

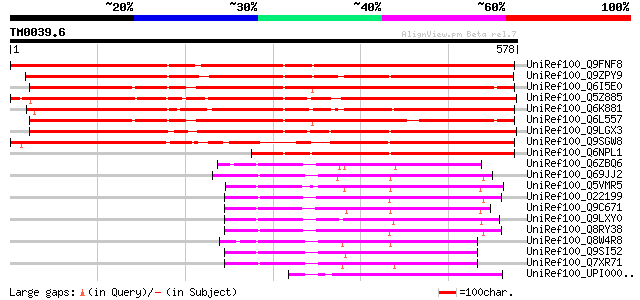
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0039.6
(578 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FNF8 Ubiquitin [Arabidopsis thaliana] 714 0.0
UniRef100_Q9ZPY9 Expressed protein [Arabidopsis thaliana] 684 0.0
UniRef100_Q6I5E0 Putative ubiquitin [Oryza sativa] 618 e-175
UniRef100_Q5Z885 Putative ubiquitin [Oryza sativa] 591 e-167
UniRef100_Q6K881 Phosphatidylinositol 3-and 4-kinase-like [Oryza... 591 e-167
UniRef100_Q6L557 Putative ubiquitin [Oryza sativa] 577 e-163
UniRef100_Q9LGX3 Similar to Arabidopsis thaliana chromosome 2 BA... 550 e-155
UniRef100_Q9SGW8 F1N19.4 [Arabidopsis thaliana] 449 e-125
UniRef100_Q6NPL1 At1g64460 [Arabidopsis thaliana] 412 e-113
UniRef100_Q6ZBQ6 Phosphatidylinositol 3-and 4-kinase family-like... 228 3e-58
UniRef100_Q69JJ2 Phosphatidylinositol 3-and 4-kinase family-like... 213 9e-54
UniRef100_Q5VMR5 Phosphatidylinositol 3-and 4-kinase family-like... 213 2e-53
UniRef100_O22199 Hypothetical protein At2g40850 [Arabidopsis tha... 211 4e-53
UniRef100_Q9C671 Hypothetical protein F28B23.7 [Arabidopsis thal... 209 1e-52
UniRef100_Q9LXY0 Hypothetical protein T5P19_250 [Arabidopsis tha... 208 3e-52
UniRef100_Q8RY38 At2g40850/T20B5.5 [Arabidopsis thaliana] 208 4e-52
UniRef100_Q8W4R8 At1g13640/F21F23_7 [Arabidopsis thaliana] 207 5e-52
UniRef100_Q9SI52 Expressed protein [Arabidopsis thaliana] 207 7e-52
UniRef100_Q7XR71 OSJNBa0043A12.14 protein [Oryza sativa] 207 7e-52
UniRef100_UPI00002EE927 UPI00002EE927 UniRef100 entry 197 7e-49
>UniRef100_Q9FNF8 Ubiquitin [Arabidopsis thaliana]
Length = 574
Score = 714 bits (1843), Expect = 0.0
Identities = 360/577 (62%), Positives = 447/577 (77%), Gaps = 11/577 (1%)
Query: 1 MACLALRPMHEEA--FGDFTSCFSHHLRDSILIYLTVGGSVVPMHIMETDSIASMKLRIQ 58
+A +AL P EE F F +L D IL++LT+ GSV+P +ME+DSIAS+KLRIQ
Sbjct: 3 VASVALSPALEELVNFPGIIGRFGFNLDDPILVFLTIAGSVIPKRVMESDSIASVKLRIQ 62
Query: 59 TFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFG 118
+ KGFFV+KQKL+++G+E++R+ +RDYG++DG +LHLV+RLSDLQAI+VRT GKEF
Sbjct: 63 SIKGFFVKKQKLLYDGREVSRNDSQIRDYGLADGKLLHLVIRLSDLQAISVRTVDGKEFE 122
Query: 119 FYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEGEALEDQRLIEDICKDNDAVIHFLVR 178
V + RNVGYVKQQIA K + L D EL +GE L+DQRLI D+C++ D VIH L+
Sbjct: 123 LVVERSRNVGYVKQQIASKEKELGIPRDHELTLDGEELDDQRLITDLCQNGDNVIHLLIS 182
Query: 179 TSDAKVTTKPVGKDFELSIEAYTVPSFSAGQLGSVSVTNRALKRNELTREFLLEPIVVNP 238
S AKV KPVGKDFE+ IE G+ G + + +EF +EP +VNP
Sbjct: 183 KS-AKVRAKPVGKDFEVFIEDVNHKHNVDGRRG------KNISSEAKPKEFFVEPFIVNP 235
Query: 239 KIKIPQVIQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAI 298
+IK+P +++ELI T EGLEKG PI+SS+GSGGAY MQD G KYVSVFKPIDEEPMA+
Sbjct: 236 EIKLPILLKELISSTLEGLEKGNGPIRSSDGSGGAYFMQDPSGHKYVSVFKPIDEEPMAV 295
Query: 299 NNPRGLPLSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTV 358
NNP G P+SVDGEG LKKGT+VG+GA+REVAAYILD+P GPR++ + D+ GFAGVPPT
Sbjct: 296 NNPHGQPVSVDGEG-LKKGTQVGEGAIREVAAYILDYPMTGPRTFPH-DQTGFAGVPPTT 353
Query: 359 MVKCLHKGFHHPEGYNSASDNVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRL 418
MVKCLHK F+HP GY+ + +N K+GSLQMF+ N+GSCEDMG VFPV++VHKISVLDIRL
Sbjct: 354 MVKCLHKDFNHPNGYSFSPENTKIGSLQMFVSNVGSCEDMGYRVFPVDQVHKISVLDIRL 413
Query: 419 ANADRHAGNILVAKDGENGSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIEY 478
ANADRHAGNILV++DG++G VL PIDHGYC P FEDCTF+WLYWPQAKEPYS +T+EY
Sbjct: 414 ANADRHAGNILVSRDGKDGQMVLTPIDHGYCFPNKFEDCTFEWLYWPQAKEPYSSETLEY 473
Query: 479 IKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLKKK 538
IKSLD E+DI+LL+FHGWE+PP C R+LRISTMLLKKG+ KGL PFTIGSIMCRETLK++
Sbjct: 474 IKSLDPEKDIELLRFHGWEIPPSCTRVLRISTMLLKKGSAKGLTPFTIGSIMCRETLKEE 533
Query: 539 SVIEQIIEEAEEGALPGTSEASFLDLVSVIMDSHLEE 575
SVIEQII +AE T+E F+ VS IMD+ L++
Sbjct: 534 SVIEQIIHDAEAIVPTETTEDEFISTVSAIMDNRLDQ 570
>UniRef100_Q9ZPY9 Expressed protein [Arabidopsis thaliana]
Length = 566
Score = 684 bits (1765), Expect = 0.0
Identities = 343/556 (61%), Positives = 433/556 (77%), Gaps = 19/556 (3%)
Query: 19 SCFSHHLRDSILIYLTVGGSVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELA 78
SC + D+I+IYLT+ GSV+PM ++E+DSI S+KLRIQ+++GF VR QKLVF G+ELA
Sbjct: 25 SCLDSYPDDTIMIYLTLPGSVIPMRVLESDSIESVKLRIQSYRGFVVRNQKLVFGGRELA 84
Query: 79 RDKLCVRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKG 138
R +RDYGVS+GN+LHLVL+LSDLQ + V+T CGK F+V + RN+GYVK+QI++K
Sbjct: 85 RSNSNMRDYGVSEGNILHLVLKLSDLQVLDVKTTCGKHCRFHVERGRNIGYVKKQISKKR 144
Query: 139 QGLLDLADQELVWEGEALEDQRLIEDICKDNDAVIHFLVRTSDAKVTTKPVGKDFELSIE 198
+D +QE+++EGE LEDQ LI DIC+++D+V+H LVR S AKV KPV K+FELSI
Sbjct: 145 GDFVDPDEQEILYEGEKLEDQSLINDICRNDDSVLHLLVRRS-AKVRVKPVEKNFELSIV 203
Query: 199 AYTVPSFSAGQLGSVSVTNRALKRNELTREFLLEPIVVNPKIKIPQVIQELIKITSEGLE 258
A + S+ ++ LEP+VVN K K+P V++++I+ S+GL+
Sbjct: 204 APQAKDKKGREAKSIVPP----------KKLSLEPVVVNSKAKVPLVVKDMIQSASDGLK 253
Query: 259 KGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLPLSVDGEGGLKKGT 318
G P++SSEG+GGAY MQ G K+V VFKPIDEEPMA NNP+GLPLS +GEG LKKGT
Sbjct: 254 SGNSPVRSSEGTGGAYFMQGPSGNKFVGVFKPIDEEPMAENNPQGLPLSPNGEG-LKKGT 312
Query: 319 RVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHKGFHHPEGYNSASD 378
+VG+GALREVAAYILDHPK G +S +E GFAGVPPT M++CLH GF+HP+G +
Sbjct: 313 KVGEGALREVAAYILDHPKSGNKSMFG-EEIGFAGVPPTAMIECLHPGFNHPKGIKT--- 368
Query: 379 NVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLANADRHAGNILVAKDGENGS 438
K+GSLQMF +N GSCEDMGP FPVEEVHKISVLDIRLANADRH GNIL+ KD E+G
Sbjct: 369 --KIGSLQMFTENDGSCEDMGPLSFPVEEVHKISVLDIRLANADRHGGNILMTKD-ESGK 425
Query: 439 TVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIEYIKSLDAEEDIKLLKFHGWEL 498
VL+PIDHGYCLPESFEDCTF+WLYWPQA++PYS +T EYI+SLDAEEDI LLKFHGW++
Sbjct: 426 LVLVPIDHGYCLPESFEDCTFEWLYWPQARKPYSAETQEYIRSLDAEEDIDLLKFHGWKM 485
Query: 499 PPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLKKKSVIEQIIEEAEEGALPGTSE 558
P E A+ LRISTMLLKKG E+GL F IG+IMCRETL KKS++E+++EEA+E LPGTSE
Sbjct: 486 PAETAQTLRISTMLLKKGVERGLTAFEIGTIMCRETLSKKSLVEEMVEEAQEAVLPGTSE 545
Query: 559 ASFLDLVSVIMDSHLE 574
A+FL+ +S +MD HL+
Sbjct: 546 AAFLEALSDVMDYHLD 561
>UniRef100_Q6I5E0 Putative ubiquitin [Oryza sativa]
Length = 586
Score = 618 bits (1594), Expect = e-175
Identities = 322/557 (57%), Positives = 396/557 (70%), Gaps = 21/557 (3%)
Query: 23 HHLRDSILIYLTVGG-SVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDK 81
H +SILIYLT G S++PM +M +DSIAS+KLR+QT KGF VRKQKLVF+G+ELAR+
Sbjct: 34 HCAAESILIYLTAPGLSMMPMRVMASDSIASVKLRVQTSKGFVVRKQKLVFDGRELARND 93
Query: 82 LCVRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGL 141
+ DYGVS GNVLHLV+R+SDL+ ITV+T G +F F V R VGYVKQQIA+
Sbjct: 94 SRIMDYGVSHGNVLHLVIRISDLRLITVQTVHGNKFRFRVEPGRTVGYVKQQIAKNSTH- 152
Query: 142 LDLADQELVWEGEALEDQRLIEDICKDNDAVIHFLVRTSDAKVTTKPVGKDFELSIEAYT 201
D LV +GE L+D LI D+C+ + AVIH LV S AK+ +PV +DFE+SI A
Sbjct: 153 -DDDHHSLVLQGEVLDDAHLIHDVCRTDGAVIHLLVHRS-AKLAARPVDRDFEVSIVARN 210
Query: 202 VPSFSAGQLGSVSVTNRALKRNELTREFLLEPIVVNPKIKIPQVIQELIKITSEGLEKGG 261
+ + A L R+F +EP++VNPK +P VI+ L+ G+EKG
Sbjct: 211 ----------RNAAADAAQPTLHLQRDFAIEPVIVNPKAALPPVIENLVGAVLAGMEKGN 260
Query: 262 KPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLPLSVDGEGGLKKGTRVG 321
PI SSEG+GGAY MQD+ G ++V+VFKP+DEEPMA NNPRGLP S GEG LKKGTRVG
Sbjct: 261 APIMSSEGTGGAYFMQDASGQEHVAVFKPVDEEPMAANNPRGLPPSPTGEG-LKKGTRVG 319
Query: 322 QGALREVAAYILDHPKKGPRSY---HNSDEKGFAGVPPTVMVKCLHKGFHHPEGYNSASD 378
+GA+REVAAYILDHP G RS+ H S GFAGV PT +V+C+H+ F P
Sbjct: 320 EGAIREVAAYILDHPPGGRRSFAGHHGSATVGFAGVAPTALVRCMHRSFKQPAASEQGPP 379
Query: 379 NVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLANADRHAGNILVAKDGENGS 438
K+GSLQ F+KN GSCEDMGP FPV EVHKI VLDIRLANADRHAGNIL +D +
Sbjct: 380 LFKVGSLQAFVKNSGSCEDMGPRAFPVHEVHKICVLDIRLANADRHAGNILTCRDEQGHG 439
Query: 439 TVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIEYIKSLDAEEDIKLLKFHGWEL 498
L+PIDHGYCLPESFEDCTF+WLYWPQ +EP+S +T+EYI+SLDAEEDI +L+FHGWE+
Sbjct: 440 LTLVPIDHGYCLPESFEDCTFEWLYWPQCREPFSEETVEYIRSLDAEEDIAILRFHGWEM 499
Query: 499 PPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLKKKSVIEQIIEEAEEGALPGTSE 558
P +C R+LR++TMLLKKG + GL F +GSI+CRETL K+SVIE+II E E+ E
Sbjct: 500 PAKCERVLRVTTMLLKKGVDSGLAAFDMGSILCRETLTKESVIEEIIREVEDDV---GDE 556
Query: 559 ASFLDLVSVIMDSHLEE 575
A+FL VS MD L E
Sbjct: 557 AAFLQSVSQSMDRRLGE 573
>UniRef100_Q5Z885 Putative ubiquitin [Oryza sativa]
Length = 568
Score = 591 bits (1524), Expect = e-167
Identities = 320/588 (54%), Positives = 406/588 (68%), Gaps = 36/588 (6%)
Query: 2 ACLALRPMHEEAFGDFTSCFS----------HHLRDSILIYLTVGGSV-VPMHIMETDSI 50
A +A+ PM EE F CF L+DSI+I+L V G+ +PM ++ ++SI
Sbjct: 4 AAIAISPMVEE-FALLPICFDGSRSPHCLSGSQLQDSIIIFLAVPGAPPMPMSVLGSESI 62
Query: 51 ASMKLRIQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGNVLHLVLRLSDLQAITVR 110
AS+KLRIQ FKGF V KQ+LV +G ELAR+ V+DYG++DGNVLHLV+RL+DL+ I +
Sbjct: 63 ASVKLRIQRFKGFVVNKQRLVLDGHELARNNCHVKDYGLADGNVLHLVIRLADLRLINIE 122
Query: 111 TECGKEFGFYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEGEALEDQRLIEDICKDND 170
T GK+F F V + RNV Y+K ++A +G L D +L +G+ LED +LI DI K +D
Sbjct: 123 TTSGKKFQFQVDQSRNVKYLKSKLAVEGDEDLG-EDHKLECDGKELEDHQLIADISKKDD 181
Query: 171 AVIHFLVRTSDAKVTTKPVGKDFELSIEAYTVPSFSAGQLGSVSVTNRALKRNELTREFL 230
AVIH +R AK+ T+ V KD +++ P N A R L
Sbjct: 182 AVIHLFIR-KPAKLRTQQVDKDTVVTV---VTPQEKENLQNEAHAVNPAKPAG--ARPAL 235
Query: 231 LEPIVVNPKIKIPQVIQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKP 290
+EPI+VN K+K+ + +I GLE G P+ S+EGSGG Y MQD+ G K ++VFKP
Sbjct: 236 VEPIIVNHKVKLSLEVMRMISSAIAGLENGYLPVMSAEGSGGVYFMQDASGEKNIAVFKP 295
Query: 291 IDEEPMAINNPRGLPLSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEK- 349
DEEPMA NNPRGLP+S DGEG +K+GT VG+GA REVAAYILDHP H S+E+
Sbjct: 296 RDEEPMAKNNPRGLPVSTDGEG-MKRGTLVGEGAFREVAAYILDHPIGD----HESEERI 350
Query: 350 GFAGVPPTVMVKCLHKGFHHPEGYNSASDNVKLGSLQMFMKNIGSCEDMGPSVFPVEEVH 409
GF+GVPPT +V+ LH+G + K+GSLQMF++N GSCEDMGP FPV+EVH
Sbjct: 351 GFSGVPPTALVRSLHRG-----------KSFKIGSLQMFIQNNGSCEDMGPRAFPVKEVH 399
Query: 410 KISVLDIRLANADRHAGNILVAKDGENGSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKE 469
KI+VLD+RLANADRHAGNILV KD E G+ L+PIDHGYCLPE FEDCTF+WLYWPQA+E
Sbjct: 400 KIAVLDLRLANADRHAGNILVCKDEEGGNYKLVPIDHGYCLPEKFEDCTFEWLYWPQARE 459
Query: 470 PYSPDTIEYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSI 529
P+S +TI YIKSLDAEEDIKLLKFHGWEL CAR+L ISTMLLKKGA +GL P+ IG I
Sbjct: 460 PFSDETIAYIKSLDAEEDIKLLKFHGWELSARCARVLCISTMLLKKGAARGLTPYDIGRI 519
Query: 530 MCRETLKKKSVIEQIIEEAEEGALPGTSEASFLDLVSVIMDSHLEEPF 577
+CRET+ + S IE I++EAE LPG+SE FL+ VS I+D HL++ F
Sbjct: 520 LCRETVNRDSEIEDIVQEAEGHVLPGSSEVIFLETVSEIIDRHLDKKF 567
>UniRef100_Q6K881 Phosphatidylinositol 3-and 4-kinase-like [Oryza sativa]
Length = 565
Score = 591 bits (1524), Expect = e-167
Identities = 320/564 (56%), Positives = 402/564 (70%), Gaps = 31/564 (5%)
Query: 20 CFSHHLR------DSILIYLTVGG-SVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVF 72
CF+ H DSI+I+L + G + +PM ++ +DS+AS+KLRIQ FKGF KQ+LVF
Sbjct: 17 CFAPHGEPRLQQLDSIVIFLAMPGVAPMPMRVLHSDSVASVKLRIQQFKGFVTTKQRLVF 76
Query: 73 EGKELARDKLCVRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQ 132
G EL+ + VRDYG++DGNVLHLV+RL+DL+AI++ T GK+F F V NVGY+K
Sbjct: 77 SGHELSLNNSHVRDYGLTDGNVLHLVVRLADLRAISIETANGKKFQFQVESCCNVGYLKD 136
Query: 133 QI-ARKGQGLLDLADQELVWEGEALEDQRLIEDICKDNDAVIHFLVRTSDAKVTTKPVGK 191
++ A GQ L L DQ LV++GE LED +LI DI K AVIH +R AKV T+ K
Sbjct: 137 KLSAESGQQLGSLKDQRLVFDGEELEDNQLIADISKKGAAVIHLFIRRP-AKVQTQQGDK 195
Query: 192 DFELSIEAYTVPSFSAGQLGSVSVTNRALKRNELTREFLLEPIVVNPKIKIPQVIQELIK 251
E + T Q ++++ A + +EPI+ N K+K+ + E+I
Sbjct: 196 --ETVVTVVTPKDNDNLQTDALNLAKPAKGKPAP-----VEPIIANGKVKLSPAVMEMIY 248
Query: 252 ITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLPLSVDGE 311
T G+E G P+ S+EGSGG Y M+DS G V+VFKPIDEEPMA NNPRGLPLS DGE
Sbjct: 249 STISGIENGYLPVMSTEGSGGVYFMKDSSGESNVAVFKPIDEEPMAKNNPRGLPLSTDGE 308
Query: 312 GGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHKGFHHPE 371
G LK+GTRVG+GALREVAAYILDHP G +S D GF+GVPPT +V+C H G +
Sbjct: 309 G-LKRGTRVGEGALREVAAYILDHPVYGCKS---CDVPGFSGVPPTALVRCFHMG----K 360
Query: 372 GYNSASDNVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLANADRHAGNILVA 431
G N K+GSLQ+F+ N GSCEDMGP FPV+EV KI++LDIRLANADRHAGNILV
Sbjct: 361 GSN------KVGSLQLFVDNNGSCEDMGPRAFPVKEVQKIAILDIRLANADRHAGNILVC 414
Query: 432 KDGENGSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIEYIKSLDAEEDIKLL 491
+DGE+ LIPIDHGYCLPE FEDCTF+WLYWPQA+EP+ P+T YI SLDA++DI LL
Sbjct: 415 QDGED-HLKLIPIDHGYCLPEKFEDCTFEWLYWPQAREPFGPETAAYIGSLDADKDIALL 473
Query: 492 KFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLKKKSVIEQIIEEAEEG 551
KFHGW L P+CAR+LRISTMLLKKGAE+GL P+ IGSI+CR+T+KK+S IE IIEEAE+
Sbjct: 474 KFHGWALSPQCARVLRISTMLLKKGAERGLTPYDIGSILCRQTVKKESEIEAIIEEAEDA 533
Query: 552 ALPGTSEASFLDLVSVIMDSHLEE 575
LPGTSE +FL+ +S IMD HL++
Sbjct: 534 ILPGTSEETFLETISEIMDFHLDK 557
>UniRef100_Q6L557 Putative ubiquitin [Oryza sativa]
Length = 612
Score = 577 bits (1486), Expect = e-163
Identities = 310/557 (55%), Positives = 383/557 (68%), Gaps = 34/557 (6%)
Query: 23 HHLRDSILIYLTVGG-SVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDK 81
H +SILIYLT G S++PM +M +DSIAS+KLR+QT KGF VRKQKLVF+G+ELAR+
Sbjct: 34 HCAAESILIYLTAPGLSMMPMRVMASDSIASVKLRVQTSKGFVVRKQKLVFDGRELARND 93
Query: 82 LCVRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGL 141
+ DYGVS GNVLHLV+R+SDL+ ITV+T G +F F V R VGYVKQQIA+
Sbjct: 94 SRIMDYGVSHGNVLHLVIRISDLRLITVQTVHGNKFRFRVEPGRTVGYVKQQIAKNSTH- 152
Query: 142 LDLADQELVWEGEALEDQRLIEDICKDNDAVIHFLVRTSDAKVTTKPVGKDFELSIEAYT 201
D LV +GE L+D LI D+C+ + AVIH LV S AK+ +PV +DFE+SI A
Sbjct: 153 -DDDHHSLVLQGEVLDDAHLIHDVCRTDGAVIHLLVHRS-AKLAARPVDRDFEVSIVARN 210
Query: 202 VPSFSAGQLGSVSVTNRALKRNELTREFLLEPIVVNPKIKIPQVIQELIKITSEGLEKGG 261
+ + A L R+F +EP++VNPK +P VI+ L+ G+EKG
Sbjct: 211 ----------RNAAADAAQPTLHLQRDFAIEPVIVNPKAALPPVIENLVGAVLAGMEKGN 260
Query: 262 KPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLPLSVDGEGGLKKGTRVG 321
PI SSEG+GGAY MQD+ G ++V+VFKP+DEEPMA NNPRGLP S GEG LKKGTRVG
Sbjct: 261 APIMSSEGTGGAYFMQDASGQEHVAVFKPVDEEPMAANNPRGLPPSPTGEG-LKKGTRVG 319
Query: 322 QGALREVAAYILDHPKKGPRSY---HNSDEKGFAGVPPTVMVKCLHKGFHHPEGYNSASD 378
+GA+REVAAYILDHP G RS+ H S GFAGV PT +V+C+H+ F P
Sbjct: 320 EGAIREVAAYILDHPPGGRRSFAGHHGSATVGFAGVAPTALVRCMHRSFKQPAASEQGPP 379
Query: 379 NVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLANADRHAGNILVAKDGENGS 438
K+GSLQ F+KN GSCEDMGP FPV EVHKI VLDIRLANADRHAGNIL +D +
Sbjct: 380 LFKVGSLQAFVKNSGSCEDMGPRAFPVHEVHKICVLDIRLANADRHAGNILTCRDEQGHG 439
Query: 439 TVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIEYIKSLDAEEDIKLLKFHGWEL 498
L+PIDHGYCLPES +EP+S +T+EYI+SLDAEEDI +L+FHGWE+
Sbjct: 440 LTLVPIDHGYCLPES-------------CREPFSEETVEYIRSLDAEEDIAILRFHGWEM 486
Query: 499 PPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLKKKSVIEQIIEEAEEGALPGTSE 558
P +C R+LR++TMLLKKG + GL F +GSI+CRETL K+SVIE+II E E+ E
Sbjct: 487 PAKCERVLRVTTMLLKKGVDSGLAAFDMGSILCRETLTKESVIEEIIREVEDDV---GDE 543
Query: 559 ASFLDLVSVIMDSHLEE 575
A+FL VS MD L E
Sbjct: 544 AAFLQSVSQSMDRRLGE 560
>UniRef100_Q9LGX3 Similar to Arabidopsis thaliana chromosome 2 BAC clone F13A10
[Oryza sativa]
Length = 588
Score = 550 bits (1418), Expect = e-155
Identities = 289/556 (51%), Positives = 380/556 (67%), Gaps = 21/556 (3%)
Query: 23 HHLRDSILIYLTVGG-SVVPMHIMETDSIASMKLRIQTFKGFFVRKQKLVFEGKELARDK 81
HH + L+YL V G + M ++E+DS+A++KLRIQ KGF R Q+LVFEG+EL+R+
Sbjct: 52 HHTPEVFLLYLAVPGVPLAKMQVLESDSVAAVKLRIQNSKGFVARNQRLVFEGRELSRND 111
Query: 82 LCVRDYGVSDGNVLHLVLRLSDLQAITVRTECGKEFGFYVGKKRNVGYVKQQIARKGQGL 141
+RDYGV G+VLHLV+RLSD + VRT G++F F V ++RN Y+KQ+I+R +
Sbjct: 112 SHIRDYGVRYGSVLHLVIRLSDPRRTAVRTVYGRKFKFQVDQRRNARYMKQEISRNVESP 171
Query: 142 LDLADQELVWEGEALEDQRLIEDICKDNDAVIHFLVRTSDAKVTTKPVGKDFELSIEAYT 201
+ + + GE L++ LI IC+ N + FL S+ G + E S E +
Sbjct: 172 NGIGESMTLVNGEKLDESTLISTICETNTSDTDFLANKSE-----NFNGNEIEESFEQLS 226
Query: 202 VPSFSAGQLGSVSVTNRALKRNELTREFLLEPIVVNPKIKIPQVIQELIKITSEGLEKGG 261
+ S + N + + L+EP++VNP + + I +I+ T GLE
Sbjct: 227 ISS---------DIGNNLQFDDAKEKYPLIEPVLVNPSVTLTPKITGMIEATLAGLEMEH 277
Query: 262 KPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLPLSVDGEGGLKKGTRVG 321
P+ SSEG+GG Y M DS G +YV+VFKPI+EEPMA +NP G PLS DGEG LK+GTRVG
Sbjct: 278 TPVMSSEGTGGVYFMLDSSGQEYVAVFKPINEEPMAKDNPNGYPLSSDGEG-LKRGTRVG 336
Query: 322 QGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHKGFHHPEGYNSASDNVK 381
+GA REVAAYILDHP G Y SDE GFAGVPPTV+V+CL+ G+ Y+ A K
Sbjct: 337 EGAFREVAAYILDHPISG---YRVSDELGFAGVPPTVLVRCLN-GYVDQTKYDCAEKEPK 392
Query: 382 LGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLANADRHAGNILVAKDGENGSTVL 441
+GSLQMF+KN GSCE+ GP FPV+EVHKI+VLD+RLAN DRH GNIL+ KD ENG L
Sbjct: 393 IGSLQMFVKNSGSCEEFGPRAFPVQEVHKIAVLDMRLANTDRHGGNILIRKD-ENGQIEL 451
Query: 442 IPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIEYIKSLDAEEDIKLLKFHGWELPPE 501
IPIDHGYCLPESFEDCTFDWLYWPQA++P++ +T++YIKSLD EEDIKLLK +G E +
Sbjct: 452 IPIDHGYCLPESFEDCTFDWLYWPQARQPFNVETLDYIKSLDEEEDIKLLKLNGCEPSSK 511
Query: 502 CARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLKKKSVIEQIIEEAEEGALPGTSEASF 561
C R+ R+STM+LKKGA +GL P+ IG+++CRE + KS IE+I+EEAE LPG E +F
Sbjct: 512 CVRVFRLSTMMLKKGAVRGLTPYEIGNMLCRENITTKSKIEEIVEEAEHVVLPGIGEKAF 571
Query: 562 LDLVSVIMDSHLEEPF 577
++ +S IMD +L E F
Sbjct: 572 MEAISGIMDRYLNELF 587
>UniRef100_Q9SGW8 F1N19.4 [Arabidopsis thaliana]
Length = 505
Score = 449 bits (1156), Expect = e-125
Identities = 262/581 (45%), Positives = 356/581 (61%), Gaps = 87/581 (14%)
Query: 1 MACLALRPMHEE---AFGDF-TSCFSHHLRDSILIYLTVGGSVVPMHIMETDSIASMKLR 56
+A +AL P+H A G F S +H+ S+L++L+V GS +PM I+E+DSIA +KLR
Sbjct: 3 VADVALSPIHRGSAFAVGGFGQSTTTHYSVKSVLVFLSVSGSTMPMLILESDSIAEVKLR 62
Query: 57 IQTFKGFFVRKQKLVFEGKELARDKLCVRDYGVSDGNVLHLVLRLSDLQAITVRTECGKE 116
IQT GF VR+QKLVF G+ELAR+ V+DYGV+ G+VLHLVL+L D +TV T CGK
Sbjct: 63 IQTCNGFRVRRQKLVFSGRELARNASRVKDYGVTGGSVLHLVLKLYDPLLVTVITTCGKV 122
Query: 117 FGFYVGKKRNVGYVKQQIARKGQGLLDLADQELVWEGEALEDQRLIEDICKDNDAVIHFL 176
F F+V ++RNVGY+K++I+++G+G ++ DQE++++GE L+D R+I+ ICKD ++VIH L
Sbjct: 123 FQFHVDRRRNVGYLKKRISKEGKGFPEVDDQEILFKGEKLDDNRIIDGICKDGNSVIHLL 182
Query: 177 VRTSDAKVTTKPVGKDFELSIEAYTVPSFSAGQLGSVSVTNRALKRNELTREFLLEPIVV 236
V+ K V ++ + + + S+G L VSV + EP+ V
Sbjct: 183 VK----KSVEDTVKREEDTATGGTYLMQDSSG-LNYVSV----------FKPMDEEPMAV 227
Query: 237 NPKIKIPQVIQELIKITSEGLEKGGKPIQSSEGSGGAYLM-QDSLGLKYVSVFKPIDEEP 295
N ++P + +GL++G + + + AYL+ GL+ VS
Sbjct: 228 NNPQQLP------VSSDGQGLKRGTRVGEGATREVAAYLLDHPKSGLRSVS--------- 272
Query: 296 MAINNPRGLPLSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVP 355
+EV + P RS H
Sbjct: 273 ------------------------------KEVMGFAGVPPTAMVRSSH----------- 291
Query: 356 PTVMVKCLHKGFHHPEGYNS-ASDNVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVL 414
K +++P G++S A+ + K+GSLQMFMKN GSCED+GP FPVEEVHKI V
Sbjct: 292 ---------KVYNYPNGFSSCATKDAKVGSLQMFMKNNGSCEDIGPGAFPVEEVHKICVF 342
Query: 415 DIRLANADRHAGNILVAKDGENGSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPD 474
DIR+ANADRHAGNIL K E G T+LIPIDHGYCLPE+FEDCTF+WLYWPQAK P+S D
Sbjct: 343 DIRMANADRHAGNILTGK-SEEGKTLLIPIDHGYCLPENFEDCTFEWLYWPQAKLPFSAD 401
Query: 475 TIEYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRET 534
TI+YI SLD+E+DI LL+ HGW +P +R LRISTMLLKKG E+ L P+ IGS+MCRET
Sbjct: 402 TIDYINSLDSEQDIALLQLHGWNVPEAVSRTLRISTMLLKKGVERNLTPYQIGSVMCRET 461
Query: 535 LKKKSVIEQIIEEAEEGALPGTSEASFLDLVSVIMDSHLEE 575
+ K S IE+I+ EA LP +SEA+FL+ VSV MD L+E
Sbjct: 462 VNKDSAIEEIVREAHNSVLPASSEATFLEAVSVAMDRRLDE 502
>UniRef100_Q6NPL1 At1g64460 [Arabidopsis thaliana]
Length = 301
Score = 412 bits (1059), Expect = e-113
Identities = 201/301 (66%), Positives = 242/301 (79%), Gaps = 4/301 (1%)
Query: 276 MQDSLGLKYVSVFKPIDEEPMAINNPRGLPLSVDGEGGLKKGTRVGQGALREVAAYILDH 335
MQDS GL YVSVFKP+DEEPMA+NNP+ LP+S DG+G LK+GTRVG+GA REVAAY+LDH
Sbjct: 1 MQDSSGLNYVSVFKPMDEEPMAVNNPQQLPVSSDGQG-LKRGTRVGEGATREVAAYLLDH 59
Query: 336 PKKGPRSYHNSDEKGFAGVPPTVMVKCLHKGFHHPEGYNS-ASDNVKLGSLQMFMKNIGS 394
PK G RS + + GFAGVPPT MV+ HK +++P G++S A+ + K+GSLQMFMKN GS
Sbjct: 60 PKSGLRSV-SKEVMGFAGVPPTAMVRSSHKVYNYPNGFSSCATKDAKVGSLQMFMKNNGS 118
Query: 395 CEDMGPSVFPVEEVHKISVLDIRLANADRHAGNILVAKDGENGSTVLIPIDHGYCLPESF 454
CED+GP FPVEEVHKI V DIR+ANADRHAGNIL K E G T+LIPIDHGYCLPE+F
Sbjct: 119 CEDIGPGAFPVEEVHKICVFDIRMANADRHAGNILTGKS-EEGKTLLIPIDHGYCLPENF 177
Query: 455 EDCTFDWLYWPQAKEPYSPDTIEYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLK 514
EDCTF+WLYWPQAK P+S DTI+YI SLD+E+DI LL+ HGW +P +R LRISTMLLK
Sbjct: 178 EDCTFEWLYWPQAKLPFSADTIDYINSLDSEQDIALLQLHGWNVPEAVSRTLRISTMLLK 237
Query: 515 KGAEKGLPPFTIGSIMCRETLKKKSVIEQIIEEAEEGALPGTSEASFLDLVSVIMDSHLE 574
KG E+ L P+ IGS+MCRET+ K S IE+I+ EA LP +SEA+FL+ VSV MD L+
Sbjct: 238 KGVERNLTPYQIGSVMCRETVNKDSAIEEIVREAHNSVLPASSEATFLEAVSVAMDRRLD 297
Query: 575 E 575
E
Sbjct: 298 E 298
>UniRef100_Q6ZBQ6 Phosphatidylinositol 3-and 4-kinase family-like [Oryza sativa]
Length = 633
Score = 228 bits (582), Expect = 3e-58
Identities = 133/315 (42%), Positives = 182/315 (57%), Gaps = 31/315 (9%)
Query: 237 NPKIKIPQVIQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPM 296
+P ++ Q++ ++++ G+EKG +P+ S G GGAY +D G ++ ++ KP DEEP
Sbjct: 129 SPSARMKQLVDDIVR----GIEKGIEPVAISSGMGGAYYFRDMWG-EHAAIVKPTDEEPF 183
Query: 297 AINNPRGLPLSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPP 356
NNP+G G GLKK RVG+ REVAAY+LDH K FA VPP
Sbjct: 184 GPNNPKGFVGKSLGLPGLKKSVRVGETGSREVAAYLLDH-------------KNFANVPP 230
Query: 357 TVMVKCLHKGFHHPEGYN---SASDNV-----KLGSLQMFMKNIGSCEDMGPSVFPVEEV 408
T++VK H FH EG + +SDN KL SLQ F+ + D G S FPV V
Sbjct: 231 TMLVKITHSVFHMNEGVDYKTKSSDNKTQAFSKLASLQEFIPHDYDASDHGTSSFPVSAV 290
Query: 409 HKISVLDIRLANADRHAGNILVAKDGENGS-----TVLIPIDHGYCLPESFEDCTFDWLY 463
H+I +LDIR+ N DRHAGNILV K + S T LIPIDHG CLPES ED F+W++
Sbjct: 291 HRIGILDIRIFNTDRHAGNILVRKLYNDASRFETQTELIPIDHGLCLPESLEDPYFEWIH 350
Query: 464 WPQAKEPYSPDTIEYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPP 523
WPQA P+S + +EYI +LD +D ++L+ + R+L +ST LK+ A G
Sbjct: 351 WPQASIPFSEEDLEYITNLDPIKDAEMLRMELHTIHEASLRVLVLSTTFLKEAAACGFCL 410
Query: 524 FTIGSIMCRETLKKK 538
IG +M R+ +K+
Sbjct: 411 SEIGEMMSRQFTRKE 425
>UniRef100_Q69JJ2 Phosphatidylinositol 3-and 4-kinase family-like [Oryza sativa]
Length = 660
Score = 213 bits (543), Expect = 9e-54
Identities = 131/335 (39%), Positives = 182/335 (54%), Gaps = 30/335 (8%)
Query: 232 EPIVVNPKIKIPQVIQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPI 291
EPI + + +++L K E + G P+ + G GGAY ++ G + V++ KP
Sbjct: 132 EPIEILGCLSPSSRMKQLAKDVVEAIRNGVDPVPVNSGMGGAYYFKNIYGER-VAIVKPT 190
Query: 292 DEEPMAINNPRGLPLSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGF 351
DEEP A NNP+G G GLK+ VG+ LREVAAY+LDH F
Sbjct: 191 DEEPFAPNNPKGFVGKTLGLPGLKRSVPVGETGLREVAAYLLDHDN-------------F 237
Query: 352 AGVPPTVMVKCLHKGFHHPEG--------YNSASDNVKLGSLQMFMKNIGSCEDMGPSVF 403
A VPPT++VK H F+ + +N KL SLQ F+ + D G S F
Sbjct: 238 ANVPPTMLVKITHSVFNVNDTVSCKSKVFHNKLQAVSKLASLQQFIAHDYDASDHGTSSF 297
Query: 404 PVEEVHKISVLDIRLANADRHAGNILVAK-----DGENGSTVLIPIDHGYCLPESFEDCT 458
PV VH+I +LDIR+ N DRHAGN+LV K D T LIPIDHG CLPES ED
Sbjct: 298 PVSAVHRIGILDIRIFNTDRHAGNLLVRKLGPGPDNFGVQTELIPIDHGLCLPESLEDPY 357
Query: 459 FDWLYWPQAKEPYSPDTIEYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAE 518
F+W++WPQA P++ + +EYI +LD +D ++L+ + C R+L +ST+ LK+ A
Sbjct: 358 FEWIHWPQASIPFTEEELEYIANLDPVKDAEMLRLELPFIRGACLRVLVLSTIFLKEAAA 417
Query: 519 KGLPPFTIGSIMCRETLKKK---SVIEQIIEEAEE 550
GL IG +M R+ K+ S +E + EA +
Sbjct: 418 FGLCLSEIGEMMSRQFTGKEEEPSELELLCMEARK 452
>UniRef100_Q5VMR5 Phosphatidylinositol 3-and 4-kinase family-like [Oryza sativa]
Length = 700
Score = 213 bits (541), Expect = 2e-53
Identities = 132/333 (39%), Positives = 184/333 (54%), Gaps = 30/333 (9%)
Query: 247 QELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLPL 306
++L+K + + G PI + G GGAY ++S G + ++ KP DEEP A NNP+G
Sbjct: 198 KQLVKDVARAIRNGVDPIPVNSGLGGAYYFRNSKG-ENAAIVKPNDEEPFAPNNPKGFTG 256
Query: 307 SVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHKG 366
G+ GLK+ RVG+ REVAAY+LD Y NS A VPPTV+VK H
Sbjct: 257 KALGQPGLKRSVRVGETGFREVAAYLLD--------YDNS-----ANVPPTVLVKISHPV 303
Query: 367 FHHPEGYNSASDNV---------KLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIR 417
F+ E +SA+ K+ S Q F+ + D G S FPV VH+I +LDIR
Sbjct: 304 FNVNECVSSANMKASKDYPGAVSKIASFQQFIPHDFDASDHGTSSFPVSAVHRIGILDIR 363
Query: 418 LANADRHAGNILVAK---DGENGS-TVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSP 473
+ N DRHAGN+LV K G+ G+ T LIPIDHG CLPE ED F+W++WPQA P+S
Sbjct: 364 IFNTDRHAGNLLVRKLTGPGKFGNQTELIPIDHGLCLPECLEDPYFEWIHWPQASIPFSD 423
Query: 474 DTIEYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRE 533
D ++YI +LD +D +L+ + C R+L +ST+ LK+ GL IG +M RE
Sbjct: 424 DELDYIANLDPMKDADMLRMELPMIHEACLRVLILSTIFLKEATSFGLCLAEIGEMMSRE 483
Query: 534 TL---KKKSVIEQIIEEAEEGALPGTSEASFLD 563
+ S +E + EA A+ ++ +D
Sbjct: 484 FTGMEDQPSELEVVCMEARRLAIEREESSTEID 516
>UniRef100_O22199 Hypothetical protein At2g40850 [Arabidopsis thaliana]
Length = 561
Score = 211 bits (538), Expect = 4e-53
Identities = 122/324 (37%), Positives = 182/324 (55%), Gaps = 23/324 (7%)
Query: 246 IQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLP 305
++ L+ + + G +P+ G GGAYL+Q G ++V KP+DEEP+A NNP+
Sbjct: 107 VRALVAEVTMAMVSGAQPLLLPSGMGGAYLLQTGKGHN-IAVAKPVDEEPLAFNNPKKSG 165
Query: 306 LSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHK 365
+ G+ G+K VG+ +RE+AAY+LD+ +GF+GVPPT +V H
Sbjct: 166 NLMLGQPGMKHSIPVGETGIRELAAYLLDY-------------QGFSGVPPTALVSISHV 212
Query: 366 GFHHPEGYNSASDNVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLANADRHA 425
FH + ++ +S K+ SLQ F+ + ++GP F VH+I +LD+RL N DRHA
Sbjct: 213 PFHVSDAFSFSSMPYKVASLQRFVGHDFDAGELGPGSFTATSVHRIGILDVRLLNLDRHA 272
Query: 426 GNILVA----KDGEN--GSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIEYI 479
GN+LV K+ N G+ L+PIDHG CLPE +D F+WL WPQA P+S ++YI
Sbjct: 273 GNMLVKRCDKKEAYNRLGTAELVPIDHGLCLPECLDDPYFEWLNWPQALVPFSDTELDYI 332
Query: 480 KSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLKKK- 538
+LD +D +LL+ LP R+L + T+ LK+ A GL IG M R+ K +
Sbjct: 333 SNLDPFKDAELLRTELHSLPESAIRVLVVCTVFLKQAAAAGLCLAEIGEKMTRDFSKGEE 392
Query: 539 --SVIEQIIEEAEEGALPGTSEAS 560
S++E + +A+ TSE S
Sbjct: 393 SFSLLETLCTKAKASVFGKTSEDS 416
>UniRef100_Q9C671 Hypothetical protein F28B23.7 [Arabidopsis thaliana]
Length = 630
Score = 209 bits (533), Expect = 1e-52
Identities = 123/314 (39%), Positives = 180/314 (57%), Gaps = 25/314 (7%)
Query: 246 IQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLP 305
++++ K ++ ++KG P+ + G GGAY ++S G + V++ KP DEEP A NNP+G
Sbjct: 148 VRQMAKDITKAVKKGIDPVAVNSGLGGAYYFKNSRG-ESVAIVKPTDEEPYAPNNPKGFV 206
Query: 306 LSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHK 365
G+ GLK+ RVG+ REVAAY+LD ++ FA VPPT +VK H
Sbjct: 207 GKALGQPGLKRSVRVGETGYREVAAYLLD-------------KEHFANVPPTALVKITHS 253
Query: 366 GFHHPEGYNSASDNVKL-----GSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLAN 420
F+ +G ++ K+ SLQ F+ + + G S FPV VH+I +LDIR+ N
Sbjct: 254 IFNVNDGVKASKPMEKMLVSKIASLQQFIPHDYDASEHGTSNFPVSAVHRIGILDIRILN 313
Query: 421 ADRHAGNILVAK---DGENGSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIE 477
DRH+GN+LV K DG G L+PIDHG CLPE+ ED F+W++WPQA P+S D ++
Sbjct: 314 TDRHSGNLLVKKLDGDGMFGQVELVPIDHGLCLPETLEDPYFEWIHWPQASIPFSEDELK 373
Query: 478 YIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETL-- 535
YI +LD D ++L+ + R+L + T+ LK+ A GL IG +M RE
Sbjct: 374 YIANLDPLGDCEMLRRELPMVREASLRVLVLCTIFLKEAAANGLCLAEIGEMMTREVRPG 433
Query: 536 -KKKSVIEQIIEEA 548
++ S IE + EA
Sbjct: 434 DEEPSEIEVVCLEA 447
>UniRef100_Q9LXY0 Hypothetical protein T5P19_250 [Arabidopsis thaliana]
Length = 533
Score = 208 bits (530), Expect = 3e-52
Identities = 118/325 (36%), Positives = 181/325 (55%), Gaps = 29/325 (8%)
Query: 246 IQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLP 305
++ L+ + + G +P+ G GGAYL+Q G ++V KP+DEEP+A NNP+G
Sbjct: 87 VRSLVAEVTIAIVSGAQPLLLPSGLGGAYLLQTEKG-NNIAVAKPVDEEPLAFNNPKGSG 145
Query: 306 LSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHK 365
G+ G+K+ RVG+ +RE+AAY+LDH +GF+ VPPT +V+ H
Sbjct: 146 GLTLGQPGMKRSIRVGESGIRELAAYLLDH-------------QGFSSVPPTALVRISHV 192
Query: 366 GFHHPEGYNSASDNVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLANADRHA 425
FH ++A K+ SLQ F+ + ++GP F V VH+I +LD+R+ N DRHA
Sbjct: 193 PFHDRGSDHAA---YKVASLQRFVGHDFDAGELGPGSFTVVSVHRIGILDVRVLNLDRHA 249
Query: 426 GNILVAKDGEN---------GSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTI 476
GN+LV K + G+ L+PIDHG CLPE +D F+WL WPQA P++ +
Sbjct: 250 GNMLVKKIHDQDETTCSNGVGAAELVPIDHGLCLPECLDDPYFEWLNWPQASVPFTDIEL 309
Query: 477 EYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLK 536
+YI +LD +D +LL+ + R+L + T+ LK+ A GL IG M R+ +
Sbjct: 310 QYISNLDPFKDAELLRTELDSIQESSLRVLIVCTIFLKEAAAAGLSLAEIGEKMTRDICR 369
Query: 537 ---KKSVIEQIIEEAEEGALPGTSE 558
SV+E + +A+ A+ G+ +
Sbjct: 370 GEESSSVLEILCNKAKASAVSGSDD 394
>UniRef100_Q8RY38 At2g40850/T20B5.5 [Arabidopsis thaliana]
Length = 560
Score = 208 bits (529), Expect = 4e-52
Identities = 120/324 (37%), Positives = 181/324 (55%), Gaps = 23/324 (7%)
Query: 246 IQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLP 305
++ L+ + + G +P+ G GGAYL+Q G ++V KP+DEEP+A NNP+
Sbjct: 107 VRALVAEVTMAMVSGAQPLLLPSGMGGAYLLQTGKGHN-IAVAKPVDEEPLAFNNPKKSG 165
Query: 306 LSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHK 365
+ G+ G+K VG+ +RE+AAY+LD+ +GF+GVPPT +V H
Sbjct: 166 NLMLGQPGMKHSIPVGETGIRELAAYLLDY-------------QGFSGVPPTALVSISHV 212
Query: 366 GFHHPEGYNSASDNVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLANADRHA 425
FH + ++ +S K+ S Q F+ + ++GP F VH+I +LD+RL N DRHA
Sbjct: 213 PFHVSDAFSFSSMPYKVASSQRFVGHDFDAGELGPGSFTATSVHRIGILDVRLLNLDRHA 272
Query: 426 GNILVA----KDGEN--GSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIEYI 479
GN+LV K+ N G+ L+PIDHG CLPE +D F+WL WPQA P+S ++YI
Sbjct: 273 GNMLVKRCDKKEAYNRLGTAELVPIDHGLCLPECLDDPYFEWLNWPQALVPFSDTELDYI 332
Query: 480 KSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLKKK- 538
+LD +D +LL+ LP ++L + T+ LK+ A GL IG M R+ K +
Sbjct: 333 SNLDPFKDAELLRTELHSLPESAIKVLVVCTVFLKQAAAAGLCLAEIGEKMTRDFSKGEE 392
Query: 539 --SVIEQIIEEAEEGALPGTSEAS 560
S++E + +A+ TSE S
Sbjct: 393 SFSLLETLCTKAKASVFGKTSEDS 416
>UniRef100_Q8W4R8 At1g13640/F21F23_7 [Arabidopsis thaliana]
Length = 622
Score = 207 bits (528), Expect = 5e-52
Identities = 121/306 (39%), Positives = 168/306 (54%), Gaps = 30/306 (9%)
Query: 240 IKIPQVIQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAIN 299
+ + Q +++K G+E PI + G GGAY +D G + V++ KP DEEP A N
Sbjct: 142 LSLKQTANDIVKAMKMGVE----PIPVNGGLGGAYYFRDEKG-QSVAIVKPTDEEPFAPN 196
Query: 300 NPRGLPLSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVM 359
NP+G G+ GLK RVG+ REVAAY+LD+ FA VPPT +
Sbjct: 197 NPKGFVGKALGQPGLKPSVRVGETGFREVAAYLLDYDH-------------FANVPPTAL 243
Query: 360 VKCLHKGFHHPEGYNSASD-------NVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKIS 412
VK H F+ +G + + K+ S Q F+ + D G S FPV VH+I
Sbjct: 244 VKITHSVFNVNDGMDGNKSREKKKLVSSKIASFQKFVPHDFDASDHGTSSFPVASVHRIG 303
Query: 413 VLDIRLANADRHAGNILVAK-----DGENGSTVLIPIDHGYCLPESFEDCTFDWLYWPQA 467
+LDIR+ N DRH GN+LV K G G LIPIDHG CLPE+ ED F+W++WPQA
Sbjct: 304 ILDIRILNTDRHGGNLLVKKLDDGGVGRFGQVELIPIDHGLCLPETLEDPYFEWIHWPQA 363
Query: 468 KEPYSPDTIEYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIG 527
P+S + ++YI+SLD +D ++L+ + C R+L + T+ LK+ A GL IG
Sbjct: 364 SIPFSEEELDYIQSLDPVKDCEMLRRELPMIREACLRVLVLCTVFLKEAAVFGLCLAEIG 423
Query: 528 SIMCRE 533
+M RE
Sbjct: 424 EMMTRE 429
>UniRef100_Q9SI52 Expressed protein [Arabidopsis thaliana]
Length = 650
Score = 207 bits (527), Expect = 7e-52
Identities = 117/297 (39%), Positives = 167/297 (55%), Gaps = 22/297 (7%)
Query: 245 VIQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGL 304
+++ ++K + ++ G +P+ G GGAY ++ G + V++ KP DEEP A NNP+G
Sbjct: 151 IVKHMVKDIVKAMKMGVEPLPVHSGLGGAYYFRNKRG-ESVAIVKPTDEEPFAPNNPKGF 209
Query: 305 PLSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLH 364
G+ GLK RVG+ REVAAY+LD+ + FA VPPT +VK H
Sbjct: 210 VGKALGQPGLKSSVRVGETGFREVAAYLLDYGR-------------FANVPPTALVKITH 256
Query: 365 KGFHHPEGYNSASDNVK-----LGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLA 419
F+ +G K + S Q F+ + D G S FPV VH+I +LDIR+
Sbjct: 257 SVFNVNDGVKGNKPREKKLVSKIASFQKFVAHDFDASDHGTSSFPVTSVHRIGILDIRIF 316
Query: 420 NADRHAGNILVAK-DGEN--GSTVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTI 476
N DRH GN+LV K DG G LIPIDHG CLPE+ ED F+W++WPQA P+S + +
Sbjct: 317 NTDRHGGNLLVKKLDGVGMFGQVELIPIDHGLCLPETLEDPYFEWIHWPQASLPFSDEEV 376
Query: 477 EYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRE 533
+YI+SLD +D +L+ + C R+L + T+ LK+ + GL IG +M RE
Sbjct: 377 DYIQSLDPVKDCDMLRRELPMIREACLRVLVLCTIFLKEASAYGLCLAEIGEMMTRE 433
>UniRef100_Q7XR71 OSJNBa0043A12.14 protein [Oryza sativa]
Length = 605
Score = 207 bits (527), Expect = 7e-52
Identities = 121/301 (40%), Positives = 163/301 (53%), Gaps = 27/301 (8%)
Query: 246 IQELIKITSEGLEKGGKPIQSSEGSGGAYLMQDSLGLKYVSVFKPIDEEPMAINNPRGLP 305
++ L+ + G P+ G GG+Y ++ G + V++ KP DEEP A NNP+G
Sbjct: 134 VKRLVDDVVTAIRSGVDPVPIGSGLGGSYYFRNISGDR-VAIVKPTDEEPFAPNNPKGFV 192
Query: 306 LSVDGEGGLKKGTRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHK 365
G+ GLKK RVG+ REVAAY+LDH FA VPPT +VK H
Sbjct: 193 GRALGQPGLKKSVRVGETGFREVAAYLLDHDN-------------FANVPPTALVKITHS 239
Query: 366 GFHHPEGYNSASD--------NVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIR 417
FH N S + K+ S Q F+ + D G S FPV VH+I +LDIR
Sbjct: 240 IFHINCPVNGGSPAHDQKQQVSSKIASFQQFIAHDFDASDHGTSSFPVAAVHRIGILDIR 299
Query: 418 LANADRHAGNILVAK-DGENG----STVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYS 472
+ N DRHAGN+LV K DG G T L PIDHG CLPE+ ED F+W++W Q+ P+S
Sbjct: 300 IFNTDRHAGNVLVRKLDGGTGRFGCQTELFPIDHGLCLPENLEDPYFEWIHWAQSSIPFS 359
Query: 473 PDTIEYIKSLDAEEDIKLLKFHGWELPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCR 532
+ +EYIK+LD D+ +L+ + C R+L + T+ LK+ A GL IG +M R
Sbjct: 360 EEELEYIKNLDPMRDVAMLRRELPIIREACLRVLVLCTIFLKEAAASGLCLAEIGEMMTR 419
Query: 533 E 533
E
Sbjct: 420 E 420
>UniRef100_UPI00002EE927 UPI00002EE927 UniRef100 entry
Length = 271
Score = 197 bits (501), Expect = 7e-49
Identities = 109/246 (44%), Positives = 146/246 (59%), Gaps = 21/246 (8%)
Query: 318 TRVGQGALREVAAYILDHPKKGPRSYHNSDEKGFAGVPPTVMVKCLHKGFHHPEGYNSAS 377
T+ G+GA REVAAY+LDH FAGVP T +V H +G
Sbjct: 1 TKAGEGATREVAAYLLDHGS-------------FAGVPATSLV-------HLTDGNGQDD 40
Query: 378 DNVKLGSLQMFMKNIGSCEDMGPSVFPVEEVHKISVLDIRLANADRHAGNILVAKDGENG 437
D+ KLGSLQ +++N E GPS+FP +EVH+I+VLDIRLAN DR+AGNIL D E
Sbjct: 41 DDGKLGSLQEYVENTAEAEGYGPSMFPTDEVHRITVLDIRLANTDRNAGNILCRSDEEGK 100
Query: 438 STVLIPIDHGYCLPESFEDCTFDWLYWPQAKEPYSPDTIEYIKSLDAEEDIKLLKFHGWE 497
LIPIDHGY LP + ED F+W +WPQA PY D EY++ LDA+ DI+ L+ + E
Sbjct: 101 VVSLIPIDHGYALPHTLEDVCFEWEFWPQASIPYGEDVKEYVELLDADADIEYLRENDIE 160
Query: 498 LPPECARLLRISTMLLKKGAEKGLPPFTIGSIMCRETLKKKSVIEQIIEEAEEGAL-PGT 556
L R+LR+ T+LLK+ ++ I S++ R+ + +KS IE+I +A A+ G
Sbjct: 161 LQASSERVLRVCTLLLKESLKRDFTAANIASMLSRKMINRKSDIEKIAAQAASTAIAEGR 220
Query: 557 SEASFL 562
SE L
Sbjct: 221 SERGLL 226
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.138 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 991,350,826
Number of Sequences: 2790947
Number of extensions: 43758065
Number of successful extensions: 98679
Number of sequences better than 10.0: 647
Number of HSP's better than 10.0 without gapping: 65
Number of HSP's successfully gapped in prelim test: 582
Number of HSP's that attempted gapping in prelim test: 96499
Number of HSP's gapped (non-prelim): 1453
length of query: 578
length of database: 848,049,833
effective HSP length: 133
effective length of query: 445
effective length of database: 476,853,882
effective search space: 212199977490
effective search space used: 212199977490
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0039.6


