

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
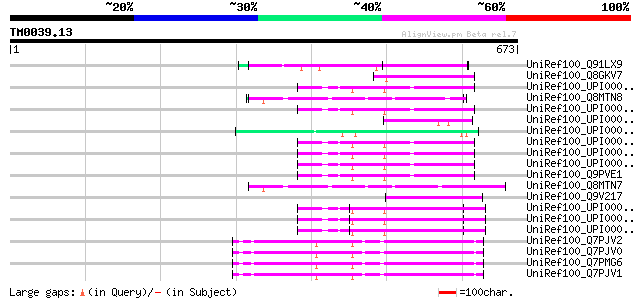
Query= TM0039.13
(673 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q91LX9 ORF73 [Human herpesvirus 8] 66 3e-09
UniRef100_Q8GKV7 M protein precursor [Streptococcus pyogenes] 59 5e-07
UniRef100_UPI0000438933 UPI0000438933 UniRef100 entry 57 1e-06
UniRef100_Q8MTN8 Glutamic acid-rich protein cNBL1500 [Trichinell... 57 2e-06
UniRef100_UPI0000439A62 UPI0000439A62 UniRef100 entry 55 5e-06
UniRef100_UPI000049A5A8 UPI000049A5A8 UniRef100 entry 55 5e-06
UniRef100_UPI0000432993 UPI0000432993 UniRef100 entry 55 9e-06
UniRef100_UPI0000437483 UPI0000437483 UniRef100 entry 54 1e-05
UniRef100_UPI0000437482 UPI0000437482 UniRef100 entry 54 1e-05
UniRef100_UPI0000437480 UPI0000437480 UniRef100 entry 54 1e-05
UniRef100_Q9PVE1 Ventricular myosin heavy chain [Brachydanio rerio] 54 1e-05
UniRef100_Q8MTN7 Glutamic acid-rich protein cNBL1700 [Trichinell... 54 1e-05
UniRef100_Q9V217 Hypothetical protein [Pyrococcus abyssi] 54 1e-05
UniRef100_UPI000036153E UPI000036153E UniRef100 entry 54 2e-05
UniRef100_UPI000036153D UPI000036153D UniRef100 entry 54 2e-05
UniRef100_UPI000036153C UPI000036153C UniRef100 entry 54 2e-05
UniRef100_Q7PJV2 ENSANGP00000024129 [Anopheles gambiae str. PEST] 54 2e-05
UniRef100_Q7PJV0 ENSANGP00000024621 [Anopheles gambiae str. PEST] 54 2e-05
UniRef100_Q7PMG6 ENSANGP00000012555 [Anopheles gambiae str. PEST] 54 2e-05
UniRef100_Q7PJV1 ENSANGP00000023782 [Anopheles gambiae str. PEST] 54 2e-05
>UniRef100_Q91LX9 ORF73 [Human herpesvirus 8]
Length = 1003
Score = 66.2 bits (160), Expect = 3e-09
Identities = 60/297 (20%), Positives = 127/297 (42%), Gaps = 6/297 (2%)
Query: 317 EDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVGGTSGSAKNVSLEADHGSDP 376
E+ + DEE + E + + EE + + + ED+ E G G+ K +S+++
Sbjct: 375 EEDEEEDEEEEDEEEEEEDEEDDDDEDNEDEEEDKKEDEEDGGDGN-KTLSIQSSQQQQE 433
Query: 377 VQGKGRSVSE----EKDTRGPASKKRRVE-DHQDHADGEVQGSEPERAEPVTVVPPSPER 431
Q + E E + P + ++ E Q+ E Q EP++ EP P +
Sbjct: 434 PQQQEPQQQEPQQQEPQQQEPLQEPQQQEPQQQEPQQQEPQQQEPQQQEPQQQEPQQQDE 493
Query: 432 LSIWGPRAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGS 491
+ ++ Q D+ + Q+ ++ + + + + Q
Sbjct: 494 QQQDEQQQDEQQQDEQQQDEQEQQDEQQQDEQQQDEQQQQDEQEQQEEQEQQEEQEQQEE 553
Query: 492 TIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAV 551
ELEE ++ E+ +Q + +ELEE +EE + L+ + E E+ ++++++ +
Sbjct: 554 QEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQE 613
Query: 552 REDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKL 608
E+ E L+ E + E L+E E ++ ++ ++ELE + LE +++L
Sbjct: 614 LEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQEL 670
Score = 52.4 bits (124), Expect = 4e-05
Identities = 64/324 (19%), Positives = 126/324 (38%), Gaps = 26/324 (8%)
Query: 304 ADIDRTLLDALRVEDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVGGTSGSA 363
+ +D+ D E+ Q TDEE D+E D EE E+ + D ED +
Sbjct: 319 SQVDKDDNDNKDDEEEQETDEE-DEEDDEEDDEEDDEEDDEEDDEED----DEEDDEEDD 373
Query: 364 KNVSLEADHGSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQDHADG-------------E 410
+ E D + + + ++ D ++ + ED +D DG E
Sbjct: 374 EEEDEEEDEEEEDEEEEEEDEEDDDDEDNEDEEEDKKEDEEDGGDGNKTLSIQSSQQQQE 433
Query: 411 VQGSEPERAEPVTVVPPSPERLSIWGPRAGNCREHLAQIDDLVLTENDQKYLGDREPAGL 470
Q EP++ EP P E L P+ ++ Q + E Q+ +EP
Sbjct: 434 PQQQEPQQQEPQQQEPQQQEPLQ--EPQQQEPQQQEPQQQEPQQQEPQQQEPQQQEPQQQ 491
Query: 471 LNYVLRASLKTASAV------RYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAI 524
+ Q + E+ +++ ++ D+ +E +E
Sbjct: 492 DEQQQDEQQQDEQQQDEQQQDEQEQQDEQQQDEQQQDEQQQQDEQEQQEEQEQQEEQEQQ 551
Query: 525 EELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKER 584
EE + L+ + E E+ ++++++ + E+ E L+ E + E L+E E ++
Sbjct: 552 EEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQE 611
Query: 585 EQSSAVKKELENRVSSLEADKKKL 608
++ ++ELE + LE +++L
Sbjct: 612 QELEEQEQELEEQEQELEEQEQEL 635
Score = 50.1 bits (118), Expect = 2e-04
Identities = 28/113 (24%), Positives = 60/113 (52%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
ELEE ++ E+ +Q + +ELEE +EE + L+ + E E+ ++++++ + E+
Sbjct: 585 ELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEE 644
Query: 555 LEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKK 607
E L+ E + E L+E E ++ ++ ++ELE + LE +++
Sbjct: 645 QEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQE 697
Score = 47.8 bits (112), Expect = 0.001
Identities = 27/111 (24%), Positives = 58/111 (51%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
ELEE ++ E+ +Q + +ELEE +EE + L+ + E E+ ++++++ + E+
Sbjct: 592 ELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEE 651
Query: 555 LEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADK 605
E L+ E + E L+E E ++ ++ ++ELE + E ++
Sbjct: 652 QEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQEQELEE 702
Score = 46.2 bits (108), Expect = 0.003
Identities = 29/114 (25%), Positives = 60/114 (52%), Gaps = 1/114 (0%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
ELEE ++ E+ +Q + +ELEE +EE + L+ + E E+ ++++++ + E+
Sbjct: 606 ELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEE 665
Query: 555 LEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKL 608
E L+ E + E L+E E L+E+EQ +++ E E + +++
Sbjct: 666 QEQELEEQEQELEEQEQELEEQEQE-LEEQEQEQELEEVEEQEQEQEEQELEEV 718
Score = 44.3 bits (103), Expect = 0.012
Identities = 26/101 (25%), Positives = 53/101 (51%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
ELEE ++ E+ +Q + +ELEE +EE + L+ + E E+ ++++++ + E+
Sbjct: 634 ELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEE 693
Query: 555 LEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELE 595
E + E + + +E E +E+EQ ++ELE
Sbjct: 694 QEQEQELEEVEEQEQEQEEQELEEVEEQEQEQEEQEEQELE 734
Score = 43.9 bits (102), Expect = 0.016
Identities = 28/113 (24%), Positives = 59/113 (51%), Gaps = 1/113 (0%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
ELEE ++ E+ +Q + +ELEE +EE + L+ + E E+ ++++++ + E+
Sbjct: 620 ELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEE 679
Query: 555 LEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKK 607
E L+ E + E + +E E +E+EQ +E+E + E +++
Sbjct: 680 QEQELEEQEQELEE-QEQEQELEEVEEQEQEQEEQELEEVEEQEQEQEEQEEQ 731
Score = 43.5 bits (101), Expect = 0.020
Identities = 50/290 (17%), Positives = 112/290 (38%), Gaps = 13/290 (4%)
Query: 320 QSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVGGTSGSAKNVSLEADHGSDPVQG 379
Q ++ +Q+ D Q +E + Q Q + E + + + E + + Q
Sbjct: 486 QEPQQQDEQQQDEQQQDEQQQDEQQQDEQEQQDEQQQDEQQQDEQQQQDEQEQQEEQEQQ 545
Query: 380 KGRSVSEEKDTRGPASKKRRVEDHQD--HADGEVQGSEPERAEPVTVVPPSPERLSIWGP 437
+ + EE++ ++ E Q+ + E++ E E E + + L
Sbjct: 546 EEQEQQEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEE--- 602
Query: 438 RAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIPELE 497
+E + + L E +Q+ + L + ELE
Sbjct: 603 -----QEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQELE 657
Query: 498 ELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEG 557
E ++ E+ +Q + +ELEE +EE + L+ + E E + + Q+ + ++LE
Sbjct: 658 EQEQELEEQEQELEEQEQELEEQEQELEEQEQELEEQEQEQELEEVEEQEQEQEEQELEE 717
Query: 558 RLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKK 607
+ + + E L+E E +E ++ V+++ E + +E +++
Sbjct: 718 VEEQEQEQEEQEEQELEEVEE---QEEQELEEVEEQEEQELEEVEEQEQQ 764
>UniRef100_Q8GKV7 M protein precursor [Streptococcus pyogenes]
Length = 227
Score = 58.9 bits (141), Expect = 5e-07
Identities = 41/146 (28%), Positives = 76/146 (51%), Gaps = 11/146 (7%)
Query: 483 SAVRYLQGSTIPELEE-----------LREKTEKDDQLISSMSKELEEGCSAIEELSKAL 531
SAVR + + +LEE L+ EK ++ I+S+++E+EE E+L++ +
Sbjct: 22 SAVRITRNTPRQKLEEEYHRLDTENHTLKHDKEKLEKNITSLTQEIEENKLKTEKLTQEI 81
Query: 532 DNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVK 591
+ KL+ E+L ++++D A LE LK AE K + L+ E+ +++ ++
Sbjct: 82 EENKLKTEELTQEIEDKRAKLSKLESDLKTAEEKVQHSKEYLELVESGHADYHKRTESLI 141
Query: 592 KELENRVSSLEADKKKLAATVSKINK 617
KE V L ++ LA TV+K ++
Sbjct: 142 KEKTMEVEKLTSEINTLAQTVNKADQ 167
>UniRef100_UPI0000438933 UPI0000438933 UniRef100 entry
Length = 1910
Score = 57.4 bits (137), Expect = 1e-06
Identities = 64/252 (25%), Positives = 106/252 (41%), Gaps = 27/252 (10%)
Query: 382 RSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRAGN 441
R EE+ +KKR++ED ++ E +T+ E+ + + N
Sbjct: 931 RLEDEEEMNAELVAKKRKLEDECSELKKDIDDLE------LTLAKVEKEKHATEN-KVKN 983
Query: 442 CREHLAQIDDLV---------LTENDQKYLGD-REPAGLLNYVLRASLKTASAVRYLQGS 491
E +A +D+++ L E Q+ L D + +N + +A K V L+GS
Sbjct: 984 LTEEMAALDEIIAKLTKEKKALQEAHQQTLDDLQSEEDKVNTLTKAKAKLEQQVDDLEGS 1043
Query: 492 TIPE------LEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKV 545
E LE + K E D +L +LE +EE K D E QL K+
Sbjct: 1044 LEQEKKLRMDLERAKRKLEGDLKLTQESIMDLENDKQQMEEKLKKKD---FEISQLNSKI 1100
Query: 546 QDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADK 605
+D A+ L+ +LK + + E L+ AA K +Q + + +ELE + LE
Sbjct: 1101 EDEQALGAQLQKKLKELQARIEELEEELEAERAARAKVEKQRADLSRELEEIIERLEEAG 1160
Query: 606 KKLAATVSKINK 617
AA + ++NK
Sbjct: 1161 GATAAQI-EMNK 1171
Score = 39.7 bits (91), Expect = 0.29
Identities = 34/127 (26%), Positives = 59/127 (45%), Gaps = 3/127 (2%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
ELE R K ++ + +S+ELEE IE L +A + E KK+ + +R D
Sbjct: 1127 ELEAERAARAKVEKQRADLSRELEE---IIERLEEAGGATAAQIEMNKKREAEFQKLRRD 1183
Query: 555 LEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVSK 614
LE E A T + ++ A ++ + VK++LE S L+ + + + + +
Sbjct: 1184 LEEATLQHEATAATLRKKHADSVADLGEQIDNLQRVKQKLEKEKSELKLELDDVVSNMEQ 1243
Query: 615 INKASFN 621
I K+ N
Sbjct: 1244 IVKSKSN 1250
Score = 37.7 bits (86), Expect = 1.1
Identities = 43/145 (29%), Positives = 72/145 (49%), Gaps = 28/145 (19%)
Query: 500 REKTEKDD-QLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRE----- 553
++K E D+ QL + + + ++E +A E+ KA+ + + E+LKK+ QD A E
Sbjct: 1707 KKKLEGDNTQLQTEVEEAVQECRNAEEKAKKAITDAAMMAEELKKE-QDTSAHLERMKKN 1765
Query: 554 ------DLEGRLKAAEGKAETGA--------ARLKEAE-AAWLKEREQSSAVK--KELEN 596
DL+ RL AE A G AR++E E L++++ S +VK ++ E
Sbjct: 1766 MEQTIKDLQHRLDEAEQIAMKGGKKQVQKLEARVRELENEVELEQKKASESVKGIRKYER 1825
Query: 597 RVSSL----EADKKKLAATVSKINK 617
R+ L E D+K LA ++K
Sbjct: 1826 RIKELTYQTEEDRKNLARLQDLVDK 1850
Score = 35.0 bits (79), Expect = 7.2
Identities = 38/164 (23%), Positives = 71/164 (43%), Gaps = 23/164 (14%)
Query: 459 QKYLGDREPAGLLNYVLRASL--KTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKE 516
QK D E A L + A+L K A +V L G I L+ +++K EK+ S + E
Sbjct: 1178 QKLRRDLEEATLQHEATAATLRKKHADSVADL-GEQIDNLQRVKQKLEKEK---SELKLE 1233
Query: 517 LEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEA 576
L++ S +E++ K+ N+ + + LE ++ KAE G + +
Sbjct: 1234 LDDVVSNMEQIVKSKSNL--------------EKMCRTLEDQMSEYRTKAEEGQRTINDF 1279
Query: 577 EAAWLKEREQSSAVKKELENR---VSSLEADKKKLAATVSKINK 617
K + ++ + ++LE + VS L K+ + + +
Sbjct: 1280 TMQKAKLQTENGELSRQLEEKDSLVSQLTRGKQSYTQQIEDLKR 1323
>UniRef100_Q8MTN8 Glutamic acid-rich protein cNBL1500 [Trichinella spiralis]
Length = 498
Score = 57.0 bits (136), Expect = 2e-06
Identities = 60/294 (20%), Positives = 125/294 (42%), Gaps = 23/294 (7%)
Query: 317 EDGQSTDEEVDQETDVAQG----EEGHAENQPQPDPEDRGGAEVGGTSGSAKNVSLEADH 372
+D ++ E +E DV+Q EE A + + PE++ E G+S + D
Sbjct: 121 KDEDKSESEASEEKDVSQEQNSKEEKGASEEDEDTPEEQNSKEENGSSEED-----DEDA 175
Query: 373 GSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERL 432
+ + + SEEK+T K E+ ++ DG E + +E T + E
Sbjct: 176 SEEQASNEEKEASEEKNTVSEERKGASEEEDEEKDDGHESEVESQASEEQT----TEEGA 231
Query: 433 SIWGPRAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGST 492
S E ++ ++ ++ +++ G+ + + + + +AS + S
Sbjct: 232 SEEEDEESASEEQTSEGEEKGASQEEEEDEGNEQESEVES---QASEEQTSEEEESASEE 288
Query: 493 IPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVR 552
E E +E+T ++++ S+ +E EE S EE + + + + E + K++ + +
Sbjct: 289 EDEENESKEQTTEEEE--SASEEEDEESASEREEKNASQEEEEDEGNESKEQTTEEEESA 346
Query: 553 EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKK 606
+ E A+EG+ + + +E E EQ S V+ + +S E +K+
Sbjct: 347 SEEEDEESASEGEEKNASQEEEEDEG-----NEQESEVESQASEEQTSEEEEKE 395
Score = 49.3 bits (116), Expect = 4e-04
Identities = 61/290 (21%), Positives = 112/290 (38%), Gaps = 41/290 (14%)
Query: 315 RVEDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVGGTSGSAKNVSLEADHGS 374
+ + Q+T+E +E D EE +E Q E E G+ + +E+
Sbjct: 220 QASEEQTTEEGASEEED----EESASEEQTSEGEEKGASQEEEEDEGNEQESEVESQASE 275
Query: 375 DPVQGKGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSI 434
+ + S SEE+D SK++ E+ + ++ E + S ER E +
Sbjct: 276 EQTSEEEESASEEEDEENE-SKEQTTEEEESASEEEDEESASEREEKNASQEEEEDE--- 331
Query: 435 WGPRAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIP 494
+E + ++ E D++ + E K AS
Sbjct: 332 ----GNESKEQTTEEEESASEEEDEESASEGEE------------KNAS----------- 364
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
+ EE E E++ ++ S S+E E S+ D EQ ++ ++G + ED
Sbjct: 365 QEEEEDEGNEQESEVESQASEEQTSEEEEKEGASQEEDEENESEEQTSEEEEEGASEEED 424
Query: 555 LEGRL--KAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLE 602
E + +E + E GA++ +E + +E EQ S V+ + +S E
Sbjct: 425 EESAFEEQTSEEEEEKGASQEEEED----EENEQESEVESQASEEQTSEE 470
Score = 40.8 bits (94), Expect = 0.13
Identities = 58/292 (19%), Positives = 108/292 (36%), Gaps = 12/292 (4%)
Query: 320 QSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVGGTSGSAKNVSLEADHGSDPVQG 379
+ ++ VD+E ++ E G +Q + E + S S + + + +
Sbjct: 86 KESENGVDKEKPTSKEESGEKTSQEKESEEKSSQEKDEDKSESEASEEKDVSQEQNSKEE 145
Query: 380 KGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRA 439
KG S +E SK+ +D D + + E E S E+ ++ R
Sbjct: 146 KGASEEDEDTPEEQNSKEENGSSEEDDEDASEEQASNEEKE------ASEEKNTVSEERK 199
Query: 440 GNCREHLAQIDDLVLTENDQKYLGDR---EPAGLLNYVLRASLKTASAVRYLQGSTIPEL 496
G E + DD +E + + ++ E A AS + S S E
Sbjct: 200 GASEEEDEEKDDGHESEVESQASEEQTTEEGASEEEDEESASEEQTSEGEEKGASQEEEE 259
Query: 497 EELRE-KTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAV--RE 553
+E E ++E + Q + E EE S E+ E E+ + +D ++ RE
Sbjct: 260 DEGNEQESEVESQASEEQTSEEEESASEEEDEENESKEQTTEEEESASEEEDEESASERE 319
Query: 554 DLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADK 605
+ + E + + E E + +E ++ SA + E +N E D+
Sbjct: 320 EKNASQEEEEDEGNESKEQTTEEEESASEEEDEESASEGEEKNASQEEEEDE 371
>UniRef100_UPI0000439A62 UPI0000439A62 UniRef100 entry
Length = 1932
Score = 55.5 bits (132), Expect = 5e-06
Identities = 63/252 (25%), Positives = 106/252 (42%), Gaps = 27/252 (10%)
Query: 382 RSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRAGN 441
R EE+ +KKR++ED ++ E +T+ E+ + + N
Sbjct: 927 RLEDEEEMNAELTAKKRKLEDECSELKKDIDDLE------LTLAKVEKEKHATEN-KVKN 979
Query: 442 CREHLAQIDDLV---------LTENDQKYLGD-REPAGLLNYVLRASLKTASAVRYLQGS 491
E +A +D+++ L E Q+ L D + +N + +A K V L+GS
Sbjct: 980 LTEEMAALDEIIARLTKEKKALQEAHQQTLDDLQSEEDKVNTLTKAKAKLEQQVDDLEGS 1039
Query: 492 TIPE------LEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKV 545
E LE ++ K E D +L +LE +EE K D E QL ++
Sbjct: 1040 LEQEKKLRMDLERVKRKLEGDLKLTQESVMDLENDKQQLEERLKKKD---FEISQLTSRI 1096
Query: 546 QDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADK 605
+D A+ L+ +LK + + E L+ AA K +Q + + +ELE LE
Sbjct: 1097 EDEQAMATQLQKKLKELQARIEELEEELEAERAARAKVEKQRADLSRELEEISERLEEAG 1156
Query: 606 KKLAATVSKINK 617
AA + ++NK
Sbjct: 1157 GATAAQI-EMNK 1167
Score = 38.1 bits (87), Expect = 0.86
Identities = 32/127 (25%), Positives = 57/127 (44%), Gaps = 3/127 (2%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
ELE R K ++ + +S+ELEE +EE A + E KK+ + +R D
Sbjct: 1123 ELEAERAARAKVEKQRADLSRELEEISERLEEAGGAT---AAQIEMNKKREAEFQKLRRD 1179
Query: 555 LEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVSK 614
LE E A T + ++ + ++ + VK++LE S L + + + + +
Sbjct: 1180 LEEATLQHEATASTLRKKHADSVSDLGEQIDNLQRVKQKLEKEKSELRLELDDVVSNMEQ 1239
Query: 615 INKASFN 621
+ KA N
Sbjct: 1240 LAKAKAN 1246
Score = 37.0 bits (84), Expect = 1.9
Identities = 36/184 (19%), Positives = 80/184 (42%), Gaps = 3/184 (1%)
Query: 451 DLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLI 510
D VL E QKY + R+ ++ ++ LE ++ + + + I
Sbjct: 1452 DKVLAEWKQKYEESQSELESSQKEARSLSTELFKLKNSYEESLDHLESMKRENKNLQEEI 1511
Query: 511 SSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGA 570
+ +++++ E I EL K ++ E +++ +++ + E EG++ A+ +
Sbjct: 1512 ADLTEQIGESGKNIHELEKMRKQLEQEKAEIQAALEEAEGSLEHEEGKILRAQLEFSQIK 1571
Query: 571 ARLKEAEAAWLKEREQSSA-VKKELENRVSSLEAD--KKKLAATVSKINKASFNNAVSQL 627
A ++ A +E EQ+ ++ ++ SSLE++ + A + K + N QL
Sbjct: 1572 ADIERKLAEKDEEMEQAKRNQQRMIDTLQSSLESETRSRNEALRLKKKMEGDLNEMEIQL 1631
Query: 628 AVVN 631
+ N
Sbjct: 1632 SQAN 1635
Score = 36.6 bits (83), Expect = 2.5
Identities = 40/123 (32%), Positives = 65/123 (52%), Gaps = 12/123 (9%)
Query: 500 REKTEKDD-QLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGR 558
++K E D+ QL + + + ++E +A E+ KA+ + + E+LKK+ QD A LE
Sbjct: 1729 KKKLEGDNTQLQTEVEEAVQECRNAEEKAKKAITDAAMMAEELKKE-QDTSA---HLERM 1784
Query: 559 LKAAEGKAETGAARLKEAEAAWL---KEREQSSAVK-KELENRVSSLEADKKKLAATVSK 614
K E + RL EAE + K++ Q V+ +ELEN V E ++KK + +V
Sbjct: 1785 KKNMEQTIKDLQHRLDEAEQIAMKGGKKQVQKLEVRVRELENEV---ELEQKKASESVKG 1841
Query: 615 INK 617
I K
Sbjct: 1842 IRK 1844
Score = 35.8 bits (81), Expect = 4.2
Identities = 40/169 (23%), Positives = 79/169 (46%), Gaps = 13/169 (7%)
Query: 459 QKYLGDREPAGLLNYVLRASL--KTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKE 516
QK D E A L + ++L K A +V L G I L+ +++K EK+ S + E
Sbjct: 1174 QKLRRDLEEATLQHEATASTLRKKHADSVSDL-GEQIDNLQRVKQKLEKEK---SELRLE 1229
Query: 517 LEEGCSAIEELSKALDNMK-----LENEQLKKKVQDGDAVREDLEGRLKAAEGKAETG-- 569
L++ S +E+L+KA N++ LE++ + + + + R + ++ A + E G
Sbjct: 1230 LDDVVSNMEQLAKAKANLEKMCRTLEDQMSEYRTKYEEGQRSINDFTMQKARLQTENGEL 1289
Query: 570 AARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVSKINKA 618
+L+E ++ + + +++E+ LE + K A + A
Sbjct: 1290 TRQLEEKDSLVSQLTRSKQSYTQQIEDLKRQLEEEVKAKNALAHAVQSA 1338
Score = 35.0 bits (79), Expect = 7.2
Identities = 39/169 (23%), Positives = 72/169 (42%), Gaps = 28/169 (16%)
Query: 440 GNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIPELEEL 499
G+ ++ Q+DD + +D L+ ++ L + + EL L
Sbjct: 1650 GHLKDAQMQLDDALRANDD----------------LKENIAIVERRNNLLQAELDELRSL 1693
Query: 500 REKTEKDDQLISSMSKELEEGCSAIEELSKALDNM--KLENEQLKKKVQDGDAVREDLEG 557
E+TE+ +L ++ E + + +L N KLE + + + + +AV+E
Sbjct: 1694 VEQTERGRKLAEQELMDVSERVQLLHSQNTSLLNQKKKLEGDNTQLQTEVEEAVQECRNA 1753
Query: 558 RLKAAEGKAETGAARLKEAEAAWLKEREQSSA----VKKELENRVSSLE 602
KA KA T AA + E LK+ + +SA +KK +E + L+
Sbjct: 1754 EEKAK--KAITDAAMMAEE----LKKEQDTSAHLERMKKNMEQTIKDLQ 1796
>UniRef100_UPI000049A5A8 UPI000049A5A8 UniRef100 entry
Length = 863
Score = 55.5 bits (132), Expect = 5e-06
Identities = 38/129 (29%), Positives = 64/129 (49%), Gaps = 11/129 (8%)
Query: 497 EELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLE 556
EEL ++ E+ + I + KE EE +EEL +N K + E+L+KKV D + +L+
Sbjct: 378 EELEKEKEEQAKKIEEIQKEKEEQTKKVEELEGEKNNEKQKVEELEKKVNDSEKENNELK 437
Query: 557 GRLKAAEGKAE-------TGAARLKEAEAAWL----KEREQSSAVKKELENRVSSLEADK 605
G+LK + K E G+ L + + + KE+E S K+L+ ++SS E +
Sbjct: 438 GQLKDLQKKLEETEKNAAAGSEELLKQKNEEIDNIKKEKEVLSKENKQLKEQISSAEENS 497
Query: 606 KKLAATVSK 614
+ K
Sbjct: 498 NSIIENEKK 506
Score = 47.0 bits (110), Expect = 0.002
Identities = 73/329 (22%), Positives = 145/329 (43%), Gaps = 53/329 (16%)
Query: 311 LDALRVEDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVGGTSGSAKNVSLEA 370
++AL+ E + ++ V +E D+ Q EE + + Q + ++ A + + + + E
Sbjct: 309 IEALKAEKDKEIEDAV-KEKDI-QIEELNKKVQEETKEKEEAKASLAISVAAEATLKAEV 366
Query: 371 DHGSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPE 430
+ ++ KG + +EK+ + A K ++ ++ +V+ E E+ V +
Sbjct: 367 EKKDQELKNKGEELEKEKEEQ--AKKIEEIQKEKEEQTKKVEELEGEKNNEKQKVEELEK 424
Query: 431 RLSIWGPRAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQG 490
+++ + N E Q+ DL QK L + E K A+A G
Sbjct: 425 KVND-SEKENN--ELKGQLKDL------QKKLEETE-------------KNAAA-----G 457
Query: 491 STIPELEELREKTE------KDDQLISSMSKELEEGCSAIEELSKAL--------DNMKL 536
S E L++K E K+ +++S +K+L+E S+ EE S ++ +++K
Sbjct: 458 SE----ELLKQKNEEIDNIKKEKEVLSKENKQLKEQISSAEENSNSIIENEKKEKEDLKH 513
Query: 537 ENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELEN 596
+NE+LK+++++ ++E+ + + K + K +E K++ SS+ +E EN
Sbjct: 514 QNEELKQQIEE---LKEENNKKERELAEKEVVIVSLQKSSEEVNKKDKSSSSSSDEE-EN 569
Query: 597 RVSSLEADKKKLAATVSKINKASFNNAVS 625
KKL SK N NN S
Sbjct: 570 EKKENGKLIKKLMIRYSKYNNKYENNIPS 598
Score = 37.7 bits (86), Expect = 1.1
Identities = 22/120 (18%), Positives = 58/120 (48%), Gaps = 1/120 (0%)
Query: 497 EELREKTEKDDQLISSMSKELEE-GCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDL 555
+ L EK +K + ++++ ++E +E+ A+ ++ E+L KKVQ+ +E+
Sbjct: 289 QALAEKKQKHHKKVAALKAQIEALKAEKDKEIEDAVKEKDIQIEELNKKVQEETKEKEEA 348
Query: 556 EGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVSKI 615
+ L + T A +++ + + E+ K+E ++ ++ +K++ V ++
Sbjct: 349 KASLAISVAAEATLKAEVEKKDQELKNKGEELEKEKEEQAKKIEEIQKEKEEQTKKVEEL 408
>UniRef100_UPI0000432993 UPI0000432993 UniRef100 entry
Length = 795
Score = 54.7 bits (130), Expect = 9e-06
Identities = 75/362 (20%), Positives = 145/362 (39%), Gaps = 42/362 (11%)
Query: 300 LRGMADIDRTLLDALRVEDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVGGT 359
LRG D + L ++ E+ DE+ +A+ + + + Q D + ++
Sbjct: 381 LRGEGDSLASELTNVKDENSALKDEKDQLNKQLAENKTENERLKKQNDELESENTKIKKE 440
Query: 360 SGSAKNVSLEADHGSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERA 419
S KN + ++ ++ + + E+ + + K R E + A+ ++Q EP+ +
Sbjct: 441 LESCKNENNNLKEENNKLKEELEKLGEQLKSLNDETNKLRRELKE--AEDKIQILEPQLS 498
Query: 420 EPVTVVPPSPERLSIWGPRAG--------------NCREHLAQIDDLVLTEN-------- 457
+ S L++ A N R L ++D VL N
Sbjct: 499 RARSENEKSQNELAVLRNEANELKVKLDREMLDNTNMRNALKILEDQVLDLNKKLDNCRE 558
Query: 458 ----------DQKYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIPELEELREKTEKDD 507
D K GD + A L N L A K +R E+++L ++
Sbjct: 559 ENDALKEENKDLKTKGDVQIASL-NSELEALKKELEKLRADNSKYKSEIDDLGKQLASAK 617
Query: 508 QLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAE 567
++ +E+ +A L LD M EN++LK++V + ED+E ++K+ E +
Sbjct: 618 NELNDCREEIVVLKNANSALRSDLDKMGTENDRLKREVDESRKKLEDMEAKVKSLENQLS 677
Query: 568 TGAARLKEAEAAWLKEREQSSAVKKELENRV---SSLEAD----KKKLAATVSKINKASF 620
+A +E + RE + ++ ELE + ++E + K++L A ++NK
Sbjct: 678 NLSAEKEELVKELYRTREDLNNLRNELEKQTGVKDTMEKESTNLKEELKALKEELNKTRD 737
Query: 621 NN 622
N
Sbjct: 738 EN 739
Score = 44.7 bits (104), Expect = 0.009
Identities = 34/148 (22%), Positives = 70/148 (46%), Gaps = 21/148 (14%)
Query: 491 STIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNM--------------KL 536
+T+ EL+++R + ++ + +ELE G I++L + +M K
Sbjct: 212 NTVNELDKMRSENADLLSELNRLKQELESGRKEIDQLKSEIGSMKDALGKCVDEIEKLKT 271
Query: 537 ENEQLKKKVQDGDAVRE-------DLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSA 589
EN+ LK +VQ ++ R+ DL+ ++ + K + +L EA+ R +
Sbjct: 272 ENKDLKSEVQGLESERDRLTNEVADLKPKISELQEKLTDASKKLDEAKTEDSDLRAEVDR 331
Query: 590 VKKELENRVSSLEADKKKLAATVSKINK 617
+KKELE+ ++ K ++ + + +NK
Sbjct: 332 LKKELESAGKEIDQLKAEMNSLKNGLNK 359
Score = 43.1 bits (100), Expect = 0.027
Identities = 32/139 (23%), Positives = 65/139 (46%), Gaps = 11/139 (7%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
+++ L ++ EKD + I + E+ A+++ ++ +K+ENE+LKK+ +A +
Sbjct: 135 KIDNLEKELEKDKKEIEQLKLEISSLKDALDKCVDEMEKLKVENEKLKKEGMKVEATWLE 194
Query: 555 LEGRLKAAEGKAETGAA-------RLKEAEAAWLKE----REQSSAVKKELENRVSSLEA 603
LKA + E A +++ A L E +++ + +KE++ S + +
Sbjct: 195 ENVNLKAKNTELEENLANTVNELDKMRSENADLLSELNRLKQELESGRKEIDQLKSEIGS 254
Query: 604 DKKKLAATVSKINKASFNN 622
K L V +I K N
Sbjct: 255 MKDALGKCVDEIEKLKTEN 273
Score = 43.1 bits (100), Expect = 0.027
Identities = 42/142 (29%), Positives = 66/142 (45%), Gaps = 26/142 (18%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
E E L+ + +K D+ + + E E L + D MKLENE LK+ V+ A+++D
Sbjct: 2 ENENLKTELDKADEQLDKLKTERNE-------LQRNFDTMKLENETLKENVK---ALKDD 51
Query: 555 LEGRLKAAEGKAETGAA-----RLKEAEAAWLKE-----REQSSAVKKE---LENRVSSL 601
LE + + G A LK+AE L++ + ++ +KKE L R S L
Sbjct: 52 LEESKREVDEMKAVGDALKDKEELKDAEFRELQQNMQNLKTENGELKKENDDLRTRSSEL 111
Query: 602 EADKKKLAATVSKINKASFNNA 623
E KL +++K NA
Sbjct: 112 E---HKLDNVKKELDKVESENA 130
Score = 42.4 bits (98), Expect = 0.045
Identities = 40/175 (22%), Positives = 87/175 (48%), Gaps = 15/175 (8%)
Query: 457 NDQKYLGD--REPAGLLNYVLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMS 514
++ K +GD ++ L + R + ++ G E ++LR ++ + + + ++
Sbjct: 60 DEMKAVGDALKDKEELKDAEFRELQQNMQNLKTENGELKKENDDLRTRSSELEHKLDNVK 119
Query: 515 KELEEGCSAIEELSKALDNMKLENEQLKKKVQ----DGDAVREDLEGRLKAAEGKAETGA 570
KEL++ S +L +DN++ E E+ KK+++ + ++++ L+ + E K +
Sbjct: 120 KELDKVESENADLRAKIDNLEKELEKDKKEIEQLKLEISSLKDALDKCVDEME-KLKVEN 178
Query: 571 ARLK----EAEAAWLKEREQSSAVKKELE----NRVSSLEADKKKLAATVSKINK 617
+LK + EA WL+E A ELE N V+ L+ + + A +S++N+
Sbjct: 179 EKLKKEGMKVEATWLEENVNLKAKNTELEENLANTVNELDKMRSENADLLSELNR 233
Score = 37.7 bits (86), Expect = 1.1
Identities = 28/123 (22%), Positives = 57/123 (45%), Gaps = 7/123 (5%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
+LEE + + ++ + ++ + E + EL + + N+K EN +LKK+ +D
Sbjct: 51 DLEESKREVDEMKAVGDALKDKEELKDAEFRELQQNMQNLKTENGELKKE-------NDD 103
Query: 555 LEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATVSK 614
L R E K + L + E+ R + ++KELE +E K ++++
Sbjct: 104 LRTRSSELEHKLDNVKKELDKVESENADLRAKIDNLEKELEKDKKEIEQLKLEISSLKDA 163
Query: 615 INK 617
++K
Sbjct: 164 LDK 166
Score = 36.2 bits (82), Expect = 3.2
Identities = 76/381 (19%), Positives = 151/381 (38%), Gaps = 41/381 (10%)
Query: 284 RCYDLCSYSVRRNAKLLRGMADIDRTLLDALRVEDGQSTDEEVDQETDVAQGEEGHAENQ 343
+C D N KL + ++ T L+ ++T+ E + V + ++ +EN
Sbjct: 166 KCVDEMEKLKVENEKLKKEGMKVEATWLEENVNLKAKNTELEENLANTVNELDKMRSENA 225
Query: 344 PQPDPEDRGGAEVGGTSGSAKNVSLEADHGSDPVQGKGRSVSE----EKDTRGPASKKRR 399
+R E+ SG + L+++ GS G+ V E + + + S+ +
Sbjct: 226 DLLSELNRLKQEL--ESGRKEIDQLKSEIGSMK-DALGKCVDEIEKLKTENKDLKSEVQG 282
Query: 400 VEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRAGNCREHL-----------AQ 448
+E +D EV +P+ +E + + ++L + R + +
Sbjct: 283 LESERDRLTNEVADLKPKISELQEKLTDASKKLDEAKTEDSDLRAEVDRLKKELESAGKE 342
Query: 449 IDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQG---STIPELEELREKTEK 505
ID L N K G + + + + + S V L+G S EL ++++
Sbjct: 343 IDQLKAEMNSLKN-GLNKCVEEMEKLTNENSELKSQVHGLRGEGDSLASELTNVKDENSA 401
Query: 506 DDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKV-----------QDGDAVRED 554
++K+L E + E L K D ++ EN ++KK++ ++ + ++E+
Sbjct: 402 LKDEKDQLNKQLAENKTENERLKKQNDELESENTKIKKELESCKNENNNLKEENNKLKEE 461
Query: 555 LE---GRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSL-----EADKK 606
LE +LK+ + LKEAE Q S + E E + L EA++
Sbjct: 462 LEKLGEQLKSLNDETNKLRRELKEAEDKIQILEPQLSRARSENEKSQNELAVLRNEANEL 521
Query: 607 KLAATVSKINKASFNNAVSQL 627
K+ ++ + NA+ L
Sbjct: 522 KVKLDREMLDNTNMRNALKIL 542
>UniRef100_UPI0000437483 UPI0000437483 UniRef100 entry
Length = 1783
Score = 54.3 bits (129), Expect = 1e-05
Identities = 64/252 (25%), Positives = 104/252 (40%), Gaps = 27/252 (10%)
Query: 382 RSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRAGN 441
R EE+ +KKR++ED ++ E +T+ E+ + + N
Sbjct: 849 RLEDEEEMNAELTAKKRKLEDECSELKKDIDDLE------LTLAKVEKEKHATEN-KVKN 901
Query: 442 CREHLAQIDDLV---------LTENDQKYLGD-REPAGLLNYVLRASLKTASAVRYLQGS 491
E +A +DD++ L E Q+ L D + +N + +A K V L+GS
Sbjct: 902 LTEEMAALDDIIAKLTKEKKALQEAHQQTLDDLQSEEDKVNTLTKAKAKLEQQVDDLEGS 961
Query: 492 TIPE------LEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKV 545
E LE + K E D +L +LE +EE K D E QL K+
Sbjct: 962 LEQEKKLRMDLERAKRKLEGDLKLTQESLMDLENDKQQLEERLKKKD---FEISQLNGKI 1018
Query: 546 QDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADK 605
+D + L+ +LK + + E L+ AA K +Q + + +ELE LE
Sbjct: 1019 EDEQTICIQLQKKLKELQARIEELEEELEAERAARAKVEKQRADLARELEEISERLEEAG 1078
Query: 606 KKLAATVSKINK 617
AA + ++NK
Sbjct: 1079 GATAAQI-EMNK 1089
Score = 45.8 bits (107), Expect = 0.004
Identities = 33/142 (23%), Positives = 71/142 (49%), Gaps = 3/142 (2%)
Query: 493 IPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVR 552
+ LE ++ + + + IS ++++L EG +I EL K ++ E +++ +++ +A
Sbjct: 1416 LDHLETMKRENKNLQEEISDLTEQLGEGGKSIHELEKMRKQLEQEKSEIQSALEEAEASL 1475
Query: 553 EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSA-VKKELENRVSSLEAD--KKKLA 609
E EG++ A+ + A ++ A +E EQS +++ ++ SSLE++ + A
Sbjct: 1476 EHEEGKILRAQLEFSQIKADIERKLAEKDEEMEQSKRNLQRTIDTLQSSLESETRSRNEA 1535
Query: 610 ATVSKINKASFNNAVSQLAVVN 631
+ K + N QL+ N
Sbjct: 1536 LRIKKKMEGDLNEMEIQLSQAN 1557
Score = 40.0 bits (92), Expect = 0.23
Identities = 37/136 (27%), Positives = 72/136 (52%), Gaps = 14/136 (10%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQD------- 547
+ EE + + E + +S EL + ++ EE L+ MK EN+ L++++ D
Sbjct: 1383 KFEESQAELESSQKEARCLSTELFKLKNSYEEALDHLETMKRENKNLQEEISDLTEQLGE 1442
Query: 548 -GDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEAD-K 605
G ++ E + R + + K+E +A L+EAEA+ E E+ ++ +LE S ++AD +
Sbjct: 1443 GGKSIHELEKMRKQLEQEKSEIQSA-LEEAEAS--LEHEEGKILRAQLE--FSQIKADIE 1497
Query: 606 KKLAATVSKINKASFN 621
+KLA ++ ++ N
Sbjct: 1498 RKLAEKDEEMEQSKRN 1513
Score = 38.5 bits (88), Expect = 0.65
Identities = 37/140 (26%), Positives = 66/140 (46%), Gaps = 5/140 (3%)
Query: 479 LKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLEN 538
L T+ V+ L L + ++ QL + + + ++E +A E+ KA+ + +
Sbjct: 1631 LDTSERVQLLHSQNTSLLNQKKKLETDISQLQTEVEEAVQECRNAEEKAKKAITDAAMMA 1690
Query: 539 EQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLK-EREQSSAVKKELENR 597
E+LKK+ QD A LE K E + RL EAE +K ++Q ++ +
Sbjct: 1691 EELKKE-QDTSA---HLERMKKNMEQTIKDLQHRLDEAEQIAMKGGKKQVQKLEARVREL 1746
Query: 598 VSSLEADKKKLAATVSKINK 617
S +E+++KK + V I K
Sbjct: 1747 ESEVESEQKKSSEAVKGIRK 1766
Score = 34.7 bits (78), Expect = 9.5
Identities = 38/150 (25%), Positives = 67/150 (44%), Gaps = 12/150 (8%)
Query: 488 LQGSTIPELEELREKTEKDDQLISSM------SKELEEGCSAIEELSKALD--NMKLENE 539
LQ E + L + E+ DQLI + +KEL E EE++ L KLE+E
Sbjct: 811 LQLQVQAEQDNLCDAEERCDQLIKNKIQLEAKAKELTERLEDEEEMNAELTAKKRKLEDE 870
Query: 540 --QLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENR 597
+LKK + D + +E A E K + + + K ++ A+++ +
Sbjct: 871 CSELKKDIDDLELTLAKVEKEKHATENKVKNLTEEMAALDDIIAKLTKEKKALQEAHQQT 930
Query: 598 VSSLEADKKKLAATVSKINKASFNNAVSQL 627
+ L++++ K+ T++K KA V L
Sbjct: 931 LDDLQSEEDKV-NTLTKA-KAKLEQQVDDL 958
>UniRef100_UPI0000437482 UPI0000437482 UniRef100 entry
Length = 1911
Score = 54.3 bits (129), Expect = 1e-05
Identities = 64/252 (25%), Positives = 104/252 (40%), Gaps = 27/252 (10%)
Query: 382 RSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRAGN 441
R EE+ +KKR++ED ++ E +T+ E+ + + N
Sbjct: 911 RLEDEEEMNAELTAKKRKLEDECSELKKDIDDLE------LTLAKVEKEKHATEN-KVKN 963
Query: 442 CREHLAQIDDLV---------LTENDQKYLGD-REPAGLLNYVLRASLKTASAVRYLQGS 491
E +A +DD++ L E Q+ L D + +N + +A K V L+GS
Sbjct: 964 LTEEMAALDDIIAKLTKEKKALQEAHQQTLDDLQSEEDKVNTLTKAKAKLEQQVDDLEGS 1023
Query: 492 TIPE------LEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKV 545
E LE + K E D +L +LE +EE K D E QL K+
Sbjct: 1024 LEQEKKLRMDLERAKRKLEGDLKLTQESLMDLENDKQQLEERLKKKD---FEISQLNGKI 1080
Query: 546 QDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADK 605
+D + L+ +LK + + E L+ AA K +Q + + +ELE LE
Sbjct: 1081 EDEQTICIQLQKKLKELQARIEELEEELEAERAARAKVEKQRADLARELEEISERLEEAG 1140
Query: 606 KKLAATVSKINK 617
AA + ++NK
Sbjct: 1141 GATAAQI-EMNK 1151
Score = 45.8 bits (107), Expect = 0.004
Identities = 33/142 (23%), Positives = 71/142 (49%), Gaps = 3/142 (2%)
Query: 493 IPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVR 552
+ LE ++ + + + IS ++++L EG +I EL K ++ E +++ +++ +A
Sbjct: 1478 LDHLETMKRENKNLQEEISDLTEQLGEGGKSIHELEKMRKQLEQEKSEIQSALEEAEASL 1537
Query: 553 EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSA-VKKELENRVSSLEAD--KKKLA 609
E EG++ A+ + A ++ A +E EQS +++ ++ SSLE++ + A
Sbjct: 1538 EHEEGKILRAQLEFSQIKADIERKLAEKDEEMEQSKRNLQRTIDTLQSSLESETRSRNEA 1597
Query: 610 ATVSKINKASFNNAVSQLAVVN 631
+ K + N QL+ N
Sbjct: 1598 LRIKKKMEGDLNEMEIQLSQAN 1619
Score = 41.2 bits (95), Expect = 0.10
Identities = 47/165 (28%), Positives = 77/165 (46%), Gaps = 27/165 (16%)
Query: 479 LKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLEN 538
L T+ V+ L L + ++ QL + + + ++E +A E+ KA+ + +
Sbjct: 1693 LDTSERVQLLHSQNTSLLNQKKKLETDISQLQTEVEEAVQECRNAEEKAKKAITDAAMMA 1752
Query: 539 EQLKKKVQDGDAVRE-----------DLEGRLKAAEGKAETGA--------ARLKEAEAA 579
E+LKK+ QD A E DL+ RL AE A G AR++E E+
Sbjct: 1753 EELKKE-QDTSAHLERMKKNMEQTIKDLQHRLDEAEQIAMKGGKKQVQKLEARVRELESE 1811
Query: 580 WLKEREQSS-AVK--KELENRVSSL----EADKKKLAATVSKINK 617
E+++SS AVK ++ E R+ L E D+K LA ++K
Sbjct: 1812 VESEQKKSSEAVKGIRKYERRIKELTYQTEEDRKNLARLQDLVDK 1856
Score = 40.0 bits (92), Expect = 0.23
Identities = 37/136 (27%), Positives = 72/136 (52%), Gaps = 14/136 (10%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQD------- 547
+ EE + + E + +S EL + ++ EE L+ MK EN+ L++++ D
Sbjct: 1445 KFEESQAELESSQKEARCLSTELFKLKNSYEEALDHLETMKRENKNLQEEISDLTEQLGE 1504
Query: 548 -GDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEAD-K 605
G ++ E + R + + K+E +A L+EAEA+ E E+ ++ +LE S ++AD +
Sbjct: 1505 GGKSIHELEKMRKQLEQEKSEIQSA-LEEAEAS--LEHEEGKILRAQLE--FSQIKADIE 1559
Query: 606 KKLAATVSKINKASFN 621
+KLA ++ ++ N
Sbjct: 1560 RKLAEKDEEMEQSKRN 1575
Score = 34.7 bits (78), Expect = 9.5
Identities = 38/150 (25%), Positives = 67/150 (44%), Gaps = 12/150 (8%)
Query: 488 LQGSTIPELEELREKTEKDDQLISSM------SKELEEGCSAIEELSKALD--NMKLENE 539
LQ E + L + E+ DQLI + +KEL E EE++ L KLE+E
Sbjct: 873 LQLQVQAEQDNLCDAEERCDQLIKNKIQLEAKAKELTERLEDEEEMNAELTAKKRKLEDE 932
Query: 540 --QLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENR 597
+LKK + D + +E A E K + + + K ++ A+++ +
Sbjct: 933 CSELKKDIDDLELTLAKVEKEKHATENKVKNLTEEMAALDDIIAKLTKEKKALQEAHQQT 992
Query: 598 VSSLEADKKKLAATVSKINKASFNNAVSQL 627
+ L++++ K+ T++K KA V L
Sbjct: 993 LDDLQSEEDKV-NTLTKA-KAKLEQQVDDL 1020
>UniRef100_UPI0000437480 UPI0000437480 UniRef100 entry
Length = 1929
Score = 54.3 bits (129), Expect = 1e-05
Identities = 64/252 (25%), Positives = 104/252 (40%), Gaps = 27/252 (10%)
Query: 382 RSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRAGN 441
R EE+ +KKR++ED ++ E +T+ E+ + + N
Sbjct: 923 RLEDEEEMNAELTAKKRKLEDECSELKKDIDDLE------LTLAKVEKEKHATEN-KVKN 975
Query: 442 CREHLAQIDDLV---------LTENDQKYLGD-REPAGLLNYVLRASLKTASAVRYLQGS 491
E +A +DD++ L E Q+ L D + +N + +A K V L+GS
Sbjct: 976 LTEEMAALDDIIAKLTKEKKALQEAHQQTLDDLQSEEDKVNTLTKAKAKLEQQVDDLEGS 1035
Query: 492 TIPE------LEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKV 545
E LE + K E D +L +LE +EE K D E QL K+
Sbjct: 1036 LEQEKKLRMDLERAKRKLEGDLKLTQESLMDLENDKQQLEERLKKKD---FEISQLNGKI 1092
Query: 546 QDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADK 605
+D + L+ +LK + + E L+ AA K +Q + + +ELE LE
Sbjct: 1093 EDEQTICIQLQKKLKELQARIEELEEELEAERAARAKVEKQRADLARELEEISERLEEAG 1152
Query: 606 KKLAATVSKINK 617
AA + ++NK
Sbjct: 1153 GATAAQI-EMNK 1163
Score = 45.8 bits (107), Expect = 0.004
Identities = 33/142 (23%), Positives = 71/142 (49%), Gaps = 3/142 (2%)
Query: 493 IPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVR 552
+ LE ++ + + + IS ++++L EG +I EL K ++ E +++ +++ +A
Sbjct: 1490 LDHLETMKRENKNLQEEISDLTEQLGEGGKSIHELEKMRKQLEQEKSEIQSALEEAEASL 1549
Query: 553 EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSA-VKKELENRVSSLEAD--KKKLA 609
E EG++ A+ + A ++ A +E EQS +++ ++ SSLE++ + A
Sbjct: 1550 EHEEGKILRAQLEFSQIKADIERKLAEKDEEMEQSKRNLQRTIDTLQSSLESETRSRNEA 1609
Query: 610 ATVSKINKASFNNAVSQLAVVN 631
+ K + N QL+ N
Sbjct: 1610 LRIKKKMEGDLNEMEIQLSQAN 1631
Score = 41.2 bits (95), Expect = 0.10
Identities = 47/165 (28%), Positives = 77/165 (46%), Gaps = 27/165 (16%)
Query: 479 LKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLEN 538
L T+ V+ L L + ++ QL + + + ++E +A E+ KA+ + +
Sbjct: 1705 LDTSERVQLLHSQNTSLLNQKKKLETDISQLQTEVEEAVQECRNAEEKAKKAITDAAMMA 1764
Query: 539 EQLKKKVQDGDAVRE-----------DLEGRLKAAEGKAETGA--------ARLKEAEAA 579
E+LKK+ QD A E DL+ RL AE A G AR++E E+
Sbjct: 1765 EELKKE-QDTSAHLERMKKNMEQTIKDLQHRLDEAEQIAMKGGKKQVQKLEARVRELESE 1823
Query: 580 WLKEREQSS-AVK--KELENRVSSL----EADKKKLAATVSKINK 617
E+++SS AVK ++ E R+ L E D+K LA ++K
Sbjct: 1824 VESEQKKSSEAVKGIRKYERRIKELTYQTEEDRKNLARLQDLVDK 1868
Score = 40.0 bits (92), Expect = 0.23
Identities = 37/136 (27%), Positives = 72/136 (52%), Gaps = 14/136 (10%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQD------- 547
+ EE + + E + +S EL + ++ EE L+ MK EN+ L++++ D
Sbjct: 1457 KFEESQAELESSQKEARCLSTELFKLKNSYEEALDHLETMKRENKNLQEEISDLTEQLGE 1516
Query: 548 -GDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEAD-K 605
G ++ E + R + + K+E +A L+EAEA+ E E+ ++ +LE S ++AD +
Sbjct: 1517 GGKSIHELEKMRKQLEQEKSEIQSA-LEEAEAS--LEHEEGKILRAQLE--FSQIKADIE 1571
Query: 606 KKLAATVSKINKASFN 621
+KLA ++ ++ N
Sbjct: 1572 RKLAEKDEEMEQSKRN 1587
Score = 34.7 bits (78), Expect = 9.5
Identities = 38/150 (25%), Positives = 67/150 (44%), Gaps = 12/150 (8%)
Query: 488 LQGSTIPELEELREKTEKDDQLISSM------SKELEEGCSAIEELSKALD--NMKLENE 539
LQ E + L + E+ DQLI + +KEL E EE++ L KLE+E
Sbjct: 885 LQLQVQAEQDNLCDAEERCDQLIKNKIQLEAKAKELTERLEDEEEMNAELTAKKRKLEDE 944
Query: 540 --QLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENR 597
+LKK + D + +E A E K + + + K ++ A+++ +
Sbjct: 945 CSELKKDIDDLELTLAKVEKEKHATENKVKNLTEEMAALDDIIAKLTKEKKALQEAHQQT 1004
Query: 598 VSSLEADKKKLAATVSKINKASFNNAVSQL 627
+ L++++ K+ T++K KA V L
Sbjct: 1005 LDDLQSEEDKV-NTLTKA-KAKLEQQVDDL 1032
>UniRef100_Q9PVE1 Ventricular myosin heavy chain [Brachydanio rerio]
Length = 1938
Score = 54.3 bits (129), Expect = 1e-05
Identities = 64/252 (25%), Positives = 104/252 (40%), Gaps = 27/252 (10%)
Query: 382 RSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRAGN 441
R EE+ +KKR++ED ++ E +T+ E+ + + N
Sbjct: 927 RLEDEEEMNAELTAKKRKLEDECSELKKDIDDLE------LTLAKVEKEKHATEN-KVKN 979
Query: 442 CREHLAQIDDLV---------LTENDQKYLGD-REPAGLLNYVLRASLKTASAVRYLQGS 491
E +A +DD++ L E Q+ L D + +N + +A K V L+GS
Sbjct: 980 LTEEMAALDDIIAKLTKEKKALQEAHQQTLDDLQSEEDKVNTLTKAKAKLEQQVDDLEGS 1039
Query: 492 TIPE------LEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKV 545
E LE + K E D +L +LE +EE K D E QL K+
Sbjct: 1040 LEQEKKLRMDLERAKRKLEGDLKLTQESLMDLENDKQQLEERLKKKD---FEISQLNGKI 1096
Query: 546 QDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADK 605
+D + L+ +LK + + E L+ AA K +Q + + +ELE LE
Sbjct: 1097 EDEQTICIQLQKKLKELQARIEELEEELEAERAARAKVEKQRADLARELEEVSERLEEAG 1156
Query: 606 KKLAATVSKINK 617
AA + ++NK
Sbjct: 1157 GATAAQI-EMNK 1167
Score = 45.8 bits (107), Expect = 0.004
Identities = 33/142 (23%), Positives = 71/142 (49%), Gaps = 3/142 (2%)
Query: 493 IPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVR 552
+ LE ++ + + + IS ++++L EG +I EL K ++ E +++ +++ +A
Sbjct: 1494 LDHLETMKRENKNLQEEISDLTEQLGEGGKSIHELEKMRKQLEQEKSEIQSALEEAEASL 1553
Query: 553 EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSA-VKKELENRVSSLEAD--KKKLA 609
E EG++ A+ + A ++ A +E EQS +++ ++ SSLE++ + A
Sbjct: 1554 EHEEGKILRAQLEFSQIKADIERKLAEKDEEMEQSKRNLQRTIDTLQSSLESETRSRNEA 1613
Query: 610 ATVSKINKASFNNAVSQLAVVN 631
+ K + N QL+ N
Sbjct: 1614 LRIKKKMEGDLNEMEIQLSQAN 1635
Score = 40.0 bits (92), Expect = 0.23
Identities = 37/136 (27%), Positives = 72/136 (52%), Gaps = 14/136 (10%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQD------- 547
+ EE + + E + +S EL + ++ EE L+ MK EN+ L++++ D
Sbjct: 1461 KFEESQAELESSQKEARCLSTELFKLKNSYEEALDHLETMKRENKNLQEEISDLTEQLGE 1520
Query: 548 -GDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEAD-K 605
G ++ E + R + + K+E +A L+EAEA+ E E+ ++ +LE S ++AD +
Sbjct: 1521 GGKSIHELEKMRKQLEQEKSEIQSA-LEEAEAS--LEHEEGKILRAQLE--FSQIKADIE 1575
Query: 606 KKLAATVSKINKASFN 621
+KLA ++ ++ N
Sbjct: 1576 RKLAEKDEEMEQSKRN 1591
Score = 38.5 bits (88), Expect = 0.65
Identities = 45/165 (27%), Positives = 76/165 (45%), Gaps = 27/165 (16%)
Query: 479 LKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLEN 538
L T+ V+ L L + ++ QL + + + ++E +A E+ KA+ + +
Sbjct: 1709 LDTSERVQLLHSQNTSLLNQKKKLETDISQLQTEVEEAVQECRNAEEKAKKAITDAAMMA 1768
Query: 539 EQLKKKVQDGDAVRE-----------DLEGRLKAAEGKAETGA--------ARLKEAEAA 579
E+LKK+ QD A E DL+ RL AE A G AR++E E
Sbjct: 1769 EELKKE-QDTSAHLERMKKNMEQTIKDLQHRLDEAEQIAMKGGKKQVQKLEARVRELECE 1827
Query: 580 WLKEREQSS-AVK--KELENRVSSL----EADKKKLAATVSKINK 617
E+++SS +VK ++ E R+ L E D+K +A ++K
Sbjct: 1828 VEAEQKRSSESVKGIRKYERRIKELTYQTEEDRKNIARLQDLVDK 1872
Score = 34.7 bits (78), Expect = 9.5
Identities = 38/150 (25%), Positives = 67/150 (44%), Gaps = 12/150 (8%)
Query: 488 LQGSTIPELEELREKTEKDDQLISSM------SKELEEGCSAIEELSKALD--NMKLENE 539
LQ E + L + E+ DQLI + +KEL E EE++ L KLE+E
Sbjct: 889 LQLQVQAEQDNLCDAEERCDQLIKNKIQLEAKAKELTERLEDEEEMNAELTAKKRKLEDE 948
Query: 540 --QLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENR 597
+LKK + D + +E A E K + + + K ++ A+++ +
Sbjct: 949 CSELKKDIDDLELTLAKVEKEKHATENKVKNLTEEMAALDDIIAKLTKEKKALQEAHQQT 1008
Query: 598 VSSLEADKKKLAATVSKINKASFNNAVSQL 627
+ L++++ K+ T++K KA V L
Sbjct: 1009 LDDLQSEEDKV-NTLTKA-KAKLEQQVDDL 1036
>UniRef100_Q8MTN7 Glutamic acid-rich protein cNBL1700 [Trichinella spiralis]
Length = 571
Score = 54.3 bits (129), Expect = 1e-05
Identities = 67/347 (19%), Positives = 146/347 (41%), Gaps = 23/347 (6%)
Query: 317 EDGQSTDEEVDQETDVAQG----EEGHAENQPQPDPEDRGGAEVGGTSGSAKNVSLEADH 372
+D ++ E +E DV+Q EE A + + PE++ E G+S + D
Sbjct: 121 KDEDKSESEASEEKDVSQEQNSKEEKGASEEDEDTPEEQNSKEENGSSEED-----DEDA 175
Query: 373 GSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERL 432
+ + + SEEK+T K E+ ++ DG E + +E T + E
Sbjct: 176 SEEQASNEEKEASEEKNTVSEERKGASEEEDEEKDDGHESEVESQASEEQT----TEEGA 231
Query: 433 SIWGPRAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGST 492
S E ++ ++ ++ +++ G+ + + + + +AS + S
Sbjct: 232 SEEEDEESASEEQTSEGEEKGASQEEEEDEGNEQESEVES---QASEEQTSEEEESASEE 288
Query: 493 IPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVR 552
E E +E+T ++++ S+ +E EE S EE + + + + E + K++ + +
Sbjct: 289 EDEENESKEQTTEEEE--SASEEEDEESASEREEKNASQEEEEDEGNESKEQTTEEEESA 346
Query: 553 EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAA-T 611
+ E +E + G +E A+ +E ++ + + E+E++ S + +++ A+
Sbjct: 347 SEEEDEESVSEEQTSEG----EEKGASQEEEEDEGNDQESEVESQASEEQTSEEEGASEE 402
Query: 612 VSKINKASFNNAVSQLAVVNPGLETAPVGYRKVVSGGTDRDCGFPRE 658
+ N++ + A E+A G K S + D G +E
Sbjct: 403 EDEENESEEQTTEEESASEEEDEESASEGEEKNASQEEEEDEGNEQE 449
Score = 47.0 bits (110), Expect = 0.002
Identities = 68/303 (22%), Positives = 120/303 (39%), Gaps = 26/303 (8%)
Query: 317 EDGQSTDEEVD----QETDV---AQGEEGHAENQPQPDPEDRGGAEVGGTSGSAKNVSLE 369
E G S +EE D QE++V A E+ E + + ED T+ ++ S E
Sbjct: 250 EKGASQEEEEDEGNEQESEVESQASEEQTSEEEESASEEEDEENESKEQTTEEEESASEE 309
Query: 370 ADHGSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSP 429
D S + + + EE++ G SK++ E+ + ++ E + S E
Sbjct: 310 EDEESASEREEKNASQEEEEDEGNESKEQTTEEEESASEEEDEESVSEEQ-----TSEGE 364
Query: 430 ERLSIWGPRAGNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQ 489
E+ + + ++++ E + G E N + + SA
Sbjct: 365 EKGASQEEEEDEGNDQESEVESQASEEQTSEEEGASEEEDEENESEEQTTEEESASEEED 424
Query: 490 GSTIPELEEL-----REKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLEN---EQL 541
+ E EE E+ E ++Q S+ EE S EE A EN EQ
Sbjct: 425 EESASEGEEKNASQEEEEDEGNEQESEVESQASEEQTSEEEEKEGASQEEDEENESEEQT 484
Query: 542 KKKVQDGDAVREDLEGRL--KAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVS 599
++ ++G + ED E + +E + E GA++ +E + +E EQ S V+ + +
Sbjct: 485 SEEEEEGASEEEDEESAFEEQTSEEEEEKGASQEEEED----EENEQESEVESQASEEQT 540
Query: 600 SLE 602
S E
Sbjct: 541 SEE 543
Score = 39.7 bits (91), Expect = 0.29
Identities = 59/285 (20%), Positives = 111/285 (38%), Gaps = 19/285 (6%)
Query: 320 QSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAEVGGTSGSAKNVSLEADHGSDPVQG 379
+ ++ VD+E ++ E G +Q + E + S S + + + +
Sbjct: 86 KESENGVDKEKPTSKEESGEKTSQEKESEEKSSQEKDEDKSESEASEEKDVSQEQNSKEE 145
Query: 380 KGRSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRA 439
KG S +E SK+ +D D + + E E S E+ ++ R
Sbjct: 146 KGASEEDEDTPEEQNSKEENGSSEEDDEDASEEQASNEEKE------ASEEKNTVSEERK 199
Query: 440 GNCREHLAQIDDLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGST--IPELE 497
G E + DD E++ + E ++AS + +G + E
Sbjct: 200 GASEEEDEEKDD--GHESEVESQASEEQTTEEGASEEEDEESASEEQTSEGEEKGASQEE 257
Query: 498 ELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEG 557
E E E++ ++ S S+E EE A + ENE ++ ++ ++ E+ E
Sbjct: 258 EEDEGNEQESEVESQASEE-----QTSEEEESASEEEDEENESKEQTTEEEESASEE-ED 311
Query: 558 RLKAAEGKAETGAARLKEAEAAWLKER---EQSSAVKKELENRVS 599
A+E + + + +E E KE+ E+ SA ++E E VS
Sbjct: 312 EESASEREEKNASQEEEEDEGNESKEQTTEEEESASEEEDEESVS 356
>UniRef100_Q9V217 Hypothetical protein [Pyrococcus abyssi]
Length = 281
Score = 54.3 bits (129), Expect = 1e-05
Identities = 44/133 (33%), Positives = 66/133 (49%), Gaps = 5/133 (3%)
Query: 500 REKTEKDDQLISSMSKELEEGCSAIEELS-KALDNMKLENEQLKKKVQDGDAVREDLEGR 558
RE+ EK + + +E EE + IE ++L+ K E E+LK KV+ + +++LE +
Sbjct: 126 REEYEKLLKEYEKLKQEFEEVKAKIEAAELESLEKAKKEIEELKGKVEKLEQEKKELEKK 185
Query: 559 LKAAEGKAETGAARLKEAEAAWLKEREQSSAVKK--ELENRVSSLEADKKKLAATVSKI- 615
LK +E K A+ K AE K RE VK+ ELE +VS LE + V +
Sbjct: 186 LKESEVKLMEYEAKAKRAEELEAKLREYEEKVKREEELERKVSELERSLNEYETKVKSLE 245
Query: 616 -NKASFNNAVSQL 627
K N V +L
Sbjct: 246 KKKEELENKVKEL 258
Score = 46.6 bits (109), Expect = 0.002
Identities = 38/143 (26%), Positives = 67/143 (46%), Gaps = 25/143 (17%)
Query: 493 IPELEELREKTEKDDQLISSMSKE-LEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAV 551
+ E E+L+++ E+ I + E LE+ IEEL ++ ++ E ++L+KK+++ +
Sbjct: 133 LKEYEKLKQEFEEVKAKIEAAELESLEKAKKEIEELKGKVEKLEQEKKELEKKLKESEVK 192
Query: 552 REDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAV--------------------K 591
+ E + K +AE A+L+E E +E E V K
Sbjct: 193 LMEYEAKAK----RAEELEAKLREYEEKVKREEELERKVSELERSLNEYETKVKSLEKKK 248
Query: 592 KELENRVSSLEADKKKLAATVSK 614
+ELEN+V LE + KL + K
Sbjct: 249 EELENKVKELEEEVNKLKEGIGK 271
Score = 37.7 bits (86), Expect = 1.1
Identities = 38/155 (24%), Positives = 71/155 (45%), Gaps = 20/155 (12%)
Query: 465 REPAGLLNYVLRASLKTASAVRYLQGSTIPELEELREKTEKD-----------DQLISSM 513
++PA L +V ++ + R G T+ E+ +TE+ QL+
Sbjct: 37 KDPAERLTWVDSLAVAAGAIAREKAGMTVSEIARELGRTEQTIRKHLKGESRAGQLVRET 96
Query: 514 SKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARL 573
+ +++G ++EL K ++ +E +K+ V RE+ E LK E +
Sbjct: 97 YELIKQG--KLDELIKTIE--MIEKGGIKEVV-----AREEYEKLLKEYEKLKQEFEEVK 147
Query: 574 KEAEAAWLKEREQSSAVKKELENRVSSLEADKKKL 608
+ EAA L+ E++ +EL+ +V LE +KK+L
Sbjct: 148 AKIEAAELESLEKAKKEIEELKGKVEKLEQEKKEL 182
Score = 37.0 bits (84), Expect = 1.9
Identities = 33/115 (28%), Positives = 55/115 (47%), Gaps = 15/115 (13%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEG----------CSAIEELSKAL----DNMKLENEQ 540
E+EEL+ K EK +Q + K+L+E EEL L + +K E E+
Sbjct: 164 EIEELKGKVEKLEQEKKELEKKLKESEVKLMEYEAKAKRAEELEAKLREYEEKVKRE-EE 222
Query: 541 LKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELE 595
L++KV + + + E ++K+ E K E ++KE E K +E K+ L+
Sbjct: 223 LERKVSELERSLNEYETKVKSLEKKKEELENKVKELEEEVNKLKEGIGKAKEILD 277
>UniRef100_UPI000036153E UPI000036153E UniRef100 entry
Length = 1857
Score = 53.9 bits (128), Expect = 2e-05
Identities = 60/237 (25%), Positives = 98/237 (41%), Gaps = 26/237 (10%)
Query: 382 RSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRAGN 441
R EE+ +KKR++ED ++ E +T+ E+ + + N
Sbjct: 852 RLEDEEEMNAELTAKKRKLEDECSELKKDIDDLE------LTLAKVEKEKHATEN-KVKN 904
Query: 442 CREHLAQIDDLV---------LTENDQKYLGD-REPAGLLNYVLRASLKTASAVRYLQGS 491
E +A +D+++ L E Q+ L D + +N + +A K V L+GS
Sbjct: 905 LTEEMAALDEIIAKLTKEKKALQEAHQQTLDDLQSEEDKVNTLTKAKTKLEQQVDDLEGS 964
Query: 492 TIPE------LEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKV 545
E LE + K E D +L +LE +EE K D E QL K+
Sbjct: 965 LEQEKKIRMDLERAKRKLEGDLKLTQETVMDLENDKQQLEERLKKKD---FEISQLNGKI 1021
Query: 546 QDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLE 602
+D A+ L+ +LK + + E L+ AA K +Q + + +ELE LE
Sbjct: 1022 EDEQAIIIQLQKKLKELQARVEELEEELEAERAARAKVEKQRADLARELEEISERLE 1078
Score = 49.7 bits (117), Expect = 3e-04
Identities = 44/184 (23%), Positives = 85/184 (45%), Gaps = 3/184 (1%)
Query: 451 DLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLI 510
D VL+E QKY + RA ++ ++ LE ++ + + + I
Sbjct: 1382 DKVLSEWKQKYEESQCELEGSQKEARALSTELFKLKNSYEESLDHLETMKRENKNLQEEI 1441
Query: 511 SSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGA 570
S +S++L EG +I EL K ++ E +++ +++ +A E EG++ A+ +
Sbjct: 1442 SDLSEQLSEGGKSIHELEKLRKQLEQEKNEIQSALEEAEASLEHEEGKILRAQLEFNQVK 1501
Query: 571 ARLKEAEAAWLKEREQSSA-VKKELENRVSSLEAD--KKKLAATVSKINKASFNNAVSQL 627
A ++ A +E EQS +++ ++ SSLEA+ + A + K + N QL
Sbjct: 1502 ADIERKLAEKDEEMEQSKRNLQRTIDTLQSSLEAECRSRNEALRLKKKMEGDLNEMEIQL 1561
Query: 628 AVVN 631
+ N
Sbjct: 1562 SQAN 1565
Score = 38.9 bits (89), Expect = 0.50
Identities = 37/159 (23%), Positives = 70/159 (43%), Gaps = 32/159 (20%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
E+ +L K E + +I + K+L+E + +EEL + L+ + +++K+ D E+
Sbjct: 1013 EISQLNGKIEDEQAIIIQLQKKLKELQARVEELEEELEAERAARAKVEKQRADLARELEE 1072
Query: 555 LEGRLKAAEG----------KAETGAARLK----------EAEAAWLKEREQSSA----- 589
+ RL+ A G K E +L+ EA AA L++++ S
Sbjct: 1073 ISERLEEAGGATTLQIEMNKKREAEFLKLRRDLEEATLQHEATAAALRKKQADSVADLGE 1132
Query: 590 -------VKKELENRVSSLEADKKKLAATVSKINKASFN 621
VK++LE S L + + + + +I K+ N
Sbjct: 1133 QIDNLQRVKQKLEKEKSELRLELDDVVSNMEQIVKSKTN 1171
Score = 35.4 bits (80), Expect = 5.5
Identities = 31/120 (25%), Positives = 48/120 (39%), Gaps = 7/120 (5%)
Query: 493 IPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVR 552
I +LE K EK+ + K L E +A++E+ L K ++ ++ D
Sbjct: 881 IDDLELTLAKVEKEKHATENKVKNLTEEMAALDEIIAKLTKEKKALQEAHQQTLDDLQSE 940
Query: 553 EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATV 612
ED L A+ K E L+ + EQ ++ +LE LE D K TV
Sbjct: 941 EDKVNTLTKAKTKLEQQVDDLEGS-------LEQEKKIRMDLERAKRKLEGDLKLTQETV 993
>UniRef100_UPI000036153D UPI000036153D UniRef100 entry
Length = 1832
Score = 53.9 bits (128), Expect = 2e-05
Identities = 60/237 (25%), Positives = 98/237 (41%), Gaps = 26/237 (10%)
Query: 382 RSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRAGN 441
R EE+ +KKR++ED ++ E +T+ E+ + + N
Sbjct: 827 RLEDEEEMNAELTAKKRKLEDECSELKKDIDDLE------LTLAKVEKEKHATEN-KVKN 879
Query: 442 CREHLAQIDDLV---------LTENDQKYLGD-REPAGLLNYVLRASLKTASAVRYLQGS 491
E +A +D+++ L E Q+ L D + +N + +A K V L+GS
Sbjct: 880 LTEEMAALDEIIAKLTKEKKALQEAHQQTLDDLQSEEDKVNTLTKAKTKLEQQVDDLEGS 939
Query: 492 TIPE------LEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKV 545
E LE + K E D +L +LE +EE K D E QL K+
Sbjct: 940 LEQEKKIRMDLERAKRKLEGDLKLTQETVMDLENDKQQLEERLKKKD---FEISQLNGKI 996
Query: 546 QDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLE 602
+D A+ L+ +LK + + E L+ AA K +Q + + +ELE LE
Sbjct: 997 EDEQAIIIQLQKKLKELQARVEELEEELEAERAARAKVEKQRADLARELEEISERLE 1053
Score = 49.7 bits (117), Expect = 3e-04
Identities = 44/184 (23%), Positives = 85/184 (45%), Gaps = 3/184 (1%)
Query: 451 DLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLI 510
D VL+E QKY + RA ++ ++ LE ++ + + + I
Sbjct: 1357 DKVLSEWKQKYEESQCELEGSQKEARALSTELFKLKNSYEESLDHLETMKRENKNLQEEI 1416
Query: 511 SSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGA 570
S +S++L EG +I EL K ++ E +++ +++ +A E EG++ A+ +
Sbjct: 1417 SDLSEQLSEGGKSIHELEKLRKQLEQEKNEIQSALEEAEASLEHEEGKILRAQLEFNQVK 1476
Query: 571 ARLKEAEAAWLKEREQSSA-VKKELENRVSSLEAD--KKKLAATVSKINKASFNNAVSQL 627
A ++ A +E EQS +++ ++ SSLEA+ + A + K + N QL
Sbjct: 1477 ADIERKLAEKDEEMEQSKRNLQRTIDTLQSSLEAECRSRNEALRLKKKMEGDLNEMEIQL 1536
Query: 628 AVVN 631
+ N
Sbjct: 1537 SQAN 1540
Score = 38.9 bits (89), Expect = 0.50
Identities = 37/159 (23%), Positives = 70/159 (43%), Gaps = 32/159 (20%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
E+ +L K E + +I + K+L+E + +EEL + L+ + +++K+ D E+
Sbjct: 988 EISQLNGKIEDEQAIIIQLQKKLKELQARVEELEEELEAERAARAKVEKQRADLARELEE 1047
Query: 555 LEGRLKAAEG----------KAETGAARLK----------EAEAAWLKEREQSSA----- 589
+ RL+ A G K E +L+ EA AA L++++ S
Sbjct: 1048 ISERLEEAGGATTLQIEMNKKREAEFLKLRRDLEEATLQHEATAAALRKKQADSVADLGE 1107
Query: 590 -------VKKELENRVSSLEADKKKLAATVSKINKASFN 621
VK++LE S L + + + + +I K+ N
Sbjct: 1108 QIDNLQRVKQKLEKEKSELRLELDDVVSNMEQIVKSKTN 1146
Score = 35.4 bits (80), Expect = 5.5
Identities = 31/120 (25%), Positives = 48/120 (39%), Gaps = 7/120 (5%)
Query: 493 IPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVR 552
I +LE K EK+ + K L E +A++E+ L K ++ ++ D
Sbjct: 856 IDDLELTLAKVEKEKHATENKVKNLTEEMAALDEIIAKLTKEKKALQEAHQQTLDDLQSE 915
Query: 553 EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATV 612
ED L A+ K E L+ + EQ ++ +LE LE D K TV
Sbjct: 916 EDKVNTLTKAKTKLEQQVDDLEGS-------LEQEKKIRMDLERAKRKLEGDLKLTQETV 968
>UniRef100_UPI000036153C UPI000036153C UniRef100 entry
Length = 1880
Score = 53.9 bits (128), Expect = 2e-05
Identities = 60/237 (25%), Positives = 98/237 (41%), Gaps = 26/237 (10%)
Query: 382 RSVSEEKDTRGPASKKRRVEDHQDHADGEVQGSEPERAEPVTVVPPSPERLSIWGPRAGN 441
R EE+ +KKR++ED ++ E +T+ E+ + + N
Sbjct: 870 RLEDEEEMNAELTAKKRKLEDECSELKKDIDDLE------LTLAKVEKEKHATEN-KVKN 922
Query: 442 CREHLAQIDDLV---------LTENDQKYLGD-REPAGLLNYVLRASLKTASAVRYLQGS 491
E +A +D+++ L E Q+ L D + +N + +A K V L+GS
Sbjct: 923 LTEEMAALDEIIAKLTKEKKALQEAHQQTLDDLQSEEDKVNTLTKAKTKLEQQVDDLEGS 982
Query: 492 TIPE------LEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKV 545
E LE + K E D +L +LE +EE K D E QL K+
Sbjct: 983 LEQEKKIRMDLERAKRKLEGDLKLTQETVMDLENDKQQLEERLKKKD---FEISQLNGKI 1039
Query: 546 QDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLE 602
+D A+ L+ +LK + + E L+ AA K +Q + + +ELE LE
Sbjct: 1040 EDEQAIIIQLQKKLKELQARVEELEEELEAERAARAKVEKQRADLARELEEISERLE 1096
Score = 49.7 bits (117), Expect = 3e-04
Identities = 44/184 (23%), Positives = 85/184 (45%), Gaps = 3/184 (1%)
Query: 451 DLVLTENDQKYLGDREPAGLLNYVLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLI 510
D VL+E QKY + RA ++ ++ LE ++ + + + I
Sbjct: 1400 DKVLSEWKQKYEESQCELEGSQKEARALSTELFKLKNSYEESLDHLETMKRENKNLQEEI 1459
Query: 511 SSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGA 570
S +S++L EG +I EL K ++ E +++ +++ +A E EG++ A+ +
Sbjct: 1460 SDLSEQLSEGGKSIHELEKLRKQLEQEKNEIQSALEEAEASLEHEEGKILRAQLEFNQVK 1519
Query: 571 ARLKEAEAAWLKEREQSSA-VKKELENRVSSLEAD--KKKLAATVSKINKASFNNAVSQL 627
A ++ A +E EQS +++ ++ SSLEA+ + A + K + N QL
Sbjct: 1520 ADIERKLAEKDEEMEQSKRNLQRTIDTLQSSLEAECRSRNEALRLKKKMEGDLNEMEIQL 1579
Query: 628 AVVN 631
+ N
Sbjct: 1580 SQAN 1583
Score = 38.9 bits (89), Expect = 0.50
Identities = 37/159 (23%), Positives = 70/159 (43%), Gaps = 32/159 (20%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVRED 554
E+ +L K E + +I + K+L+E + +EEL + L+ + +++K+ D E+
Sbjct: 1031 EISQLNGKIEDEQAIIIQLQKKLKELQARVEELEEELEAERAARAKVEKQRADLARELEE 1090
Query: 555 LEGRLKAAEG----------KAETGAARLK----------EAEAAWLKEREQSSA----- 589
+ RL+ A G K E +L+ EA AA L++++ S
Sbjct: 1091 ISERLEEAGGATTLQIEMNKKREAEFLKLRRDLEEATLQHEATAAALRKKQADSVADLGE 1150
Query: 590 -------VKKELENRVSSLEADKKKLAATVSKINKASFN 621
VK++LE S L + + + + +I K+ N
Sbjct: 1151 QIDNLQRVKQKLEKEKSELRLELDDVVSNMEQIVKSKTN 1189
Score = 35.4 bits (80), Expect = 5.5
Identities = 31/120 (25%), Positives = 48/120 (39%), Gaps = 7/120 (5%)
Query: 493 IPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDAVR 552
I +LE K EK+ + K L E +A++E+ L K ++ ++ D
Sbjct: 899 IDDLELTLAKVEKEKHATENKVKNLTEEMAALDEIIAKLTKEKKALQEAHQQTLDDLQSE 958
Query: 553 EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELENRVSSLEADKKKLAATV 612
ED L A+ K E L+ + EQ ++ +LE LE D K TV
Sbjct: 959 EDKVNTLTKAKTKLEQQVDDLEGS-------LEQEKKIRMDLERAKRKLEGDLKLTQETV 1011
>UniRef100_Q7PJV2 ENSANGP00000024129 [Anopheles gambiae str. PEST]
Length = 1937
Score = 53.5 bits (127), Expect = 2e-05
Identities = 77/342 (22%), Positives = 140/342 (40%), Gaps = 23/342 (6%)
Query: 296 NAKLLRGMADIDRTLLDALRVEDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAE 355
N+KLL LLD+L E G +E ++ ++ ENQ + E E
Sbjct: 877 NSKLLAEKT----ALLDSLSGEKGAL--QEYQEKAAKLTAQKNDLENQLRDTQERLAQEE 930
Query: 356 VGGTSGSAKNVSLEADHGSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQD---HADGEVQ 412
LE + GS + + +K + ASK ++ + D H D +
Sbjct: 931 DARNQLFQTKKKLEQEIGSQKKDAEDLELQIQKIEQDKASKDHQIRNLNDEIAHQDELIN 990
Query: 413 GSEPERAEPVTVVPPSPERLSIWGPRAGNCREHLAQIDDLV-----LTENDQKYLGDREP 467
E+ V + E L + + + A+++ + E ++K GD E
Sbjct: 991 KLNKEKKMQGEVNQKTAEELQAAEDKVNHLNKVKAKLEQTLDELEDSLEREKKLRGDVEK 1050
Query: 468 AGLLNYVLRASLK-TASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEE 526
A + LK T AV L+ + +EL + + D+ IS++S +LE+ S + +
Sbjct: 1051 A---KRKVEGDLKLTQEAVADLERNK----KELEQTVLRKDKEISALSAKLEDEQSLVGK 1103
Query: 527 LSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQ 586
L K + ++ E+L+++V+ R E + + E RL+EA A + E
Sbjct: 1104 LQKQIKELQARIEELEEEVEAERQARAKAEKQRADLARELEELGERLEEAGGATSAQIEL 1163
Query: 587 SSAVKKELENRVSSLEADKKKLAATVSKINKASFNNAVSQLA 628
+ + EL LE + T++ + K N+AV+++A
Sbjct: 1164 NKKREAELAKLRRDLEEANIQHEGTLANLRK-KHNDAVAEMA 1204
Score = 37.0 bits (84), Expect = 1.9
Identities = 35/140 (25%), Positives = 67/140 (47%), Gaps = 9/140 (6%)
Query: 474 VLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDN 533
+ A+ K A + G ++++L + + + + S EL A EE + L+
Sbjct: 1438 IANAAEKKQKAFDKIIGEWKLKVDDLAAELDASQKECRNYSTELFRLKGAYEEGQEQLEA 1497
Query: 534 MKLENEQLKKKVQD-----GDAVR--EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQ 586
++ EN+ L +V+D G+ R ++E K E + + A L+EAEAA E+E+
Sbjct: 1498 VRRENKNLADEVKDLLDQIGEGGRNIHEIEKSRKRLEAEKDELQAALEEAEAA--LEQEE 1555
Query: 587 SSAVKKELENRVSSLEADKK 606
+ ++ +LE E D++
Sbjct: 1556 NKVLRAQLELSQVRQEIDRR 1575
Score = 35.8 bits (81), Expect = 4.2
Identities = 28/143 (19%), Positives = 60/143 (41%), Gaps = 18/143 (12%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDA---- 550
+LE +R + + + + ++ EG I E+ K+ ++ E ++L+ +++ +A
Sbjct: 1494 QLEAVRRENKNLADEVKDLLDQIGEGGRNIHEIEKSRKRLEAEKDELQAALEEAEAALEQ 1553
Query: 551 --------------VREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELEN 596
VR++++ R++ E + E + A + E + K E
Sbjct: 1554 EENKVLRAQLELSQVRQEIDRRIQEKEEEFENTRKNHQRALDSMQASLEAEAKGKAEALR 1613
Query: 597 RVSSLEADKKKLAATVSKINKAS 619
LEAD +L + NKA+
Sbjct: 1614 MKKKLEADINELEIALDHANKAN 1636
>UniRef100_Q7PJV0 ENSANGP00000024621 [Anopheles gambiae str. PEST]
Length = 1937
Score = 53.5 bits (127), Expect = 2e-05
Identities = 77/342 (22%), Positives = 140/342 (40%), Gaps = 23/342 (6%)
Query: 296 NAKLLRGMADIDRTLLDALRVEDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAE 355
N+KLL LLD+L E G +E ++ ++ ENQ + E E
Sbjct: 877 NSKLLAEKT----ALLDSLSGEKGAL--QEYQEKAAKLTAQKNDLENQLRDTQERLAQEE 930
Query: 356 VGGTSGSAKNVSLEADHGSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQD---HADGEVQ 412
LE + GS + + +K + ASK ++ + D H D +
Sbjct: 931 DARNQLFQTKKKLEQEIGSQKKDAEDLELQIQKIEQDKASKDHQIRNLNDEIAHQDELIN 990
Query: 413 GSEPERAEPVTVVPPSPERLSIWGPRAGNCREHLAQIDDLV-----LTENDQKYLGDREP 467
E+ V + E L + + + A+++ + E ++K GD E
Sbjct: 991 KLNKEKKMQGEVNQKTAEELQAAEDKVNHLNKVKAKLEQTLDELEDSLEREKKLRGDVEK 1050
Query: 468 AGLLNYVLRASLK-TASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEE 526
A + LK T AV L+ + +EL + + D+ IS++S +LE+ S + +
Sbjct: 1051 A---KRKVEGDLKLTQEAVADLERNK----KELEQTVLRKDKEISALSAKLEDEQSLVGK 1103
Query: 527 LSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQ 586
L K + ++ E+L+++V+ R E + + E RL+EA A + E
Sbjct: 1104 LQKQIKELQARIEELEEEVEAERQARAKAEKQRADLARELEELGERLEEAGGATSAQIEL 1163
Query: 587 SSAVKKELENRVSSLEADKKKLAATVSKINKASFNNAVSQLA 628
+ + EL LE + T++ + K N+AV+++A
Sbjct: 1164 NKKREAELAKLRRDLEEANIQHEGTLANLRK-KHNDAVAEMA 1204
Score = 37.0 bits (84), Expect = 1.9
Identities = 35/140 (25%), Positives = 67/140 (47%), Gaps = 9/140 (6%)
Query: 474 VLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDN 533
+ A+ K A + G ++++L + + + + S EL A EE + L+
Sbjct: 1438 IANAAEKKQKAFDKIIGEWKLKVDDLAAELDASQKECRNYSTELFRLKGAYEEGQEQLEA 1497
Query: 534 MKLENEQLKKKVQD-----GDAVR--EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQ 586
++ EN+ L +V+D G+ R ++E K E + + A L+EAEAA E+E+
Sbjct: 1498 VRRENKNLADEVKDLLDQIGEGGRNIHEIEKSRKRLEAEKDELQAALEEAEAA--LEQEE 1555
Query: 587 SSAVKKELENRVSSLEADKK 606
+ ++ +LE E D++
Sbjct: 1556 NKVLRAQLELSQVRQEIDRR 1575
Score = 35.8 bits (81), Expect = 4.2
Identities = 28/143 (19%), Positives = 60/143 (41%), Gaps = 18/143 (12%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDA---- 550
+LE +R + + + + ++ EG I E+ K+ ++ E ++L+ +++ +A
Sbjct: 1494 QLEAVRRENKNLADEVKDLLDQIGEGGRNIHEIEKSRKRLEAEKDELQAALEEAEAALEQ 1553
Query: 551 --------------VREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELEN 596
VR++++ R++ E + E + A + E + K E
Sbjct: 1554 EENKVLRAQLELSQVRQEIDRRIQEKEEEFENTRKNHQRALDSMQASLEAEAKGKAEALR 1613
Query: 597 RVSSLEADKKKLAATVSKINKAS 619
LEAD +L + NKA+
Sbjct: 1614 MKKKLEADINELEIALDHANKAN 1636
>UniRef100_Q7PMG6 ENSANGP00000012555 [Anopheles gambiae str. PEST]
Length = 1943
Score = 53.5 bits (127), Expect = 2e-05
Identities = 77/342 (22%), Positives = 140/342 (40%), Gaps = 23/342 (6%)
Query: 296 NAKLLRGMADIDRTLLDALRVEDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAE 355
N+KLL LLD+L E G +E ++ ++ ENQ + E E
Sbjct: 877 NSKLLAEKT----ALLDSLSGEKGAL--QEYQEKAAKLTAQKNDLENQLRDTQERLAQEE 930
Query: 356 VGGTSGSAKNVSLEADHGSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQD---HADGEVQ 412
LE + GS + + +K + ASK ++ + D H D +
Sbjct: 931 DARNQLFQTKKKLEQEIGSQKKDAEDLELQIQKIEQDKASKDHQIRNLNDEIAHQDELIN 990
Query: 413 GSEPERAEPVTVVPPSPERLSIWGPRAGNCREHLAQIDDLV-----LTENDQKYLGDREP 467
E+ V + E L + + + A+++ + E ++K GD E
Sbjct: 991 KLNKEKKMQGEVNQKTAEELQAAEDKVNHLNKVKAKLEQTLDELEDSLEREKKLRGDVEK 1050
Query: 468 AGLLNYVLRASLK-TASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEE 526
A + LK T AV L+ + +EL + + D+ IS++S +LE+ S + +
Sbjct: 1051 A---KRKVEGDLKLTQEAVADLERNK----KELEQTVLRKDKEISALSAKLEDEQSLVGK 1103
Query: 527 LSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQ 586
L K + ++ E+L+++V+ R E + + E RL+EA A + E
Sbjct: 1104 LQKQIKELQARIEELEEEVEAERQARAKAEKQRADLARELEELGERLEEAGGATSAQIEL 1163
Query: 587 SSAVKKELENRVSSLEADKKKLAATVSKINKASFNNAVSQLA 628
+ + EL LE + T++ + K N+AV+++A
Sbjct: 1164 NKKREAELAKLRRDLEEANIQHEGTLANLRK-KHNDAVAEMA 1204
Score = 37.0 bits (84), Expect = 1.9
Identities = 35/140 (25%), Positives = 67/140 (47%), Gaps = 9/140 (6%)
Query: 474 VLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDN 533
+ A+ K A + G ++++L + + + + S EL A EE + L+
Sbjct: 1438 IANAAEKKQKAFDKIIGEWKLKVDDLAAELDASQKECRNYSTELFRLKGAYEEGQEQLEA 1497
Query: 534 MKLENEQLKKKVQD-----GDAVR--EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQ 586
++ EN+ L +V+D G+ R ++E K E + + A L+EAEAA E+E+
Sbjct: 1498 VRRENKNLADEVKDLLDQIGEGGRNIHEIEKSRKRLEAEKDELQAALEEAEAA--LEQEE 1555
Query: 587 SSAVKKELENRVSSLEADKK 606
+ ++ +LE E D++
Sbjct: 1556 NKVLRAQLELSQVRQEIDRR 1575
Score = 35.8 bits (81), Expect = 4.2
Identities = 28/143 (19%), Positives = 60/143 (41%), Gaps = 18/143 (12%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDA---- 550
+LE +R + + + + ++ EG I E+ K+ ++ E ++L+ +++ +A
Sbjct: 1494 QLEAVRRENKNLADEVKDLLDQIGEGGRNIHEIEKSRKRLEAEKDELQAALEEAEAALEQ 1553
Query: 551 --------------VREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELEN 596
VR++++ R++ E + E + A + E + K E
Sbjct: 1554 EENKVLRAQLELSQVRQEIDRRIQEKEEEFENTRKNHQRALDSMQASLEAEAKGKAEALR 1613
Query: 597 RVSSLEADKKKLAATVSKINKAS 619
LEAD +L + NKA+
Sbjct: 1614 MKKKLEADINELEIALDHANKAN 1636
>UniRef100_Q7PJV1 ENSANGP00000023782 [Anopheles gambiae str. PEST]
Length = 1943
Score = 53.5 bits (127), Expect = 2e-05
Identities = 77/342 (22%), Positives = 140/342 (40%), Gaps = 23/342 (6%)
Query: 296 NAKLLRGMADIDRTLLDALRVEDGQSTDEEVDQETDVAQGEEGHAENQPQPDPEDRGGAE 355
N+KLL LLD+L E G +E ++ ++ ENQ + E E
Sbjct: 877 NSKLLAEKT----ALLDSLSGEKGAL--QEYQEKAAKLTAQKNDLENQLRDTQERLAQEE 930
Query: 356 VGGTSGSAKNVSLEADHGSDPVQGKGRSVSEEKDTRGPASKKRRVEDHQD---HADGEVQ 412
LE + GS + + +K + ASK ++ + D H D +
Sbjct: 931 DARNQLFQTKKKLEQEIGSQKKDAEDLELQIQKIEQDKASKDHQIRNLNDEIAHQDELIN 990
Query: 413 GSEPERAEPVTVVPPSPERLSIWGPRAGNCREHLAQIDDLV-----LTENDQKYLGDREP 467
E+ V + E L + + + A+++ + E ++K GD E
Sbjct: 991 KLNKEKKMQGEVNQKTAEELQAAEDKVNHLNKVKAKLEQTLDELEDSLEREKKLRGDVEK 1050
Query: 468 AGLLNYVLRASLK-TASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEE 526
A + LK T AV L+ + +EL + + D+ IS++S +LE+ S + +
Sbjct: 1051 A---KRKVEGDLKLTQEAVADLERNK----KELEQTVLRKDKEISALSAKLEDEQSLVGK 1103
Query: 527 LSKALDNMKLENEQLKKKVQDGDAVREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQ 586
L K + ++ E+L+++V+ R E + + E RL+EA A + E
Sbjct: 1104 LQKQIKELQARIEELEEEVEAERQARAKAEKQRADLARELEELGERLEEAGGATSAQIEL 1163
Query: 587 SSAVKKELENRVSSLEADKKKLAATVSKINKASFNNAVSQLA 628
+ + EL LE + T++ + K N+AV+++A
Sbjct: 1164 NKKREAELAKLRRDLEEANIQHEGTLANLRK-KHNDAVAEMA 1204
Score = 37.0 bits (84), Expect = 1.9
Identities = 35/140 (25%), Positives = 67/140 (47%), Gaps = 9/140 (6%)
Query: 474 VLRASLKTASAVRYLQGSTIPELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDN 533
+ A+ K A + G ++++L + + + + S EL A EE + L+
Sbjct: 1438 IANAAEKKQKAFDKIIGEWKLKVDDLAAELDASQKECRNYSTELFRLKGAYEEGQEQLEA 1497
Query: 534 MKLENEQLKKKVQD-----GDAVR--EDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQ 586
++ EN+ L +V+D G+ R ++E K E + + A L+EAEAA E+E+
Sbjct: 1498 VRRENKNLADEVKDLLDQIGEGGRNIHEIEKSRKRLEAEKDELQAALEEAEAA--LEQEE 1555
Query: 587 SSAVKKELENRVSSLEADKK 606
+ ++ +LE E D++
Sbjct: 1556 NKVLRAQLELSQVRQEIDRR 1575
Score = 35.8 bits (81), Expect = 4.2
Identities = 28/143 (19%), Positives = 60/143 (41%), Gaps = 18/143 (12%)
Query: 495 ELEELREKTEKDDQLISSMSKELEEGCSAIEELSKALDNMKLENEQLKKKVQDGDA---- 550
+LE +R + + + + ++ EG I E+ K+ ++ E ++L+ +++ +A
Sbjct: 1494 QLEAVRRENKNLADEVKDLLDQIGEGGRNIHEIEKSRKRLEAEKDELQAALEEAEAALEQ 1553
Query: 551 --------------VREDLEGRLKAAEGKAETGAARLKEAEAAWLKEREQSSAVKKELEN 596
VR++++ R++ E + E + A + E + K E
Sbjct: 1554 EENKVLRAQLELSQVRQEIDRRIQEKEEEFENTRKNHQRALDSMQASLEAEAKGKAEALR 1613
Query: 597 RVSSLEADKKKLAATVSKINKAS 619
LEAD +L + NKA+
Sbjct: 1614 MKKKLEADINELEIALDHANKAN 1636
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,204,532,927
Number of Sequences: 2790947
Number of extensions: 54746922
Number of successful extensions: 209102
Number of sequences better than 10.0: 5273
Number of HSP's better than 10.0 without gapping: 777
Number of HSP's successfully gapped in prelim test: 4710
Number of HSP's that attempted gapping in prelim test: 182163
Number of HSP's gapped (non-prelim): 23309
length of query: 673
length of database: 848,049,833
effective HSP length: 134
effective length of query: 539
effective length of database: 474,062,935
effective search space: 255519921965
effective search space used: 255519921965
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0039.13


