

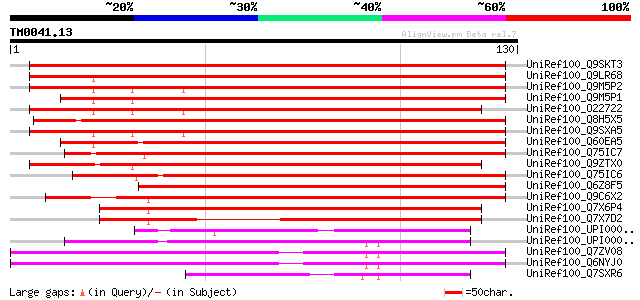
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0041.13
(130 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SKT3 Putative secretory carrier-associated membrane ... 191 2e-48
UniRef100_Q9LR68 F21B7.17 [Arabidopsis thaliana] 173 7e-43
UniRef100_Q9M5P2 Secretory carrier membrane protein [Arabidopsis... 167 6e-41
UniRef100_Q9M5P1 Secretory carrier membrane protein [Oryza sativa] 162 2e-39
UniRef100_O22722 F11P17.4 protein [Arabidopsis thaliana] 161 3e-39
UniRef100_Q8H5X5 Putative secretory carrier membrane protein [Or... 160 6e-39
UniRef100_Q9SXA5 T28P6.15 protein [Arabidopsis thaliana] 154 4e-37
UniRef100_Q60EA5 Putative secretory carrier membrane protein [Or... 149 2e-35
UniRef100_Q75IC7 Putative secretory carrier membrane protein [Or... 145 2e-34
UniRef100_Q9ZTX0 Similarity to SCAMP37 [Pisum sativum] 143 7e-34
UniRef100_Q75IC6 Putative secretory carrier membrane protein [Or... 134 5e-31
UniRef100_Q6Z8F5 Putative secretory carrier membrane protein [Or... 123 1e-27
UniRef100_Q9C6X2 Secretory carrier membrane protein, putative [A... 122 1e-27
UniRef100_Q7X6P4 OSJNBa0006A01.21 protein [Oryza sativa] 122 2e-27
UniRef100_Q7X7D2 OSJNBa0093F12.6 protein [Oryza sativa] 92 3e-18
UniRef100_UPI00003C1C31 UPI00003C1C31 UniRef100 entry 53 1e-06
UniRef100_UPI000031902F UPI000031902F UniRef100 entry 49 2e-05
UniRef100_Q7ZV08 Secretory carrier membrane protein 2, isoform 1... 49 2e-05
UniRef100_Q6NYJ0 Secretory carrier membrane protein 2, isoform 2... 49 2e-05
UniRef100_Q7SXR6 Scamp2l protein [Brachydanio rerio] 47 7e-05
>UniRef100_Q9SKT3 Putative secretory carrier-associated membrane protein [Arabidopsis
thaliana]
Length = 282
Score = 191 bits (486), Expect = 2e-48
Identities = 91/122 (74%), Positives = 107/122 (87%)
Query: 6 EMEPARNSGSVPLATSKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRRE 65
E+ P N SVP ATSKL PLPPEPYDRGAT+DIPLD+ KD+KAKEKEL+ +EAELKRRE
Sbjct: 13 EINPFANPTSVPAATSKLSPLPPEPYDRGATMDIPLDSGKDLKAKEKELREKEAELKRRE 72
Query: 66 QELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFLF 125
QE+KRKEDAIA+AGIVIEEKNWPPFFP+IHH+IS EIP+HLQRIQYVAF + LG++ L
Sbjct: 73 QEIKRKEDAIAQAGIVIEEKNWPPFFPLIHHDISNEIPIHLQRIQYVAFTSMLGLVVCLL 132
Query: 126 FS 127
++
Sbjct: 133 WN 134
>UniRef100_Q9LR68 F21B7.17 [Arabidopsis thaliana]
Length = 283
Score = 173 bits (439), Expect = 7e-43
Identities = 84/123 (68%), Positives = 102/123 (82%), Gaps = 1/123 (0%)
Query: 6 EMEPARNSGSVPLAT-SKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRR 64
E+ P N SVP A+ S LKPLPPEPYDRGAT+DIPLD+ D++AKE ELQA+E ELKR+
Sbjct: 13 EINPFANHTSVPPASNSYLKPLPPEPYDRGATVDIPLDSGNDLRAKEMELQAKENELKRK 72
Query: 65 EQELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFL 124
EQELKR+EDAIAR G+VIEEKNWP FFP+IHH+I EIP+HLQ+IQYVAF T LG++ L
Sbjct: 73 EQELKRREDAIARTGVVIEEKNWPEFFPLIHHDIPNEIPIHLQKIQYVAFTTLLGLVGCL 132
Query: 125 FFS 127
++
Sbjct: 133 LWN 135
>UniRef100_Q9M5P2 Secretory carrier membrane protein [Arabidopsis thaliana]
Length = 289
Score = 167 bits (422), Expect = 6e-41
Identities = 84/127 (66%), Positives = 104/127 (81%), Gaps = 5/127 (3%)
Query: 6 EMEPARNSGSVPLAT-SKLKPLPPEP--YDRGATIDIPLDT--TKDIKAKEKELQAREAE 60
E+ P N+ VP A+ S+L PLPPEP +D G T+DIPLD T+D+K KEKELQA+EAE
Sbjct: 15 EVNPFANARGVPPASNSRLSPLPPEPVGFDYGRTVDIPLDRAGTQDLKKKEKELQAKEAE 74
Query: 61 LKRREQELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGM 120
LKRREQ+LKRKEDA ARAGIVIE KNWPPFFP+IHH+I+ EIP+HLQR+QYV F T+LG+
Sbjct: 75 LKRREQDLKRKEDAAARAGIVIEVKNWPPFFPLIHHDIANEIPVHLQRLQYVTFATYLGL 134
Query: 121 LKFLFFS 127
+ LF++
Sbjct: 135 VLCLFWN 141
>UniRef100_Q9M5P1 Secretory carrier membrane protein [Oryza sativa]
Length = 286
Score = 162 bits (409), Expect = 2e-39
Identities = 80/118 (67%), Positives = 100/118 (83%), Gaps = 4/118 (3%)
Query: 14 GSVPLAT-SKLKPLPPEP---YDRGATIDIPLDTTKDIKAKEKELQAREAELKRREQELK 69
GSVP A+ S++ PLP EP DRGAT+DIPLD+TKD+K KEKELQA+EAEL +RE EL+
Sbjct: 21 GSVPPASNSRMPPLPHEPGFYNDRGATVDIPLDSTKDMKKKEKELQAKEAELNKRESELR 80
Query: 70 RKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFLFFS 127
R+E+A +RAGIVIEEKNWPPFFPIIHH+IS EIP+HLQR+QY+AF + LG+ LF++
Sbjct: 81 RREEAASRAGIVIEEKNWPPFFPIIHHDISNEIPIHLQRMQYLAFSSLLGLAACLFWN 138
>UniRef100_O22722 F11P17.4 protein [Arabidopsis thaliana]
Length = 234
Score = 161 bits (407), Expect = 3e-39
Identities = 81/121 (66%), Positives = 99/121 (80%), Gaps = 5/121 (4%)
Query: 6 EMEPARNSGSVPLAT-SKLKPLPPEP--YDRGATIDIPLDT--TKDIKAKEKELQAREAE 60
E+ P N+ VP A+ S+L PLPPEP +D G T+DIPLD T+D+K KEKELQA+EAE
Sbjct: 15 EVNPFANARGVPPASNSRLSPLPPEPVGFDYGRTVDIPLDRAGTQDLKKKEKELQAKEAE 74
Query: 61 LKRREQELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGM 120
LKRREQ+LKRKEDA ARAGIVIE KNWPPFFP+IHH+I+ EIP+HLQR+QYV F T+L +
Sbjct: 75 LKRREQDLKRKEDAAARAGIVIEVKNWPPFFPLIHHDIANEIPVHLQRLQYVTFATYLDV 134
Query: 121 L 121
+
Sbjct: 135 I 135
>UniRef100_Q8H5X5 Putative secretory carrier membrane protein [Oryza sativa]
Length = 306
Score = 160 bits (405), Expect = 6e-39
Identities = 79/121 (65%), Positives = 99/121 (81%), Gaps = 1/121 (0%)
Query: 7 MEPARNSGSVPLATSKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRREQ 66
M RN SV + S+L PLPPEP GAT+DIPLD++KD+K +EKELQAREAEL +RE+
Sbjct: 39 MPNPRNVPSVS-SNSRLSPLPPEPAAFGATVDIPLDSSKDLKNREKELQAREAELNKREK 97
Query: 67 ELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFLFF 126
ELKR+E+A ARAGIVIEEKNWPPF P+IHH+I+ EIP HLQR+QYVAF ++LG+ LF+
Sbjct: 98 ELKRREEAAARAGIVIEEKNWPPFLPLIHHDITNEIPSHLQRMQYVAFASFLGLACCLFW 157
Query: 127 S 127
+
Sbjct: 158 N 158
>UniRef100_Q9SXA5 T28P6.15 protein [Arabidopsis thaliana]
Length = 288
Score = 154 bits (389), Expect = 4e-37
Identities = 78/129 (60%), Positives = 101/129 (77%), Gaps = 7/129 (5%)
Query: 6 EMEPARNSGSVPLAT-SKLKPLPPEP--YDRGATIDIPLDT----TKDIKAKEKELQARE 58
E+ P N GSVP A+ S+L PLPPEP + G T+DIPLD +D+K KEKELQA+E
Sbjct: 15 EVNPFANPGSVPAASNSRLSPLPPEPAGFGYGRTVDIPLDRPGSGAQDLKKKEKELQAKE 74
Query: 59 AELKRREQELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWL 118
A+L+RREQ+LKRK+DA ARAGIVIE KNWP FFP+IHH+I+ EI + LQR+QY+AF T+L
Sbjct: 75 ADLRRREQDLKRKQDAAARAGIVIEAKNWPTFFPLIHHDIANEILVRLQRLQYIAFATYL 134
Query: 119 GMLKFLFFS 127
G++ LF++
Sbjct: 135 GLVLALFWN 143
>UniRef100_Q60EA5 Putative secretory carrier membrane protein [Oryza sativa]
Length = 282
Score = 149 bits (375), Expect = 2e-35
Identities = 75/115 (65%), Positives = 94/115 (81%), Gaps = 2/115 (1%)
Query: 14 GSVPLAT-SKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRKE 72
GSVP A S+L PL EP D +DIPLD++KD+K KEKELQA EAEL +RE+ELKRKE
Sbjct: 21 GSVPPANNSRLPPLSHEPADF-YNVDIPLDSSKDLKKKEKELQAMEAELNKRERELKRKE 79
Query: 73 DAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFLFFS 127
+A A+AGIVIE+KNWPPFFP+IHH IS EIP+HLQR+QY+AF ++LG+ LF++
Sbjct: 80 EAAAQAGIVIEDKNWPPFFPLIHHNISNEIPIHLQRMQYLAFSSFLGLAACLFWN 134
>UniRef100_Q75IC7 Putative secretory carrier membrane protein [Oryza sativa]
Length = 313
Score = 145 bits (366), Expect = 2e-34
Identities = 72/117 (61%), Positives = 92/117 (78%), Gaps = 5/117 (4%)
Query: 15 SVPLATSKLKPLPPEPYDR----GATIDIPLDTTKDIKAKEKELQAREAELKRREQELKR 70
S P AT L PLPPEP D +DIP+DT+KD+K +EKEL A+EAEL RRE+E+KR
Sbjct: 50 SAPPATH-LSPLPPEPADFYNDFSTPVDIPMDTSKDMKTREKELLAKEAELNRREKEIKR 108
Query: 71 KEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFLFFS 127
+E+A ARAGIV+E+KNWPPFFPIIH++I EIP+HLQR QYVAF + LG++ LF++
Sbjct: 109 REEAAARAGIVLEDKNWPPFFPIIHNDIGNEIPVHLQRTQYVAFASLLGLVLCLFWN 165
>UniRef100_Q9ZTX0 Similarity to SCAMP37 [Pisum sativum]
Length = 289
Score = 143 bits (361), Expect = 7e-34
Identities = 71/122 (58%), Positives = 93/122 (76%), Gaps = 7/122 (5%)
Query: 6 EMEPARNSGSVPLATSKLKPLPPEP------YDRGATIDIPLDTTKDIKAKEKELQAREA 59
++ P N S AT+ +P P P Y G T+DIPLDT+ D K KE++LQA+EA
Sbjct: 14 QVNPFSNPRSAASATNS-RPAPLNPDRADYNYGFGPTVDIPLDTSTDGKKKERDLQAKEA 72
Query: 60 ELKRREQELKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLG 119
EL++REQE++RKE+AIARAGIVIEEKNWPPFFPIIHH+I+ EIP+HL+ +QYVAF + LG
Sbjct: 73 ELRKREQEVRRKEEAIARAGIVIEEKNWPPFFPIIHHDITNEIPIHLRTLQYVAFFSLLG 132
Query: 120 ML 121
++
Sbjct: 133 LV 134
>UniRef100_Q75IC6 Putative secretory carrier membrane protein [Oryza sativa]
Length = 283
Score = 134 bits (337), Expect = 5e-31
Identities = 66/115 (57%), Positives = 89/115 (77%), Gaps = 5/115 (4%)
Query: 17 PLATSKLKPLPPEPY----DRGATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRKE 72
P + + PLP EP D GA++D PLD+ K +K KE+EL A+EAEL +REQELKR+E
Sbjct: 24 PFSHPRPTPLPHEPVAFYNDPGASVD-PLDSKKGLKKKERELLAKEAELNKREQELKRRE 82
Query: 73 DAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFLFFS 127
+A+ARAG+ IE KNWPPFFP+IH +IS +IP+HLQR+QYVAF + LG++ LF++
Sbjct: 83 EALARAGVFIEPKNWPPFFPVIHVDISNDIPVHLQRVQYVAFASLLGLVICLFWN 137
>UniRef100_Q6Z8F5 Putative secretory carrier membrane protein [Oryza sativa]
Length = 281
Score = 123 bits (308), Expect = 1e-27
Identities = 54/94 (57%), Positives = 75/94 (79%)
Query: 34 GATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRKEDAIARAGIVIEEKNWPPFFPI 93
GAT+DIPLD D K K KEL EA+LKRRE +++R+E+A+ AG+ +EEKNWPPFFPI
Sbjct: 58 GATVDIPLDNMSDSKGKGKELLQWEADLKRREADIRRREEALKSAGVPMEEKNWPPFFPI 117
Query: 94 IHHEISKEIPLHLQRIQYVAFITWLGMLKFLFFS 127
IHH+I+ EIP + Q++QY+AF +WLG++ LF++
Sbjct: 118 IHHDIANEIPANAQKLQYLAFASWLGIVLCLFWN 151
>UniRef100_Q9C6X2 Secretory carrier membrane protein, putative [Arabidopsis thaliana]
Length = 264
Score = 122 bits (307), Expect = 1e-27
Identities = 57/120 (47%), Positives = 86/120 (71%), Gaps = 8/120 (6%)
Query: 10 ARNSGSVPLATSKLKPLPPEPYDRG--ATIDIPLDTTKDIKAKEKELQAREAELKRREQE 67
++ G VP A+ P Y + AT+DIPLD D K+++L EAEL+++E +
Sbjct: 21 SKGGGRVPAASR------PVEYGQSLDATVDIPLDNMNDSSQKQRKLADWEAELRKKEMD 74
Query: 68 LKRKEDAIARAGIVIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGMLKFLFFS 127
+KR+E+AIA+ G+ I++KNWPPFFPIIHH+I+KEIP+H Q++QY+AF +WLG++ L F+
Sbjct: 75 IKRREEAIAKFGVQIDDKNWPPFFPIIHHDIAKEIPVHAQKLQYLAFASWLGIVLCLVFN 134
>UniRef100_Q7X6P4 OSJNBa0006A01.21 protein [Oryza sativa]
Length = 273
Score = 122 bits (305), Expect = 2e-27
Identities = 53/101 (52%), Positives = 78/101 (76%), Gaps = 3/101 (2%)
Query: 24 KPLPPEPYDRG---ATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRKEDAIARAGI 80
+P P + G AT+D+PLDT D K+K +EL + E +LKRRE ++KR+E+A+ AG+
Sbjct: 37 RPTEPAGFGAGRGDATVDVPLDTMGDSKSKARELSSWETDLKRREADIKRREEALRNAGV 96
Query: 81 VIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGML 121
+E+KNWPPFFPIIHH+I+ EIP +LQ++QY+AF +WLG++
Sbjct: 97 PMEDKNWPPFFPIIHHDIANEIPANLQKLQYLAFASWLGIV 137
>UniRef100_Q7X7D2 OSJNBa0093F12.6 protein [Oryza sativa]
Length = 245
Score = 91.7 bits (226), Expect = 3e-18
Identities = 44/101 (43%), Positives = 63/101 (61%), Gaps = 24/101 (23%)
Query: 24 KPLPPEPYDRG---ATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRKEDAIARAGI 80
+P P + G AT+D+PLDT DIK R+E+A+ AG+
Sbjct: 37 RPTEPAGFGAGRGDATVDVPLDTMGDIK---------------------RREEALRNAGV 75
Query: 81 VIEEKNWPPFFPIIHHEISKEIPLHLQRIQYVAFITWLGML 121
+E+KNWPPFFPIIHH+I+ EIP +LQ++QY+AF +WLG++
Sbjct: 76 PMEDKNWPPFFPIIHHDIANEIPANLQKLQYLAFASWLGIV 116
>UniRef100_UPI00003C1C31 UPI00003C1C31 UniRef100 entry
Length = 289
Score = 53.1 bits (126), Expect = 1e-06
Identities = 31/87 (35%), Positives = 49/87 (55%), Gaps = 8/87 (9%)
Query: 33 RGATIDIPLDTTKDIKAKE-KELQAREAELKRREQELKRKEDAIARAGIVIEEKNWPPFF 91
+G+T+D + ++A +EL+ RE EL RRE++L + I + G NWP F+
Sbjct: 47 KGSTVD---EIPSRVEANRMEELRRREEELARRERDLDNRAAHIQKHG----RNNWPFFY 99
Query: 92 PIIHHEISKEIPLHLQRIQYVAFITWL 118
P+I+H+I+ EIP Q I + WL
Sbjct: 100 PLIYHDIAAEIPPDSQAIMENLYKLWL 126
>UniRef100_UPI000031902F UPI000031902F UniRef100 entry
Length = 416
Score = 49.3 bits (116), Expect = 2e-05
Identities = 29/110 (26%), Positives = 52/110 (46%), Gaps = 8/110 (7%)
Query: 15 SVPLATSKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAELKRREQELKRKEDA 74
S P+ + +P +P + T+D + EL R+ EL+++ EL R+E
Sbjct: 55 STPVPSQNTQPAIMKPTEEPPAYS--QQQTQDQARAQAELLKRQEELEKKAAELDRRERE 112
Query: 75 IARAGIVIEEKNWPPF---FPI---IHHEISKEIPLHLQRIQYVAFITWL 118
+ G + NWPP FP+ +H+IS +IP+ Q+ + + W+
Sbjct: 113 LQSHGAAGRKNNWPPLPEKFPVGPCFYHDISVDIPVEFQKTVKIMYNLWI 162
>UniRef100_Q7ZV08 Secretory carrier membrane protein 2, isoform 1 [Brachydanio rerio]
Length = 326
Score = 49.3 bits (116), Expect = 2e-05
Identities = 33/133 (24%), Positives = 62/133 (45%), Gaps = 12/133 (9%)
Query: 1 MDFFSEMEPARNSGSVPLATSKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAE 60
++ FS ++P +++ +P +P + + +++EL+ + AE
Sbjct: 34 IESFSPFPDGNQGSTIPASSAPTQPAVLQPSVEPSPQATAAAAQASLIKQQEELERKAAE 93
Query: 61 LKRREQELKRKEDAIARAGIVIEEKNWPPF---FPI---IHHEISKEIPLHLQRIQYVAF 114
L+RREQEL+ +R ++ NWPP FPI + + S+EIP QR+ + +
Sbjct: 94 LERREQELQ------SRGAPTGKQNNWPPLPKRFPIQPCFYQDFSEEIPQEHQRLCKMIY 147
Query: 115 ITWLGMLKFLFFS 127
W+ LF +
Sbjct: 148 YLWMWECVTLFLN 160
>UniRef100_Q6NYJ0 Secretory carrier membrane protein 2, isoform 2 [Brachydanio rerio]
Length = 246
Score = 49.3 bits (116), Expect = 2e-05
Identities = 33/133 (24%), Positives = 62/133 (45%), Gaps = 12/133 (9%)
Query: 1 MDFFSEMEPARNSGSVPLATSKLKPLPPEPYDRGATIDIPLDTTKDIKAKEKELQAREAE 60
++ FS ++P +++ +P +P + + +++EL+ + AE
Sbjct: 34 IESFSPFPDGNQGSTIPASSAPTQPAVLQPSVEPSPQATAAAAQASLIKQQEELERKAAE 93
Query: 61 LKRREQELKRKEDAIARAGIVIEEKNWPPF---FPI---IHHEISKEIPLHLQRIQYVAF 114
L+RREQEL+ +R ++ NWPP FPI + + S+EIP QR+ + +
Sbjct: 94 LERREQELQ------SRGAPTGKQNNWPPLPKRFPIQPCFYQDFSEEIPQEHQRLCKMIY 147
Query: 115 ITWLGMLKFLFFS 127
W+ LF +
Sbjct: 148 YLWMWECVTLFLN 160
>UniRef100_Q7SXR6 Scamp2l protein [Brachydanio rerio]
Length = 325
Score = 47.4 bits (111), Expect = 7e-05
Identities = 25/79 (31%), Positives = 46/79 (57%), Gaps = 12/79 (15%)
Query: 46 DIKAKEKELQAREAELKRREQELKRKEDAIARAGIVIEEKNWPP---FFPI---IHHEIS 99
++ ++ EL+ + AEL+R+EQEL+ ++ A + E NWPP F PI + + +
Sbjct: 80 ELLRQQAELERKAAELERKEQELRDRDAARGK------ENNWPPLPKFLPIKPCFYQDFN 133
Query: 100 KEIPLHLQRIQYVAFITWL 118
+EIP+ QR+ + + W+
Sbjct: 134 EEIPVEYQRVCKMMYYLWM 152
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.138 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 224,713,710
Number of Sequences: 2790947
Number of extensions: 9181576
Number of successful extensions: 68280
Number of sequences better than 10.0: 553
Number of HSP's better than 10.0 without gapping: 283
Number of HSP's successfully gapped in prelim test: 271
Number of HSP's that attempted gapping in prelim test: 65980
Number of HSP's gapped (non-prelim): 2008
length of query: 130
length of database: 848,049,833
effective HSP length: 106
effective length of query: 24
effective length of database: 552,209,451
effective search space: 13253026824
effective search space used: 13253026824
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0041.13


