

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
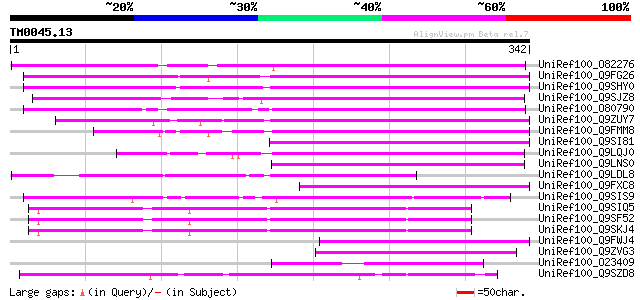
Query= TM0045.13
(342 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O82276 Putative non-LTR retroelement reverse transcrip... 199 7e-50
UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like... 191 2e-47
UniRef100_Q9SHY0 F1E22.12 [Arabidopsis thaliana] 184 2e-45
UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcrip... 181 3e-44
UniRef100_O80790 Putative non-LTR retroelement reverse transcrip... 166 9e-40
UniRef100_Q9ZUY7 Putative reverse transcriptase [Arabidopsis tha... 164 3e-39
UniRef100_Q9FMM8 Non-LTR retroelement reverse transcriptase-like... 149 8e-35
UniRef100_Q9SI81 F23N19.5 [Arabidopsis thaliana] 142 1e-32
UniRef100_Q9LQJ0 F28G4.15 protein [Arabidopsis thaliana] 141 2e-32
UniRef100_Q9LNS0 F1L3.4 [Arabidopsis thaliana] 137 3e-31
UniRef100_Q9LDL8 Hypothetical protein AT4g08830 [Arabidopsis tha... 121 2e-26
UniRef100_Q9FXC8 F12A21.24 [Arabidopsis thaliana] 102 2e-20
UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcrip... 99 2e-19
UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcrip... 99 2e-19
UniRef100_Q9SF52 Putative non-LTR reverse transcriptase [Arabido... 98 3e-19
UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcrip... 97 6e-19
UniRef100_Q9FWJ4 Hypothetical protein F21N10.9 [Arabidopsis thal... 89 2e-16
UniRef100_Q9ZVG3 T2P11.14 protein [Arabidopsis thaliana] 87 7e-16
UniRef100_O23409 SIMILARITY to probable splicing factor CEPRP21 ... 87 7e-16
UniRef100_Q9SZD8 Hypothetical protein F19B15.120 [Arabidopsis th... 86 2e-15
>UniRef100_O82276 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1231
Score = 199 bits (507), Expect = 7e-50
Identities = 117/341 (34%), Positives = 167/341 (48%), Gaps = 12/341 (3%)
Query: 2 NPGLSWHHIWKWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHL 61
N G ++ IWK P +V F+W V N I N R R ++ +++C +CN ETI+H+
Sbjct: 898 NMGSFFNRIWKLITPERVRVFIWLVSQNVIMTNVERVRRHLSENAICSVCNGAEETILHV 957
Query: 62 LRDCACVRPLWQQLVPGHLFQAFIASDLCPWLLGQLRSSLLAPIGGGVVPDFGLLLWNIW 121
LRDC + P+W++L+P F + L WL + P+ G FG+ +W W
Sbjct: 958 LRDCPAMEPIWRRLLPLRRHHEFFSQSLLEWLFTNMD-----PVKGIWPTLFGMGIWWAW 1012
Query: 122 TSRSCFVFQGVPFDLHKVLTSGLRQQLDYVHCRNMMQNPALPRRASYHTVQ--VGWSPPP 179
R C VF K+ L+ D + A+ R + V+ + W P
Sbjct: 1013 KWRCCDVFGE-----RKICRDRLKFIKDMAEEVRRVHVGAVGNRPNGVRVERMIRWQVPS 1067
Query: 180 PAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLA 239
WVK TDG+ RG LAA GG R+ G WL GF N+G+ AELWG + + +A
Sbjct: 1068 DGWVKITTDGASRGNHGLAAAGGAIRNGQGEWLGGFALNIGSCAAPLAELWGAYYGLLIA 1127
Query: 240 WDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSV 299
WD+G R+ ++ D + V ++ G HP ++ + + RDW + H REAN +
Sbjct: 1128 WDKGFRRVELDLDCKLVVGFLSTGVSNAHPLSFLVRLCQGFFTRDWLVRVSHVYREANRL 1187
Query: 300 ADELANMAHDLDFGIHILDYPPPRVSNLLLFDVMGGSLPRA 340
AD LAN A L G+H D P V LLL DV G PRA
Sbjct: 1188 ADGLANYAFTLPLGLHCFDACPEGVRLLLLADVNGTEFPRA 1228
>UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like [Arabidopsis
thaliana]
Length = 676
Score = 191 bits (486), Expect = 2e-47
Identities = 115/336 (34%), Positives = 164/336 (48%), Gaps = 10/336 (2%)
Query: 10 IWKWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCACVR 69
+W+ P + FLW V N + N R R MA +CPLC SE++IH+LRDC +
Sbjct: 347 VWRVMVPERARIFLWLVGNQVVLTNAERVRRHMADSDVCPLCKGASESLIHVLRDCPAMM 406
Query: 70 PLWQQLVPGHLFQAFIASDLCPWLLGQLRSSLLAPIGGGVVPDFGLLLWNIWTSRSCFVF 129
+W ++VP + F + L W+ G L+ + F L +W W R +VF
Sbjct: 407 GIWMRVVPVMEQRRFFETSLLEWMYGNLKERSDSERRSWPTL-FALTVWWGWKWRCGYVF 465
Query: 130 --QGVPFDLHKVLTSGLRQ-QLDYVHCRNMMQNPALPRRASYHTVQVGWSPPPPAWVKCN 186
D K L S + + + ++ + L R + W P WV N
Sbjct: 466 GEDSRCRDRVKFLKSAVAEVEAAHLAANGDAREDVLVER------MIAWRKPAEGWVTMN 519
Query: 187 TDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPR 246
TDG+ G A GGV RD +G WL GF N+G + AELWG++ + +AW+RG R
Sbjct: 520 TDGASHGNPGQATAGGVIRDEHGSWLVGFALNIGVCSAPLAELWGVYYGLVVAWERGWRR 579
Query: 247 LVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELANM 306
+ +E DS++ V + G HP ++ + +DW + H REAN +AD LAN
Sbjct: 580 VRLEVDSALVVGFLQSGIGDSHPLAFLVRLCHGFISKDWIVRITHVYREANRLADGLANY 639
Query: 307 AHDLDFGIHILDYPPPRVSNLLLFDVMGGSLPRATR 342
A L FG +LD P VS++LL DVMG S PR R
Sbjct: 640 AFTLPFGFLLLDSCPEHVSSILLEDVMGTSFPRHVR 675
>UniRef100_Q9SHY0 F1E22.12 [Arabidopsis thaliana]
Length = 1055
Score = 184 bits (468), Expect = 2e-45
Identities = 110/335 (32%), Positives = 159/335 (46%), Gaps = 8/335 (2%)
Query: 10 IWKWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCACVR 69
+WK P +V FLW V N + R R ++ ++C +C E+++H+LRDC
Sbjct: 400 LWKVRVPERVKTFLWLVGNQAVMTEEERHRRHLSASNVCQVCKGGVESMLHVLRDCPAQL 459
Query: 70 PLWQQLVPGHLFQAFIASDLCPWLLGQLRS-SLLAPIGGGVVPDFGLLLWNIWTSRSCFV 128
+W ++VP Q F + L WL L S I + F +++W W R +
Sbjct: 460 GIWVRVVPQRRQQGFFSKSLFEWLYDNLGDRSGCEDIPWSTI--FAVIIWWGWKWRCGNI 517
Query: 129 FQGVPFDLHKV-LTSGLRQQLDYVHCRNMMQNPALPRRASYHTVQVGWSPPPPAWVKCNT 187
F +V ++ H N++ PR +GW P WVK NT
Sbjct: 518 FGENTKCRDRVKFVKEWAVEVYRAHSGNVLVGITQPRVER----MIGWVSPCVGWVKVNT 573
Query: 188 DGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPRL 247
DG+ RG LA+ GGV RD G W GF N+G + AELWG++ + AW++ VPR+
Sbjct: 574 DGASRGNPGLASAGGVLRDCTGAWCGGFSLNIGRCSAPQAELWGVYYGLYFAWEKKVPRV 633
Query: 248 VVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELANMA 307
+E DS V V + G HP ++ + +DW + VH REAN +AD LAN A
Sbjct: 634 ELEVDSEVIVGFLKTGISDSHPLSFLVRLCHGFLQKDWLVRIVHVYREANRLADGLANYA 693
Query: 308 HDLDFGIHILDYPPPRVSNLLLFDVMGGSLPRATR 342
L G H D P +S+LL D +G + PR R
Sbjct: 694 FSLSLGFHSFDLVPDAMSSLLREDTLGSTRPRRVR 728
>UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 321
Score = 181 bits (458), Expect = 3e-44
Identities = 108/332 (32%), Positives = 158/332 (47%), Gaps = 25/332 (7%)
Query: 16 PFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCACVRPLWQQL 75
P +V FLW V+ I N R+R ++ +C +C ETI+H+LRDC + +W +L
Sbjct: 3 PERVRVFLWLVVQQVIITNVERYRRHLSDTRVCQICQGGEETILHVLRDCPAMAGIWSRL 62
Query: 76 VPGHLFQAFIASDLCPWLLGQLRSSLLAPIGGGVVPDFGLLLWNIWTSRSCFVFQGVPFD 135
VP + F + L W+ LR G F + +W W R +F G
Sbjct: 63 VPRDQIRQFFTASLLEWIYKNLRER------GSWPTVFVMAVWWGWKWRCGNIFGG---- 112
Query: 136 LHKVLTSGLRQQLDYVHCRNMMQNPALPR--------RASYHTVQVGWSPPPPAWVKCNT 187
R ++ ++ +++ + A+ R S V W P WVK NT
Sbjct: 113 -----NGKCRDRVKFI--KDLAEEVAIANAFVKGNEVRVSRVERLVSWVSPEDGWVKLNT 165
Query: 188 DGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPRL 247
DG+ RG A GGV RD G W+ GF N+G + AELWG++ + +AW RG R+
Sbjct: 166 DGASRGNPGFATAGGVLRDHNGAWIGGFAVNIGVCSAPLAELWGVYYGLFIAWGRGARRV 225
Query: 248 VVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELANMA 307
+E DS + V + G HP ++ + + W + H REAN +AD LAN A
Sbjct: 226 ELEVDSKMVVGFLTTGIADSHPLSFLLRLCYDFLSKGWIVRISHVYREANRLADGLANYA 285
Query: 308 HDLDFGIHILDYPPPRVSNLLLFDVMGGSLPR 339
L G+H+L+ P VS++LL DV G S PR
Sbjct: 286 FSLSLGLHLLESRPDVVSSILLDDVAGVSYPR 317
>UniRef100_O80790 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 970
Score = 166 bits (420), Expect = 9e-40
Identities = 105/332 (31%), Positives = 155/332 (46%), Gaps = 12/332 (3%)
Query: 10 IWKWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCACVR 69
IWK P +V F+W V + I N R R ++ + C +CN E+I+H+LRDC +
Sbjct: 647 IWKLVAPERVRVFIWLVSHMVIMTNVERVRRHLSDIATCSVCNGADESILHVLRDCPAMT 706
Query: 70 PLWQQLVPGHLFQAFIASDLCPWLLGQLRSSLLAPIGGGVVPDFGLLLWNIWTSRSCFVF 129
P+WQ+L+P F + WL L P G F + +W W R VF
Sbjct: 707 PIWQRLLPQRRQNEFFSQ--FEWLFTNLD-----PAKGDWPTLFSMGIWWAWKWRCGDVF 759
Query: 130 QGVPFDLHKV-LTSGLRQQLDYVHCRNMMQNPALPRRASYHTVQVGWSPPPPAWVKCNTD 188
++ + +++ H + + +RA + + W P WVK TD
Sbjct: 760 GERKLCRDRLKFIKDIAEEVRKAHVGTLNNHV---KRARVERM-IRWKAPSDRWVKLTTD 815
Query: 189 GSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPRLV 248
G+ RG LAA G + G WL GF N+G+ + AELWG + + +AWD+G R+
Sbjct: 816 GASRGHQGLAAASGAILNLQGEWLGGFALNIGSCDAPLAELWGAYYGLLIAWDKGFRRVE 875
Query: 249 VESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELANMAH 308
+ DS + V ++ G HP ++ + + RDW + H REAN +AD LAN A
Sbjct: 876 LNLDSELVVGFLSTGISKAHPLSFLVRLCQGFFTRDWLVRVSHVYREANRLADGLANYAF 935
Query: 309 DLDFGIHILDYPPPRVSNLLLFDVMGGSLPRA 340
L G H + P V L+ D G S PRA
Sbjct: 936 FLPLGFHCFEICPEDVLLFLVEDANGTSFPRA 967
>UniRef100_Q9ZUY7 Putative reverse transcriptase [Arabidopsis thaliana]
Length = 314
Score = 164 bits (415), Expect = 3e-39
Identities = 103/317 (32%), Positives = 146/317 (45%), Gaps = 13/317 (4%)
Query: 31 IWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCACVRPLWQQLVPGHLFQAFIASDLC 90
+ N R R ++ +C +C +TIIH+LRDC + +W +LVP + F L
Sbjct: 5 LMTNAERRRRHLSDSDICQICKGAEKTIIHILRDCPAMEGIWIRLVPAGKRREFFTQSLL 64
Query: 91 PWL---LGQLRSSLLAPIGGGVVPDFGLLLWNIWTSR--SCFVFQGVPFDLHKVLTSGLR 145
WL LG R + + F L +W W R + F Q D + L L
Sbjct: 65 EWLFANLGDRRKTCESTWS----TLFALSIWWAWKWRCGNIFGVQDKCRDRVRFL-KDLA 119
Query: 146 QQLDYVHCRNMMQNPALPRRASYHTVQVGWSPPPPAWVKCNTDGSVRGTSNLAACGGVCR 205
++ H + R + WS P W K NTDG+ RG LA+ GGV R
Sbjct: 120 RETSMAHVIVRTLSGGHGERVER---LIAWSKPEEGWWKLNTDGASRGNPGLASAGGVLR 176
Query: 206 DSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPRLVVESDSSVAVKLINEGSD 265
D G W GF N+G + AELWG++ + +AW+R V RL +E DS + V + G +
Sbjct: 177 DEEGAWRGGFALNIGVCSAPLAELWGVYYGLYIAWERRVTRLEIEVDSEIVVGFLKIGIN 236
Query: 266 VLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELANMAHDLDFGIHILDYPPPRVS 325
+HP ++ + RDW + H REAN +AD LAN A L G H L P +
Sbjct: 237 EVHPLSFLVRLCHDFISRDWRVRISHVYREANRLADGLANYAFSLPLGFHSLSLVPDSLR 296
Query: 326 NLLLFDVMGGSLPRATR 342
+LL D G ++ R R
Sbjct: 297 FILLDDTSGATVSRQVR 313
>UniRef100_Q9FMM8 Non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 308
Score = 149 bits (377), Expect = 8e-35
Identities = 96/296 (32%), Positives = 137/296 (45%), Gaps = 22/296 (7%)
Query: 56 ETIIHLLRDCACVRPLWQQLVPGHLFQAFIASDLCPWLLGQL--RSSLLAPIGGGVVPDF 113
E+++H+ RDC +W + VP Q F + L WL L RSS I + F
Sbjct: 25 ESVLHVFRDCPAQLGIWVRFVPRRRQQGFFSKSLFEWLYDNLCDRSSC-EDIPWSTI--F 81
Query: 114 GLLLWNIWTSRSCFVF-------QGVPFDLHKVLTSGLRQQLDYVHCRNMMQNPALPRRA 166
+++W W R +F V F V+ ++ H N + PR
Sbjct: 82 AVIIWWGWKWRCSNIFGENTKCRDRVKFVKEWVV------EVYRAHLGNALVGSTQPRVE 135
Query: 167 SYHTVQVGWSPPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLW 226
+GW P WVK NTDG+ RG LA+ GGV RD G W GF N+G +
Sbjct: 136 RL----IGWVLPCVGWVKVNTDGASRGNPGLASAGGVLRDCEGAWCGGFSLNIGRCSAQH 191
Query: 227 AELWGIFSVIQLAWDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWD 286
AELWG++ + AW++ VPR+ +E DS V + G HP ++ + +DW
Sbjct: 192 AELWGVYYGLYFAWEKKVPRVELEVDSEAIVGFLKTGISDSHPLSFLVRLCHNFLQKDWL 251
Query: 287 FQCVHACREANSVADELANMAHDLDFGIHILDYPPPRVSNLLLFDVMGGSLPRATR 342
+ V+ REAN +AD LAN L G H D+ P +S+LL D +G + R R
Sbjct: 252 VRIVYVYREANCLADGLANYTILLSLGFHSFDFVPDAMSSLLKEDTLGSTQLRRVR 307
>UniRef100_Q9SI81 F23N19.5 [Arabidopsis thaliana]
Length = 233
Score = 142 bits (359), Expect = 1e-32
Identities = 71/171 (41%), Positives = 92/171 (53%)
Query: 172 QVGWSPPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWG 231
QV WS P W K NTDG+ G LA GG R+ YG W FGF N+G AELWG
Sbjct: 62 QVRWSKPSLGWCKLNTDGASHGNPGLATAGGALRNEYGEWCFGFALNIGRCLAPLAELWG 121
Query: 232 IFSVIQLAWDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVH 291
++ + +AWDRG+ RL +E DS + V + G HP ++ + RDW + H
Sbjct: 122 VYYGLFMAWDRGITRLELEVDSEMVVGFLRTGIGSSHPLSFLVRMCHGFLSRDWIVRIGH 181
Query: 292 ACREANSVADELANMAHDLDFGIHILDYPPPRVSNLLLFDVMGGSLPRATR 342
REAN +AD LAN A DL G H PP + ++L D +G S+PR R
Sbjct: 182 VYREANRLADGLANYAFDLPLGYHAFASPPNSLDSILRDDELGISVPRLVR 232
>UniRef100_Q9LQJ0 F28G4.15 protein [Arabidopsis thaliana]
Length = 272
Score = 141 bits (356), Expect = 2e-32
Identities = 89/277 (32%), Positives = 126/277 (45%), Gaps = 20/277 (7%)
Query: 71 LWQQLVPGHLFQAFIASDLCPWLLGQLRSSLLAPIGGGVVPDFGLLLWNIWTSRSCFVFQ 130
+W +L+P +F + L W+ L + G F +W W R
Sbjct: 4 IWTRLLPVRRLSSFFSKSLLEWIYANLGEEIEIN-GCPWAVTFSQAIWWGWKWR-----Y 57
Query: 131 GVPFDLHKVLTSGLR----QQLD----YVHCRNMMQNPALPRRASYHTVQVGWSPPPPAW 182
G F +K +R + LD +VH + A R + WSPP W
Sbjct: 58 GNIFGENKKCRDRVRFIKDRALDVWKAHVHKMGVTTRTAREERL------IAWSPPRVGW 111
Query: 183 VKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDR 242
K NTDG+ RG LA GGV RD G W +GF N+G + AELWG + + +AW+R
Sbjct: 112 FKLNTDGASRGNPRLATAGGVVRDGDGNWCYGFSLNIGICSAPLAELWGAYYGLNIAWER 171
Query: 243 GVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADE 302
GV +L +E DS + V + G D HP ++ +DW + H REAN +AD
Sbjct: 172 GVTQLEMEIDSEMVVGFLRTGIDDSHPLSFLVRLCHGLLSKDWSVRISHVYREANRLADG 231
Query: 303 LANMAHDLDFGIHILDYPPPRVSNLLLFDVMGGSLPR 339
LAN A L G H+ + P V +++ DV G + PR
Sbjct: 232 LANYAFFLPLGFHLFNSTPDIVMSIVHDDVAGSAYPR 268
>UniRef100_Q9LNS0 F1L3.4 [Arabidopsis thaliana]
Length = 253
Score = 137 bits (346), Expect = 3e-31
Identities = 67/167 (40%), Positives = 91/167 (54%)
Query: 173 VGWSPPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGI 232
+ WSPP W K NTDG+ RG LA GGV RD G W +GF N+G + AELWG
Sbjct: 83 IAWSPPRVGWFKLNTDGASRGNPRLATAGGVVRDGDGNWCYGFSLNIGICSAPLAELWGA 142
Query: 233 FSVIQLAWDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHA 292
+ + +AW+RGV +L +E DS + V + G D HP ++ +DW + H
Sbjct: 143 YYGLNIAWERGVTQLEMEIDSEMVVGFLRTGIDDSHPLSFLVRLCHGLLSKDWSVRISHV 202
Query: 293 CREANSVADELANMAHDLDFGIHILDYPPPRVSNLLLFDVMGGSLPR 339
REAN +AD LAN A L G H+ + P V +++ DV G + PR
Sbjct: 203 YREANRLADGLANYAFFLPLGFHLFNSTPDIVMSIVHDDVAGSAYPR 249
>UniRef100_Q9LDL8 Hypothetical protein AT4g08830 [Arabidopsis thaliana]
Length = 947
Score = 121 bits (304), Expect = 2e-26
Identities = 80/268 (29%), Positives = 117/268 (42%), Gaps = 22/268 (8%)
Query: 2 NPGLSWHHIWKWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHL 61
N + +W+ +V FLW + + S+C +C ETI+H+
Sbjct: 688 NMAAFFDRLWRVVALERVKTFLWHIGDT----------------SVCQVCKGGDETILHV 731
Query: 62 LRDCACVRPLWQQLVPGHLFQAFIASDLCPWLLGQLRSSLLAPIGGGVVPDFGLLLWNIW 121
L+DC + +W++LV F L WL L A G F +++W W
Sbjct: 732 LKDCPSIAGIWRRLVQVQRSYDFFNGSLFGWLYVNLGMKN-AETGYAWATLFAIVVWWSW 790
Query: 122 TSRSCFVFQGVPFDLHKV-LTSGLRQQLDYVHCRNMMQNPALPRRASYHTVQVGWSPPPP 180
R +VF V +V L ++ + H + QN L R V W PP
Sbjct: 791 KWRCGYVFGEVGKCRDRVKFFRDLAAEVSHAHAIHS-QNGGLRTRVER---LVAWKPPDG 846
Query: 181 AWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAW 240
WVK NTDG+ RG LA GGV RD G W GF ++G + AELWG++ + +AW
Sbjct: 847 EWVKLNTDGASRGNLGLATTGGVLRDGIGHWCGGFALDIGVCSAPLAELWGVYYGLYMAW 906
Query: 241 DRGVPRLVVESDSSVAVKLINEGSDVLH 268
+R R+ +E DS + V + G H
Sbjct: 907 ERRFTRVELEVDSELVVGFLTTGISDTH 934
>UniRef100_Q9FXC8 F12A21.24 [Arabidopsis thaliana]
Length = 803
Score = 102 bits (254), Expect = 2e-20
Identities = 55/151 (36%), Positives = 74/151 (48%)
Query: 192 RGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPRLVVES 251
RG LA GGV RD G W GF N+G + AELWG++ LAW + + R+ +E
Sbjct: 652 RGNPGLATAGGVIRDGAGNWCGGFALNIGRCSAPLAELWGVYYGFYLAWTKALTRVELEV 711
Query: 252 DSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELANMAHDLD 311
DS + V + G HP ++ +DW + H REAN +AD LAN A L
Sbjct: 712 DSELVVGFLKTGIGDQHPLSFLVRLCHGLLSKDWIVRITHVYREANRLADGLANYAFSLP 771
Query: 312 FGIHILDYPPPRVSNLLLFDVMGGSLPRATR 342
G H L P + +L D +G + PR R
Sbjct: 772 LGFHSLIDVPDDLEVILHEDSLGSTRPRRVR 802
>UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1715
Score = 98.6 bits (244), Expect = 2e-19
Identities = 86/335 (25%), Positives = 146/335 (42%), Gaps = 26/335 (7%)
Query: 10 IWKWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCACVR 69
IW+ K+ +F+WR L+ + + + D C C ETI H++ C+ +
Sbjct: 1388 IWRLKITPKIKHFIWRCLSGALSTTTQLRNRNIPADPTCQRCCNADETINHIIFTCSYAQ 1447
Query: 70 PLWQQL-VPGH---LFQAFIASDLCPWLLGQLRSSLLAPIGGGVVPDFGLLLWNIWTSRS 125
+W+ G F + ++ L G+ +L PI G++P + ++W +W SR+
Sbjct: 1448 VVWRSANFSGSNRLCFTDNLEENIRLILQGKKNQNL--PILNGLMPFW--IMWRLWKSRN 1503
Query: 126 CFVFQGVPFDLHKVLTSGLRQQLDYVHCRNMMQNPALPRRASYHTVQVG---------WS 176
++FQ + KV ++ ++V M+ + A+ S++T Q WS
Sbjct: 1504 EYLFQQLDRFPWKVAQKAEQEATEWV--ETMVNDTAI----SHNTAQSNDRPLSRSKQWS 1557
Query: 177 PPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTS-NVLWAELWGIFSV 235
PP ++KCN D + + G + RD GR L C L S + L AE G
Sbjct: 1558 SPPEGFLKCNFDSGYVQGRDYTSTGWILRDCNGRVLHSGCAKLQQSYSALQAEALGFLHA 1617
Query: 236 IQLAWDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACRE 295
+Q+ W RG + E D+ LIN+ D H ++ IR W + + RE
Sbjct: 1618 LQMVWIRGYCYVWFEGDNLELTNLINKTED-HHLLETLLYDIRFWMTKLPFSSIGYVNRE 1676
Query: 296 ANSVADELANMAHDLDFGIHILDYPPPRVSNLLLF 330
N AD+L A+ + ++ + PPR L L+
Sbjct: 1677 RNLAADKLTKYANSMS-SLYETFHVPPRWLQLYLY 1710
>UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1524
Score = 98.6 bits (244), Expect = 2e-19
Identities = 78/300 (26%), Positives = 125/300 (41%), Gaps = 15/300 (5%)
Query: 13 WDGPF--KVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCACVRP 70
W+ P K+ +FLWR L+ + R GM +D +CP C+ E+E+I H L C
Sbjct: 1203 WNLPIMPKLKHFLWRALSQALATTERLTTRGMRIDPICPRCHRENESINHALFTCPFATM 1262
Query: 71 LWQQLVPGHLFQAFIASDLCPWLLGQLRSSLLAPIGGGVVPDFGLLL-----WNIWTSRS 125
W + +++D + S++L + + DF LL W IW +R+
Sbjct: 1263 AWWLSDSSLIRNQLMSND-----FEENISNILNFVQDTTMSDFHKLLPVWLIWRIWKARN 1317
Query: 126 CFVFQGVPFDLHKVLTSGLRQQLDYVHCRNMMQNPALPRRASYHTVQVGWSPPPPAWVKC 185
VF K + S + D+++ + P R ++ W PP +VKC
Sbjct: 1318 NVVFNKFRESPSKTVLSAKAETHDWLNATQSHKKTPSPTRQIAEN-KIEWRNPPATYVKC 1376
Query: 186 NTDGSVRGTSNLAACGGVCRDSYGRWL-FGFCRNLGTSNVLWAELWGIFSVIQLAWDRGV 244
N D A G + R+ YG + +G + TSN L AE + + +Q W RG
Sbjct: 1377 NFDAGFDVQKLEATGGWIIRNHYGTPISWGSMKLAHTSNPLEAETKALLAALQQTWIRGY 1436
Query: 245 PRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELA 304
++ +E D + LIN G + I W ++ Q R+ N +A LA
Sbjct: 1437 TQVFMEGDCQTLINLIN-GISFHSSLANHLEDISFWANKFASIQFGFIRRKGNKLAHVLA 1495
>UniRef100_Q9SF52 Putative non-LTR reverse transcriptase [Arabidopsis thaliana]
Length = 484
Score = 98.2 bits (243), Expect = 3e-19
Identities = 78/300 (26%), Positives = 125/300 (41%), Gaps = 15/300 (5%)
Query: 13 WDGPF--KVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCACVRP 70
W+ P K+ +FLWR L+ + R GM +D CP C+ E+E+I H L C
Sbjct: 163 WNLPIMPKLKHFLWRALSQALATTERLTTRGMRIDPSCPRCHRENESINHALFTCPFATM 222
Query: 71 LWQQLVPGHLFQAFIASDLCPWLLGQLRSSLLAPIGGGVVPDFGLLL-----WNIWTSRS 125
W+ + +++D + S++L + + DF LL W IW +R+
Sbjct: 223 AWRLSDSSLIRNQLMSND-----FEENISNILNFVQDTTMSDFHKLLPVWLIWRIWKARN 277
Query: 126 CFVFQGVPFDLHKVLTSGLRQQLDYVHCRNMMQNPALPRRASYHTVQVGWSPPPPAWVKC 185
VF K + S + D+++ + P R ++ W PP +VKC
Sbjct: 278 NVVFNKFRESPSKTVLSAKAETHDWLNATQSHKKTPSPTRQIAEN-KIEWRNPPATYVKC 336
Query: 186 NTDGSVRGTSNLAACGGVCRDSYGRWL-FGFCRNLGTSNVLWAELWGIFSVIQLAWDRGV 244
N D A G + R+ YG + +G + TSN L AE + + +Q W RG
Sbjct: 337 NFDAGFDVQKLEATGGWIIRNHYGTPISWGSMKLAHTSNPLEAETKALLAALQQTWIRGY 396
Query: 245 PRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELA 304
++ +E D + LIN G + I W ++ Q R+ N +A LA
Sbjct: 397 TQVFMEGDCQTLINLIN-GISFHSSLANHLEDISFWANKFASIQFGFIRRKGNKLAHVLA 455
>UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1750
Score = 97.1 bits (240), Expect = 6e-19
Identities = 77/300 (25%), Positives = 125/300 (41%), Gaps = 15/300 (5%)
Query: 13 WDGPF--KVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCACVRP 70
W+ P K+ +FLWR L+ + R GM +D CP C+ E+E+I H L C
Sbjct: 1429 WNLPIMPKLKHFLWRALSQALATTERLTTRGMRIDPSCPRCHRENESINHALFTCPFATM 1488
Query: 71 LWQQLVPGHLFQAFIASDLCPWLLGQLRSSLLAPIGGGVVPDFGLLL-----WNIWTSRS 125
W+ + +++D + S++L + + DF LL W IW +R+
Sbjct: 1489 AWRLSDSSLIRNQLMSND-----FEENISNILNFVQDTTMSDFHKLLPVWLIWRIWKARN 1543
Query: 126 CFVFQGVPFDLHKVLTSGLRQQLDYVHCRNMMQNPALPRRASYHTVQVGWSPPPPAWVKC 185
VF K + S + D+++ + P R ++ W PP +VKC
Sbjct: 1544 NVVFNKFRESPSKTVLSAKAETHDWLNATQSHKKTPSPTRQIAEN-KIEWRNPPATYVKC 1602
Query: 186 NTDGSVRGTSNLAACGGVCRDSYGRWL-FGFCRNLGTSNVLWAELWGIFSVIQLAWDRGV 244
N D A G + R+ YG + +G + TSN L AE + + +Q W RG
Sbjct: 1603 NFDAGFDVQKLEATGGWIIRNHYGTPISWGSMKLAHTSNPLEAETKALLAALQQTWIRGY 1662
Query: 245 PRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELA 304
++ +E D + LIN G + I W ++ Q ++ N +A LA
Sbjct: 1663 TQVFMEGDCQTLINLIN-GISFHSSLANHLEDISFWANKFASIQFGFIRKKGNKLAHVLA 1721
>UniRef100_Q9FWJ4 Hypothetical protein F21N10.9 [Arabidopsis thaliana]
Length = 256
Score = 88.6 bits (218), Expect = 2e-16
Identities = 47/138 (34%), Positives = 68/138 (49%)
Query: 205 RDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPRLVVESDSSVAVKLINEGS 264
R+ G W GF ++G + AELWG++ + +AW++ V R+ +E DS + V + G
Sbjct: 37 REHQGNWCGGFSVSIGICSAPQAELWGVYYGLYIAWEKWVTRVELEVDSEIVVGFLKTGI 96
Query: 265 DVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELANMAHDLDFGIHILDYPPPRV 324
HP + + +DW + H REAN +AD LAN A L G H P V
Sbjct: 97 GDSHPLSFLARLCYGFISKDWIVRISHVYREANRLADGLANYAFTLPLGFHFFHSSPDVV 156
Query: 325 SNLLLFDVMGGSLPRATR 342
++ L D G S PR R
Sbjct: 157 DSVRLEDTSGTSCPRLVR 174
>UniRef100_Q9ZVG3 T2P11.14 protein [Arabidopsis thaliana]
Length = 158
Score = 87.0 bits (214), Expect = 7e-16
Identities = 46/133 (34%), Positives = 65/133 (48%)
Query: 202 GVCRDSYGRWLFGFCRNLGTSNVLWAELWGIFSVIQLAWDRGVPRLVVESDSSVAVKLIN 261
G RD YG W F NLG AELWG++ + +AW++G+ RL +E DS + +
Sbjct: 17 GALRDEYGDWRGSFALNLGRCTAPLAELWGVYYGLVIAWEKGITRLELEVDSKLVAGFLT 76
Query: 262 EGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREANSVADELANMAHDLDFGIHILDYPP 321
G + H ++ + +DW + H REAN ADELAN A L G D
Sbjct: 77 TGIEDSHLLSFLVRLCYGFSSKDWIVRVSHVYREANRFADELANYAFSLLLGFLSFDSGL 136
Query: 322 PRVSNLLLFDVMG 334
P V +++ D G
Sbjct: 137 PFVDSIMREDAAG 149
>UniRef100_O23409 SIMILARITY to probable splicing factor CEPRP21 [Arabidopsis
thaliana]
Length = 559
Score = 87.0 bits (214), Expect = 7e-16
Identities = 48/140 (34%), Positives = 65/140 (46%), Gaps = 14/140 (10%)
Query: 173 VGWSPPPPAWVKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNLGTSNVLWAELWGI 232
+ W+ PP WVK +TDG+ RG AA GGV RD G W+ GF L
Sbjct: 26 IAWTKPPEGWVKVSTDGASRGNPGPAAAGGVIRDEDGLWVGGFALQL------------- 72
Query: 233 FSVIQLAWDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHA 292
+ ++L W R+ +E DS + V + G HP ++ +DW + H
Sbjct: 73 -AFVRLRWQSCGGRVELEVDSELVVGFLQSGISEAHPLAFLVRLCHGLLSKDWLVRVSHV 131
Query: 293 CREANSVADELANMAHDLDF 312
REAN +AD LAN A L F
Sbjct: 132 YREANRLADGLANYAFSLQF 151
>UniRef100_Q9SZD8 Hypothetical protein F19B15.120 [Arabidopsis thaliana]
Length = 575
Score = 85.5 bits (210), Expect = 2e-15
Identities = 82/324 (25%), Positives = 139/324 (42%), Gaps = 26/324 (8%)
Query: 7 WHHIWKWDGPFKVANFLWRVLNNGIWVNYRRWRAGMALDSLCPLCNEESETIIHLLRDCA 66
+ IWK K+ +FLW+ L+N + V ++ +S C C ET+ HLL C
Sbjct: 254 YQKIWKSQTSPKIQHFLWKCLSNSLPVAGALAYRHLSKESACIRCPSCKETVNHLLFKCT 313
Query: 67 CVRPLWQ-QLVPGHLFQAFIASDLCP--WLLGQLRSSLLAPIGGGVVPDFGLLLWNIWTS 123
R W +P L + S W+ + +VP LLW +W +
Sbjct: 314 FARLTWAISSIPIPLGGEWADSIYVNLYWVFNLGNGNPQWEKASQLVP---WLLWRLWKN 370
Query: 124 RSCFVFQGVPFDLHKVLTSGLRQQLDYVHCRNMMQNPALPRRASYHTVQVG-WSPPPPAW 182
R+ VF+G F+ +VL R + D R + + + + G W PPP W
Sbjct: 371 RNELVFRGREFNAQEVLR---RAEDDLEEWRIRTEAESCGTKPQVNRSSCGRWRPPPHQW 427
Query: 183 VKCNTDGSVRGTSNLAACGGVCRDSYGRWLFGFCRNL-GTSNVLWAEL----WGIFSVIQ 237
VKCNTD + + G V R+ G + R L +VL AEL W + S+ +
Sbjct: 428 VKCNTDATWNRDNERCGIGWVLRNEKGEVKWMGARALPKLKSVLEAELEAMRWAVLSLSR 487
Query: 238 LAWDRGVPRLVVESDSSVAVKLINEGSDVLHPYGGMISQIRQWKDRDWDFQCVHACREAN 297
++ ++ ESDS V ++++N ++ I +++ + + + V RE N
Sbjct: 488 FQYN----YVIFESDSQVLIEILN-NDEIWPSLKPTIQDLQRLLSQFTEVKFVFIPREGN 542
Query: 298 SVADELANMAHDLDFGIHILDYPP 321
++A+ +A + + L+Y P
Sbjct: 543 TLAERVARES------LSFLNYDP 560
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.141 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 654,835,447
Number of Sequences: 2790947
Number of extensions: 27134125
Number of successful extensions: 76829
Number of sequences better than 10.0: 261
Number of HSP's better than 10.0 without gapping: 122
Number of HSP's successfully gapped in prelim test: 139
Number of HSP's that attempted gapping in prelim test: 76512
Number of HSP's gapped (non-prelim): 316
length of query: 342
length of database: 848,049,833
effective HSP length: 128
effective length of query: 214
effective length of database: 490,808,617
effective search space: 105033044038
effective search space used: 105033044038
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0045.13


