

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0041.3
(261 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
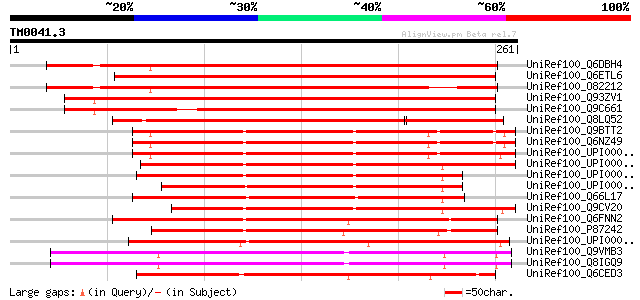
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6DBH4 At2g23820 [Arabidopsis thaliana] 324 2e-87
UniRef100_Q6ETL6 Putative metal-dependent phosphohydrolase HD do... 305 6e-82
UniRef100_O82212 Hypothetical protein At2g23820 [Arabidopsis tha... 289 4e-77
UniRef100_Q93ZV1 Hypothetical protein At1g26160 [Arabidopsis tha... 282 7e-75
UniRef100_Q9C661 Hypothetical protein F28B23.16 [Arabidopsis tha... 258 1e-67
UniRef100_Q8LQ52 Metal-dependent phosphohydrolase HD domain-cont... 214 1e-54
UniRef100_Q9BTT2 C6orf74 protein [Homo sapiens] 174 2e-42
UniRef100_Q6NZ49 Hypothetical protein [Homo sapiens] 174 2e-42
UniRef100_UPI000036D95F UPI000036D95F UniRef100 entry 174 2e-42
UniRef100_UPI00003ACF75 UPI00003ACF75 UniRef100 entry 170 4e-41
UniRef100_UPI00001CE6A7 UPI00001CE6A7 UniRef100 entry 167 3e-40
UniRef100_UPI0000438DFE UPI0000438DFE UniRef100 entry 166 7e-40
UniRef100_Q66L17 MGC85244 protein [Xenopus laevis] 164 2e-39
UniRef100_Q9CV20 Mus musculus adult male tongue cDNA, RIKEN full... 162 6e-39
UniRef100_Q6FNN2 Candida glabrata strain CBS138 chromosome J com... 162 6e-39
UniRef100_P87242 SPCC4G3.17 protein [Schizosaccharomyces pombe] 161 1e-38
UniRef100_UPI000042E2B5 UPI000042E2B5 UniRef100 entry 160 4e-38
UniRef100_Q9VMB3 CG11050-PA, isoform A [Drosophila melanogaster] 159 5e-38
UniRef100_Q8IGQ9 RE44531p [Drosophila melanogaster] 158 1e-37
UniRef100_Q6CED3 Similar to sp|P38331 Saccharomyces cerevisiae Y... 155 1e-36
>UniRef100_Q6DBH4 At2g23820 [Arabidopsis thaliana]
Length = 257
Score = 324 bits (830), Expect = 2e-87
Identities = 165/238 (69%), Positives = 188/238 (78%), Gaps = 9/238 (3%)
Query: 20 SLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPS------AS 73
S + + +RF SP+R H S SP S + S P +A PS AS
Sbjct: 21 SFTRSHIRFTYSAAGASSPNRAIH---CMASDSPQSGDGSVSSPPNVAAVPSSSSSSSAS 77
Query: 74 SAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMA 133
SAIDFLS+C RLKTT RAGWI++DV+DPESIADHMYRM LMALI++DIPGV+RDKC+KMA
Sbjct: 78 SAIDFLSLCTRLKTTPRAGWIKRDVKDPESIADHMYRMGLMALISSDIPGVNRDKCMKMA 137
Query: 134 IVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSS 193
IVHDIAEAIVGDITP+ G+ KEEKNR E EAL+HMCK+LGGG RAKE+AELW EYE NSS
Sbjct: 138 IVHDIAEAIVGDITPSCGISKEEKNRRESEALEHMCKLLGGGERAKEIAELWREYEENSS 197
Query: 194 PEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRKK 251
PEAK VKD DKVE+ILQALEYE +QGKDL+EFFQSTAGKFQT IGKAWASEIVSRR+K
Sbjct: 198 PEAKVVKDFDKVELILQALEYEQDQGKDLEEFFQSTAGKFQTNIGKAWASEIVSRRRK 255
>UniRef100_Q6ETL6 Putative metal-dependent phosphohydrolase HD domain-containing
protein [Oryza sativa]
Length = 228
Score = 305 bits (782), Expect = 6e-82
Identities = 145/197 (73%), Positives = 177/197 (89%), Gaps = 1/197 (0%)
Query: 55 SSSVTTSPPSAASATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLM 114
++S+++S PS + A P+++SAIDFL++C+RLKTTKRAGW+R+ VQ PES+ADHMYRM +M
Sbjct: 32 AASMSSSSPSPSPAAPASASAIDFLTLCYRLKTTKRAGWVRRGVQGPESVADHMYRMGVM 91
Query: 115 ALIAADIP-GVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLG 173
AL+AAD+P GV+RD+CVKMAIVHDIAEAIVGDITP+DGVPKEEK+R EQEALDHMC +LG
Sbjct: 92 ALVAADLPSGVNRDRCVKMAIVHDIAEAIVGDITPSDGVPKEEKSRREQEALDHMCSLLG 151
Query: 174 GGSRAKEVAELWAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGKF 233
GG RA+E+ ELW EYE N++ EAK VKD DKVEMILQALEYE EQG DL+EFFQSTAGKF
Sbjct: 152 GGPRAEEIRELWMEYEQNATLEAKVVKDFDKVEMILQALEYEKEQGLDLEEFFQSTAGKF 211
Query: 234 QTEIGKAWASEIVSRRK 250
QT++GKAWA+E+ SRRK
Sbjct: 212 QTDVGKAWAAEVASRRK 228
>UniRef100_O82212 Hypothetical protein At2g23820 [Arabidopsis thaliana]
Length = 243
Score = 289 bits (740), Expect = 4e-77
Identities = 153/238 (64%), Positives = 174/238 (72%), Gaps = 23/238 (9%)
Query: 20 SLSSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPS------AS 73
S + + +RF SP+R H S SP S + S P +A PS AS
Sbjct: 21 SFTRSHIRFTYSAAGASSPNRAIH---CMASDSPQSGDGSVSSPPNVAAVPSSSSSSSAS 77
Query: 74 SAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMA 133
SAIDFLS+C RLKTT RAGWI++DV+DPESIADHMYRM LMALI++DIPGV+RDKC+KMA
Sbjct: 78 SAIDFLSLCTRLKTTPRAGWIKRDVKDPESIADHMYRMGLMALISSDIPGVNRDKCMKMA 137
Query: 134 IVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSS 193
IVHDIAEAIVGDITP+ G+ KEEKNR E EAL+HMCK+LGGG RAKE+AELW EYE NSS
Sbjct: 138 IVHDIAEAIVGDITPSCGISKEEKNRRESEALEHMCKLLGGGERAKEIAELWREYEENSS 197
Query: 194 PEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRKK 251
PEAK VKD DKVE+ILQALEYE GKFQT IGKAWASEIVSRR+K
Sbjct: 198 PEAKVVKDFDKVELILQALEYEQ--------------GKFQTNIGKAWASEIVSRRRK 241
>UniRef100_Q93ZV1 Hypothetical protein At1g26160 [Arabidopsis thaliana]
Length = 258
Score = 282 bits (721), Expect = 7e-75
Identities = 147/230 (63%), Positives = 170/230 (73%), Gaps = 8/230 (3%)
Query: 29 KSVTLSPHSPSRVF--------HMTTAAGSSSPPSSSVTTSPPSAASATPSASSAIDFLS 80
+S+ S HS SR F T + P S +S S S SS+IDFL+
Sbjct: 21 RSILASFHSSSRNFLFLGKPTPSSTIVSVRCQKPVSDGVSSMESMNHVASSVSSSIDFLT 80
Query: 81 ICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMAIVHDIAE 140
+CHRLKTTKR GWI + + PESIADHMYRM+LMALIA D+ GVDR++C+KMAIVHDIAE
Sbjct: 81 LCHRLKTTKRKGWINQGINGPESIADHMYRMALMALIAGDLTGVDRERCIKMAIVHDIAE 140
Query: 141 AIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSSPEAKFVK 200
AIVGDITP+DGVPKEEK+R E AL MC+VLGGG RA+E+ ELW EYE N+S EA VK
Sbjct: 141 AIVGDITPSDGVPKEEKSRRETAALKEMCEVLGGGLRAEEITELWLEYENNASLEANIVK 200
Query: 201 DLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRK 250
D DKVEMILQALEYE E GK LDEFF STAGKFQTEIGK+WA+EI +RRK
Sbjct: 201 DFDKVEMILQALEYEAEHGKVLDEFFISTAGKFQTEIGKSWAAEINARRK 250
>UniRef100_Q9C661 Hypothetical protein F28B23.16 [Arabidopsis thaliana]
Length = 248
Score = 258 bits (659), Expect = 1e-67
Identities = 140/230 (60%), Positives = 162/230 (69%), Gaps = 18/230 (7%)
Query: 29 KSVTLSPHSPSRVF--------HMTTAAGSSSPPSSSVTTSPPSAASATPSASSAIDFLS 80
+S+ S HS SR F T + P S +S S S SS+IDFL+
Sbjct: 21 RSILASFHSSSRNFLFLGKPTPSSTIVSVRCQKPVSDGVSSMESMNHVASSVSSSIDFLT 80
Query: 81 ICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMAIVHDIAE 140
+CHRLK + PESIADHMYRM+LMALIA D+ GVDR++C+KMAIVHDIAE
Sbjct: 81 LCHRLK----------GINGPESIADHMYRMALMALIAGDLTGVDRERCIKMAIVHDIAE 130
Query: 141 AIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSSPEAKFVK 200
AIVGDITP+DGVPKEEK+R E AL MC+VLGGG RA+E+ ELW EYE N+S EA VK
Sbjct: 131 AIVGDITPSDGVPKEEKSRRETAALKEMCEVLGGGLRAEEITELWLEYENNASLEANIVK 190
Query: 201 DLDKVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRK 250
D DKVEMILQALEYE E GK LDEFF STAGKFQTEIGK+WA+EI +RRK
Sbjct: 191 DFDKVEMILQALEYEAEHGKVLDEFFISTAGKFQTEIGKSWAAEINARRK 240
>UniRef100_Q8LQ52 Metal-dependent phosphohydrolase HD domain-containing protein- like
[Oryza sativa]
Length = 461
Score = 214 bits (546), Expect = 1e-54
Identities = 103/151 (68%), Positives = 125/151 (82%), Gaps = 2/151 (1%)
Query: 54 PSSSVTTSPPSAASATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSL 113
P+ + +PPSAA++ SASSAIDFL++CHRLKTTKR GWI ++ PESIADHMYRM+L
Sbjct: 146 PADAAAAAPPSAAAS--SASSAIDFLTLCHRLKTTKRKGWINHSIKGPESIADHMYRMAL 203
Query: 114 MALIAADIPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLG 173
MALIA D+P VDR++C+K+AIVHDIAEAIVGDITP+DG+PK EK+R EQ+AL+ MC+VLG
Sbjct: 204 MALIAGDLPAVDRERCIKIAIVHDIAEAIVGDITPSDGIPKAEKSRREQKALNEMCEVLG 263
Query: 174 GGSRAKEVAELWAEYEANSSPEAKFVKDLDK 204
GG A E+ ELW EYE NSS EA VKD DK
Sbjct: 264 GGPIADEIKELWEEYENNSSIEANLVKDFDK 294
Score = 78.2 bits (191), Expect = 2e-13
Identities = 38/51 (74%), Positives = 44/51 (85%)
Query: 204 KVEMILQALEYEHEQGKDLDEFFQSTAGKFQTEIGKAWASEIVSRRKKTNG 254
KVEMILQALEYE E GK LDEFF STAGKFQTEIGK+WA+E+ +RR++ G
Sbjct: 408 KVEMILQALEYEKEHGKVLDEFFLSTAGKFQTEIGKSWAAEVNARREQRCG 458
>UniRef100_Q9BTT2 C6orf74 protein [Homo sapiens]
Length = 218
Score = 174 bits (442), Expect = 2e-42
Identities = 101/206 (49%), Positives = 138/206 (66%), Gaps = 13/206 (6%)
Query: 64 SAASATPS---ASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAAD 120
S +SAT S A S + FL + +LK R GW+ ++VQ PES++DHMYRM++MA++ D
Sbjct: 17 SVSSATFSGHGARSLLQFLRLVGQLKRVPRTGWVYRNVQRPESVSDHMYRMAVMAMVIKD 76
Query: 121 IPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKE 180
+++D+CV++A+VHD+AE IVGDI P D +PKEEK+R E+EA+ + ++L R KE
Sbjct: 77 -DRLNKDRCVRLALVHDMAECIVGDIAPADNIPKEEKHRREEEAMKQITQLLPEDLR-KE 134
Query: 181 VAELWAEYEANSSPEAKFVKDLDKVEMILQALEY---EHEQGKDLDEFFQSTAGKFQTEI 237
+ ELW EYE SS EAKFVK LD+ EMILQA EY EH+ G+ L +F+ STAGKF
Sbjct: 135 LYELWEEYETQSSAEAKFVKQLDQCEMILQASEYEDLEHKPGR-LQDFYDSTAGKFNHPE 193
Query: 238 GKAWASEIVSRRKKTN---GSSHPHS 260
SE+ + R TN +S PHS
Sbjct: 194 IVQLVSELEAER-STNIAAAASEPHS 218
>UniRef100_Q6NZ49 Hypothetical protein [Homo sapiens]
Length = 204
Score = 174 bits (442), Expect = 2e-42
Identities = 101/206 (49%), Positives = 138/206 (66%), Gaps = 13/206 (6%)
Query: 64 SAASATPS---ASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAAD 120
S +SAT S A S + FL + +LK R GW+ ++VQ PES++DHMYRM++MA++ D
Sbjct: 3 SVSSATFSGHGARSLLQFLRLVGQLKRVPRTGWVYRNVQRPESVSDHMYRMAVMAMVIKD 62
Query: 121 IPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKE 180
+++D+CV++A+VHD+AE IVGDI P D +PKEEK+R E+EA+ + ++L R KE
Sbjct: 63 -DRLNKDRCVRLALVHDMAECIVGDIAPADNIPKEEKHRREEEAMKQITQLLPEDLR-KE 120
Query: 181 VAELWAEYEANSSPEAKFVKDLDKVEMILQALEY---EHEQGKDLDEFFQSTAGKFQTEI 237
+ ELW EYE SS EAKFVK LD+ EMILQA EY EH+ G+ L +F+ STAGKF
Sbjct: 121 LYELWEEYETRSSAEAKFVKQLDQCEMILQASEYEDLEHKPGR-LQDFYDSTAGKFNHPE 179
Query: 238 GKAWASEIVSRRKKTN---GSSHPHS 260
SE+ + R TN +S PHS
Sbjct: 180 IVQLVSELEAER-STNIAAAASEPHS 204
>UniRef100_UPI000036D95F UPI000036D95F UniRef100 entry
Length = 218
Score = 174 bits (441), Expect = 2e-42
Identities = 99/205 (48%), Positives = 136/205 (66%), Gaps = 11/205 (5%)
Query: 64 SAASATPS---ASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAAD 120
S +SAT S A S + FL + +LK R GW+ ++VQ PES++DHMYRM++MA++ D
Sbjct: 17 SVSSATFSGHGARSLLQFLRLVGQLKRVPRTGWVYRNVQRPESVSDHMYRMAVMAMVIKD 76
Query: 121 IPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKE 180
+++D+CV++A+VHD+AE IVGDI P D +PKEEK+R E+EA+ + ++L R KE
Sbjct: 77 -DRLNKDRCVRLALVHDMAECIVGDIAPADNIPKEEKHRREEEAMKQITQLLPEDLR-KE 134
Query: 181 VAELWAEYEANSSPEAKFVKDLDKVEMILQALEY---EHEQGKDLDEFFQSTAGKFQTEI 237
+ ELW EYE SS EAKFVK LD+ EMILQA EY EH+ G+ L +F+ STAGKF
Sbjct: 135 LYELWEEYETQSSAEAKFVKQLDQCEMILQASEYEDLEHKPGR-LQDFYDSTAGKFNHPE 193
Query: 238 GKAWASEIVSRRKK--TNGSSHPHS 260
SE+ + R +S PHS
Sbjct: 194 IVQLVSELEAERNANIAAAASEPHS 218
>UniRef100_UPI00003ACF75 UPI00003ACF75 UniRef100 entry
Length = 194
Score = 170 bits (430), Expect = 4e-41
Identities = 89/196 (45%), Positives = 127/196 (64%), Gaps = 5/196 (2%)
Query: 68 ATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRD 127
A P + FL + +LK R GW+ ++V+ PES++DHMYRM++MAL+ D +++D
Sbjct: 1 AGPGGGGMLPFLRLLGQLKRVPRTGWVYRNVEKPESVSDHMYRMAVMALVTED-KSLNKD 59
Query: 128 KCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAE 187
+C+++A+VHD+AE IVGDI P D + KEEK+R E+ A+ + ++L + KE+ ELW E
Sbjct: 60 RCIRLALVHDMAECIVGDIAPADNISKEEKHRREEAAMRQLTQLLSEDLK-KEIYELWEE 118
Query: 188 YEANSSPEAKFVKDLDKVEMILQALEYEHEQGKD--LDEFFQSTAGKF-QTEIGKAWASE 244
YE + EAKFVK LD+ EMILQALEYE + L +F+ STAGKF EI + +
Sbjct: 119 YENQCTAEAKFVKQLDQCEMILQALEYEELENTPGRLQDFYDSTAGKFIHPEILQLVSLI 178
Query: 245 IVSRRKKTNGSSHPHS 260
R KK + HPHS
Sbjct: 179 NTERNKKIAATCHPHS 194
>UniRef100_UPI00001CE6A7 UPI00001CE6A7 UniRef100 entry
Length = 199
Score = 167 bits (422), Expect = 3e-40
Identities = 85/171 (49%), Positives = 122/171 (70%), Gaps = 5/171 (2%)
Query: 66 ASATPSASSAI-DFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGV 124
ASAT + + + FL + +LK R GW+ ++V+ PES++DHMYRM++MA++ D +
Sbjct: 2 ASATSNCQAGLLRFLRLVGQLKRVPRTGWVYRNVEKPESVSDHMYRMAVMAMVTRD-DRL 60
Query: 125 DRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAEL 184
++D+C+++A+VHD+AE IVGDI P D +PKEEK+R E+EA+ + ++L R KE+ EL
Sbjct: 61 NKDRCIRLALVHDMAECIVGDIAPADNIPKEEKHRREEEAMKEITQLLPEDLR-KELYEL 119
Query: 185 WAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKD--LDEFFQSTAGKF 233
W EYE SS EA+FVK LD+ EMILQA EYE + K L +F+ STAGKF
Sbjct: 120 WEEYETQSSEEARFVKQLDQCEMILQASEYEDMENKPGRLQDFYDSTAGKF 170
>UniRef100_UPI0000438DFE UPI0000438DFE UniRef100 entry
Length = 193
Score = 166 bits (419), Expect = 7e-40
Identities = 83/157 (52%), Positives = 113/157 (71%), Gaps = 4/157 (2%)
Query: 79 LSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMAIVHDI 138
+ + +LK R GW+ ++++ PES++DHMYRMS+MAL DI V++++C+K+A+VHD+
Sbjct: 1 MKLVGQLKRVPRTGWVYRNIKQPESVSDHMYRMSMMALTIQDI-SVNKERCMKLALVHDL 59
Query: 139 AEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSSPEAKF 198
AE IVGDI P D V K EK+R E++A+ H+ +L G R KE+ LW EYE SSPEAK
Sbjct: 60 AECIVGDIAPADNVSKAEKHRREKDAMVHITGLLDDGLR-KEIYNLWEEYETQSSPEAKL 118
Query: 199 VKDLDKVEMILQALEYEHEQGKD--LDEFFQSTAGKF 233
VK+LD +EMI+QA EYE +GK L EFF ST GKF
Sbjct: 119 VKELDNLEMIIQAHEYEELEGKPGRLQEFFVSTEGKF 155
>UniRef100_Q66L17 MGC85244 protein [Xenopus laevis]
Length = 201
Score = 164 bits (415), Expect = 2e-39
Identities = 80/173 (46%), Positives = 122/173 (70%), Gaps = 4/173 (2%)
Query: 64 SAASATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPG 123
+A S + + S + F+ + +LK R GWI + V+ PES++DHMYRM++MA++ D
Sbjct: 3 AAGSCSSAGKSLLQFMKLVGQLKRVPRTGWIYRQVEKPESVSDHMYRMAVMAMLTEDRK- 61
Query: 124 VDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAE 183
+++D+C+++A+VHD+AE IVGDI P D + KEEK+R E+ A++H+ ++L + EV +
Sbjct: 62 LNKDRCIRLALVHDMAECIVGDIAPADNIAKEEKHRKEKAAMEHLTQLLPDNLKT-EVYD 120
Query: 184 LWAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKD--LDEFFQSTAGKFQ 234
LW EYE + EAKFVK+LD+ EMILQALEYE + + L +F+ STAGKF+
Sbjct: 121 LWEEYEHQFTAEAKFVKELDQCEMILQALEYEELEKRPGRLQDFYNSTAGKFK 173
>UniRef100_Q9CV20 Mus musculus adult male tongue cDNA, RIKEN full-length enriched
library, clone:2310057G13 product:hypothetical HD domain
containing protein, full insert sequence [Mus musculus]
Length = 182
Score = 162 bits (411), Expect = 6e-39
Identities = 86/181 (47%), Positives = 122/181 (66%), Gaps = 6/181 (3%)
Query: 84 RLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMAIVHDIAEAIV 143
+LK R GW+ ++V+ PES++DHMYRM++MA++ D +++D+C+++A+VHD+AE IV
Sbjct: 4 QLKRVPRTGWVYRNVEKPESVSDHMYRMAVMAMVTRD-DRLNKDRCIRLALVHDMAECIV 62
Query: 144 GDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEANSSPEAKFVKDLD 203
GDI P D +PKEEK+R E+EA+ + ++L R KE+ ELW EYE SS EAKFVK LD
Sbjct: 63 GDIAPADNIPKEEKHRREEEAMKQITQLLPEDLR-KELYELWEEYETQSSEEAKFVKQLD 121
Query: 204 KVEMILQALEYEHEQGKD--LDEFFQSTAGKFQTEIGKAWASEIVSRRKKT--NGSSHPH 259
+ EMILQA EYE + K L +F+ STAGKF SE+ + R + S+ P
Sbjct: 122 QCEMILQASEYEDLENKPGRLQDFYDSTAGKFSHPEIVQLVSELETERNASMATASAEPG 181
Query: 260 S 260
S
Sbjct: 182 S 182
>UniRef100_Q6FNN2 Candida glabrata strain CBS138 chromosome J complete sequence
[Candida glabrata]
Length = 209
Score = 162 bits (411), Expect = 6e-39
Identities = 80/201 (39%), Positives = 128/201 (62%), Gaps = 5/201 (2%)
Query: 54 PSSSVTTSPPSAASATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSL 113
P + + A + T +++ I L+I +LK KR GW+ V+DPESI+DHMYRM +
Sbjct: 7 PEDHIPDNVKKALAGTKNSNYVIALLNIVQQLKIQKRTGWVDHKVKDPESISDHMYRMGI 66
Query: 114 MALIAADIPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMC-KVL 172
+++ + P ++RDKCV++++VHD+AE++VGDITP D + KEEK+R E + + ++C K++
Sbjct: 67 TSMLITN-PDINRDKCVRISLVHDLAESLVGDITPNDPMTKEEKHRREYDTIKYLCDKII 125
Query: 173 G--GGSRAKEVAELWAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQGKDLDEFFQSTA 230
AKE+ E W YE SSPEA++VKD+DK EM++Q EYE LD+F+ S
Sbjct: 126 APFNSKAAKEILEDWLAYENVSSPEARYVKDIDKFEMLVQCFEYEQSNNIRLDQFW-SAK 184
Query: 231 GKFQTEIGKAWASEIVSRRKK 251
+T W ++++ R++
Sbjct: 185 DAIKTPEVSEWCNDLIKMREE 205
>UniRef100_P87242 SPCC4G3.17 protein [Schizosaccharomyces pombe]
Length = 198
Score = 161 bits (408), Expect = 1e-38
Identities = 85/184 (46%), Positives = 126/184 (68%), Gaps = 9/184 (4%)
Query: 74 SAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVKMA 133
S + FL RLKTT R GW+ ++ PESIADHMYRM ++ ++ D P +++++C+K+A
Sbjct: 8 SIVPFLDCLSRLKTTPRTGWLYHGIEKPESIADHMYRMGILTMLCND-PSINKERCLKIA 66
Query: 134 IVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCK---VLGGGSRAKEVAELWAEYEA 190
+VHD+AE+IVGDITP + V KEEK+R+E EA+ + + L +A+E+ EL+ EYE+
Sbjct: 67 VVHDMAESIVGDITPHENVSKEEKHRMESEAMVSITQQLIPLNLSLQAEEIKELFLEYES 126
Query: 191 NSSPEAKFVKDLDKVEMILQALEYEHEQG--KDLDEFFQSTAGK-FQTEIGKAWASEIVS 247
S+PEAKFVKD+DK EMI Q EYE + KDL +F + AGK Q + K W ++++
Sbjct: 127 ASTPEAKFVKDIDKFEMIAQMFEYERKFNGEKDLSQF--TWAGKLIQHPLVKGWLNDVLQ 184
Query: 248 RRKK 251
R++
Sbjct: 185 EREQ 188
>UniRef100_UPI000042E2B5 UPI000042E2B5 UniRef100 entry
Length = 259
Score = 160 bits (404), Expect = 4e-38
Identities = 86/207 (41%), Positives = 131/207 (62%), Gaps = 12/207 (5%)
Query: 62 PPSAASATPSASSAIDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALI---A 118
PP S A + FL + +LK KR+GWIR+ V+ ESI+DHM RM+LMA++ +
Sbjct: 46 PPPYKSTGNEALDTLAFLHMLEQLKIQKRSGWIREGVKQAESISDHMCRMALMAMMLPNS 105
Query: 119 ADIPGVDRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRA 178
++ P +D +CV MA+VHD+AEA VGDITP +GVP K++LE++A+D + GG
Sbjct: 106 SERP-LDIPRCVMMALVHDLAEAYVGDITPVEGVPTHVKHQLEEQAMDTFLNEMLGGKGN 164
Query: 179 KEVAE----LWAEYEANSSPEAKFVKDLDKVEMILQALEYEHEQG-KDLDEFFQSTAGKF 233
K+ E LW EYEA +PE++ VKDLD++E+ LQA+EYE Q + LD FF+ +
Sbjct: 165 KDARERFRSLWDEYEARETPESRLVKDLDRIELALQAVEYERSQDIQTLDPFFKGSIPNL 224
Query: 234 QTEIGKAWASEIVSRRKK---TNGSSH 257
+ + + WA+ ++ R++ + G +H
Sbjct: 225 EHPVTRQWAATLMEERRQLWTSRGRTH 251
>UniRef100_Q9VMB3 CG11050-PA, isoform A [Drosophila melanogaster]
Length = 388
Score = 159 bits (403), Expect = 5e-38
Identities = 88/252 (34%), Positives = 142/252 (55%), Gaps = 17/252 (6%)
Query: 22 SSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSA------ 75
++T+ S+ P + +T + G + + S A+A+ SA
Sbjct: 117 TATKFAMSSLAAEPLQSQKSDEITASTGGGDVETPGLGASCHEVAAASASAGKKPCFSSG 176
Query: 76 ----IDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVK 131
+ F+ + LK TKR GW+ +DV D ESI+ HMYRMS++ + G+++ +C++
Sbjct: 177 LGEILQFMELIGNLKHTKRTGWVLRDVNDCESISGHMYRMSMLTFLLDGSEGLNQIRCME 236
Query: 132 MAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEAN 191
+A+VHD+AE++VGDITP G+ K++K +E +A++ +CK++ R K + EL+ EYE
Sbjct: 237 LALVHDLAESLVGDITPFCGISKDDKRAMEFKAMEDICKLI--EPRGKRIMELFEEYEHG 294
Query: 192 SSPEAKFVKDLDKVEMILQALEYEHEQGKDL--DEFFQSTAGKFQTEIGKAWASEIVSRR 249
+ E+KFVKDLD+++M++QA EYE L EFF ST GKF K +EI +R
Sbjct: 295 QTAESKFVKDLDRLDMVMQAFEYEKRDNCLLKHQEFFDSTEGKFNHPFVKKLVNEIYEQR 354
Query: 250 ---KKTNGSSHP 258
K G++ P
Sbjct: 355 DVLAKAKGATPP 366
>UniRef100_Q8IGQ9 RE44531p [Drosophila melanogaster]
Length = 388
Score = 158 bits (400), Expect = 1e-37
Identities = 88/252 (34%), Positives = 141/252 (55%), Gaps = 17/252 (6%)
Query: 22 SSTQLRFKSVTLSPHSPSRVFHMTTAAGSSSPPSSSVTTSPPSAASATPSASSA------ 75
++T+ S+ P + +T + G + + S A+A+ SA
Sbjct: 117 TATKFAMSSLAAEPLQSQKSDEITASTGGGDVETPGLGASCHEVAAASASAGKKPCFSSG 176
Query: 76 ----IDFLSICHRLKTTKRAGWIRKDVQDPESIADHMYRMSLMALIAADIPGVDRDKCVK 131
+ F+ + LK TKR GW+ +DV D ESI+ HMYRMS + + G+++ +C++
Sbjct: 177 LGEILQFMELIGNLKHTKRTGWVLRDVNDCESISGHMYRMSTLTFLLDGSEGLNQIRCME 236
Query: 132 MAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLGGGSRAKEVAELWAEYEAN 191
+A+VHD+AE++VGDITP G+ K++K +E +A++ +CK++ R K + EL+ EYE
Sbjct: 237 LALVHDLAESLVGDITPFCGISKDDKRAMEFKAMEDICKLI--EPRGKRIMELFEEYEHG 294
Query: 192 SSPEAKFVKDLDKVEMILQALEYEHEQGKDL--DEFFQSTAGKFQTEIGKAWASEIVSRR 249
+ E+KFVKDLD+++M++QA EYE L EFF ST GKF K +EI +R
Sbjct: 295 QTAESKFVKDLDRLDMVMQAFEYEKRDNCLLKHQEFFDSTEGKFNHPFVKKLVNEIYEQR 354
Query: 250 ---KKTNGSSHP 258
K G++ P
Sbjct: 355 DVLAKAKGATPP 366
>UniRef100_Q6CED3 Similar to sp|P38331 Saccharomyces cerevisiae YBR242w [Yarrowia
lipolytica]
Length = 242
Score = 155 bits (391), Expect = 1e-36
Identities = 86/190 (45%), Positives = 126/190 (66%), Gaps = 8/190 (4%)
Query: 66 ASATPSASSAIDFLSICHRLKTTKRAGWIR-KDVQDPESIADHMYRMSLMALIAADIPGV 124
A+A S + FL++ RLKTT R GW+R K + DPESIADH YRMS++A+++ + V
Sbjct: 45 AAAATGTESLLAFLNVVERLKTTPRTGWLRYKMIDDPESIADHQYRMSIIAMLS--LSPV 102
Query: 125 DRDKCVKMAIVHDIAEAIVGDITPTDGVPKEEKNRLEQEALDHMCKVLG--GGSRAKEVA 182
+++ CVKMA+VHD+AEAIVGDITP D + K EK+R E ++ +M ++ AKE+
Sbjct: 103 NQNTCVKMALVHDMAEAIVGDITPFDDMTKAEKSRREHSSIIYMAALVEKYNPVAAKEIV 162
Query: 183 ELWAEYEANSSPEAKFVKDLDKVEMILQALEYE--HEQGKDLDEFFQSTAGKFQTEIGKA 240
+LW +YE S+ EA+ VKD+DK E++LQ EYE H+ +DL +F+ EIGK
Sbjct: 163 DLWNQYENCSTDEARLVKDIDKFELMLQTYEYEKQHKFAEDLSQFYTLRGVIKTEEIGKL 222
Query: 241 WASEIVSRRK 250
A E++ +R+
Sbjct: 223 -ADELLRKRQ 231
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.312 0.125 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 399,648,172
Number of Sequences: 2790947
Number of extensions: 15277229
Number of successful extensions: 105347
Number of sequences better than 10.0: 643
Number of HSP's better than 10.0 without gapping: 289
Number of HSP's successfully gapped in prelim test: 367
Number of HSP's that attempted gapping in prelim test: 92656
Number of HSP's gapped (non-prelim): 7868
length of query: 261
length of database: 848,049,833
effective HSP length: 125
effective length of query: 136
effective length of database: 499,181,458
effective search space: 67888678288
effective search space used: 67888678288
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0041.3


