQR 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
PKS+KVTTAYLH IQ+GK IQPFGCLLALDEKT KVIA+SENAPEMLTMVSHAVPSVGEH
Sbjct: 61 PKSDKVTTAYLHQIQKGKFIQPFGCLLALDEKTLKVIAFSENAPEMLTMVSHAVPSVGEH 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P LGI DIRTIFT PS +ALQKALGF EV+LLNP+LVHCK SGKPFYAI+HRVTGSLII
Sbjct: 121 PVLGIGIDIRTIFTGPSGAALQKALGFGEVSLLNPVLVHCKNSGKPFYAIVHRVTGSLII 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM
Sbjct: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
YKFH+DDHGEV++EITKPGLEPYLGLHYPATDIPQA+RFLFMKNKVRMI DC AK VKV
Sbjct: 241 GYKFHDDDHGEVVSEITKPGLEPYLGLHYPATDIPQAARFLFMKNKVRMICDCRAKHVKV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQK 360
+ DEKLPFDLTLCGSTLRAPH CHLQYM NM+SIASLVMAVVVND DE+G+ SDS Q QK
Sbjct: 301 VQDEKLPFDLTLCGSTLRAPHYCHLQYMENMNSIASLVMAVVVNDGDEEGESSDSSQSQK 360
Query: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLC 420
RKRLWGLVV HNT+PRF PFPLRYACEFLAQVFAI VNKE+ELE Q LEKNILRTQTLLC
Sbjct: 361 RKRLWGLVVSHNTTPRFAPFPLRYACEFLAQVFAILVNKELELENQFLEKNILRTQTLLC 420
Query: 421 DMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHTD 480
DMLMRDAPLGI++QSPN+MDL+KCDGAALLYKNK+ LG+ PS+ + DI SWL +YHTD
Sbjct: 421 DMLMRDAPLGIVSQSPNIMDLIKCDGAALLYKNKIHRLGMNPSDFQLHDIVSWLCEYHTD 480
Query: 481 STGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGE 540
STGLSTDSL DAGFPGAL+LGD VCGMAAVRI+ KD +FW+RSHTAAE+RWGGAKHEPGE
Sbjct: 481 STGLSTDSLYDAGFPGALALGDAVCGMAAVRISDKDWLFWYRSHTAAEVRWGGAKHEPGE 540
Query: 541 QDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAID 600
+DDG+KMHPRSSFK FLEVV+ RS PWKDYEMD IHSLQLILRNAFKD D+++ NT +I
Sbjct: 541 KDDGRKMHPRSSFKGFLEVVKTRSIPWKDYEMDRIHSLQLILRNAFKDADAVNSNTISIH 600
Query: 601 TRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAI 660
T+L+DLKI+GMQELEAVT+EMVRLIETA+VPI AVD+DG VNGWN K+AELTGLPV EAI
Sbjct: 601 TKLNDLKIDGMQELEAVTAEMVRLIETASVPIFAVDVDGQVNGWNTKVAELTGLPVDEAI 660
Query: 661 GKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDL 720
GKHLLTLVED S D V KML+LAL G+EE+NV+FEIKTHG +S PISL+VNACAS+D+
Sbjct: 661 GKHLLTLVEDSSVDTVNKMLELALQGQEERNVEFEIKTHGPSRDSSPISLIVNACASKDV 720
Query: 721 RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNPA 780
R++VVGVCF+AQDIT QK++MDKFTRIEGDY+AI+QNP+PLIPPIFGTD+FGWC EWN A
Sbjct: 721 RDSVVGVCFIAQDITGQKSIMDKFTRIEGDYRAIIQNPHPLIPPIFGTDQFGWCSEWNSA 780
Query: 781 MTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGF 840
MT LTGW+R++VMDKMLLGEVFGT A CRLKNQEAFVNFG++LN A+TG E+EK+ FGF
Sbjct: 781 MTMLTGWRRDDVMDKMLLGEVFGTQAACCRLKNQEAFVNFGVILNNAITGQESEKIPFGF 840
Query: 841 FARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALT 900
FAR GKYVECLL VSK+LD EG VTG+FCFLQLAS ELQQALH+Q LSEQTALKRLK L
Sbjct: 841 FARYGKYVECLLCVSKRLDKEGAVTGLFCFLQLASHELQQALHVQRLSEQTALKRLKVLA 900
Query: 901 YMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYL 960
Y++RQIRNPLSGI+FSRK LEGT LG EQK ++HTSAQCQRQL KILDD+DLDSI++GYL
Sbjct: 901 YIRRQIRNPLSGIIFSRKMLEGTSLGEEQKNILHTSAQCQRQLDKILDDTDLDSIIEGYL 960
Query: 961 DLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLL 1020
DLEM EF L +VL+ S+SQ+M +S+ + I I ND+ E+++ E LYGDS RLQQVLA+FLL
Sbjct: 961 DLEMLEFKLHEVLVASISQVMMKSNGKNIMISNDMVEDLLNETLYGDSPRLQQVLANFLL 1020
Query: 1021 ISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLESE 1080
+S+N TP+GG++ ++ LTK+++G+SV LA LE I H G GVPE LL+QMFG++ SE
Sbjct: 1021 VSVNSTPSGGKLSISGKLTKDRIGESVQLALLEFRIRHTGGGVPEELLSQMFGSEADASE 1080
Query: 1081 EGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
EGISLL+SRKL+KLM+G+V+YLREAG+S+FI+SVELA A K
Sbjct: 1081 EGISLLVSRKLVKLMNGEVQYLREAGRSTFIISVELAVATK 1121
>UniRef100_P06592 Phytochrome A [Cucurbita pepo]
Length = 1124
Score = 1786 bits (4626), Expect = 0.0
Identities = 880/1121 (78%), Positives = 997/1121 (88%), Gaps = 1/1121 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MS+SRPSQSS+NSGRSRHS R+IAQT+VDA + A+FEESG+SFDYSSSVR + D Q
Sbjct: 1 MSTSRPSQSSSNSGRSRHSTRIIAQTSVDANVQADFEESGNSFDYSSSVRVTSDVSGDQQ 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
P+S+KVTTAYLHHIQ+GKLIQPFGCLLALD+KT KVIAYSENAPEMLTMVSHAVPS+G++
Sbjct: 61 PRSDKVTTAYLHHIQKGKLIQPFGCLLALDDKTFKVIAYSENAPEMLTMVSHAVPSMGDY 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P LGI TD+RTIFTAPSASAL KALGF EVTLLNPILVHCKTSGKPFYAI+HRVTGSLII
Sbjct: 121 PVLGIGTDVRTIFTAPSASALLKALGFGEVTLLNPILVHCKTSGKPFYAIVHRVTGSLII 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYE P+TAAGALQSYKLAAKAITRLQSLPSGSM RLCDTMVQEVFELTGYDRVM
Sbjct: 181 DFEPVKPYEGPVTAAGALQSYKLAAKAITRLQSLPSGSMARLCDTMVQEVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFH+DDHGEVI+E+ KPGL+PYLGLHYPATDIPQA+RFLFMKNKVRMIVDC AK +KV
Sbjct: 241 AYKFHDDDHGEVISEVAKPGLQPYLGLHYPATDIPQAARFLFMKNKVRMIVDCRAKHLKV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQK 360
L DEKL FDLTLCGSTLRAPHSCHLQYM NM+SIASLVMAVVVN+ DE+ +G ++Q QK
Sbjct: 301 LQDEKLQFDLTLCGSTLRAPHSCHLQYMENMNSIASLVMAVVVNEGDEENEGP-ALQQQK 359
Query: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLC 420
RKRLWGLVVCHN+SPRFVPFPLRYACEFLAQVFAIHVNKE+ELE QI+EKNILRTQTLLC
Sbjct: 360 RKRLWGLVVCHNSSPRFVPFPLRYACEFLAQVFAIHVNKELELENQIIEKNILRTQTLLC 419
Query: 421 DMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHTD 480
DMLMRDAPLGI+++SPN+MDLVK DGAALLYK K+W LG+TP++ + DIASWLS+YH D
Sbjct: 420 DMLMRDAPLGIVSRSPNIMDLVKSDGAALLYKKKIWRLGLTPNDFQLLDIASWLSEYHMD 479
Query: 481 STGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGE 540
STGLSTDSL DAG+PGA++LGD VCGMAAVRIT D++FWFRSHTA+EIRWGGAKHE G+
Sbjct: 480 STGLSTDSLYDAGYPGAIALGDEVCGMAAVRITNNDMIFWFRSHTASEIRWGGAKHEHGQ 539
Query: 541 QDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAID 600
+DD +KMHPRSSFKAFLEVV+ RS PWKDYEMDAIHSLQLILRN FKDTD+ +IN +I
Sbjct: 540 KDDARKMHPRSSFKAFLEVVKTRSLPWKDYEMDAIHSLQLILRNTFKDTDATEINRKSIQ 599
Query: 601 TRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAI 660
T L DLKIEG QELE+VTSEMVRLIETATVPILAVD+DGL+NGWN KIAELTGLPV +AI
Sbjct: 600 TTLGDLKIEGRQELESVTSEMVRLIETATVPILAVDLDGLINGWNTKIAELTGLPVDKAI 659
Query: 661 GKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDL 720
GKHLLTLVED S + V+KML LAL G+EE+NVQFEIKTHGS +E G ISLVVNACASRDL
Sbjct: 660 GKHLLTLVEDSSVEVVRKMLFLALQGQEEQNVQFEIKTHGSHIEVGSISLVVNACASRDL 719
Query: 721 RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNPA 780
RENVVGV FVAQDIT QK VMDKFTR+EGDYKAIVQNPNPLIPPIFG+DEFGWC EWNPA
Sbjct: 720 RENVVGVFFVAQDITGQKMVMDKFTRLEGDYKAIVQNPNPLIPPIFGSDEFGWCSEWNPA 779
Query: 781 MTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGF 840
M KLTGW REEV+DKMLLGEVFG H + CRLKNQEAFVN GIVLN AM G + EK FGF
Sbjct: 780 MAKLTGWSREEVIDKMLLGEVFGVHKSCCRLKNQEAFVNLGIVLNNAMCGQDPEKASFGF 839
Query: 841 FARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALT 900
AR+G YVECLL V+K LD +G VTG FCFLQL S ELQQAL+IQ L EQTALKRL+AL
Sbjct: 840 LARNGMYVECLLCVNKILDKDGAVTGFFCFLQLPSHELQQALNIQRLCEQTALKRLRALG 899
Query: 901 YMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYL 960
Y+KRQI+NPLSGI+FSR+ LE T+LG+EQK L+ TS CQ+Q+SK+LD+SD+D I+DG++
Sbjct: 900 YIKRQIQNPLSGIIFSRRLLERTELGVEQKELLRTSGLCQKQISKVLDESDIDKIIDGFI 959
Query: 961 DLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLL 1020
DLEM EFTL +VL+ S+SQ+M + +GI+IVN+ EE M E LYGDSLRLQQVLADFLL
Sbjct: 960 DLEMDEFTLHEVLMVSISQVMLKIKGKGIQIVNETPEEAMSETLYGDSLRLQQVLADFLL 1019
Query: 1021 ISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLESE 1080
IS++ P+GGQ+ ++ +TK QLGKSVHL +LE IT+ G G+PE+LLN+MFG++ SE
Sbjct: 1020 ISVSYAPSGGQLTISTDVTKNQLGKSVHLVHLEFRITYAGGGIPESLLNEMFGSEEDASE 1079
Query: 1081 EGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
EG SLLISRKL+KLM+GDVRY+REAGKSSFI++VELAAAHK
Sbjct: 1080 EGFSLLISRKLVKLMNGDVRYMREAGKSSFIITVELAAAHK 1120
>UniRef100_Q6V7X1 Phytochrome A [Monotropastrum globosum]
Length = 1130
Score = 1776 bits (4601), Expect = 0.0
Identities = 877/1123 (78%), Positives = 993/1123 (88%), Gaps = 4/1123 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MSSSRP+QSS +SGRS+HSAR+IAQTTVDAK+HA+FEESG SFDYS+SVR +GT D Q
Sbjct: 1 MSSSRPTQSSGSSGRSKHSARIIAQTTVDAKLHADFEESGGSFDYSTSVRFTGTVGGDIQ 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
P+S+KVTTAYLH IQRGKLIQPFGCLLA+DEKT KVIAYSENAPEMLTMVSHAVPSVG+H
Sbjct: 61 PRSDKVTTAYLHQIQRGKLIQPFGCLLAVDEKTFKVIAYSENAPEMLTMVSHAVPSVGDH 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P LGI TD+RTIFT PSA+ALQKA+G+ EV+LLNPILVHCKTSGKPFYAI+HRVTGSLII
Sbjct: 121 PLLGIGTDVRTIFTNPSAAALQKAMGYGEVSLLNPILVHCKTSGKPFYAIVHRVTGSLII 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSG+M LCD MVQEVFELTGYDRVM
Sbjct: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGNMRILCDAMVQEVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
YKFH+DDHGEV +E+TKPGLEPYLGLHYPATDIPQA+RFLFMKNK+RMI DCHAKQVKV
Sbjct: 241 VYKFHDDDHGEVFSELTKPGLEPYLGLHYPATDIPQAARFLFMKNKIRMICDCHAKQVKV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSV---- 356
+ D+KLP DLTLCGSTLRAPHSCHLQYM NM+SIASLVM+VVVN+ DE+GDG S
Sbjct: 301 IQDDKLPIDLTLCGSTLRAPHSCHLQYMENMNSIASLVMSVVVNEGDEEGDGGGSSVSSN 360
Query: 357 QPQKRKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQ 416
Q QK KRLWGL+VCHNT+PRFVPFPLRYACEFLAQVF IHVNKE+ELE QI EKNILRTQ
Sbjct: 361 QQQKIKRLWGLLVCHNTTPRFVPFPLRYACEFLAQVFTIHVNKELELENQIHEKNILRTQ 420
Query: 417 TLLCDMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSK 476
TLLCDMLMRDAPLGI++QSPN+MDLVKCDGA LLYK+K + +G TP++ +RDI WLS+
Sbjct: 421 TLLCDMLMRDAPLGIVSQSPNVMDLVKCDGAVLLYKDKTYRMGTTPTDFQLRDIVYWLSE 480
Query: 477 YHTDSTGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKH 536
YHTDSTGLSTDSL DAG+PGAL+ GD VCGMAAV+IT D++FWF++ TAAEI+WGGAKH
Sbjct: 481 YHTDSTGLSTDSLYDAGYPGALAFGDGVCGMAAVKITSNDMLFWFKAQTAAEIQWGGAKH 540
Query: 537 EPGEQDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINT 596
E GE+DDG+KMHPRSSFKAFLEVV+ RS PWKDYEMDAIHSLQLILRNAFKDT +MD T
Sbjct: 541 ESGERDDGRKMHPRSSFKAFLEVVKTRSLPWKDYEMDAIHSLQLILRNAFKDTKAMDATT 600
Query: 597 TAIDTRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPV 656
I TRL DLKIEGM+ELEAVTSEMVRLIETATVPILAVD+DGLVNGWN+KIAELTGLPV
Sbjct: 601 DVIHTRLHDLKIEGMEELEAVTSEMVRLIETATVPILAVDVDGLVNGWNLKIAELTGLPV 660
Query: 657 GEAIGKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACA 716
+AIG+ LL+LVED ST VKKMLDLAL G+EE+N+QFE+KT S+ +SGPISLVVNACA
Sbjct: 661 DKAIGRDLLSLVEDSSTGIVKKMLDLALQGKEEQNIQFELKTDESRRDSGPISLVVNACA 720
Query: 717 SRDLRENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCE 776
SRD ENVVGVCFVAQDIT KTVMDKFTRIEGDYKAIVQNPNPLIPPI GTDEFGWC E
Sbjct: 721 SRDHHENVVGVCFVAQDITGHKTVMDKFTRIEGDYKAIVQNPNPLIPPILGTDEFGWCSE 780
Query: 777 WNPAMTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKV 836
WN AM K++GW RE+V++KMLLGEVFGTH+ CRLKNQEAFVN GIVLN A+TG E+EK+
Sbjct: 781 WNLAMEKISGWNREDVINKMLLGEVFGTHVVCCRLKNQEAFVNLGIVLNNAVTGRESEKI 840
Query: 837 GFGFFARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRL 896
FGFFAR+GKYVEC+L SKK+D EG VTGVFC LQLASPELQQALH+Q L+EQTALKR
Sbjct: 841 SFGFFARNGKYVECILCASKKIDGEGAVTGVFCLLQLASPELQQALHVQRLTEQTALKRF 900
Query: 897 KALTYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIM 956
K L Y++RQ R LSGI++S + +EGTDL QK+L+HTSAQCQ QL+KILDD+DLD I+
Sbjct: 901 KELAYIRRQTRASLSGIMYSWRLMEGTDLRERQKQLLHTSAQCQHQLTKILDDTDLDCII 960
Query: 957 DGYLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLA 1016
DGYLDLEM EFTL +VL S+SQ+ +S+ +GI I N + EE M LYGD+LRLQQV+A
Sbjct: 961 DGYLDLEMVEFTLYEVLSASISQVTLKSNGKGIHIANPLQEEKMPATLYGDNLRLQQVIA 1020
Query: 1017 DFLLISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDG 1076
DFL IS+N TPNGGQ+V +ASLTK++LG+SV L ++E+ ITH G GVPE LLNQMFG D
Sbjct: 1021 DFLSISVNFTPNGGQIVASASLTKDRLGQSVQLVHVEIRITHMGGGVPEGLLNQMFGGDT 1080
Query: 1077 LESEEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAA 1119
SEEGISLL+SRKL+KLM+GDV+YLREAGKS+FI+SVELAAA
Sbjct: 1081 DTSEEGISLLVSRKLVKLMNGDVQYLREAGKSTFIISVELAAA 1123
>UniRef100_Q6K0L1 PHYA4 photoreceptor [Stellaria longipes]
Length = 1122
Score = 1699 bits (4400), Expect = 0.0
Identities = 834/1119 (74%), Positives = 965/1119 (85%), Gaps = 1/1119 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
M+S SQSS NSGRS+HSAR+IAQT DAK+HA FEES + FDYSSSVR S T+ +
Sbjct: 1 MASPAQSQSSTNSGRSKHSARIIAQTIQDAKLHAEFEESSNEFDYSSSVRGS-TSGVNQL 59
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
PKS+KVT++YL IQ+GK IQ FGCLLALD+KT +VIA+SENAPEMLTMVSHAVPSVG+
Sbjct: 60 PKSDKVTSSYLLQIQKGKFIQLFGCLLALDDKTFRVIAFSENAPEMLTMVSHAVPSVGDL 119
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P +GI T+IRTIFT PSASALQKALGF +V+LLNPILVHCK SGKPFYAI+HRVT SL+I
Sbjct: 120 PVIGIGTNIRTIFTGPSASALQKALGFTDVSLLNPILVHCKNSGKPFYAIVHRVTRSLVI 179
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSG+M+RL DTMVQEVFELTGYDRVM
Sbjct: 180 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGNMDRLVDTMVQEVFELTGYDRVM 239
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFH+DDHGEV++E+TKP L+ YLGLHYPATDIPQA+RFLFMKNKVR+I DC AK V+V
Sbjct: 240 AYKFHDDDHGEVVSEVTKPNLDSYLGLHYPATDIPQAARFLFMKNKVRLICDCRAKNVRV 299
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQK 360
+ DEKL DLTLCGSTLRAPH CH QYM NM+SI SLVMAVVVND D++ GS QP K
Sbjct: 300 VQDEKLSVDLTLCGSTLRAPHGCHAQYMENMNSIGSLVMAVVVNDEDDEDGGSAPAQPHK 359
Query: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLC 420
RKRLWGLVVCH+TSPRFVPFPLRYACEFLAQVFAIHVNKE+ELE Q LEK ILRTQTLLC
Sbjct: 360 RKRLWGLVVCHHTSPRFVPFPLRYACEFLAQVFAIHVNKELELENQFLEKKILRTQTLLC 419
Query: 421 DMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHTD 480
DMLMRDAPLGI+TQ+PN+MDLVKCDGAALLY NK+W LG+TP++ +RDIA WLS+ HTD
Sbjct: 420 DMLMRDAPLGIVTQNPNVMDLVKCDGAALLYNNKIWKLGITPTDYQLRDIAGWLSRDHTD 479
Query: 481 STGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGE 540
STGLSTDSL DAG+PGA SLGD VCGMAAVRITP D++FWFRSHTAAE++WGGAKHE GE
Sbjct: 480 STGLSTDSLHDAGYPGARSLGDTVCGMAAVRITPNDMLFWFRSHTAAEVKWGGAKHETGE 539
Query: 541 QDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAID 600
+DDG KMHPR+SFKAFLEVV+ RS PWKDYEMDAIHSLQLILRNAFKD ++ D+NT+ I
Sbjct: 540 KDDGSKMHPRTSFKAFLEVVKRRSVPWKDYEMDAIHSLQLILRNAFKDVEASDLNTSVIH 599
Query: 601 TRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAI 660
+++SDL+I G++ELEAVTSEMVRLIETATVPILAVD DGLVNGWN KI+ELTG+PV EA+
Sbjct: 600 SKISDLQINGLRELEAVTSEMVRLIETATVPILAVDADGLVNGWNTKISELTGVPVAEAV 659
Query: 661 GKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDL 720
GKH+ +L E+ S D VK+ML LAL GEE+KNVQFEIK H S +SGPISLVVNACAS+D+
Sbjct: 660 GKHIASLAEESSIDNVKRMLQLALQGEEKKNVQFEIKRHQSNPDSGPISLVVNACASKDV 719
Query: 721 RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNPA 780
NVVGVC +AQDIT QKTVMDKFTRIEGDYKAI+Q+PNPLIPPIFGTDEFGWC EWNPA
Sbjct: 720 NGNVVGVCLIAQDITGQKTVMDKFTRIEGDYKAIIQSPNPLIPPIFGTDEFGWCSEWNPA 779
Query: 781 MTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGF 840
M KLTGW REEV+DKMLLGEVFGTH + CRLKNQEAFVNFGI+LN AM+G T+K+ F
Sbjct: 780 MAKLTGWSREEVIDKMLLGEVFGTHKSCCRLKNQEAFVNFGIILNGAMSGQNTDKLPIEF 839
Query: 841 FARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALT 900
F R GKY+ECLL V+KKLD +G VTGVFCFLQLAS +LQ ALHIQ L+EQ A KR KAL
Sbjct: 840 FTRFGKYIECLLCVNKKLDGDGAVTGVFCFLQLASHDLQHALHIQRLAEQAATKRAKALA 899
Query: 901 YMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYL 960
YMKRQI+NPLSGI+FS K L+GT++G +Q++++ TS +CQ QL+KILDDSDLDSI+DGY
Sbjct: 900 YMKRQIKNPLSGIMFSGKILDGTEMGEDQRQVLQTSIRCQGQLNKILDDSDLDSIIDGYC 959
Query: 961 DLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLL 1020
+LEM EFT+QD+L+ S+SQ+MA+SS +GI++ N+ E E LYGDSLRLQQ+LADFL
Sbjct: 960 ELEMVEFTVQDILVASISQVMAKSSEKGIQMSNNCTEHGFKETLYGDSLRLQQILADFLS 1019
Query: 1021 ISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLESE 1080
IS+N T GG + V LTK+++G+SV LANLE I H G G+ E LL++MF + G SE
Sbjct: 1020 ISVNFTSPGGHIGVTVRLTKDKIGESVQLANLEFRIMHTGGGISEELLSEMFESRGNASE 1079
Query: 1081 EGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAA 1119
+GISLLISRKL+KLM+GD++YLR AG +FI+ VELA A
Sbjct: 1080 DGISLLISRKLVKLMNGDIQYLRSAGTCTFIIYVELAVA 1118
>UniRef100_Q6V7X3 Phytochrome A [Cuscuta pentagona]
Length = 1119
Score = 1696 bits (4392), Expect = 0.0
Identities = 837/1126 (74%), Positives = 981/1126 (86%), Gaps = 9/1126 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MSSSRPSQSS+NS RS+HSAR+IAQT++DAK+HA FEESG SFDYSSS+R + + +
Sbjct: 1 MSSSRPSQSSSNSARSKHSARIIAQTSIDAKLHAEFEESGDSFDYSSSIRVTSVNTGEQK 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
P+S+KVTTAYLH IQ+ K IQPFGCLLALDEKT +VIA+SENAP+MLTMVSHAVPSVG+
Sbjct: 61 PRSDKVTTAYLHQIQKAKFIQPFGCLLALDEKTFRVIAFSENAPDMLTMVSHAVPSVGDL 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P LGI TDIRTIFTAPS +ALQKALGF EV+LLNPILVHCKTSGKPFYAI+HRVTGSLI+
Sbjct: 121 PVLGIGTDIRTIFTAPSGAALQKALGFGEVSLLNPILVHCKTSGKPFYAIVHRVTGSLIV 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYE PMTAAGALQSYKLAAKAI RLQSLPSGS+ER CDT+VQEVFELTGYDRVM
Sbjct: 181 DFEPVKPYEAPMTAAGALQSYKLAAKAIARLQSLPSGSLERFCDTIVQEVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFH+DDHGEV++EITKPGLEPYLGLHYPATDIPQA+RFLFMKNKVRMI DC AK VKV
Sbjct: 241 AYKFHDDDHGEVVSEITKPGLEPYLGLHYPATDIPQAARFLFMKNKVRMICDCQAKHVKV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQK 360
+ DEKL FDLTLCGSTLRAPH+CHLQYM NM+SIASLVMA+VVND D++ + S K
Sbjct: 301 VQDEKLLFDLTLCGSTLRAPHTCHLQYMENMNSIASLVMAIVVNDGDDEEEEERSGSG-K 359
Query: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLC 420
RKRLWGLVVCHNT+PRFVPFPLRYACEFLAQVFAIHVNKE+ELE QI+EKNILRTQTLLC
Sbjct: 360 RKRLWGLVVCHNTTPRFVPFPLRYACEFLAQVFAIHVNKELELENQIVEKNILRTQTLLC 419
Query: 421 DMLMRDAPLGILTQS--PNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYH 478
D+L+RDA LGI++QS PN+MDLVKCDGA LLYK+K+ LG+TP++ ++DI L+++H
Sbjct: 420 DILLRDAVLGIVSQSQSPNMMDLVKCDGAVLLYKSKIHRLGITPTDFQLQDIVYRLNEHH 479
Query: 479 TDSTGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEP 538
DSTGLSTDSL DAGFPGALSLG +CGMA+VRI+ KD +FWFRSHTA+E+RWGG KHEP
Sbjct: 480 MDSTGLSTDSLYDAGFPGALSLG--LCGMASVRISEKDWLFWFRSHTASEVRWGGVKHEP 537
Query: 539 GEQDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTA 598
DDG+KMHPRSSFKAFLEVV RS PWKDYEMD IHSLQLI+RNAF + ++ + T
Sbjct: 538 ---DDGRKMHPRSSFKAFLEVVETRSLPWKDYEMDGIHSLQLIMRNAFFN-EADTVATNV 593
Query: 599 IDTRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGE 658
I +L+DL+I+G+QELEAVTSEMVRLIETA VPI+AV +DGLVNGWN KIAELTGL V E
Sbjct: 594 IHAKLNDLRIDGLQELEAVTSEMVRLIETAMVPIIAVGVDGLVNGWNTKIAELTGLSVDE 653
Query: 659 AIGKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASR 718
AIG HLLTLVED S VKKML+LAL GEEEKNVQFEI THG + E GPISLVVNACASR
Sbjct: 654 AIGNHLLTLVEDSSVHTVKKMLNLALQGEEEKNVQFEIMTHGIRSECGPISLVVNACASR 713
Query: 719 DLRENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWN 778
D++E+VVGVCF+AQDIT QKTVMDKFTRIEGDY+AI+QNPNPLIPPIFGTDEFGWC EWN
Sbjct: 714 DVQESVVGVCFIAQDITGQKTVMDKFTRIEGDYRAIIQNPNPLIPPIFGTDEFGWCSEWN 773
Query: 779 PAMTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGF 838
AMTKL+GW+R+EV+DKM+LGEVFGT A CRLK+ EAFV G+VLN A+TG E++K F
Sbjct: 774 SAMTKLSGWRRDEVIDKMVLGEVFGTQKACCRLKSHEAFVTLGVVLNNAITGHESDKTVF 833
Query: 839 GFFARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKA 898
GF R+GKYVECLLSV+K+L+ +G V G+FCFLQLAS ELQQALH Q LSEQTA KRLK
Sbjct: 834 GFCTRNGKYVECLLSVTKRLNQDGAVIGLFCFLQLASQELQQALHFQKLSEQTATKRLKV 893
Query: 899 LTYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDG 958
L Y+++Q++NPLSGI+FSRK LEGT+LG +Q+ ++HTSAQCQ+QLSK+LDD+DLD I++G
Sbjct: 894 LAYLRKQVKNPLSGIMFSRKMLEGTELGNDQQNILHTSAQCQQQLSKVLDDTDLDCIIEG 953
Query: 959 YLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADF 1018
YLDLEM EF L +VL+ S+SQ+M +S+ + +R++NDVAE ++ E LYGDSLRLQQVLA+F
Sbjct: 954 YLDLEMVEFKLDEVLLASISQVMTKSNGKSLRVINDVAENVLCETLYGDSLRLQQVLAEF 1013
Query: 1019 LLISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLE 1078
L +++N TP+GGQ+ V++SLTK+ LG+SV LA+LE +TH G GVPE LL QMFG+D
Sbjct: 1014 LSVAVNFTPSGGQLAVSSSLTKDHLGQSVQLAHLEFRVTHSGGGVPEELLTQMFGSDVDA 1073
Query: 1079 SEEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHKLKA 1124
EEGISLL+SR L+KLM+GDV+Y REAG+S+FI+SVELA A K +A
Sbjct: 1074 LEEGISLLVSRNLVKLMNGDVQYHREAGRSAFIISVELAVATKPRA 1119
>UniRef100_Q6K0L2 PHYA3 photoreceptor [Stellaria longipes]
Length = 1123
Score = 1692 bits (4383), Expect = 0.0
Identities = 832/1120 (74%), Positives = 963/1120 (85%), Gaps = 2/1120 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
M+S SQSS NSGRS+HSAR+IAQT DAK+HA FEES + FDYSSSVR S T+ +
Sbjct: 1 MASPAQSQSSTNSGRSKHSARIIAQTIQDAKLHAEFEESSNEFDYSSSVRGS-TSGVNQL 59
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
P+S+KVT++YL IQ+GK IQPFGCLLALD+KT +VIA+SENAPEMLTMVSHAVPSVG+
Sbjct: 60 PQSDKVTSSYLLQIQKGKFIQPFGCLLALDDKTFRVIAFSENAPEMLTMVSHAVPSVGDL 119
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P +GI TDIRTIFT PSASALQKALGF +V+LLNPILVHCK SGKPFYAI+HRVT SL+I
Sbjct: 120 PVIGIGTDIRTIFTDPSASALQKALGFTDVSLLNPILVHCKNSGKPFYAIVHRVTRSLVI 179
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSG+M+RL DTMVQEVFELTGYDRVM
Sbjct: 180 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGNMDRLVDTMVQEVFELTGYDRVM 239
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFH+DDHGEV++E+TKP L+ YLGLHYPATDIPQA+RFLFMKNKVR+I DC AK V+V
Sbjct: 240 AYKFHDDDHGEVVSEVTKPNLDSYLGLHYPATDIPQAARFLFMKNKVRLICDCRAKNVRV 299
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQK 360
+ DEKL DLTLCGSTLRAPH CH QYM NM+SI SLVMAVVVND D++ GS QP K
Sbjct: 300 VQDEKLSVDLTLCGSTLRAPHGCHAQYMENMNSIGSLVMAVVVNDEDDEDGGSAPAQPHK 359
Query: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLC 420
RKRLWGLVVCH+TSPRFVPFPLRYACEFLAQVFAIHVNKE+ELE Q LEK ILRTQTLLC
Sbjct: 360 RKRLWGLVVCHHTSPRFVPFPLRYACEFLAQVFAIHVNKELELESQFLEKKILRTQTLLC 419
Query: 421 DMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHTD 480
DMLMRDAPLGI+TQ+PN+MDLVKCDGAALLY NK+W LG++P++ +RDIA WLS+ HTD
Sbjct: 420 DMLMRDAPLGIVTQNPNVMDLVKCDGAALLYNNKIWKLGISPTDYQLRDIAGWLSRDHTD 479
Query: 481 STGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGE 540
STGLSTDSL DAG+PGA SLGD VCGMAAVRIT +++FWFRSHTAAE++WGGAKHE GE
Sbjct: 480 STGLSTDSLHDAGYPGARSLGDTVCGMAAVRITLNNMLFWFRSHTAAEVKWGGAKHETGE 539
Query: 541 QDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAID 600
+DDG KMHPR+SFKAFLEVV+ RS PWKDYEMDAIHSLQLILRNAFKD ++ D+NT+ I
Sbjct: 540 KDDGSKMHPRTSFKAFLEVVKRRSVPWKDYEMDAIHSLQLILRNAFKDVEASDLNTSVIH 599
Query: 601 TRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAI 660
+++SDL+I G++ELEAVTSEMVRLIETATVPILAVD DGLVNGWN KI ELTG+PV EA+
Sbjct: 600 SKISDLQINGLRELEAVTSEMVRLIETATVPILAVDADGLVNGWNTKIFELTGVPVAEAV 659
Query: 661 GKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDL 720
GKH+ +L E+ S D VK+ML LAL GEE+KNVQFEIK H S +S PISLVVNACAS+D+
Sbjct: 660 GKHIASLAEESSIDNVKRMLQLALQGEEKKNVQFEIKRHQSNPDSSPISLVVNACASKDV 719
Query: 721 RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNPA 780
NVVGVC + QDIT QKTVMDKFTRIEGDYKAI+Q+PNPLIPPIFGTDEFGWC EWNPA
Sbjct: 720 NGNVVGVCLITQDITGQKTVMDKFTRIEGDYKAIIQSPNPLIPPIFGTDEFGWCSEWNPA 779
Query: 781 MTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGF 840
M KLTGW REEV+DKMLLGEVFGT + CRLKNQEAFVNFGI+LN AM+G T+K+ F
Sbjct: 780 MAKLTGWSREEVIDKMLLGEVFGTQKSCCRLKNQEAFVNFGIILNGAMSGQNTDKLPIEF 839
Query: 841 FARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALT 900
F R GKY+ECLL V+KKLD +G VTGVFCFLQLAS +LQ ALHIQ L+EQ A KR KAL
Sbjct: 840 FTRFGKYIECLLCVNKKLDGDGAVTGVFCFLQLASHDLQHALHIQRLAEQAATKRAKALA 899
Query: 901 YMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYL 960
YMKRQI+NPLSGI+FS K L+GT++G +Q++++ TS +CQ QL+KILDDSDLDSI+DGY
Sbjct: 900 YMKRQIKNPLSGIMFSGKILDGTEMGEDQRQVLQTSIRCQGQLNKILDDSDLDSIIDGYC 959
Query: 961 DLEMAEFTLQDVLITSLSQIMARSSARGIRIVND-VAEEIMVEILYGDSLRLQQVLADFL 1019
+LEM EFT+QD+L+ S Q+MA+S+ +GI+I ND E + E LYGDSLRLQQ+LADFL
Sbjct: 960 ELEMVEFTVQDILVASTCQVMAKSNEKGIQIANDSTTEHGLKETLYGDSLRLQQILADFL 1019
Query: 1020 LISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLES 1079
IS+N TP GG V + LTK+++G+S+ ANLE I+H G G+ E LL+QMF N G S
Sbjct: 1020 WISVNFTPAGGNVGIKVRLTKDKIGESIQHANLEFRISHTGGGISEELLSQMFENQGEVS 1079
Query: 1080 EEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAA 1119
EEGISLL+SRK++KLM+GDV+YLR AG S+FI+SVELA A
Sbjct: 1080 EEGISLLVSRKIVKLMNGDVQYLRSAGSSTFIISVELAIA 1119
>UniRef100_Q6V7X2 Phytochrome A [Orobanche minor]
Length = 1123
Score = 1678 bits (4346), Expect = 0.0
Identities = 824/1122 (73%), Positives = 972/1122 (86%), Gaps = 3/1122 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
M+SS+P +SS NS +SR SAR+IAQT++DAK+ A+FEESGSSFDYS+SVR + +
Sbjct: 1 MASSQPGRSSTNSAQSRQSARIIAQTSIDAKLDADFEESGSSFDYSTSVRVTNYPAGLSE 60
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
P+S+KVTTAYLH IQ+GKLIQ FGCLLALDEKT +VIAYSENAPEMLTMVSHAVPSVG+H
Sbjct: 61 PRSDKVTTAYLHQIQKGKLIQQFGCLLALDEKTFRVIAYSENAPEMLTMVSHAVPSVGDH 120
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P LGI +DIRTIFTAPSA+ALQKALGF EV+LLNPILVHCKTSGKPFYAIIHRVTGSLII
Sbjct: 121 PLLGIGSDIRTIFTAPSAAALQKALGFGEVSLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKP+EVPMTAAGALQSYKLAAKAI LQ+LP GS+ERLCDTMVQ+VFELTGYDRVM
Sbjct: 181 DFEPVKPHEVPMTAAGALQSYKLAAKAIACLQALPGGSIERLCDTMVQQVFELTGYDRVM 240
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
YKFHEDDHGEV EITKPGLEPY+GLHYPATDIPQA+RFLFMKNKVRMI DC A VKV
Sbjct: 241 IYKFHEDDHGEVFTEITKPGLEPYVGLHYPATDIPQAARFLFMKNKVRMICDCRANHVKV 300
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQK 360
+ D+ LPFDLTLCGSTLRAPH CH QYM NM+SIASLVM+VVVN+ DEDG S S P K
Sbjct: 301 VQDDNLPFDLTLCGSTLRAPHGCHSQYMENMNSIASLVMSVVVNEGDEDGPDSSS-GPYK 359
Query: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLC 420
RKRLWGLVVCHNT PRF+PFPLRYACEFL QVF+IHVNKE+ELE Q+LEKNILRTQTLLC
Sbjct: 360 RKRLWGLVVCHNTCPRFIPFPLRYACEFLVQVFSIHVNKELELENQMLEKNILRTQTLLC 419
Query: 421 DMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHTD 480
D+L+RD PLGI++QSPN+MDLVKCDGA LL+K + LG+TP++ IRDI SWL +YH D
Sbjct: 420 DLLLRDVPLGIVSQSPNVMDLVKCDGAILLHKRTKYRLGLTPTDFQIRDIVSWLDEYHQD 479
Query: 481 STGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGE 540
STGLSTDSL DAGFPGAL+LG+ +CGMAAV+IT +D +FWFRSHTAAEIRWGGAKHE
Sbjct: 480 STGLSTDSLYDAGFPGALALGNALCGMAAVKITDEDWLFWFRSHTAAEIRWGGAKHELEA 539
Query: 541 QDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAID 600
+DDG+KMHPRSSF+AFLEVV+ RS PWKDYEMD IHSLQLILRNA+K+++ D+ + I
Sbjct: 540 KDDGRKMHPRSSFRAFLEVVKTRSLPWKDYEMDGIHSLQLILRNAYKESEEKDLESREIH 599
Query: 601 TRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAI 660
RL++L+I+G++E+EAVTSEMVRLIETATVPI +V +DGLVNGWN KI++LTGL V EAI
Sbjct: 600 ARLNELQIDGVKEIEAVTSEMVRLIETATVPIFSVGVDGLVNGWNTKISDLTGLSVVEAI 659
Query: 661 GKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDL 720
G H L LVED S D V KML LAL G+EE +VQFEIKTHG + ESGPISL+VNACAS+D+
Sbjct: 660 GMHFLALVEDSSADTVSKMLGLALQGKEEHDVQFEIKTHGQRSESGPISLIVNACASKDV 719
Query: 721 RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNPA 780
+ENVVGVCF+AQDIT QK++MDKFTRIEGDY++I+QNPNPLIPPIFGTDEFGWC EWN A
Sbjct: 720 KENVVGVCFIAQDITTQKSMMDKFTRIEGDYRSIIQNPNPLIPPIFGTDEFGWCSEWNAA 779
Query: 781 MTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGF 840
M KL+GW RE V+DKMLLGEVFG + A CRLKNQEA+VN G+VLN +TG E+ KV FGF
Sbjct: 780 MIKLSGWGREAVIDKMLLGEVFGLNKACCRLKNQEAYVNLGVVLNNTVTGQESGKVSFGF 839
Query: 841 FARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALT 900
F+RSGKYV CLL VSKK+D EG VTG+FCFLQLASPELQQALHIQ +SEQTA KRL+ L
Sbjct: 840 FSRSGKYVACLLCVSKKVDSEGSVTGLFCFLQLASPELQQALHIQRISEQTASKRLRVLA 899
Query: 901 YMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYL 960
Y++R+IR+PLSGI+FSRK +EGTDL EQK +V TS CQ Q++KIL+D+DLD I++GYL
Sbjct: 900 YIRREIRSPLSGIIFSRKLMEGTDLNDEQKNIVRTSLHCQSQMNKILEDTDLDHIIEGYL 959
Query: 961 DLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLL 1020
DLEM EF L +VLI S+SQ++++S+ +GI+IV+++A + E LYGDSLRLQQVLA FLL
Sbjct: 960 DLEMVEFKLHEVLIASISQVISKSNGKGIKIVDNLAPNLSNETLYGDSLRLQQVLAAFLL 1019
Query: 1021 ISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLE-S 1079
I+++ TP+GGQ+ VAA+L K+ +G+ V L LE ITHGG GVP+ +LNQMFG++ + S
Sbjct: 1020 IAVDSTPSGGQLGVAATLAKDSIGEFVQLGRLECRITHGG-GVPQEILNQMFGDEPTDAS 1078
Query: 1080 EEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
E+GISL ISRKL+KLM GD++YLREAG+S+FI+SVE+A ++K
Sbjct: 1079 EDGISLFISRKLVKLMKGDIQYLREAGRSTFIISVEIAISNK 1120
>UniRef100_Q717V8 Phytochrome A1 [Stellaria longipes]
Length = 1122
Score = 1667 bits (4317), Expect = 0.0
Identities = 824/1119 (73%), Positives = 949/1119 (84%), Gaps = 1/1119 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
M+S SQSS NSGRS+HSAR+IAQT DAK HA FEES + FDYSSSVR S T+ +
Sbjct: 1 MASRAQSQSSTNSGRSKHSARIIAQTIQDAKFHAEFEESSNEFDYSSSVRGS-TSGVNQL 59
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
PKS+KVT++YL IQ+GK IQPFGCLLALD+KT +VIA+SENAPEMLTMVSHAVPSVG+
Sbjct: 60 PKSDKVTSSYLLQIQKGKFIQPFGCLLALDDKTFRVIAFSENAPEMLTMVSHAVPSVGDL 119
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P +GI TDIRTIFT PSASALQKALGF +V+LLNPILVHCK SGKPFYAI+HRVT SL+I
Sbjct: 120 PVIGIGTDIRTIFTGPSASALQKALGFTDVSLLNPILVHCKNSGKPFYAIVHRVTRSLVI 179
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSG+M RL DTMVQEVFELTGYDRVM
Sbjct: 180 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGNMVRLVDTMVQEVFELTGYDRVM 239
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFH+DDHGEV++E+TKP L+ YLGLHYPATDIPQA+RFLFMKNKVR+I DC AK V+V
Sbjct: 240 AYKFHDDDHGEVVSEVTKPNLDSYLGLHYPATDIPQAARFLFMKNKVRLICDCRAKNVRV 299
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQK 360
+ DEKL DLTLCGSTLRAPH CH QYM NM+SI SLVMAVVVND D++ GS QP K
Sbjct: 300 VQDEKLSVDLTLCGSTLRAPHGCHAQYMENMNSIGSLVMAVVVNDEDDEDGGSAPAQPHK 359
Query: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLC 420
RKRLWGLVVCH+TSPRFVPFPLRYACEFLAQVFAIHVNKE+ELE Q LEK ILRTQTLLC
Sbjct: 360 RKRLWGLVVCHHTSPRFVPFPLRYACEFLAQVFAIHVNKELELENQFLEKKILRTQTLLC 419
Query: 421 DMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHTD 480
DML+RDAPLGI+T SPN+MDLVKCDGAALLY NKVW LG TP++ +++I WLS+ H D
Sbjct: 420 DMLIRDAPLGIVTHSPNIMDLVKCDGAALLYNNKVWRLGSTPTDYQLQEIGGWLSRDHMD 479
Query: 481 STGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGE 540
STGLSTDSL DAG+P AL LGD VCGMAAV IT D++FWF SHTAAEI+WGGAKHE GE
Sbjct: 480 STGLSTDSLYDAGYPAALELGDSVCGMAAVSITVNDMLFWFTSHTAAEIKWGGAKHEAGE 539
Query: 541 QDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTTAID 600
+DDG KMHPRSSFKAFLEVV+ RS PWKDYEMDAIHSLQLILRNAFKD ++ D+NT+ I
Sbjct: 540 KDDGSKMHPRSSFKAFLEVVKRRSVPWKDYEMDAIHSLQLILRNAFKDGEAADLNTSVIH 599
Query: 601 TRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVGEAI 660
+++SDL+I G++ELEAVTSEMVRLIETATVPI AVD DGLVNGWN KI ELTG+PV EA+
Sbjct: 600 SKISDLQISGLKELEAVTSEMVRLIETATVPIFAVDSDGLVNGWNTKIYELTGIPVEEAV 659
Query: 661 GKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACASRDL 720
GKH+ LVED S D VK+ML AL GEE+KNVQFE+K H S +SGPISL+VNACAS+D+
Sbjct: 660 GKHIAALVEDSSIDNVKQMLQSALQGEEKKNVQFEVKRHHSIPDSGPISLIVNACASKDV 719
Query: 721 RENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEWNPA 780
NVVGVC +AQDIT QKTVMDKF RIEGDYKAI+Q+PNPLIPPIFGTDEFGWC EWNPA
Sbjct: 720 NGNVVGVCLIAQDITGQKTVMDKFLRIEGDYKAIIQSPNPLIPPIFGTDEFGWCSEWNPA 779
Query: 781 MTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVGFGF 840
M KLTGW REEV+DKMLLGEVFG H + CRLKNQEAFVN G++LN AM+G EK+ GF
Sbjct: 780 MAKLTGWTREEVIDKMLLGEVFGMHKSCCRLKNQEAFVNLGVLLNGAMSGQNIEKLSIGF 839
Query: 841 FARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLKALT 900
F RSGKY+ECLL V+KKL+ EG VTGVFCFLQLAS +LQ ALHIQ L+EQ A KR L
Sbjct: 840 FTRSGKYIECLLCVNKKLNGEGDVTGVFCFLQLASHDLQHALHIQRLAEQAATKRANVLA 899
Query: 901 YMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMDGYL 960
YMKRQI+NPL+GI+FS K L+GT++ +Q+ ++ TSA+CQ QL+KILDDSDLDSI+DGY
Sbjct: 900 YMKRQIKNPLAGIIFSGKILDGTNVDEKQRLVLQTSARCQGQLNKILDDSDLDSIIDGYC 959
Query: 961 DLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLADFLL 1020
+LEM EF +QD+L+ S+SQ+MA+SS +GI++ N+ E E LYGDSLRLQQ+LADFL
Sbjct: 960 ELEMVEFAVQDILVASISQVMAKSSEKGIQMSNNCTEHGFKETLYGDSLRLQQILADFLS 1019
Query: 1021 ISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGLESE 1080
IS+N T GG + V LTK+++G+SV LANLE I H G G+ E LL++MF + G SE
Sbjct: 1020 ISVNFTSPGGHIGVTVRLTKDKIGESVQLANLEFRIMHTGGGISEELLSEMFESRGNASE 1079
Query: 1081 EGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAA 1119
+GISLLISRKL+KLM+GD++YLR AG S+FI+SVELA A
Sbjct: 1080 DGISLLISRKLVKLMNGDIQYLRSAGTSTFIISVELAVA 1118
>UniRef100_P55141 Phytochrome A [Petroselinum crispum]
Length = 1129
Score = 1657 bits (4290), Expect = 0.0
Identities = 818/1128 (72%), Positives = 958/1128 (84%), Gaps = 9/1128 (0%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MSSSRP+ SS+N GR+ +ARV+ TT+DAKIHA+FEESG+SFDYSSSVR + +
Sbjct: 1 MSSSRPANSSSNPGRANQNARVVL-TTLDAKIHADFEESGNSFDYSSSVRVTSAVGENSS 59
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
+SNK+TTAYLHHIQ+GKLIQP GCLLA+DEK+ K++AYSENAPEMLTMVSHAVPSVGEH
Sbjct: 60 IQSNKLTTAYLHHIQKGKLIQPVGCLLAVDEKSFKIMAYSENAPEMLTMVSHAVPSVGEH 119
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P LGI TD+RTIFTAPSA+ALQKA+GF ++ LLNPILVHCKTSGKPFYAI HRVTGSLII
Sbjct: 120 PVLGIGTDVRTIFTAPSAAALQKAVGFTDINLLNPILVHCKTSGKPFYAIAHRVTGSLII 179
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKPYEVPMTAAGALQSYKLA+KA+ RLQ+LP GSMERLCDTMVQEVFELTGYDRVM
Sbjct: 180 DFEPVKPYEVPMTAAGALQSYKLASKAVNRLQALPGGSMERLCDTMVQEVFELTGYDRVM 239
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
AYKFH+DDHGEV AE+TKPGLEPY GLHYPATD+PQA+RFLF+KNKVRMI DC A V
Sbjct: 240 AYKFHDDDHGEVTAEVTKPGLEPYFGLHYPATDVPQAARFLFLKNKVRMICDCRANSAPV 299
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDGDGSDSVQPQK 360
L DEKLPF+LTLCGSTLRAPHSCHLQYM NM+SIASLVMAVV+ND+DE + SD K
Sbjct: 300 LQDEKLPFELTLCGSTLRAPHSCHLQYMENMNSIASLVMAVVINDSDEVVESSDR-NSVK 358
Query: 361 RKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTLLC 420
K+LWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHV+KE+ELE QI+EKNILRTQTLLC
Sbjct: 359 SKKLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVSKELELENQIVEKNILRTQTLLC 418
Query: 421 DMLMRDAPLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKYHTD 480
D+LMRDAPLGI++QSPN+MDLVKCDGAALLYKNKV+ LG TPS+ +RDI SWL++YHTD
Sbjct: 419 DLLMRDAPLGIVSQSPNMMDLVKCDGAALLYKNKVYRLGATPSDYQLRDIVSWLTEYHTD 478
Query: 481 STGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHEPGE 540
STGLSTDSL DAG+PGAL+LGD+VCGMA V+IT D++FWFRSH A IRWGGAK EP E
Sbjct: 479 STGLSTDSLYDAGYPGALALGDVVCGMAVVKITSHDMLFWFRSHAAGHIRWGGAKAEPDE 538
Query: 541 QDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFK-------DTDSMD 593
DG+KMHPRSSFKAFLEVV+ RS+ WK++EMDAIHSLQLILR A D +
Sbjct: 539 NHDGRKMHPRSSFKAFLEVVKTRSTTWKEFEMDAIHSLQLILRKALSVEKAVAAQGDEIR 598
Query: 594 INTTAIDTRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTG 653
NT I T+L+DLKIEG+QELEAVTSEMVRLIETATVPI AVD D +VNGWN KIAELTG
Sbjct: 599 SNTDVIHTKLNDLKIEGIQELEAVTSEMVRLIETATVPIFAVDADEIVNGWNTKIAELTG 658
Query: 654 LPVGEAIGKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVN 713
LPV +A+GKHLLTLVED S V +L LAL G+EE+ + FE KT+GS+ +S PI++VVN
Sbjct: 659 LPVDQAMGKHLLTLVEDSSVGTVVFLLALALQGKEEQGIPFEFKTYGSREDSVPITVVVN 718
Query: 714 ACASRDLRENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGW 773
ACA+R L +NVVGVCFVAQD+T+QKT+MDKFTRI+GDYKAIVQNPNPLIPPIFGTDEFGW
Sbjct: 719 ACATRGLHDNVVGVCFVAQDVTSQKTIMDKFTRIQGDYKAIVQNPNPLIPPIFGTDEFGW 778
Query: 774 CCEWNPAMTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSET 833
C EWN AMT+L+GW+RE+VM+KMLLGE+FG + C LK++EAFVN G+VLN A+TG +
Sbjct: 779 CSEWNQAMTELSGWRREDVMNKMLLGEIFGIQTSCCHLKSKEAFVNLGVVLNNALTGQIS 838
Query: 834 EKVGFGFFARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTAL 893
EK+ F FFA GKYVECLL SKKL EG VTG+FCFLQLAS ELQQALHIQ L+EQTA+
Sbjct: 839 EKICFSFFATDGKYVECLLCASKKLHGEGTVTGIFCFLQLASQELQQALHIQRLTEQTAM 898
Query: 894 KRLKALTYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLD 953
KRLK L+Y++RQ +NPL GI F R+ LE +G EQ +L TS CQR ++KILDD+DLD
Sbjct: 899 KRLKTLSYLRRQAKNPLCGINFVREKLEEIGMGEEQTKLFRTSVHCQRHVNKILDDTDLD 958
Query: 954 SIMDGYLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQ 1013
SI+DGYLDLEM+EF L DV + S SQ+ RS+ + I++V++ +EE+M E LYGDSLRLQ+
Sbjct: 959 SIIDGYLDLEMSEFRLHDVYVASRSQVSMRSNGKAIQVVDNFSEEMMSETLYGDSLRLQK 1018
Query: 1014 VLADFLLISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFG 1073
VLADF+ + +N TP GG + ++ +LT++ LG+SV L +LE ITH G+GVPE ++QMFG
Sbjct: 1019 VLADFMSVCVNLTPVGGHLGISVTLTEDNLGQSVQLVHLEFRITHTGAGVPEEAVSQMFG 1078
Query: 1074 NDGLESEEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
+D SEEGISLLISRKL+KLM+GDV YLREAGKS+FI++VELAAA K
Sbjct: 1079 SDSETSEEGISLLISRKLVKLMNGDVHYLREAGKSTFIITVELAAASK 1126
>UniRef100_Q6VAP1 Phytochrome A [Cyrtosia septentrionalis]
Length = 1118
Score = 1522 bits (3941), Expect = 0.0
Identities = 757/1124 (67%), Positives = 918/1124 (81%), Gaps = 18/1124 (1%)
Query: 1 MSSSRPSQSSNNSGRSRHSARVIAQTTVDAKIHANFEESGSSFDYSSSVRASGTADADHQ 60
MS SRPSQSS+ S RSR SAR+IAQTTVDAK+ A F+ G+ FDYS S+RA +
Sbjct: 1 MSFSRPSQSSSASSRSRQSARIIAQTTVDAKLDAEFDAMGTCFDYSQSIRAP-----PDE 55
Query: 61 PKSNKVTTAYLHHIQRGKLIQPFGCLLALDEKTCKVIAYSENAPEMLTMVSHAVPSVGEH 120
+S KVT AYL HIQRGKLIQPFGCLLALDEKT KV+A+SENAPEMLTMVS VPSVG+H
Sbjct: 56 QRSEKVT-AYLQHIQRGKLIQPFGCLLALDEKTFKVLAFSENAPEMLTMVSFTVPSVGDH 114
Query: 121 PALGIDTDIRTIFTAPSASALQKALGFAEVTLLNPILVHCKTSGKPFYAIIHRVTGSLII 180
P + I TD+R +FT+PS +ALQKALGFAEV+LLNPILVHCK+SG+PFYAI+HRVTG LI+
Sbjct: 115 PTIVIGTDVRNLFTSPSTAALQKALGFAEVSLLNPILVHCKSSGRPFYAIVHRVTGCLIV 174
Query: 181 DFEPVKPYEVPMTAAGALQSYKLAAKAITRLQSLPSGSMERLCDTMVQEVFELTGYDRVM 240
DFEPVKP +VPMTAAGALQSYKLAAKAI++LQSLPSGSME+LC+T+++EVFELTGYDRVM
Sbjct: 175 DFEPVKPNDVPMTAAGALQSYKLAAKAISKLQSLPSGSMEKLCNTVIEEVFELTGYDRVM 234
Query: 241 AYKFHEDDHGEVIAEITKPGLEPYLGLHYPATDIPQASRFLFMKNKVRMIVDCHAKQVKV 300
YKFHEDDHGEV AEITKPGLE Y GLHYPATDIPQA+RFLFMKNKVRMI DCHAK VKV
Sbjct: 235 VYKFHEDDHGEVFAEITKPGLESYFGLHYPATDIPQAARFLFMKNKVRMICDCHAKHVKV 294
Query: 301 LIDEKLPFDLTLCGSTLRAPHSCHLQYMANMDSIASLVMAVVVNDNDEDG--DGSDSVQP 358
D+KLPFD++ CGSTLRAPHSCHLQYM NM+SIASLVMAVVVN+ +EDG + ++ P
Sbjct: 295 YQDKKLPFDISFCGSTLRAPHSCHLQYMENMNSIASLVMAVVVNEGEEDGGNEAEENSPP 354
Query: 359 QKRKRLWGLVVCHNTSPRFVPFPLRYACEFLAQVFAIHVNKEIELECQILEKNILRTQTL 418
+RKRLWGLVVCHN SPRFVPFPLRYACEFL QVFAIHVNKE ELE + EK I+RTQT+
Sbjct: 355 HRRKRLWGLVVCHNESPRFVPFPLRYACEFLMQVFAIHVNKEFELENLVKEKKIMRTQTM 414
Query: 419 LCDMLMRD-APLGILTQSPNLMDLVKCDGAALLYKNKVWMLGVTPSELHIRDIASWLSKY 477
LCDML+R+ PLGI+TQ+PN+MDLVKCDGAA LY++K+W LGVTPSE I DI WLS
Sbjct: 415 LCDMLLREFVPLGIITQTPNIMDLVKCDGAAFLYQDKIWRLGVTPSEPQIYDIVHWLSAC 474
Query: 478 HTDSTGLSTDSLSDAGFPGALSLGDLVCGMAAVRITPKDVVFWFRSHTAAEIRWGGAKHE 537
H DSTGLSTD+L +AG+PG SLGD+VCGMA RIT KD++FWFRS AA IRWGGAKH+
Sbjct: 475 HMDSTGLSTDNLHEAGYPGISSLGDVVCGMAVARITSKDMLFWFRSPAAASIRWGGAKHD 534
Query: 538 PGEQDDGKKMHPRSSFKAFLEVVRARSSPWKDYEMDAIHSLQLILRNAFKDTDSMDINTT 597
++DDG++MHPRSSFKAFLEV + RS PW D+EM+AIHSLQLILR+ + N
Sbjct: 535 AADKDDGRRMHPRSSFKAFLEVAKVRSLPWGDHEMNAIHSLQLILRDTLNGIE----NKA 590
Query: 598 AIDTRLSDLKIEGMQELEAVTSEMVRLIETATVPILAVDIDGLVNGWNIKIAELTGLPVG 657
ID +L++LK+EGM +E VT+EMVRLIETATVPILAVD DGL+NGWN+KIA+LTGL
Sbjct: 591 IIDPQLNELKLEGM--VEVVTNEMVRLIETATVPILAVDADGLINGWNMKIAQLTGLSDD 648
Query: 658 EAIGKHLLTLVEDCSTDRVKKMLDLALSGEEEKNVQFEIKTHGSKMESGPISLVVNACAS 717
EA GKHLLT+VED S D VK+ML LAL G EE+NVQF++KT G + + GP+ LVVNAC S
Sbjct: 649 EARGKHLLTIVEDSSIDVVKRMLLLALQGIEEQNVQFQVKTSGVRRDDGPLILVVNACVS 708
Query: 718 RDLRENVVGVCFVAQDITAQKTVMDKFTRIEGDYKAIVQNPNPLIPPIFGTDEFGWCCEW 777
RD+ NVVG CFVAQD+T QK ++DKFT+IEGDYKAIVQNP PLIPPIFGTDEFGWC EW
Sbjct: 709 RDIHSNVVGACFVAQDVTGQKFILDKFTKIEGDYKAIVQNPCPLIPPIFGTDEFGWCSEW 768
Query: 778 NPAMTKLTGWKREEVMDKMLLGEVFGTHMAACRLKNQEAFVNFGIVLNKAMTGSETEKVG 837
N AMTKL+GWKR+EVMDKMLLGEVFG + + CR+K+Q+A VNF I+++ A +G ETEK
Sbjct: 769 NLAMTKLSGWKRDEVMDKMLLGEVFGINTSCCRMKSQDALVNFSILISNAFSGQETEKSP 828
Query: 838 FGFFARSGKYVECLLSVSKKLDVEGLVTGVFCFLQLASPELQQALHIQHLSEQTALKRLK 897
F F RSGK+V+CLLSVS+K+DVEG +TG+FCF+ ELQQ+ Q L +Q +KR+K
Sbjct: 829 FSFMNRSGKHVDCLLSVSRKVDVEGNLTGIFCFVLATGHELQQS---QPLVQQETVKRMK 885
Query: 898 ALTYMKRQIRNPLSGIVFSRKTLEGTDLGIEQKRLVHTSAQCQRQLSKILDDSDLDSIMD 957
AL Y++ +IRNPLSGI+++RK L GT+L EQ L+ T A+C QL++IL+D +L+ IM+
Sbjct: 886 ALAYIRNEIRNPLSGIMYTRKMLVGTNLDEEQSLLLSTGAKCHNQLNRILEDLNLEDIMN 945
Query: 958 GYLDLEMAEFTLQDVLITSLSQIMARSSARGIRIVNDVAEEIMVEILYGDSLRLQQVLAD 1017
L+LEM EF L+DV++T++SQ+M S +G+ +V D+ + ++ E +YGDSLRLQQ+ AD
Sbjct: 946 SCLELEMNEFNLKDVVLTAVSQVMLPSEGKGVNVVYDLPDGLLSEYVYGDSLRLQQIXAD 1005
Query: 1018 FLLISINCTPNGGQVVVAASLTKEQLGKSVHLANLELSITHGGSGVPEALLNQMFGNDGL 1077
FLL+ + +P+G QV + A+L K LGKS+ L ++E+ ITH G+GV E LL++MFG+
Sbjct: 1006 FLLVCVKYSPDGAQVEITANLKKNTLGKSLQLIHVEIRITHAGNGVAEELLSEMFGSKEE 1065
Query: 1078 ESEEGISLLISRKLLKLMSGDVRYLREAGKSSFILSVELAAAHK 1121
SEEG+SL++ RKLL+LM+GDV YLREA KS FILS ELA A K
Sbjct: 1066 TSEEGMSLMVCRKLLRLMNGDVCYLREANKSVFILSAELACASK 1109
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,759,508,731
Number of Sequences: 2790947
Number of extensions: 72991202
Number of successful extensions: 177136
Number of sequences better than 10.0: 2307
Number of HSP's better than 10.0 without gapping: 972
Number of HSP's successfully gapped in prelim test: 1336
Number of HSP's that attempted gapping in prelim test: 173061
Number of HSP's gapped (non-prelim): 2868
length of query: 1124
length of database: 848,049,833
effective HSP length: 138
effective length of query: 986
effective length of database: 462,899,147
effective search space: 456418558942
effective search space used: 456418558942
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0048.6


