

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
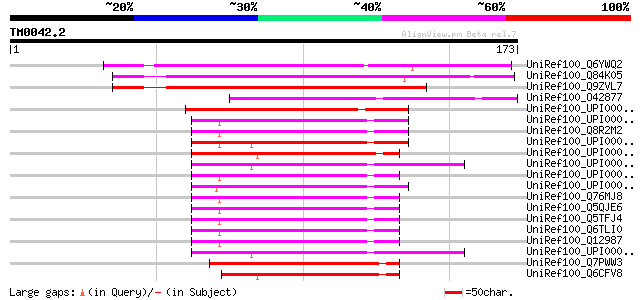
Query= TM0042.2
(173 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6YWQ2 Acidic 82 kDa protein-like [Oryza sativa] 139 4e-32
UniRef100_Q84K05 Hypothetical protein At1g54770 [Arabidopsis tha... 118 7e-26
UniRef100_Q9ZVL7 T22H22.18 [Arabidopsis thaliana] 107 1e-22
UniRef100_O42877 SPAC3G9.15c protein [Schizosaccharomyces pombe] 67 1e-10
UniRef100_UPI00002F0191 UPI00002F0191 UniRef100 entry 64 2e-09
UniRef100_UPI00001CEF34 UPI00001CEF34 UniRef100 entry 57 3e-07
UniRef100_Q8R2M2 Acidic 82 kDa protein mRNA [Mus musculus] 56 3e-07
UniRef100_UPI0000247594 UPI0000247594 UniRef100 entry 56 4e-07
UniRef100_UPI00004329F6 UPI00004329F6 UniRef100 entry 56 4e-07
UniRef100_UPI000035F520 UPI000035F520 UniRef100 entry 55 1e-06
UniRef100_UPI00003681D6 UPI00003681D6 UniRef100 entry 55 1e-06
UniRef100_UPI00003AEA11 UPI00003AEA11 UniRef100 entry 55 1e-06
UniRef100_Q76MJ8 TdT interacting factor 2 [Homo sapiens] 55 1e-06
UniRef100_Q5QJE6 LPTS-RP2 [Homo sapiens] 55 1e-06
UniRef100_Q5TFJ4 Acidic 82 kDa protein mRNA [Homo sapiens] 55 1e-06
UniRef100_Q6TLI0 Estrogen receptor binding protein [Homo sapiens] 55 1e-06
UniRef100_Q12987 Acidic 82 kDa protein [Homo sapiens] 55 1e-06
UniRef100_UPI00002BB7BA UPI00002BB7BA UniRef100 entry 52 5e-06
UniRef100_Q7PWW3 ENSANGP00000016794 [Anopheles gambiae str. PEST] 52 5e-06
UniRef100_Q6CFV8 Similarities with tr|Q8R2M2 Mus musculus Simila... 51 1e-05
>UniRef100_Q6YWQ2 Acidic 82 kDa protein-like [Oryza sativa]
Length = 195
Score = 139 bits (349), Expect = 4e-32
Identities = 80/174 (45%), Positives = 99/174 (55%), Gaps = 39/174 (22%)
Query: 33 RDDGSHSTSQAESSTITLWKHNSELVHALFVPPNEPTKLNKLLREQVKDTTGRIWFDMPA 92
+ D S+S+A+ S LWK SELV LFVPP +P K NKL R+ VKDT+G+ WFDMPA
Sbjct: 25 KKDTGASSSRAQGS---LWKPASELVDGLFVPPRDPRKANKLARKNVKDTSGKGWFDMPA 81
Query: 93 QTINPELQKDETSLQLRAAIDPKRHYKKGNSKSKALPKYFQASS---------------- 136
TI PEL+KD LQLR +DPKRH+K+ KSKALPKYFQ +
Sbjct: 82 PTITPELKKDLEILQLRHVMDPKRHFKRA-GKSKALPKYFQVGTVIEPASEFFSSRLTKR 140
Query: 137 -------------------GARKVREIEEQNQPAGNEKWKVKGGNSRKRAKERR 171
RKVREI+E P G++KW+ KG + KRAK+RR
Sbjct: 141 ERKTTLVDELLSDQHLKNYRMRKVREIQESRTPGGSQKWRNKGKKTLKRAKDRR 194
>UniRef100_Q84K05 Hypothetical protein At1g54770 [Arabidopsis thaliana]
Length = 189
Score = 118 bits (295), Expect = 7e-26
Identities = 72/171 (42%), Positives = 90/171 (52%), Gaps = 42/171 (24%)
Query: 36 GSHSTSQAESSTITLWKHNSELVHALFVPPNEPTKLNKLLREQVKDTTGRIWFDMPAQTI 95
GS + AESS SELV L +PPN+P K+NK++R+Q+KDTTG WFDMPA T+
Sbjct: 27 GSTRSKTAESS-------KSELVDGLCLPPNDPRKINKMIRKQLKDTTGSNWFDMPAPTM 79
Query: 96 NPELQKDETSLQLRAAIDPKRHYKKGNSKSKALPKYFQ---------------------- 133
PEL++D L+LR +DP HYKK S+SK KYFQ
Sbjct: 80 TPELKRDLQLLKLRTVMDPALHYKKSVSRSKLAEKYFQIGTVIEPAEEFYGRLTKKNRKA 139
Query: 134 ------------ASSGARKVREIEEQNQPAGNEKWKVKGGNSRKRAKERRN 172
A RKVREIEE+++ N+KW K GN K K RRN
Sbjct: 140 TLADELVSDPKTALYRKRKVREIEEKSRAVTNKKWN-KKGNQSKNTKPRRN 189
>UniRef100_Q9ZVL7 T22H22.18 [Arabidopsis thaliana]
Length = 284
Score = 107 bits (267), Expect = 1e-22
Identities = 56/107 (52%), Positives = 70/107 (65%), Gaps = 7/107 (6%)
Query: 36 GSHSTSQAESSTITLWKHNSELVHALFVPPNEPTKLNKLLREQVKDTTGRIWFDMPAQTI 95
GS + AESS SELV L +PPN+P K+NK++R+Q+KDTTG WFDMPA T+
Sbjct: 27 GSTRSKTAESS-------KSELVDGLCLPPNDPRKINKMIRKQLKDTTGSNWFDMPAPTM 79
Query: 96 NPELQKDETSLQLRAAIDPKRHYKKGNSKSKALPKYFQASSGARKVR 142
PEL++D L+LR +DP HYKK S+SK KYFQAS R R
Sbjct: 80 TPELKRDLQLLKLRTVMDPALHYKKSVSRSKLAEKYFQASFRTRSKR 126
>UniRef100_O42877 SPAC3G9.15c protein [Schizosaccharomyces pombe]
Length = 230
Score = 67.4 bits (163), Expect = 1e-10
Identities = 35/98 (35%), Positives = 54/98 (54%), Gaps = 4/98 (4%)
Query: 76 REQVKDTTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKKGNSKSKALPKYFQAS 135
++ +KDT G WFDMPA + +++D L++R A+DPKRHY++ N+KS +PKYFQ
Sbjct: 104 KKNIKDTAGSNWFDMPATELTESVKRDIQLLKMRNALDPKRHYRRENTKS--MPKYFQVG 161
Query: 136 SGARKVREIEEQNQPAGNEKWKVKGGNSRKRAKERRNY 173
S ++ P K + + ERR+Y
Sbjct: 162 SIVEGPQDFYSSRIPTRERKETIV--DELLHDSERRSY 197
>UniRef100_UPI00002F0191 UPI00002F0191 UniRef100 entry
Length = 170
Score = 63.9 bits (154), Expect = 2e-09
Identities = 31/76 (40%), Positives = 47/76 (61%), Gaps = 2/76 (2%)
Query: 61 LFVPPNEPTKLNKLLREQVKDTTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKK 120
LFV P + +K+ + +R+ + T G+ WFDM A PEL+++ L+LR A DPKR YK
Sbjct: 25 LFVAPQDRSKVRREMRKMERKTAGKGWFDMKAVEYTPELRREMRMLKLRGAYDPKRFYK- 83
Query: 121 GNSKSKALPKYFQASS 136
N+ + LP +FQ +
Sbjct: 84 -NADTSKLPTHFQVGT 98
>UniRef100_UPI00001CEF34 UPI00001CEF34 UniRef100 entry
Length = 761
Score = 56.6 bits (135), Expect = 3e-07
Identities = 31/76 (40%), Positives = 44/76 (57%), Gaps = 4/76 (5%)
Query: 63 VPPNEPTK--LNKLLREQVKDTTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKK 120
VPP +K L K R++ + T G WF M A + EL+ D +L++RA++DPKR YKK
Sbjct: 624 VPPYSESKHQLQKQRRKERQKTAGNGWFGMKAPELTDELKNDLKALKMRASMDPKRFYKK 683
Query: 121 GNSKSKALPKYFQASS 136
+ PKYFQ +
Sbjct: 684 ND--RDGFPKYFQVGT 697
>UniRef100_Q8R2M2 Acidic 82 kDa protein mRNA [Mus musculus]
Length = 758
Score = 56.2 bits (134), Expect = 3e-07
Identities = 31/76 (40%), Positives = 43/76 (55%), Gaps = 4/76 (5%)
Query: 63 VPPNEPTK--LNKLLREQVKDTTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKK 120
VPP +K L K R++ + T G WF M A + EL+ D +L++RA +DPKR YKK
Sbjct: 621 VPPYSESKHRLQKQRRKERQKTAGNGWFGMKAPELTDELKNDLRALKMRAGMDPKRFYKK 680
Query: 121 GNSKSKALPKYFQASS 136
+ PKYFQ +
Sbjct: 681 ND--RDGFPKYFQVGT 694
>UniRef100_UPI0000247594 UPI0000247594 UniRef100 entry
Length = 921
Score = 55.8 bits (133), Expect = 4e-07
Identities = 33/76 (43%), Positives = 47/76 (61%), Gaps = 4/76 (5%)
Query: 63 VPPNEPTK-LNKLLREQVKD-TTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKK 120
VPP +K KL R+ ++ TTG WF+M A + EL+ D +L++RAA+DPKR YKK
Sbjct: 778 VPPCRESKHAAKLKRKAEREKTTGDGWFNMRAPELTEELKNDLKALKMRAAMDPKRFYKK 837
Query: 121 GNSKSKALPKYFQASS 136
+ + PKYFQ +
Sbjct: 838 ND--REGFPKYFQVGT 851
>UniRef100_UPI00004329F6 UPI00004329F6 UniRef100 entry
Length = 156
Score = 55.8 bits (133), Expect = 4e-07
Identities = 30/72 (41%), Positives = 45/72 (61%), Gaps = 3/72 (4%)
Query: 63 VPPNEPTKLNKLLREQVKDTT-GRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKKG 121
VP TKL K R + + T G+ WF+M A + PE++ D +++R+ +DPKR YKK
Sbjct: 22 VPCELSTKLLKAKRRKERSKTKGKDWFNMGAPELTPEVKHDLQVIRMRSVLDPKRFYKKN 81
Query: 122 NSKSKALPKYFQ 133
+ K+ LP+YFQ
Sbjct: 82 DLKT--LPRYFQ 91
>UniRef100_UPI000035F520 UPI000035F520 UniRef100 entry
Length = 832
Score = 54.7 bits (130), Expect = 1e-06
Identities = 33/95 (34%), Positives = 48/95 (49%), Gaps = 4/95 (4%)
Query: 63 VPPNEPTKLNKLLREQVKD--TTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKK 120
VPP +K L+ +V+ TTG WF+M A ++ EL+ D L++R ++DPKR YKK
Sbjct: 690 VPPYSESKQKMKLKRRVEKEKTTGDGWFNMKAPEMSQELKGDLQVLKMRGSLDPKRFYKK 749
Query: 121 GNSKSKALPKYFQASSGARKVREIEEQNQPAGNEK 155
+ PKYFQ + + P N K
Sbjct: 750 ND--RDGFPKYFQVGTVVDNPVDFYHSQIPKKNRK 782
>UniRef100_UPI00003681D6 UPI00003681D6 UniRef100 entry
Length = 742
Score = 54.7 bits (130), Expect = 1e-06
Identities = 31/73 (42%), Positives = 43/73 (58%), Gaps = 4/73 (5%)
Query: 63 VPPNEPTK--LNKLLREQVKDTTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKK 120
VPP +K L K R++ + T G WF M A + EL+ D +L++RA++DPKR YKK
Sbjct: 619 VPPYSESKYQLQKKRRKERQKTAGDGWFGMKAPEMTNELKNDLKALKMRASMDPKRFYKK 678
Query: 121 GNSKSKALPKYFQ 133
+ PKYFQ
Sbjct: 679 ND--RDGFPKYFQ 689
>UniRef100_UPI00003AEA11 UPI00003AEA11 UniRef100 entry
Length = 686
Score = 54.7 bits (130), Expect = 1e-06
Identities = 32/76 (42%), Positives = 43/76 (56%), Gaps = 4/76 (5%)
Query: 63 VPPNEPT--KLNKLLREQVKDTTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKK 120
VPP + +L K R + + TTG WF M A I EL+ D L++RA++DPK YKK
Sbjct: 549 VPPFRESLQQLKKQRRAEREKTTGDGWFGMKAPEITSELKNDLKVLKMRASLDPKHFYKK 608
Query: 121 GNSKSKALPKYFQASS 136
+ LPKYFQ +
Sbjct: 609 ND--RDGLPKYFQVGT 622
>UniRef100_Q76MJ8 TdT interacting factor 2 [Homo sapiens]
Length = 756
Score = 54.7 bits (130), Expect = 1e-06
Identities = 31/73 (42%), Positives = 43/73 (58%), Gaps = 4/73 (5%)
Query: 63 VPPNEPTK--LNKLLREQVKDTTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKK 120
VPP +K L K R++ + T G WF M A + EL+ D +L++RA++DPKR YKK
Sbjct: 619 VPPYSESKYQLQKKRRKERQKTAGDGWFGMKAPEMTNELKNDLKALKMRASMDPKRFYKK 678
Query: 121 GNSKSKALPKYFQ 133
+ PKYFQ
Sbjct: 679 ND--RDGFPKYFQ 689
>UniRef100_Q5QJE6 LPTS-RP2 [Homo sapiens]
Length = 756
Score = 54.7 bits (130), Expect = 1e-06
Identities = 31/73 (42%), Positives = 43/73 (58%), Gaps = 4/73 (5%)
Query: 63 VPPNEPTK--LNKLLREQVKDTTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKK 120
VPP +K L K R++ + T G WF M A + EL+ D +L++RA++DPKR YKK
Sbjct: 619 VPPYSESKYQLQKKRRKERQKTAGDGWFGMKAPEMTNELKNDLKALKMRASMDPKRFYKK 678
Query: 121 GNSKSKALPKYFQ 133
+ PKYFQ
Sbjct: 679 ND--RDGFPKYFQ 689
>UniRef100_Q5TFJ4 Acidic 82 kDa protein mRNA [Homo sapiens]
Length = 810
Score = 54.7 bits (130), Expect = 1e-06
Identities = 31/73 (42%), Positives = 43/73 (58%), Gaps = 4/73 (5%)
Query: 63 VPPNEPTK--LNKLLREQVKDTTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKK 120
VPP +K L K R++ + T G WF M A + EL+ D +L++RA++DPKR YKK
Sbjct: 619 VPPYSESKYQLQKKRRKERQKTAGDGWFGMKAPEMTNELKNDLKALKMRASMDPKRFYKK 678
Query: 121 GNSKSKALPKYFQ 133
+ PKYFQ
Sbjct: 679 ND--RDGFPKYFQ 689
>UniRef100_Q6TLI0 Estrogen receptor binding protein [Homo sapiens]
Length = 756
Score = 54.7 bits (130), Expect = 1e-06
Identities = 31/73 (42%), Positives = 43/73 (58%), Gaps = 4/73 (5%)
Query: 63 VPPNEPTK--LNKLLREQVKDTTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKK 120
VPP +K L K R++ + T G WF M A + EL+ D +L++RA++DPKR YKK
Sbjct: 619 VPPYSESKYQLQKKRRKERQKTAGDGWFGMKAPEMTNELKNDLKALKMRASMDPKRFYKK 678
Query: 121 GNSKSKALPKYFQ 133
+ PKYFQ
Sbjct: 679 ND--RDGFPKYFQ 689
>UniRef100_Q12987 Acidic 82 kDa protein [Homo sapiens]
Length = 736
Score = 54.7 bits (130), Expect = 1e-06
Identities = 31/73 (42%), Positives = 43/73 (58%), Gaps = 4/73 (5%)
Query: 63 VPPNEPTK--LNKLLREQVKDTTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKK 120
VPP +K L K R++ + T G WF M A + EL+ D +L++RA++DPKR YKK
Sbjct: 619 VPPYSESKYQLQKKRRKERQKTAGDGWFGMKAPEMTNELKNDLKALKMRASMDPKRFYKK 678
Query: 121 GNSKSKALPKYFQ 133
+ PKYFQ
Sbjct: 679 ND--RDGFPKYFQ 689
>UniRef100_UPI00002BB7BA UPI00002BB7BA UniRef100 entry
Length = 1018
Score = 52.4 bits (124), Expect = 5e-06
Identities = 32/95 (33%), Positives = 48/95 (49%), Gaps = 4/95 (4%)
Query: 63 VPPNEPTKLNKLLREQVKD--TTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKK 120
VPP + +K L+ +V+ TTG WF+M A ++ EL+ D L++R ++D KR YKK
Sbjct: 919 VPPYQESKQALKLKRRVEKEKTTGDSWFNMKAPELSQELKGDLQVLKMRGSLDSKRFYKK 978
Query: 121 GNSKSKALPKYFQASSGARKVREIEEQNQPAGNEK 155
+ PKYFQ + + P N K
Sbjct: 979 ND--RDGFPKYFQVGTVVDNPVDFYHSRIPKKNRK 1011
>UniRef100_Q7PWW3 ENSANGP00000016794 [Anopheles gambiae str. PEST]
Length = 288
Score = 52.4 bits (124), Expect = 5e-06
Identities = 24/65 (36%), Positives = 43/65 (65%), Gaps = 2/65 (3%)
Query: 69 TKLNKLLREQVKDTTGRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKKGNSKSKAL 128
T + KL R + K T G+ W+ + A I+PELQ + T +++R+ +DPK+ +K+ + + +
Sbjct: 154 TAIRKLNRLERKKTKGKGWYGLAAPEISPELQNELTLMKMRSVLDPKQSFKRLDKRKQ-- 211
Query: 129 PKYFQ 133
PKYF+
Sbjct: 212 PKYFE 216
>UniRef100_Q6CFV8 Similarities with tr|Q8R2M2 Mus musculus Similar to acidic 82 kDa
protein mRNA [Yarrowia lipolytica]
Length = 321
Score = 51.2 bits (121), Expect = 1e-05
Identities = 27/62 (43%), Positives = 39/62 (62%), Gaps = 3/62 (4%)
Query: 73 KLLREQVKDTT-GRIWFDMPAQTINPELQKDETSLQLRAAIDPKRHYKKGNSKSKALPKY 131
KL +++ +D T G WF M A + PEL+ D L++R +DPKR YKK +SK PK+
Sbjct: 193 KLDKKKARDNTAGDKWFGMKAPDMTPELKMDMELLKMRNVLDPKRFYKKSDSKKD--PKF 250
Query: 132 FQ 133
F+
Sbjct: 251 FE 252
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.128 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 293,136,181
Number of Sequences: 2790947
Number of extensions: 11202469
Number of successful extensions: 24681
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 24630
Number of HSP's gapped (non-prelim): 81
length of query: 173
length of database: 848,049,833
effective HSP length: 118
effective length of query: 55
effective length of database: 518,718,087
effective search space: 28529494785
effective search space used: 28529494785
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0042.2


