

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
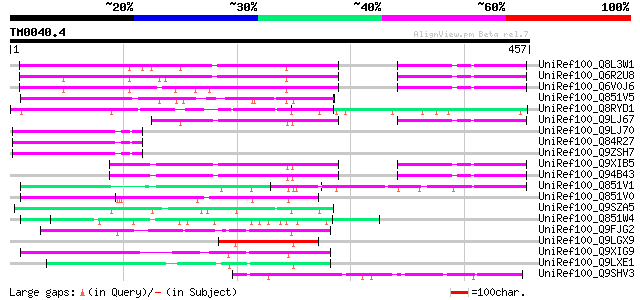
Query= TM0040.4
(457 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8L3W1 Reduced vernalization response 1 [Arabidopsis t... 138 3e-31
UniRef100_Q6R2U8 Reduced vernalization response 1 [Brassica camp... 127 9e-28
UniRef100_Q6V0J6 Reduced vernalization response 1 [Brassica camp... 125 3e-27
UniRef100_Q851V5 Putative auxin response factor [Oryza sativa] 83 1e-14
UniRef100_Q8RYD1 Auxin response factor 36 [Arabidopsis thaliana] 82 3e-14
UniRef100_Q9LJ67 Gb|AAD43153.1 [Arabidopsis thaliana] 77 1e-12
UniRef100_Q9LJ70 Gb|AAC72857.1 [Arabidopsis thaliana] 77 1e-12
UniRef100_Q84R27 Hypothetical protein At3g18960 [Arabidopsis tha... 77 1e-12
UniRef100_Q9ZSH7 T15B16.18 protein [Arabidopsis thaliana] 76 2e-12
UniRef100_Q9XIB5 F13F21.8 protein [Arabidopsis thaliana] 75 4e-12
UniRef100_Q94B43 Hypothetical protein F13F21.8 [Arabidopsis thal... 73 1e-11
UniRef100_Q851V1 Putative auxin response factor [Oryza sativa] 71 6e-11
UniRef100_Q851V0 Putative auxin response factor [Oryza sativa] 71 7e-11
UniRef100_Q9SZA5 Hypothetical protein F17M5.40 [Arabidopsis thal... 67 8e-10
UniRef100_Q851W4 Putative auxin response factor [Oryza sativa] 62 3e-08
UniRef100_Q9FJG2 Gb|AAD26962.1 [Arabidopsis thaliana] 58 5e-07
UniRef100_Q9LGX9 Hypothetical protein [Oryza sativa] 58 6e-07
UniRef100_Q9XIG9 Hypothetical protein At2g16210 [Arabidopsis tha... 54 9e-06
UniRef100_Q9LXE1 Hypothetical protein F17I14_30 [Arabidopsis tha... 54 1e-05
UniRef100_Q9SHV3 Hypothetical protein At2g24700 [Arabidopsis tha... 52 5e-05
>UniRef100_Q8L3W1 Reduced vernalization response 1 [Arabidopsis thaliana]
Length = 341
Score = 138 bits (348), Expect = 3e-31
Identities = 93/323 (28%), Positives = 151/323 (45%), Gaps = 46/323 (14%)
Query: 9 RIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMV 68
R+P+ F ++ +++ L PDG W++ K D +WFQ+GW++F YS+ G+++
Sbjct: 21 RVPDKFVSKFKDELSVAVALTVPDGHVWRVGLRKADNKIWFQDGWQEFVDRYSIRIGYLL 80
Query: 69 LFEYKDTSHFGVHIFDKSTLEIRYPSHENQDEEHNP-------DQIDDDSVEIL------ 115
+F Y+ S F V+IF+ S EI Y S D HN + ++D+ E++
Sbjct: 81 IFRYEGNSAFSVYIFNLSHSEINYHSTGLMDSAHNHFKRARLFEDLEDEDAEVIFPSSVY 140
Query: 116 -----DKIPPCKK--------------TKLKSPMSCPK-PRKKLRTSTSEDVGE----SP 151
+ P K K + P PK P+K+ R + D E +P
Sbjct: 141 PSPLPESTVPANKGYASSAIQTLFTGPVKAEEPTPTPKIPKKRGRKKKNADPEEINSSAP 200
Query: 152 ELHNLPKQVKIEDDAGRTTECLEGEDLTSKITEALNRARTFQSKFPSFMTVMKPAYV-NG 210
+ + K + A + E+ A+N A+TF+ P F V++P+Y+ G
Sbjct: 201 RDDDPENRSKFYESASARKRTVTAEER----ERAINAAKTFEPTNPFFRVVLRPSYLYRG 256
Query: 211 YSLHIPSFFAKKYLKNETDVLLQVLDGRTWSV----SFNLGKFNAGWKKFTSVNNLKVGD 266
+++PS FA+KYL + + L + W V KF+ GW +FT NNL GD
Sbjct: 257 CIMYLPSGFAEKYLSGISGFIKVQLAEKQWPVRCLYKAGRAKFSQGWYEFTLENNLGEGD 316
Query: 267 VCLFELNKSVPLSLKVLIFPLAE 289
VC+FEL ++ LKV F + E
Sbjct: 317 VCVFELLRTRDFVLKVTAFRVNE 339
Score = 58.2 bits (139), Expect = 5e-07
Identities = 39/115 (33%), Positives = 56/115 (47%), Gaps = 6/115 (5%)
Query: 342 KNAALEEANKFNSNNPFFIVNIAPDK-NRDYRPRVPKLFIRKHFNDIQQSEQTVILQCEK 400
+ A+ A F NPFF V + P R +P F K+ + I + +Q +
Sbjct: 227 RERAINAAKTFEPTNPFFRVVLRPSYLYRGCIMYLPSGFAEKYLSGISGF---IKVQLAE 283
Query: 401 KLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIYR 455
K PV+ C K G ++ S+GW EF ++ L GD CVFEL+ D VL V +R
Sbjct: 284 KQWPVR--CLYKAGRAKFSQGWYEFTLENNLGEGDVCVFELLRTRDFVLKVTAFR 336
>UniRef100_Q6R2U8 Reduced vernalization response 1 [Brassica campestris]
Length = 329
Score = 127 bits (318), Expect = 9e-28
Identities = 90/311 (28%), Positives = 146/311 (46%), Gaps = 34/311 (10%)
Query: 9 RIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGD--VWFQNGWKDFAKYYSLDHGH 66
R+P+ F R+ +++ L PDG W++ K D + +WFQ+GW++F YS+ G+
Sbjct: 21 RVPDKFVSRFKDELSVAVALTVPDGHVWRVGLRKADNNNKIWFQDGWQEFVDRYSIRIGY 80
Query: 67 MVLFEYKDTSHFGVHIFDKSTLEIRYPSHENQDE-EHNP--------DQIDDDSVEILDK 117
+++F Y+ S F V I++ EI Y S D HN + ++D+ E L
Sbjct: 81 LLIFRYEGNSAFSVCIYNLPQSEINYHSTGLMDSASHNNHFKRPRLFEDLEDEDAETLHT 140
Query: 118 IPPCKKTKLKSPM-----------SCPKP--RKKLRTSTSEDVGESPELHNLPK-QVKIE 163
++ P+ PK RKK E+V S + P+ + K
Sbjct: 141 TASAIQSFFTGPVKPEEATPTQTSKVPKKRGRKKKNADHPEEVNSSAPRDDDPESRSKFY 200
Query: 164 DDAGRTTECLEGEDLTSKITEALNRARTFQSKFPSFMTVMKPAYV-NGYSLHIPSFFAKK 222
+ A + E+ A+N A+TF+ P F V++P+Y+ G +++PS FA+K
Sbjct: 201 ESASARKRTVNAEER----ERAVNAAKTFEPTNPFFRVVLRPSYLYRGCIMYLPSGFAEK 256
Query: 223 YLKNETDVLLQVLDGRTWSV----SFNLGKFNAGWKKFTSVNNLKVGDVCLFELNKSVPL 278
YL + + L + W V KF+ GW +FT NNL GDVC+FEL ++
Sbjct: 257 YLSGISGFIKVQLGEKQWPVRCLYKAGRAKFSQGWYEFTVENNLGEGDVCVFELLRTRDF 316
Query: 279 SLKVLIFPLAE 289
LKV + + E
Sbjct: 317 VLKVTAYRVNE 327
Score = 58.9 bits (141), Expect = 3e-07
Identities = 40/115 (34%), Positives = 56/115 (47%), Gaps = 6/115 (5%)
Query: 342 KNAALEEANKFNSNNPFFIVNIAPDK-NRDYRPRVPKLFIRKHFNDIQQSEQTVILQCEK 400
+ A+ A F NPFF V + P R +P F K+ + I + +Q +
Sbjct: 215 RERAVNAAKTFEPTNPFFRVVLRPSYLYRGCIMYLPSGFAEKYLSGISGF---IKVQLGE 271
Query: 401 KLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIYR 455
K PV+ C K G ++ S+GW EF ++ L GD CVFEL+ D VL V YR
Sbjct: 272 KQWPVR--CLYKAGRAKFSQGWYEFTVENNLGEGDVCVFELLRTRDFVLKVTAYR 324
>UniRef100_Q6V0J6 Reduced vernalization response 1 [Brassica campestris]
Length = 329
Score = 125 bits (313), Expect = 3e-27
Identities = 89/310 (28%), Positives = 149/310 (47%), Gaps = 32/310 (10%)
Query: 9 RIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGD--VWFQNGWKDFAKYYSLDHGH 66
R+P+ F ++ +++ L PDG W++ K D + +WFQ+GW++F YS+ G+
Sbjct: 21 RVPDKFVSKFKDELSVAVALTVPDGHVWRVGLRKADNNNKIWFQDGWQEFVDRYSIRIGY 80
Query: 67 MVLFEYKDTSHFGVHIFDKSTLEIRYPSHENQDE-EHNP--------DQIDDDSVEILDK 117
+++F Y+ S F V I++ EI Y S D HN + ++D+ E L
Sbjct: 81 LLIFRYEGNSAFSVCIYNLPQSEINYHSTGLMDSASHNNHFKRPRLFEDLEDEDAETLHT 140
Query: 118 IPPCKKTKLKSPMSCPKPRKKLRTSTS---EDVGESPELHNLPKQVKI----EDDAGRTT 170
++ P+ KP + T TS + G + + P++V +DD +
Sbjct: 141 TASAIQSFFTGPV---KPEEATPTQTSKVPKKRGRKKKNADHPEEVNSSAPRDDDPESRS 197
Query: 171 ECLE------GEDLTSKITEALNRARTFQSKFPSFMTVMKPAYV-NGYSLHIPSFFAKKY 223
+ E G + A+N A+TF+ P F V++P+Y+ G +++PS FA+KY
Sbjct: 198 KFYESASARKGTVNAEERERAVNAAKTFEPTNPFFRVVLRPSYLYRGCIMYLPSGFAEKY 257
Query: 224 LKNETDVLLQVLDGRTWSV----SFNLGKFNAGWKKFTSVNNLKVGDVCLFELNKSVPLS 279
L + + L + W V KF+ GW +FT NNL GDVC+FEL ++
Sbjct: 258 LSGISGFIKVQLGEKQWPVRCLYKAGRAKFSQGWYEFTVENNLGEGDVCVFELLRTRDFV 317
Query: 280 LKVLIFPLAE 289
LKV + + E
Sbjct: 318 LKVTAYRVNE 327
Score = 58.9 bits (141), Expect = 3e-07
Identities = 40/115 (34%), Positives = 56/115 (47%), Gaps = 6/115 (5%)
Query: 342 KNAALEEANKFNSNNPFFIVNIAPDK-NRDYRPRVPKLFIRKHFNDIQQSEQTVILQCEK 400
+ A+ A F NPFF V + P R +P F K+ + I + +Q +
Sbjct: 215 RERAVNAAKTFEPTNPFFRVVLRPSYLYRGCIMYLPSGFAEKYLSGISGF---IKVQLGE 271
Query: 401 KLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIYR 455
K PV+ C K G ++ S+GW EF ++ L GD CVFEL+ D VL V YR
Sbjct: 272 KQWPVR--CLYKAGRAKFSQGWYEFTVENNLGEGDVCVFELLRTRDFVLKVTAYR 324
>UniRef100_Q851V5 Putative auxin response factor [Oryza sativa]
Length = 1029
Score = 83.2 bits (204), Expect = 1e-14
Identities = 79/300 (26%), Positives = 127/300 (42%), Gaps = 43/300 (14%)
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
IPN F + + G + L+ DG + + +K G + Q+GWK F + L G ++
Sbjct: 748 IPNEFLQYFRGKIPRTIKLQLRDGCTYDVQVTKNLGKISLQSGWKAFVTAHDLQMGDFLV 807
Query: 70 FEYKDTSHFGVHIFDKSTLEIRYPSHENQDEEHNPDQIDDDSVEILDKIPPCKKTKLKSP 129
F Y S V IF S E + ++ H ++ + E L +KSP
Sbjct: 808 FSYDGISKLKVLIFGPSGCEKVHSRSTLKNATHCGEKWE----EPLHISSNSHDLPVKSP 863
Query: 130 MSCPKPRKKLRTSTSE--------------DVGESPELH-NLPKQVKIEDDAGRTTECLE 174
+ K K+ +S E D+ P L+ LPK ++ D ++
Sbjct: 864 QNVSKSEKQWDSSEQENDTANIEEVALQGDDLQGHPVLNCILPKHTRLTD--------MQ 915
Query: 175 GEDLTSKITEALNRARTFQSKFPSFMTVMKPAYVNGYS--LHIPSFFAKKYLK-NETDVL 231
+ L SK+ S+ P + +++ + V+G S + I +A YL E ++
Sbjct: 916 KQQLESKV-------GAIHSEIPIYGCILRKSRVHGKSQTVDICREYADVYLPFKELNMT 968
Query: 232 LQVLDGRTWSV-----SFNLGKFNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIFP 286
LQ G+ W V + + GW +F NNL+VGD+CLFEL K S+ V I P
Sbjct: 969 LQ-RHGKNWEVLCRTKDTRTKRLSTGWSRFAQENNLQVGDICLFELLKKKEYSMNVHIIP 1027
Score = 70.9 bits (172), Expect = 7e-11
Identities = 71/300 (23%), Positives = 111/300 (36%), Gaps = 66/300 (22%)
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
IP F + + + LK +G + K + GW +FA + + G ++
Sbjct: 164 IPYKFAENFRDQIQGTIKLKARNGNTCSVLVDKCSNKLVLTKGWAEFANSHDIKMGDFLV 223
Query: 70 FEYKDTSHFGVHIFDKSTLEIRYPSHENQDEEHNPDQIDDDSVEILDKIPPCKKTKLKSP 129
F Y S F V IFD S ++ SH + + + D +EIL C L++
Sbjct: 224 FRYTGNSQFEVKIFDPSGC-VKAASHNAVNIGQHAQNMQGDPIEILS----CSDEHLRAQ 278
Query: 130 M---------------SCPKPRKKLRTSTSEDVGESP--ELHNLPKQVKIEDDAGRTTEC 172
+C K K S+SED E+P E+H ++K+E+
Sbjct: 279 SLTTERQNQPEKDVIDNCNKKMKTEHASSSEDDQETPTAEVH----RMKVEE-------- 326
Query: 173 LEGEDLTSKITEALNRARTFQSKFPSFMTVMKPAYVNGYSLH--IPSFFAKKYLKNETDV 230
R S P F+ VMK + V + I +A +Y +
Sbjct: 327 ---------------MVRAIHSNHPVFVAVMKKSNVTRQPCYVAISRKYANEYFPGGDQM 371
Query: 231 LLQVLDGRTWSVSFNLGK-----FNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIF 285
L G+ W V F + K + GW+KFT N EL PL+L + +F
Sbjct: 372 LTLQRHGKRWQVKFCISKRKLRMLSKGWRKFTRDN----------ELQHWAPLALFITLF 421
Score = 47.0 bits (110), Expect = 0.001
Identities = 19/37 (51%), Positives = 29/37 (78%)
Query: 417 RLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHI 453
RLS GW FA+++ L+VGD C+FEL+ K++ ++VHI
Sbjct: 989 RLSTGWSRFAQENNLQVGDICLFELLKKKEYSMNVHI 1025
Score = 38.9 bits (89), Expect = 0.31
Identities = 21/80 (26%), Positives = 38/80 (47%)
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
IP+ F + + G ++ L+P G + + +K + +GW+ F + L+ G ++
Sbjct: 467 IPDKFARHFKGVISKTIKLEPRSGYTFDVQVTKKLNILVLGSGWESFVNAHDLNMGDFLV 526
Query: 70 FEYKDTSHFGVHIFDKSTLE 89
F+Y V IFD S E
Sbjct: 527 FKYNGDFLLQVLIFDPSGCE 546
Score = 38.5 bits (88), Expect = 0.40
Identities = 30/93 (32%), Positives = 46/93 (49%), Gaps = 16/93 (17%)
Query: 207 YVNGYSLHIPSFFAKKYLKNE----------TDVLLQVLDGRTWSVSF--NLGKFN--AG 252
Y N + + I F + + NE + LQ+ DG T+ V NLGK + +G
Sbjct: 731 YKNFFKVMIGRFRERMIIPNEFLQYFRGKIPRTIKLQLRDGCTYDVQVTKNLGKISLQSG 790
Query: 253 WKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIF 285
WK F + ++L++GD +F + LKVLIF
Sbjct: 791 WKAFVTAHDLQMGDFLVFSYDGI--SKLKVLIF 821
>UniRef100_Q8RYD1 Auxin response factor 36 [Arabidopsis thaliana]
Length = 337
Score = 82.0 bits (201), Expect = 3e-14
Identities = 87/314 (27%), Positives = 133/314 (41%), Gaps = 52/314 (16%)
Query: 1 MLMESFVQR--IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVW-FQNGWKDFA 57
+L+ F R IP F+ + LK P G + + + D F+ GW F
Sbjct: 28 LLLPGFHNRLVIPRKFSTHCKRKLPQIVTLKSPSGVTYNVGVEEDDEKTMAFRFGWDKFV 87
Query: 58 KYYSLDHGHMVLFEYKDTSHFGVHIFDKSTL---------------EIRYPSHEN-QDEE 101
K +SL+ +++F++ S F V +FD TL E SH + EE
Sbjct: 88 KDHSLEENDLLVFKFHGVSEFEVLVFDGQTLCEKPTSYFVRKCGHAEKTKASHTGYEQEE 147
Query: 102 HNPDQIDDDSVEILDKIPPCKKTKLKSPMSCPKPRKKLRTSTSEDVGESPELHNLPKQVK 161
H ID S + L I P ++ KK+R S D NL ++
Sbjct: 148 HINSDIDTASAQ-LPVISPTSTVRVSEGKYPLSGFKKMRRELSND--------NLDQKAD 198
Query: 162 IEDDAGRTTECLEGEDLTSKITEALNRARTFQSKFPSFMTVMKPAYV-NGYSLHIP-SFF 219
+E + + + +AL+ A+ S F+ MK ++V + L IP +
Sbjct: 199 VEMISAGSNK------------KALSLAKRAISP-DGFLVFMKRSHVVSKCFLTIPYKWC 245
Query: 220 AKKYLKNETDVLLQVLDGRTWSVSFNL------GKFNAGWKKFTSVNNLKVGDVCLFE-- 271
K L +V++QV D W + FN+ G + GWKKF NNL+ GDVC+FE
Sbjct: 246 VKNMLITRQEVVMQV-DQTKWEMKFNIFGARGSGGISTGWKKFVQDNNLREGDVCVFEPA 304
Query: 272 LNKSVPLSLKVLIF 285
+++ PL L V IF
Sbjct: 305 NSETKPLHLNVYIF 318
Score = 47.4 bits (111), Expect = 9e-04
Identities = 58/244 (23%), Positives = 97/244 (38%), Gaps = 38/244 (15%)
Query: 249 FNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIFP---LAEEPHSL---PRSLVQGD 302
F GW KF ++L+ D+ +F+ + +VL+F L E+P S +
Sbjct: 79 FRFGWDKFVKDHSLEENDLLVFKFHGVS--EFEVLVFDGQTLCEKPTSYFVRKCGHAEKT 136
Query: 303 RVNHINYGRFRYTESQNVLTSKCAETIKRTSKL------------------ISPCPLKNA 344
+ +H Y + + S S I TS + +S L
Sbjct: 137 KASHTGYEQEEHINSDIDTASAQLPVISPTSTVRVSEGKYPLSGFKKMRRELSNDNLDQK 196
Query: 345 ALEEANKFNSNNPFFIVN---IAPDKNRDYRPR---VPKLFIRKHFN----DIQQSEQTV 394
A E SN + I+PD + R V K F+ + ++ + Q V
Sbjct: 197 ADVEMISAGSNKKALSLAKRAISPDGFLVFMKRSHVVSKCFLTIPYKWCVKNMLITRQEV 256
Query: 395 ILQCEKKLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPV--LDVH 452
++Q ++ +K + +G +S GW +F + + L GD CVFE N E L+V+
Sbjct: 257 VMQVDQTKWEMKFNIFGARGSGGISTGWKKFVQDNNLREGDVCVFEPANSETKPLHLNVY 316
Query: 453 IYRG 456
I+RG
Sbjct: 317 IFRG 320
>UniRef100_Q9LJ67 Gb|AAD43153.1 [Arabidopsis thaliana]
Length = 230
Score = 77.0 bits (188), Expect = 1e-12
Identities = 56/174 (32%), Positives = 87/174 (49%), Gaps = 14/174 (8%)
Query: 126 LKSPMSCPK-PRKKLRTSTSEDVGE----SPELHNLPKQVKIEDDAGRTTECLEGEDLTS 180
+K P PK P+K+ R + D E +P + + K + A + E+
Sbjct: 59 VKEPTPTPKIPKKRGRKKKNADPEEINSSAPRDDDPENRSKFYESASARKRTVTAEERE- 117
Query: 181 KITEALNRARTFQSKFPSFMTVMKPAYV-NGYSLHIPSFFAKKYLKNETDVLLQVLDGRT 239
A+N A+TF+ P F V++P+Y+ G +++PS FA+KYL + + L +
Sbjct: 118 ---RAINAAKTFEPTNPFFRVVLRPSYLYRGCIMYLPSGFAEKYLSGISGFIKVQLAEKQ 174
Query: 240 WSVS--FNLG--KFNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIFPLAE 289
W V + G KF+ GW +FT NNL GDVC+FEL ++ LKV F + E
Sbjct: 175 WPVRCLYKAGRAKFSQGWYEFTLENNLGEGDVCVFELLRTRDFVLKVTAFRVNE 228
Score = 58.2 bits (139), Expect = 5e-07
Identities = 39/115 (33%), Positives = 56/115 (47%), Gaps = 6/115 (5%)
Query: 342 KNAALEEANKFNSNNPFFIVNIAPDK-NRDYRPRVPKLFIRKHFNDIQQSEQTVILQCEK 400
+ A+ A F NPFF V + P R +P F K+ + I + +Q +
Sbjct: 116 RERAINAAKTFEPTNPFFRVVLRPSYLYRGCIMYLPSGFAEKYLSGISGF---IKVQLAE 172
Query: 401 KLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIYR 455
K PV+ C K G ++ S+GW EF ++ L GD CVFEL+ D VL V +R
Sbjct: 173 KQWPVR--CLYKAGRAKFSQGWYEFTLENNLGEGDVCVFELLRTRDFVLKVTAFR 225
>UniRef100_Q9LJ70 Gb|AAC72857.1 [Arabidopsis thaliana]
Length = 193
Score = 76.6 bits (187), Expect = 1e-12
Identities = 41/115 (35%), Positives = 61/115 (52%), Gaps = 5/115 (4%)
Query: 3 MESFVQRIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSL 62
M+ + +IP F K G ++ L+ P G I + ++WF GW +FA+ +S+
Sbjct: 39 MKDKMMKIPPRFVKLQGSKLSEVVTLETPAGFKRSIKLKRIGEEIWFHEGWSEFAEAHSI 98
Query: 63 DHGHMVLFEYKDTSHFGVHIFDKSTLEIRYPSHENQDEEHNPDQIDDDSVEILDK 117
+ GH +LFEYK+ S F V IF+ S E +YP D H D DDD ++I K
Sbjct: 99 EEGHFLLFEYKENSSFRVIIFNVSACETKYP----LDAVHIIDS-DDDIIDITGK 148
>UniRef100_Q84R27 Hypothetical protein At3g18960 [Arabidopsis thaliana]
Length = 209
Score = 76.6 bits (187), Expect = 1e-12
Identities = 41/115 (35%), Positives = 61/115 (52%), Gaps = 5/115 (4%)
Query: 3 MESFVQRIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSL 62
M+ + +IP F K G ++ L+ P G I + ++WF GW +FA+ +S+
Sbjct: 39 MKDKMMKIPPRFVKLQGSKLSEVVTLETPAGFKRSIKLKRIGEEIWFHEGWSEFAEAHSI 98
Query: 63 DHGHMVLFEYKDTSHFGVHIFDKSTLEIRYPSHENQDEEHNPDQIDDDSVEILDK 117
+ GH +LFEYK+ S F V IF+ S E +YP D H D DDD ++I K
Sbjct: 99 EEGHFLLFEYKENSSFRVIIFNVSACETKYP----LDAVHIIDS-DDDIIDITGK 148
>UniRef100_Q9ZSH7 T15B16.18 protein [Arabidopsis thaliana]
Length = 190
Score = 76.3 bits (186), Expect = 2e-12
Identities = 43/115 (37%), Positives = 58/115 (50%), Gaps = 5/115 (4%)
Query: 3 MESFVQRIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSL 62
M+ + RIP F K G ++ L P G I + ++WF GW +FA+ +S+
Sbjct: 39 MKDKMMRIPPRFVKLQGSKLSEVVTLVTPAGYKRSIKLKRIGEEIWFHEGWSEFAEAHSI 98
Query: 63 DHGHMVLFEYKDTSHFGVHIFDKSTLEIRYPSHENQDEEHNPDQIDDDSVEILDK 117
+ GH +LFEYK S F V IF+ S E YP D H D DDD +EI K
Sbjct: 99 EEGHFLLFEYKKNSSFRVIIFNASACETNYP----LDAVHIIDS-DDDVIEITGK 148
>UniRef100_Q9XIB5 F13F21.8 protein [Arabidopsis thaliana]
Length = 226
Score = 75.1 bits (183), Expect = 4e-12
Identities = 58/207 (28%), Positives = 97/207 (46%), Gaps = 14/207 (6%)
Query: 89 EIRYPSHENQDEEHNPDQIDDDSVEILDKIPPCKKTKLKSPMSCPKPRKKLRTSTSEDVG 148
++ YPS+ E N +++ K ++T P K +K + E+V
Sbjct: 26 KVIYPSNPESTEPVNKGYGGSTAIQSFFKESKAEET----PKVLKKRGRKKKNPNPEEVN 81
Query: 149 ES-PELHNLPKQVKIEDDAGRTTECLEGEDLTSKITEALNRARTFQSKFPSFMTVMKPAY 207
S P + + K + A + E+ A+N A+TF+ P F V++P+Y
Sbjct: 82 SSTPGGDDSENRSKFYESASARKRTVTAEERE----RAVNAAKTFEPTNPYFRVVLRPSY 137
Query: 208 V-NGYSLHIPSFFAKKYLKNETDVLLQVLDGRTWSVS--FNLG--KFNAGWKKFTSVNNL 262
+ G +++PS FA+KYL + + L + W V + G KF+ GW +FT NN+
Sbjct: 138 LYRGCIMYLPSGFAEKYLSGISGFIKLQLGEKQWPVRCLYKAGRAKFSQGWYEFTLENNI 197
Query: 263 KVGDVCLFELNKSVPLSLKVLIFPLAE 289
GDVC+FEL ++ L+V F + E
Sbjct: 198 GEGDVCVFELLRTRDFVLEVTAFRVNE 224
Score = 57.8 bits (138), Expect = 6e-07
Identities = 38/115 (33%), Positives = 57/115 (49%), Gaps = 6/115 (5%)
Query: 342 KNAALEEANKFNSNNPFFIVNIAPDK-NRDYRPRVPKLFIRKHFNDIQQSEQTVILQCEK 400
+ A+ A F NP+F V + P R +P F K+ + I + LQ +
Sbjct: 112 RERAVNAAKTFEPTNPYFRVVLRPSYLYRGCIMYLPSGFAEKYLSGISGF---IKLQLGE 168
Query: 401 KLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIYR 455
K PV+ C K G ++ S+GW EF ++ + GD CVFEL+ D VL+V +R
Sbjct: 169 KQWPVR--CLYKAGRAKFSQGWYEFTLENNIGEGDVCVFELLRTRDFVLEVTAFR 221
>UniRef100_Q94B43 Hypothetical protein F13F21.8 [Arabidopsis thaliana]
Length = 226
Score = 73.2 bits (178), Expect = 1e-11
Identities = 58/207 (28%), Positives = 95/207 (45%), Gaps = 14/207 (6%)
Query: 89 EIRYPSHENQDEEHNPDQIDDDSVEILDKIPPCKKTKLKSPMSCPKPRKKLRTSTSEDVG 148
++ YPS E N ++ K ++T P K +K + E+V
Sbjct: 26 KVIYPSIPESTEPVNKGYGGSTDIQSFFKESKAEET----PKVLKKRGRKKKNPNPEEVN 81
Query: 149 ES-PELHNLPKQVKIEDDAGRTTECLEGEDLTSKITEALNRARTFQSKFPSFMTVMKPAY 207
S P + + K + A + E+ A+N A+TF+ P F V++P+Y
Sbjct: 82 SSTPGGDDSENRSKFYESASARKRTVTAEERE----RAVNAAKTFEPTNPYFRVVLRPSY 137
Query: 208 V-NGYSLHIPSFFAKKYLKNETDVLLQVLDGRTWSVS--FNLG--KFNAGWKKFTSVNNL 262
+ G +++PS FA+KYL + + L + W V + G KF+ GW +FT NN+
Sbjct: 138 LYRGCIMYLPSGFAEKYLSGISGFIKLQLGEKQWPVRCLYKAGRAKFSQGWYEFTLENNI 197
Query: 263 KVGDVCLFELNKSVPLSLKVLIFPLAE 289
GDVC+FEL ++ L+V F + E
Sbjct: 198 GEGDVCVFELLRTRDFVLEVTAFRVNE 224
Score = 57.8 bits (138), Expect = 6e-07
Identities = 38/115 (33%), Positives = 57/115 (49%), Gaps = 6/115 (5%)
Query: 342 KNAALEEANKFNSNNPFFIVNIAPDK-NRDYRPRVPKLFIRKHFNDIQQSEQTVILQCEK 400
+ A+ A F NP+F V + P R +P F K+ + I + LQ +
Sbjct: 112 RERAVNAAKTFEPTNPYFRVVLRPSYLYRGCIMYLPSGFAEKYLSGISGF---IKLQLGE 168
Query: 401 KLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPVLDVHIYR 455
K PV+ C K G ++ S+GW EF ++ + GD CVFEL+ D VL+V +R
Sbjct: 169 KQWPVR--CLYKAGRAKFSQGWYEFTLENNIGEGDVCVFELLRTRDFVLEVTAFR 221
>UniRef100_Q851V1 Putative auxin response factor [Oryza sativa]
Length = 306
Score = 71.2 bits (173), Expect = 6e-11
Identities = 63/280 (22%), Positives = 111/280 (39%), Gaps = 49/280 (17%)
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
IPN F +GG + L+ G + + +K G V Q+GW + + L G ++
Sbjct: 46 IPNEFLHNFGGKIPKSIKLETRSGLTFDVQVTKNSGRVVLQSGWASYVSAHDLKIGDFLV 105
Query: 70 FEYKDTSHFGVHIFDKSTLEIRYPSHENQDEEHNPDQIDDDSVEILDKIPPCKKTKLKSP 129
F+Y S IFD S E C+K P
Sbjct: 106 FKYSGDSQLKTLIFDSSGCE-----------------------------KVCEK-----P 131
Query: 130 MSCPKPRKKLRTSTSEDVGESPELHNLPKQVKIE-DDAGRTTECLEGEDLTSKITEA--- 185
+ + S+D + + ++ +Q ++ + G E + G L S+
Sbjct: 132 VDMSGRSYDIAMRNSQDEKKKRKQRDISRQGTVKPSEEGLKAELVPGCILPSRTDLTRLQ 191
Query: 186 ----LNRARTFQSKFPSFMTVMKPAYVNGY--SLHIPSFFAKKYLKNETDVLLQVLDGRT 239
+ + + S+ P + VM + ++G ++ I +A YL E ++ G++
Sbjct: 192 KNILIEKVKAINSETPIYGYVMNNSSIHGIPCTVEISKKYADVYLPFEDGTVVLQHHGKS 251
Query: 240 WSVSFNLGKFNA-----GWKKFTSVNNLKVGDVCLFELNK 274
W+V L K N+ GW++F N L +GD+CLF+L K
Sbjct: 252 WNVRCCLTKQNSKRFLKGWRQFAGDNKLHLGDICLFDLLK 291
Score = 64.3 bits (155), Expect = 7e-09
Identities = 70/252 (27%), Positives = 114/252 (44%), Gaps = 35/252 (13%)
Query: 230 VLLQVLDGRTWSVSF--NLGK--FNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIF 285
+ L+ G T+ V N G+ +GW + S ++LK+GD +F+ S LK LIF
Sbjct: 62 IKLETRSGLTFDVQVTKNSGRVVLQSGWASYVSAHDLKIGDFLVFKY--SGDSQLKTLIF 119
Query: 286 P------LAEEPHSLP-RSLVQGDRVNHINYGRFRYTESQNVLTSKCAETIKRTSKLISP 338
+ E+P + RS R + + + + T K +E + ++L+
Sbjct: 120 DSSGCEKVCEKPVDMSGRSYDIAMRNSQDEKKKRKQRDISRQGTVKPSEEGLK-AELVPG 178
Query: 339 CPL----------KNAALEEANKFNSNNPFF--IVNIAPDKNRDYRPRVPKLFIRKHFND 386
C L KN +E+ NS P + ++N N + I K + D
Sbjct: 179 CILPSRTDLTRLQKNILIEKVKAINSETPIYGYVMN-----NSSIHGIPCTVEISKKYAD 233
Query: 387 IQQ--SEQTVILQCEKKLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELI-N 443
+ + TV+LQ K V+ C K+ R KGW +FA +KL +GD C+F+L+ +
Sbjct: 234 VYLPFEDGTVVLQHHGKSWNVRC-CLTKQNSKRFLKGWRQFAGDNKLHLGDICLFDLLKD 292
Query: 444 KEDPVLDVHIYR 455
K+ V+DVHI R
Sbjct: 293 KKKYVMDVHIIR 304
>UniRef100_Q851V0 Putative auxin response factor [Oryza sativa]
Length = 750
Score = 70.9 bits (172), Expect = 7e-11
Identities = 79/298 (26%), Positives = 123/298 (40%), Gaps = 39/298 (13%)
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTD-GDVWFQNGWKDFAKYYSLDHGHMV 68
+P F + G ++ L+ P G W I + +D G++ Q GWK+F ++ G +
Sbjct: 424 VPARFANNFNGHISEEVNLRSPSGETWSIGVANSDAGELVLQPGWKEFVDGNGIEEGDCL 483
Query: 69 LFEYKD-TSHFGVHIFDKSTLEIRYP----SH-----EN--------QDEEHNPDQIDDD 110
LF Y +S F V IFD S E P SH EN ++ P +D D
Sbjct: 484 LFRYSGVSSSFDVLIFDPSGCEKASPHFVGSHGFGRAENSAGAEQGGRNGRRTPPIVDGD 543
Query: 111 SVEILDKIPPCKKTKLKS-PMSCPKPRKKLRTSTSEDVGESPELHNLPKQVKIED----D 165
+ + +S P +C + T E+ GE ++ + + +
Sbjct: 544 NGHRHHLEMTLHRNSCRSIPRACKRSLFSDETEAKENDGEDEDVVAAAEGGRYGEYYFSR 603
Query: 166 AGRTTECLEGEDLTSKITEALNRART-FQSKFPSFMTVMKPAYVNGYS---LHIPSFFAK 221
GR E +L + E ++R Q P F+ V+ ++V + + FA
Sbjct: 604 HGRVAE----YNLREEDREEISRVPVPVQPGNPVFVQVIHSSHVRSSKYCIVGVSPEFAG 659
Query: 222 KYL-KNETDVLLQ-VLDGRTWSVSF-----NLGKFNAGWKKFTSVNNLKVGDVCLFEL 272
KYL E +V+L+ G W V F G + AGW++F N L DVCLFEL
Sbjct: 660 KYLGAVEREVVLERASRGGEWHVPFVHRQNTRGFYGAGWRQFAGDNRLVAHDVCLFEL 717
Score = 52.4 bits (124), Expect = 3e-05
Identities = 27/85 (31%), Positives = 41/85 (47%), Gaps = 1/85 (1%)
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTD-GDVWFQNGWKDFAKYYSLDHGHMV 68
+P+ F + G ++ LK P G W I + +D G++ ++GWK+F + +
Sbjct: 46 VPSRFANNFNGHISEVVNLKSPSGKTWSIGVAYSDTGELVLRSGWKEFVDANGVQENDCL 105
Query: 69 LFEYKDTSHFGVHIFDKSTLEIRYP 93
LF Y S F V IFD S E P
Sbjct: 106 LFRYSGVSSFDVLIFDPSGCEKASP 130
Score = 35.8 bits (81), Expect = 2.6
Identities = 28/93 (30%), Positives = 44/93 (47%), Gaps = 10/93 (10%)
Query: 199 FMTVMKPAYVNGYSLHIPSFFAKKYLKNETDVL-LQVLDGRTWSVSFNLGK-----FNAG 252
F+T M + S+ +PS FA + + ++V+ L+ G+TWS+ +G
Sbjct: 32 FLTYMVGDFTE--SMIVPSRFANNFNGHISEVVNLKSPSGKTWSIGVAYSDTGELVLRSG 89
Query: 253 WKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIF 285
WK+F N ++ D LF S S VLIF
Sbjct: 90 WKEFVDANGVQENDCLLFRY--SGVSSFDVLIF 120
Score = 34.3 bits (77), Expect = 7.6
Identities = 25/77 (32%), Positives = 38/77 (48%), Gaps = 7/77 (9%)
Query: 215 IPSFFAKKYLKN-ETDVLLQVLDGRTWSVSF---NLGKF--NAGWKKFTSVNNLKVGDVC 268
+P+ FA + + +V L+ G TWS+ + G+ GWK+F N ++ GD
Sbjct: 424 VPARFANNFNGHISEEVNLRSPSGETWSIGVANSDAGELVLQPGWKEFVDGNGIEEGDCL 483
Query: 269 LFELNKSVPLSLKVLIF 285
LF + V S VLIF
Sbjct: 484 LFRYS-GVSSSFDVLIF 499
>UniRef100_Q9SZA5 Hypothetical protein F17M5.40 [Arabidopsis thaliana]
Length = 461
Score = 67.4 bits (163), Expect = 8e-10
Identities = 79/346 (22%), Positives = 133/346 (37%), Gaps = 71/346 (20%)
Query: 5 SFVQRIPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVW-FQNGWKDFAKYYSLD 63
+ +Q IP F+ + LK P G + + + D F+ GW F K +SL+
Sbjct: 103 NLLQVIPRKFSTHCKRKLPQIVTLKSPSGVTYNVGVEEDDEKTMAFRFGWDKFVKDHSLE 162
Query: 64 HGHMVLFEYKDTSHFGVHIFDKSTL----------------------EIRYPSHENQDEE 101
+++F++ S F V +FD TL + S +
Sbjct: 163 ENDLLVFKFHGVSEFEVLVFDGQTLCEKPTSYFVRKCGHAEKTKGIIDFNATSSRSPKRH 222
Query: 102 HNPDQIDDDSVEILDKIPPCKKT--------------KLKSPMSCPKPRKKLRTSTSEDV 147
NPD ++ + L PP K+ +P+ T ++
Sbjct: 223 FNPDDVETTPNQQLVISPPVDNELEDLIDIDLDFDIDKILNPLLVAS-----HTGYEQEE 277
Query: 148 GESPELHNLPKQVKIEDDAG--RTTEC-------------LEGEDLTSKITEALNRARTF 192
+ ++ Q+ + R +E L ++L K + +A +
Sbjct: 278 HINSDIDTASAQLPVISPTSTVRVSEGKYPLSGFKKMRRELSNDNLDQKAAGSNKKALSL 337
Query: 193 QSKFPS---FMTVMKPAYV-NGYSLHIP-SFFAKKYLKNETDVLLQVLDGRTWSVSFNL- 246
+ S F+ MK ++V + L IP + K L +V++QV D W + FN+
Sbjct: 338 AKRAISPDGFLVFMKRSHVVSKCFLTIPYKWCVKNMLITRQEVVMQV-DQTKWEMKFNIF 396
Query: 247 -----GKFNAGWKKFTSVNNLKVGDVCLFE--LNKSVPLSLKVLIF 285
G + GWKKF NNL+ GDVC+FE +++ PL L V IF
Sbjct: 397 GARGSGGISTGWKKFVQDNNLREGDVCVFEPANSETKPLHLNVYIF 442
Score = 47.0 bits (110), Expect = 0.001
Identities = 23/69 (33%), Positives = 38/69 (54%), Gaps = 2/69 (2%)
Query: 390 SEQTVILQCEKKLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACVFELINKEDPV- 448
+ Q V++Q ++ +K + +G +S GW +F + + L GD CVFE N E
Sbjct: 376 TRQEVVMQVDQTKWEMKFNIFGARGSGGISTGWKKFVQDNNLREGDVCVFEPANSETKPL 435
Query: 449 -LDVHIYRG 456
L+V+I+RG
Sbjct: 436 HLNVYIFRG 444
>UniRef100_Q851W4 Putative auxin response factor [Oryza sativa]
Length = 545
Score = 62.4 bits (150), Expect = 3e-08
Identities = 69/336 (20%), Positives = 128/336 (37%), Gaps = 31/336 (9%)
Query: 10 IPNNFTKRYGGDMANPTFLKPPDGTDWKIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVL 69
IPN F ++GG ++ L+ P G + + SK Q GW+ F + ++ +L
Sbjct: 43 IPNKFLDQFGGKISRTVELESPKGNVYVVKVSKHMNKTVLQCGWEAFVDAHQIEENDSLL 102
Query: 70 FEYKDTSH--FGVHIFDKSTLEIRYPSHENQDEEHNPDQI---DDDSVEILDKIPPCKKT 124
F + S GV + +++ +H++ + ++ + S K T
Sbjct: 103 FRHIKNSRRASGVQERNADPIDVSSSTHDDTVQSSGGERFARSESGSDSQHSKTAKLAAT 162
Query: 125 KLKSPMSCPKPRKKLRTSTSEDVG---ESPELHNLPKQVKIEDDAGRTTECLEGEDLTSK 181
C K +S+ + P++ +P V G +L+
Sbjct: 163 CSSGGSECTGEEAKESSSSEHESSYDLVDPQIAPMPGYV-----------LSRGTNLSQA 211
Query: 182 ITEALNR-ARTFQSKFPSFMTVMKPAYVNGY--SLHIPSFFAKKYLKNETDVLLQVLDG- 237
E L+ + + + P ++T MK + VN + SL I +A Y + + + G
Sbjct: 212 HEEKLDMLVQEIRPEIPLYVTTMKHSNVNSHHASLVIAKHYACAYFPHTSQTITLKWHGK 271
Query: 238 -RTWSVSFNLGKFNAG------WKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIFPLAEE 290
R W F + K G W F N++K GD+C+F L + + L E
Sbjct: 272 NRKWHPKFYIRKDQVGYILHGRWIDFVRHNHVKEGDICIFHLKNFNGRKFRATVHLLRET 331
Query: 291 -PHSLPRSLVQGDRVNHINYGRFRYTESQNVLTSKC 325
PHS + + R + T+ + V +++C
Sbjct: 332 IPHSFGALHIPKRFESRNGRMRLKMTDDRRVSSTEC 367
Score = 47.4 bits (111), Expect = 9e-04
Identities = 55/267 (20%), Positives = 108/267 (39%), Gaps = 25/267 (9%)
Query: 37 KIYHSKTDGDVWFQNGWKDFAKYYSLDHGHMVLFEYKDTSHFGVHIFDKSTLEIRYPSHE 96
K Y K W DF ++ + G + +F K+ F F + +R
Sbjct: 278 KFYIRKDQVGYILHGRWIDFVRHNHVKEGDICIFHLKN---FNGRKFRATVHLLRETIPH 334
Query: 97 NQDEEHNPDQIDDDSVEILDKIPPCKKTKLKSPMSCPKPRKKLRTSTSEDVGESPELHN- 155
+ H P + + + + K+ ++ S C + + T+ + ++ + +N
Sbjct: 335 SFGALHIPKRFESRNGRMRLKMTDDRRV---SSTECRRGTMEPSTTNVKKEADNEQCNNG 391
Query: 156 ---LPKQVKIEDDAGRTTECLEGEDLT---SKITEALNRARTFQSKFPSFMTVMKPAYVN 209
+ + + G + L + ++ + + + QS+ P ++++M + V
Sbjct: 392 QGKRQEPLNFDVSVGSSKPYLTADRVSLTEEQFMKVEENVHSIQSEGPIYVSIMNKSNVG 451
Query: 210 GYSLHIPSF---FAKKYLKN--ETDVLLQVLDGRTWSV-----SFNLGKFNAGWKKFTSV 259
L+I + FA +YL +T LL G+ W V S + F GW+ F
Sbjct: 452 TDGLYIITLGRQFAIRYLPEGEQTLTLLTTGTGKAWQVKMRPRSGDARMFTLGWRDFVRD 511
Query: 260 NNLKVGDVCLFELNKSVP--LSLKVLI 284
N L+ D+CLF+L K+ L++KV I
Sbjct: 512 NRLQTEDICLFQLTKNSERGLAMKVHI 538
>UniRef100_Q9FJG2 Gb|AAD26962.1 [Arabidopsis thaliana]
Length = 307
Score = 58.2 bits (139), Expect = 5e-07
Identities = 65/269 (24%), Positives = 113/269 (41%), Gaps = 34/269 (12%)
Query: 28 LKPPDGTDWKIYHSKTDGDVWFQN-GWKDFAKYYSLDHGHMVLFEYKDTSHFGVHIFDKS 86
L+ P G W++ SK + ++ GW F L + F ++ F V IF+
Sbjct: 56 LRVPWGRSWQVKISKNPNFHYMEDRGWNQFVNDNGLGENEYLTFTHEANMCFNVTIFEAD 115
Query: 87 TLEIRYP-------SHENQDEEHNPDQIDDDSVEILDKIPPCKKTKLKSPMSCPKPRKKL 139
E+ P S N+ EE D K+ +++S P K
Sbjct: 116 GTEMLRPRKTITSSSGRNKREERKSIYKDVK-----------KEEEIESWSESSHPCHKT 164
Query: 140 RTSTSEDVGESPELHNLPKQVKIEDDAGRTTECLEGEDLTSKITEALNRARTFQSKFPSF 199
STS + + EL NL K+ + + +T++ + E +++ + A T S P F
Sbjct: 165 AESTSGRLTQKQEL-NLRKKEADKTEKSKTSKKKKVETVSND-----SEAGT-SSLIPEF 217
Query: 200 MTVMKPAYVNGYSLHIPSFFAKKYLKNETDVL-LQVLDGR-TWSVSFNL----GKFNAGW 253
+K +++ L IP F ++ ET + + G+ +W V++ + +F+ GW
Sbjct: 218 KLTIKKSHL--LFLGIPKKFVDMHMPTETTMFKIHYPRGKKSWDVTYVVTDVQSRFSGGW 275
Query: 254 KKFTSVNNLKVGDVCLFELNKSVPLSLKV 282
+ L VGDVC F+L K + +KV
Sbjct: 276 SRLAKELGLLVGDVCTFKLIKPTEMRVKV 304
Score = 35.8 bits (81), Expect = 2.6
Identities = 52/244 (21%), Positives = 94/244 (38%), Gaps = 33/244 (13%)
Query: 230 VLLQVLDGRTWSVSFNLGKF-----NAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKV-- 282
++L+V GR+W V + + GW +F + N L + F ++ ++ +
Sbjct: 54 MVLRVPWGRSWQVKISKNPNFHYMEDRGWNQFVNDNGLGENEYLTFTHEANMCFNVTIFE 113
Query: 283 -----LIFPLAEEPHSLPRSLVQGDRVNHINYGRFRYTESQNVLTSKCAETIKRTSKLIS 337
++ P S R+ + + + + + ES + + C +T + TS ++
Sbjct: 114 ADGTEMLRPRKTITSSSGRNKREERKSIYKDVKKEEEIESWSESSHPCHKTAESTSGRLT 173
Query: 338 PCPLKNAALEEANKFNSNNPFF---IVNIAPDKNRDYRPRVPK------------LFIRK 382
N +EA+K + + ++ D +P+ L I K
Sbjct: 174 QKQELNLRKKEADKTEKSKTSKKKKVETVSNDSEAGTSSLIPEFKLTIKKSHLLFLGIPK 233
Query: 383 HFNDIQQSEQTVILQCE----KKLCPVKLTCYPKKGPSRLSKGWVEFAKKSKLEVGDACV 438
F D+ +T + + KK V T SR S GW AK+ L VGD C
Sbjct: 234 KFVDMHMPTETTMFKIHYPRGKKSWDV--TYVVTDVQSRFSGGWSRLAKELGLLVGDVCT 291
Query: 439 FELI 442
F+LI
Sbjct: 292 FKLI 295
>UniRef100_Q9LGX9 Hypothetical protein [Oryza sativa]
Length = 524
Score = 57.8 bits (138), Expect = 6e-07
Identities = 36/96 (37%), Positives = 61/96 (63%), Gaps = 8/96 (8%)
Query: 185 ALNRARTFQSKFP----SFMTVMKPAYV-NGYSLHIPSFFAKKYL-KNETDVLLQVLDGR 238
A+ RA+ Q+K P SF+ M ++V +G+ L +P+ F KYL K++TD++L+ +G
Sbjct: 137 AMKRAQEIQTKLPAEHPSFVKHMLHSHVVSGFWLGLPAGFCNKYLPKHDTDIVLEDENGN 196
Query: 239 TWSVSFNLGK--FNAGWKKFTSVNNLKVGDVCLFEL 272
+ ++ GK +AGW+ F +++KVGDV +FEL
Sbjct: 197 NHNTNYLGGKQGLSAGWRGFAINHDIKVGDVVVFEL 232
>UniRef100_Q9XIG9 Hypothetical protein At2g16210 [Arabidopsis thaliana]
Length = 286
Score = 53.9 bits (128), Expect = 9e-06
Identities = 59/284 (20%), Positives = 117/284 (40%), Gaps = 47/284 (16%)
Query: 10 IPNNFTKRYGG-DMANPTFLKPPDGTDWKIYHSKTDGDVWFQ-NGWKDFAKYYSLDHGHM 67
+P++F++ + +++ ++ G W++ SK + + +GW+ F + +L + +
Sbjct: 36 LPHDFSRSFTDKELSRKMKIRAQWGNSWEVGISKNPRFYFMEKSGWEKFVRDNALGNSEL 95
Query: 68 VLFEYKDTSHFGVHIFDKSTLEIRYPSHENQDEEHNPDQIDDDSVEILDKIPPCKKTKLK 127
+ F +K HF V+IF E+ P + + +D E++
Sbjct: 96 LTFTHKGKMHFTVNIFKLDGKEMMQPPQSRSFFASSKQEENDIKEEVV------------ 143
Query: 128 SPMSCPKPRKKLRTSTSEDVGESPELHNLPKQVKIEDDAGRTTECLEGEDLTSKITEALN 187
S + G++ + +++ + A + E +SK TE +
Sbjct: 144 ---------------VSSNRGQTTAAESKGRKLNLGKRAAK-------ESQSSKRTEKVV 181
Query: 188 RART-----FQSKFPSFMTVMKPAYVNGYSLHIPSFFAKKYLKNETDVL-LQVLDG-RTW 240
RAR+ S +F + K Y+ L IP+ +K + +E V + +G ++W
Sbjct: 182 RARSDYAGASSSTAAAFTILFKQGYL--VFLRIPNSVSKDQVPDEKTVFKIHHPNGKKSW 239
Query: 241 SVSF--NLGKFNAGWKKFTSVNNLKVGDVCLFELNKSVPLSLKV 282
+V + G F+ GW++ L VGD C F K L L V
Sbjct: 240 NVVYLERFGAFSGGWRRVVKEYPLAVGDTCKFTFIKPKELLLVV 283
>UniRef100_Q9LXE1 Hypothetical protein F17I14_30 [Arabidopsis thaliana]
Length = 308
Score = 53.5 bits (127), Expect = 1e-05
Identities = 57/260 (21%), Positives = 100/260 (37%), Gaps = 24/260 (9%)
Query: 33 GTDWKIYHSKTDGDVWFQ-NGWKDFAKYYSLDHGHMVLFEYKDTSHFGVHIFDKSTLEIR 91
G WK+ SK + + +GW+ F +L + F +K F V IF+K E+
Sbjct: 60 GNSWKVKISKNPRFYFMEKSGWEKFVIDNALGDHEFLTFTHKGQMSFTVKIFNKDGKEMM 119
Query: 92 YPSHENQDEEHNPDQIDDDSVEILDKIPPCKKTKLKSPMSCPKPRKKLRTSTSEDVGESP 151
P + + V+ +++ ++ + P + + + G
Sbjct: 120 QPPQSRASFASSSRVKTEQDVKREEEVLVSSDSRSRGPTTAAETNRG---------GSYK 170
Query: 152 ELHNLPKQVKIEDDAGRTTECLEGEDLTSKITEALNRARTF----QSKFPSFMTVMKPAY 207
N K+ E + TE + T ++ +++ R + S F + +Y
Sbjct: 171 RKLNFGKKKAEETQTYKRTERTQNSKRTERV---VSKERVYAGEPSSSVAGFKIFISKSY 227
Query: 208 VNGYSLHIPSFFAKKYLKNETDVLLQVLDG-RTWSVSFNLGK----FNAGWKKFTSVNNL 262
+ SL IP F K +T V + DG +TW V F + + F+ GWK+ +
Sbjct: 228 IK--SLAIPKPFGNYMPKEKTRVKIHHPDGEKTWKVVFVVKERGQIFSGGWKRLCKEYPV 285
Query: 263 KVGDVCLFELNKSVPLSLKV 282
GD C F L + L L V
Sbjct: 286 VFGDTCKFTLITPLELLLVV 305
>UniRef100_Q9SHV3 Hypothetical protein At2g24700 [Arabidopsis thaliana]
Length = 899
Score = 51.6 bits (122), Expect = 5e-05
Identities = 64/269 (23%), Positives = 111/269 (40%), Gaps = 26/269 (9%)
Query: 197 PSFMTVMKPAYVNGYSLHIPSFFAKKYLKN----ETDVLLQVLDGRTWSVSFNLGKFNAG 252
P F + P + N L IP F KYL +T L TW V + + + G
Sbjct: 12 PQFFQPLLPGFTN--HLDIPVAFFLKYLVGTNVGKTAELRSDASEMTWKVKIDGRRLSNG 69
Query: 253 WKKFTSVNNLKVGDVCLFELNKSVPLSLKVLIFPLAEEPHSLPRSLVQGDRVNHINY--- 309
W+ FT ++L+VGD+ +F + + L E + ++ D++ ++++
Sbjct: 70 WEDFTIAHDLRVGDIVVFRQEGELVFHVTALGPSCCEIQYG--EDTLEEDKIGNLSFILN 127
Query: 310 --GRFRYTES--QNVLTSKCAETIKRTSKLISPCPLKNAALEEANKFNSNNPFFIVNIAP 365
FR + N+ T K T +SK S LK A E+ NS + F+ +
Sbjct: 128 IISTFRMSLRILLNLSTEKLCGTENVSSKKKS---LKREA--ESAPDNSLDSCFVATVTG 182
Query: 366 DKNRDYRPRVPKLFIRKHFNDIQQSEQTVILQCEKKLCPVKL--TCYPKKGPSRLSKGWV 423
+ +PK F N + Q V++ E + + L Y G + +GW
Sbjct: 183 SNLKRDTLYIPKEFALS--NGLMNKYQIVLMNEEGESWKIDLRREAY-NYGRFYMRRGWR 239
Query: 424 EFAKKSKLEVGDACVFELI-NKEDPVLDV 451
F + + GD F+L+ N+E P++ +
Sbjct: 240 SFCIANGKKPGDVFAFKLVKNEETPMIQL 268
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 825,279,239
Number of Sequences: 2790947
Number of extensions: 36779265
Number of successful extensions: 91190
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 57
Number of HSP's that attempted gapping in prelim test: 90814
Number of HSP's gapped (non-prelim): 360
length of query: 457
length of database: 848,049,833
effective HSP length: 131
effective length of query: 326
effective length of database: 482,435,776
effective search space: 157274062976
effective search space used: 157274062976
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0040.4


