

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0037a.11
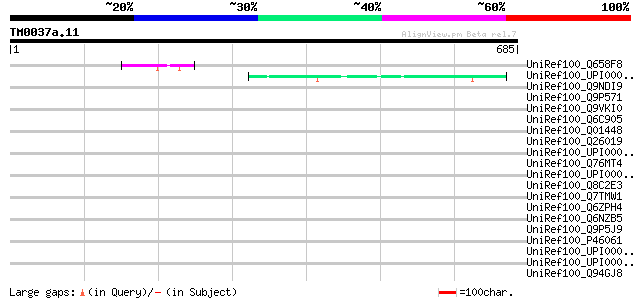
(685 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q658F8 Hypothetical protein P0015E04.9 [Oryza sativa] 51 1e-04
UniRef100_UPI00001224EC UPI00001224EC UniRef100 entry 49 4e-04
UniRef100_Q9NDI9 Merozoite surface protein 3g [Plasmodium vivax] 47 0.001
UniRef100_Q9P571 Related to actin-interacting protein AIP3 [Neur... 47 0.002
UniRef100_Q9VKI0 CG6392-PA [Drosophila melanogaster] 46 0.004
UniRef100_Q6C905 Yarrowia lipolytica chromosome D of strain CLIB... 45 0.005
UniRef100_Q01448 Histone promoter control 2 protein [Saccharomyc... 45 0.005
UniRef100_Q26019 Polymorphic antigen precursor [Plasmodium falci... 45 0.007
UniRef100_UPI00004375BD UPI00004375BD UniRef100 entry 45 0.009
UniRef100_Q76MT4 ABTAP [Rattus rattus] 45 0.009
UniRef100_UPI000023F6CB UPI000023F6CB UniRef100 entry 44 0.012
UniRef100_Q8C2E3 Mus musculus 2 days neonate thymus thymic cells... 44 0.012
UniRef100_Q7TMW1 Rangap1 protein [Mus musculus] 44 0.012
UniRef100_Q6ZPH4 MKIAA1835 protein [Mus musculus] 44 0.012
UniRef100_Q6NZB5 Rangap1 protein [Mus musculus] 44 0.012
UniRef100_Q9P5J9 Related to BCS1 protein [Neurospora crassa] 44 0.012
UniRef100_P46061 Ran GTPase-activating protein 1 [Mus musculus] 44 0.012
UniRef100_UPI000023DA4F UPI000023DA4F UniRef100 entry 44 0.016
UniRef100_UPI0000219DB0 UPI0000219DB0 UniRef100 entry 44 0.016
UniRef100_Q94GJ8 Putative retrotransposon protein [Oryza sativa] 44 0.016
>UniRef100_Q658F8 Hypothetical protein P0015E04.9 [Oryza sativa]
Length = 569
Score = 51.2 bits (121), Expect = 1e-04
Identities = 35/107 (32%), Positives = 49/107 (45%), Gaps = 10/107 (9%)
Query: 151 IKLPFSPFICHVLSFLNVAPCQLQPNAWGFLRCFEILCEHLSFTPTYP----LFFLFYKS 206
++LP PF VL+ +AP QL PNAW L F LC P P +F F+ +
Sbjct: 98 MRLPLQPFFAAVLAHFGLAPSQLTPNAWRLLAGFAALCRRAGVAPPPPPPLTVFRHFFTA 157
Query: 207 VTTKPTSVKWVPLLARKNMSR---FTALKS-HYKVWQKKYFKVMESP 249
+ P + W AR + S FT L + W K+ F ++ SP
Sbjct: 158 IGCPPGA--WYSFRARGSASTGSLFTRLSNVTMNPWWKEQFILVSSP 202
>UniRef100_UPI00001224EC UPI00001224EC UniRef100 entry
Length = 3376
Score = 49.3 bits (116), Expect = 4e-04
Identities = 79/357 (22%), Positives = 140/357 (39%), Gaps = 21/357 (5%)
Query: 323 KGKVDANPIKMKEYLAQSAAAAKKRAAETEQKKKNEGTSGSDNVRDPKRQKTSGAAGGKP 382
K + DA K + A+ AK + E ++K K E + + D K +K + A
Sbjct: 1137 KKEADAKTKKEADDKAKKDLEAKTKK-EADEKAKKEAEAKTKKEADDKARKEAEAKKEAE 1195
Query: 383 LHQSTLDSKSRPAEKKKGHDNVPPPQQDSSTL----INRPSTPFGQAGPSSAIGGEAPPP 438
+ D + A KK D+ + D + + + + EA
Sbjct: 1196 AKKEADDKAKKEAVAKKVADDKTKKEADDKARNEAEAKKEAEDKAKKEDDAKTKKEADDK 1255
Query: 439 LLNLSDPHFNGLEFMTRTFDNQIHKDISGQGPPNIASMAIHHALSAASTVAGMAQCVKEL 498
+D N + + + KD+ + A L A + + KE
Sbjct: 1256 AKKKADEKAN------KEAEAKTKKDVEAKTKKEADDKA-KKDLEAKTKKEADEKAKKE- 1307
Query: 499 IAAKNRFEKKAADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDLA-ASDLLLQKTKSL 557
A+ + +K+A K A ++AK D+E NKK A+EK K E A D ++ ++
Sbjct: 1308 --AEAKIKKEAEAKKEADDKAKKDSEAKNKK--EADEKARKEAETKKEAEDKAKKEAETK 1363
Query: 558 KEAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRI 617
KEA K + K + ++KK D+ ++SI +S + F ++ VE D S
Sbjct: 1364 KEAELGKSDETKPKKRVVKKKTDKSDSSISQISSADTSSEFTVQEKPAEKVEQLPDESTS 1423
Query: 618 GWLKEI---KDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQA 671
+K+ ++ +V + +S D P +PEE E ++ + +K D A
Sbjct: 1424 ATIKKAAQPQEPEVEKKELLSDDKKPVEGKPPKPEESDETTKKRVIKKKTQKSDSVA 1480
Score = 40.8 bits (94), Expect = 0.13
Identities = 53/252 (21%), Positives = 95/252 (37%), Gaps = 26/252 (10%)
Query: 323 KGKVDANPIKMKEYLAQSAAAAKKRAAETEQKKKNEGTSGSDNVRDPKRQKTSGAAGGKP 382
K + +A K + A++ A AKK A + +K + T + + K A K
Sbjct: 905 KKEAEAKTKKEADDKARNEAEAKKEAEDKAEKGADAKTKKEADDKAKKEAVAKKEADDKT 964
Query: 383 LHQSTLDSKSRPAEKKKGHDNVPPPQQDSSTLINRPSTPFGQAGPSSAIGGEAPPPLLNL 442
++ + ++ KK+ D V ++D + + + + I EA N
Sbjct: 965 KKEAEAEKEAEAKTKKEADDKV---KKDLEAKTKKEADEKAKKEAEAKIKKEADDKARNE 1021
Query: 443 SDPHFNGLEFMTRTFDNQIHKDISGQGPPNIASMAIHHALSAASTVAGMAQCVKELIAAK 502
++ + + D + K+ + ++ +M K+ K
Sbjct: 1022 AEAKKEAEDKAKKEADAKTKKEADDKAKKDLEAMT------------------KKEADEK 1063
Query: 503 NRFEKKAADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQKTKSLKEAIN 562
+ E +A K A E+AK +AE K K E K K +D A DL + K+ KEA
Sbjct: 1064 AKKEAEAKTKKEADEKAKKEAEAKTK--KDVEAKTKKEADDKAKKDL---EAKTKKEADE 1118
Query: 563 DKHTAVQAKYQK 574
+AK +K
Sbjct: 1119 KAKKEAEAKIKK 1130
>UniRef100_Q9NDI9 Merozoite surface protein 3g [Plasmodium vivax]
Length = 969
Score = 47.4 bits (111), Expect = 0.001
Identities = 79/357 (22%), Positives = 127/357 (35%), Gaps = 31/357 (8%)
Query: 342 AAAKKRAAETEQKK-KNEGTSGSDNVRDPKRQKTSGAAGGKPLHQSTLDSKSRPAEKKKG 400
A K + AE E KK K+E + + K+Q K + +K A+ +
Sbjct: 566 ACEKAKTAEQEAKKAKDEAVKAAKEAEEAKKQAEKAEKITKTATEEANKAKEEEAKASEA 625
Query: 401 HDNVPPPQQDSSTLINRPSTPFGQAGPSSAIGGEAPPPLLNLSDPHFNGLEFMTRTFDNQ 460
D + + F ++ P + L+ + +N
Sbjct: 626 KQEAETKAGDVDEEVYAVNVEFESVKAAAKAAAHHKVPEI---------LDKEKKNAENA 676
Query: 461 IHKDISGQGPPNIASMAIHHALSAASTVAGMAQCVKE---LIAAKNRFEKKAADYKTAYE 517
K + + + A T AG AQ E IAA EK + + ++ E
Sbjct: 677 AKKASAKATEAKTTAETATKKATEAKTAAGNAQKASENAKAIAADVLAEKASTEAQSLKE 736
Query: 518 RAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQKTKSLKEAINDKHTAVQAKYQ-KLE 576
AK A K + EEK + D AA+D Q + S +A K A QAK + LE
Sbjct: 737 EAKKLAADIKKSNVTNEEKAKR---DKAANDAAHQASLSASKAKEAKTAATQAKGEVALE 793
Query: 577 KKYDRLNASILGRASLQYAQGFLAAK----------EQINVVEPG-FDLSRIGWLKEIKD 625
KK + ++ A+ A + EQ++V G +L+ + ++
Sbjct: 794 KKKEESAKAVEAAKEAMKARDKAAFELLKLKKQDVLEQVDVSPSGNNNLNDVDEQVSLEV 853
Query: 626 GQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQAGTSQGNNANNE 682
G+ + D D PQ +E E D ED E +E Q++ T A E
Sbjct: 854 GEQENESD---DAPPQETEEVTEEGDEEDEEEMEEDEIQDESGHTEETPTEQAAEQE 907
>UniRef100_Q9P571 Related to actin-interacting protein AIP3 [Neurospora crassa]
Length = 1127
Score = 46.6 bits (109), Expect = 0.002
Identities = 37/135 (27%), Positives = 61/135 (44%), Gaps = 10/135 (7%)
Query: 558 KEAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRI 617
KE + + A+Q +Q + +R A L + LQ KE + VE G L +
Sbjct: 942 KEGVLGEVRALQPNHQSRLEAIER--AEKLRQKELQTRMVNPLTKELASFVEEG-KLKKS 998
Query: 618 GWLKEIKDGQVVGDDDISLD-------LLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQ 670
G ++E++ + DD I + L+ + DD+ E EED E+ E+ E+ E+E+ +
Sbjct: 999 GGVEEVERARKAKDDQIRREVWERMNGLIAENDDDGEDEEDDEEEEEEEEEEEGEEEEEE 1058
Query: 671 AGTSQGNNANNENLA 685
G Q E A
Sbjct: 1059 EGEEQEEGEEGEEKA 1073
>UniRef100_Q9VKI0 CG6392-PA [Drosophila melanogaster]
Length = 2013
Score = 45.8 bits (107), Expect = 0.004
Identities = 25/101 (24%), Positives = 54/101 (52%), Gaps = 3/101 (2%)
Query: 497 ELIAAKNRFEKKAADYKTAYE---RAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQK 553
E+++ +++ E + + A + TD E +K+L +++ +L E A D Q
Sbjct: 704 EIVSVRDKLESVESAFNLASSEIIQKATDCERLSKELSTSQNAFGQLQERYDALDQQWQA 763
Query: 554 TKSLKEAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQY 594
++ +++KH VQ KYQKL+++Y++L + +S ++
Sbjct: 764 QQAGITTLHEKHEHVQEKYQKLQEEYEQLESRARSASSAEF 804
>UniRef100_Q6C905 Yarrowia lipolytica chromosome D of strain CLIB99 of Yarrowia
lipolytica [Yarrowia lipolytica]
Length = 1098
Score = 45.4 bits (106), Expect = 0.005
Identities = 51/197 (25%), Positives = 87/197 (43%), Gaps = 24/197 (12%)
Query: 496 KELIAAKNRFEKKA--ADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQK 553
K + A+N+ + A A K +R K +AE KK K+ + E+ +++ K
Sbjct: 33 KNAVKAENKAKNDAREAKKKEHEQRKKEEAEAMKKKAKNFNKSKFGKKEEEKVTEVKPSK 92
Query: 554 TKSL-------------KEAINDKHTAVQAKYQKLEKKYDRLNASILG----RASLQYAQ 596
TK +++ +DK T + K + E K D L A IL L+ +
Sbjct: 93 TKGSDKTEKSKGSRTKSQDSSSDKSTKTKTKTK--EFKPDSLLAEILELGGTEEDLKLVE 150
Query: 597 GFLAAKEQINVVEP--GFDLSRIGWLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGED 654
++ + VV P G + LK +KD + GD ++ + D E E EED ++
Sbjct: 151 DLSDGEDDV-VVNPSNGSKIDSGDLLKFMKDIGLDGDVPQLVEDVKDEDVEEEQEEDDDE 209
Query: 655 GNEQHRNEDQEKEDPQA 671
G E +ED E++D ++
Sbjct: 210 GMEDDEDEDTEEDDEES 226
>UniRef100_Q01448 Histone promoter control 2 protein [Saccharomyces cerevisiae]
Length = 623
Score = 45.4 bits (106), Expect = 0.005
Identities = 75/327 (22%), Positives = 133/327 (39%), Gaps = 26/327 (7%)
Query: 258 SDNNPLFPFYWTKNPRRKISVSYDALDESEQGVADYLRTLPVISGHDLIEASKSGTLNQF 317
SD+ LF + K RKI + L ++ V + +ISG S SG +
Sbjct: 45 SDSEDLFNKFSNKKTNRKIPNIAEELAKNRNYVKGASPSPIIISGSS--STSPSGPSSSS 102
Query: 318 FTTMG--KGKVDANPIKMKEYLAQSAAAAKKRAAETEQKKKNEGTSGSDNVRDPKRQKTS 375
MG + + N +++ ++ K +TE+K++N DN P+R +S
Sbjct: 103 TNPMGIPTNRFNKNTVELYQHSPSPVMTTNK--TDTEEKRQNN--RNMDNKNTPERGSSS 158
Query: 376 GAAGGKPLHQSTLDSKSRPAEKKKGHDNVPPPQQDSSTLINR-PSTPFGQAGPSSAIGGE 434
AA K L S+L + S + K H N ++S+ N PS ++I
Sbjct: 159 FAA--KQLKISSLLTISSNEDSKTLHINDTNGNKNSNAASNNIPSAYAELHTEGNSIESL 216
Query: 435 APPPLLNLSDPHFNGLE----FMTRTFDNQIHKDIS-GQGPPN--IASMAIHHALSAAST 487
PP S P L T+ D I KD G P IA + L ST
Sbjct: 217 IKPP----SSPRNKSLTPKVILPTQNMDGTIAKDPHLGDNTPGILIAKTSSPVNLDVEST 272
Query: 488 VAGMAQCVKELIAAKNRFEKKAADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDLAAS 547
+ + K + K K A+ K + +R+ + ++ + S+++ ++ + +++
Sbjct: 273 AQSLGKFNKSTNSLKAALTKAPAE-KVSLKRSISSVTNSDSNISSSKKPTSEKAKKSSSA 331
Query: 548 DLLLQK---TKSLKEAINDKHTAVQAK 571
+L K TK+ K+A ++ + + K
Sbjct: 332 SAILPKPTTTKTSKKAASNSSDSTRKK 358
>UniRef100_Q26019 Polymorphic antigen precursor [Plasmodium falciparum]
Length = 380
Score = 45.1 bits (105), Expect = 0.007
Identities = 59/260 (22%), Positives = 106/260 (40%), Gaps = 26/260 (10%)
Query: 448 NGLEFMTRTFDNQIHKDISGQGPPNIASMAIHHALSAASTVAGMAQ-CVKELIAAK--NR 504
N +EF F KDI AS A A + A+ KE + K ++
Sbjct: 73 NSMEF-GEGFSADDQKDIEAYKKAKQASQDAEQAAKDAENASKEAEEAAKEAVNLKESDK 131
Query: 505 FEKKAADYKTAYERAKTDAETANKKLKSAEE--KCAKLTEDLAASDLLLQKTKSLKEAIN 562
KA + TA +AK ETA K AE K ++ E + +L +KTK E
Sbjct: 132 SYTKAKEACTAASKAKKAVETALKAKDDAETALKTSETPEKPSRINLFSRKTKEYAEKAK 191
Query: 563 DKHTAVQAKYQKLEK---------KYDRLNASILGRASLQYAQG-------FLAAKEQIN 606
+ + + YQK + YD + G ++ + ++++K++ N
Sbjct: 192 NAYEKAKNAYQKANQAVLKAKEASSYDYILGWEFGGGVPEHKKEENMLSHLYVSSKDKEN 251
Query: 607 VVEPGFDL--SRIGWLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQ 664
+ + D+ + +E ++ ++ ++ + D+E E EE+ E+ NE + ++Q
Sbjct: 252 ISKENDDVLDEKEEEAEETEEEELEEKNEEETESEISEDEEEEEEEEKEEENE--KKKEQ 309
Query: 665 EKEDPQAGTSQGNNANNENL 684
EKE Q + +NL
Sbjct: 310 EKEQSNENNDQKKDMEAQNL 329
>UniRef100_UPI00004375BD UPI00004375BD UniRef100 entry
Length = 1034
Score = 44.7 bits (104), Expect = 0.009
Identities = 67/302 (22%), Positives = 114/302 (37%), Gaps = 45/302 (14%)
Query: 332 KMKEYLAQSAAAAKKRAAETEQKKKNEGTSGSDNVRDPKRQKTSGAAGGKPLHQSTLDSK 391
K K +S A + +E +QK++ GT SD + + + A+ P + + +
Sbjct: 193 KEKPCKPESKDTAADQMSE-KQKEEKRGTGESDKLTKALTSEVT-ASNPSPPYDAQPSTP 250
Query: 392 SRPAEKKKGHDN------VPPPQQDSSTLINRPSTPFGQAGPSSAIGGEAPPPLLNLSDP 445
S K HD+ +PPP DS+ ++ PS+ + P+ P LN +
Sbjct: 251 SIEEALKIIHDSERPQSSLPPPSGDSAFFLHNPSSSNSASDPNE------PEDSLNKARA 304
Query: 446 HFNGLEFMTRTFDNQIHKDISGQGPPNIASMAIHHALSAASTVAGMAQCVKELIAAKNRF 505
L + D IH SG P++ L S MA + I +
Sbjct: 305 DDADLNSTSTDMDTGIHVHTSGSPCPSL--------LGTRSLATSMASSLGSAIRMTSFA 356
Query: 506 EKKAADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQKTKSLKEAINDKH 565
E+K R+ T A++T S LL + LK + +K
Sbjct: 357 EQKFRKLNHGDGRSIT----------PDTHSPARITSPRDPSHLLASEMVQLKMKLEEKR 406
Query: 566 TAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRIGWLKEIKD 625
A++A+ +K+E + R + +GR + +NVV +S +G E D
Sbjct: 407 RAIEAQKKKVEAAFTR-HRQRMGRTAF------------LNVVRRKGVVSPLGGSSEGPD 453
Query: 626 GQ 627
G+
Sbjct: 454 GR 455
>UniRef100_Q76MT4 ABTAP [Rattus rattus]
Length = 842
Score = 44.7 bits (104), Expect = 0.009
Identities = 40/169 (23%), Positives = 71/169 (41%), Gaps = 27/169 (15%)
Query: 513 KTAYERAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQKTKSLKEAINDKHTAVQAKY 572
K +++ K D+E + K +EE + +DL ++ + K D T+ K
Sbjct: 142 KNSHKSPKIDSEVSPK---DSEESLQSRKKKRDTTDLSVEASPKGKLRTKDPSTSAMVK- 197
Query: 573 QKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVV-------EPGFDLSRIGWLKEIKD 625
++++ G + + Q + AK+ + E S +G +E +
Sbjct: 198 ----------SSTVSGSKAKREKQAVIMAKDNAGRMLHEEAPEEDSDSASELGRDEE-SE 246
Query: 626 GQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQAGTS 674
G++ DD S D DDE+E EE+ EDG E+ E++E E S
Sbjct: 247 GEITSDDRASAD-----DDENEDEEEEEDGEEEEEEEEEEDESDDESDS 290
>UniRef100_UPI000023F6CB UPI000023F6CB UniRef100 entry
Length = 1493
Score = 44.3 bits (103), Expect = 0.012
Identities = 55/204 (26%), Positives = 79/204 (37%), Gaps = 39/204 (19%)
Query: 492 AQCVKELIAAKNRFEKKAADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDLAASDLLL 551
A+ KEL AA EK D ERA E ++ AEE A ++L A L
Sbjct: 1063 AKSSKELAAASRDLEKLENDINNQGERA----EELQAQVAEAEEGLATKKQELKALKAEL 1118
Query: 552 -QKTKSLKEA----------INDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLA 600
+KT L E + + A+ Q+L D+L+ +L
Sbjct: 1119 DEKTDELNETRAMEIEMRNKLEENQKALAENQQRLRHWNDKLSKIVL------------- 1165
Query: 601 AKEQINVVEPGFDLSRIGWLKEIKDGQVVGDDDISLDLLPQFDDE--SEPEEDGEDGNEQ 658
+ I+ + G K+ K Q D DI +D PQ D + PEE+GE
Sbjct: 1166 --QNIDDLTGGSTNGHSKPRKQPKPQQNDDDGDIDMDDAPQDDVDMTDAPEEEGE----- 1218
Query: 659 HRNEDQEKEDPQAGTSQGNNANNE 682
ED ++E+ + G Q N NE
Sbjct: 1219 --QEDADEEEEEEGEEQMNGQPNE 1240
>UniRef100_Q8C2E3 Mus musculus 2 days neonate thymus thymic cells cDNA, RIKEN full-
length enriched library, clone:E430025K03 product:RAN
GTPase activating protein 1, full insert sequence [Mus
musculus]
Length = 589
Score = 44.3 bits (103), Expect = 0.012
Identities = 33/117 (28%), Positives = 59/117 (50%), Gaps = 24/117 (20%)
Query: 559 EAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRIG 618
EA+ DK A+ +KL+ LN + LG EQ+ V F+++++
Sbjct: 315 EAVADK-----AELEKLD-----LNGNALGEEGC----------EQLQEVMDSFNMAKV- 353
Query: 619 WLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQAGTSQ 675
L + D + G+D+ + + D+E E EED ED +E+ +++E+E PQ G+ +
Sbjct: 354 -LASLSDDE--GEDEDEEEEGEEDDEEEEDEEDEEDDDEEEEEQEEEEEPPQRGSGE 407
>UniRef100_Q7TMW1 Rangap1 protein [Mus musculus]
Length = 589
Score = 44.3 bits (103), Expect = 0.012
Identities = 33/117 (28%), Positives = 59/117 (50%), Gaps = 24/117 (20%)
Query: 559 EAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRIG 618
EA+ DK A+ +KL+ LN + LG EQ+ V F+++++
Sbjct: 315 EAVADK-----AELEKLD-----LNGNALGEEGC----------EQLQEVMDSFNMAKV- 353
Query: 619 WLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQAGTSQ 675
L + D + G+D+ + + D+E E EED ED +E+ +++E+E PQ G+ +
Sbjct: 354 -LASLSDDE--GEDEDEEEEGEEDDEEEEDEEDEEDDDEEEEEQEEEEEPPQRGSGE 407
>UniRef100_Q6ZPH4 MKIAA1835 protein [Mus musculus]
Length = 646
Score = 44.3 bits (103), Expect = 0.012
Identities = 33/117 (28%), Positives = 59/117 (50%), Gaps = 24/117 (20%)
Query: 559 EAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRIG 618
EA+ DK A+ +KL+ LN + LG EQ+ V F+++++
Sbjct: 372 EAVADK-----AELEKLD-----LNGNALGEEGC----------EQLQEVMDSFNMAKV- 410
Query: 619 WLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQAGTSQ 675
L + D + G+D+ + + D+E E EED ED +E+ +++E+E PQ G+ +
Sbjct: 411 -LASLSDDE--GEDEDEEEEGEEDDEEEEDEEDEEDDDEEEEEQEEEEEPPQRGSGE 464
>UniRef100_Q6NZB5 Rangap1 protein [Mus musculus]
Length = 589
Score = 44.3 bits (103), Expect = 0.012
Identities = 33/117 (28%), Positives = 59/117 (50%), Gaps = 24/117 (20%)
Query: 559 EAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRIG 618
EA+ DK A+ +KL+ LN + LG EQ+ V F+++++
Sbjct: 315 EAVADK-----AELEKLD-----LNGNALGEEGC----------EQLQEVMDSFNMAKV- 353
Query: 619 WLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQAGTSQ 675
L + D + G+D+ + + D+E E EED ED +E+ +++E+E PQ G+ +
Sbjct: 354 -LASLSDDE--GEDEDEEEEGEEDDEEEEDEEDEEDDDEEEEEQEEEEEPPQRGSGE 407
>UniRef100_Q9P5J9 Related to BCS1 protein [Neurospora crassa]
Length = 779
Score = 44.3 bits (103), Expect = 0.012
Identities = 53/189 (28%), Positives = 74/189 (39%), Gaps = 24/189 (12%)
Query: 399 KGHDNVPPPQQDSSTLINRPSTPFGQAGPSSAIGGEAPPPLLNLSDPHFNGLEFMTRTFD 458
+G D P DS L PST AG ++A EA L ++ +E + D
Sbjct: 477 EGDDVDTPSDADSKDLFKTPSTSTSPAGAAAAAAAEARRELAR-NEATLKVVELADQFAD 535
Query: 459 NQIHKDISGQGPPNIASMAIHHALSAASTVAGMAQCVKELIAAKNRFEKKAADYKTAYE- 517
+ S P I + H +A + V G Q V + A K + EK D + A E
Sbjct: 536 KIPAHEFS---PAEIQGFLLKHKRNATAAVEGAEQWVVD--ARKEKMEK---DLREARER 587
Query: 518 RAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQKTKSLKEAINDKHTAVQAKYQKLEK 577
R K + A KK EEK K KT++ K + KH V +K K
Sbjct: 588 REKEEKAEAEKKGTKKEEKGGK------------DKTETKKSKKSSKHNKVTSK--KSSS 633
Query: 578 KYDRLNASI 586
K + A+I
Sbjct: 634 KKSTMKAAI 642
>UniRef100_P46061 Ran GTPase-activating protein 1 [Mus musculus]
Length = 589
Score = 44.3 bits (103), Expect = 0.012
Identities = 33/117 (28%), Positives = 59/117 (50%), Gaps = 24/117 (20%)
Query: 559 EAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINVVEPGFDLSRIG 618
EA+ DK A+ +KL+ LN + LG EQ+ V F+++++
Sbjct: 315 EAVADK-----AELEKLD-----LNGNALGEEGC----------EQLQEVMDSFNMAKV- 353
Query: 619 WLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQEKEDPQAGTSQ 675
L + D + G+D+ + + D+E E EED ED +E+ +++E+E PQ G+ +
Sbjct: 354 -LASLSDDE--GEDEDEEEEGEEDDEEEEDEEDEEDDDEEEEEQEEEEEPPQRGSGE 407
>UniRef100_UPI000023DA4F UPI000023DA4F UniRef100 entry
Length = 1075
Score = 43.9 bits (102), Expect = 0.016
Identities = 41/174 (23%), Positives = 73/174 (41%), Gaps = 11/174 (6%)
Query: 506 EKKAADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDLAASDLLLQKTKSLKEAINDKH 565
+KK A+ + +++K D E AEEKC L + L+ D ++ ++ + +
Sbjct: 891 KKKDAEVEALQQQSKDDLEQQKSMTLKAEEKCQHLEKQLSGGDAGMKDMQARLARMESEL 950
Query: 566 TAVQAKYQKLEKKYDRLNASI---LGRASLQYAQGFLAAKEQINVVEPGFD--LSRIGWL 620
+A EK +L + +A + + +E + + D L G L
Sbjct: 951 SAANKARDAAEKNLSKLKEAAKEEAAKAKSEVEKQLSELREAREMTQSELDDLLMVFGDL 1010
Query: 621 KE----IKDGQVVGDDDISLDLLPQFDDESEPEEDGE--DGNEQHRNEDQEKED 668
+E KD + + +S DD+ E EE+G DG+E N D EK++
Sbjct: 1011 EEKMTKYKDRLIELGETVSDGEDDDEDDDDEEEENGSDADGSEGSDNNDDEKDE 1064
>UniRef100_UPI0000219DB0 UPI0000219DB0 UniRef100 entry
Length = 709
Score = 43.9 bits (102), Expect = 0.016
Identities = 32/102 (31%), Positives = 53/102 (51%), Gaps = 8/102 (7%)
Query: 508 KAADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDLAA----SDLLLQKTKSLKEAIND 563
K A + E A ETA + + + K +KL EDLAA SD L Q+ ++ K++++D
Sbjct: 256 KIAKLEEDLEAANKSTETAQAEAATLKTKISKLEEDLAAAKSQSDKLTQEAEAQKKSLDD 315
Query: 564 KHTAVQAKYQKLEKKYDRLNASILG----RASLQYAQGFLAA 601
+ +QAK ++ E +L A+ +ASL+ Q L +
Sbjct: 316 ANAQIQAKTKETEDLLAKLKAAEASVAEKQASLEKTQAELTS 357
Score = 43.1 bits (100), Expect = 0.027
Identities = 51/253 (20%), Positives = 92/253 (36%), Gaps = 22/253 (8%)
Query: 341 AAAAKKRAAETEQKKKNEGTSGSDNVRDPKRQKTSGAAGGKPLHQSTLDS----KSRPAE 396
AAA K++ ++E+ K + S + + K H T + K+ A+
Sbjct: 459 AAAEAKKSLDSEKASKADANSALETQKSAVADAQKAVEAEKKAHAETKSALEAEKTAAAD 518
Query: 397 KKKGHDNVPPPQQDSSTLINRPSTPFGQAGPSSAIGGEAPPPLLNLSDPHFNGLEFMTRT 456
KK ++ Q++ + + SS G A +
Sbjct: 519 AKKAVESEKSTAQEAQKALEAEKAAHAETKKSSEAGTSAVTEAQKALES----------- 567
Query: 457 FDNQIHKDISGQGPPNIASMA-IHHALSAASTVAGMAQCVKELIAAKNRFEKKAADYKTA 515
+ H + + A++A AL A A AQ E A KKA + + A
Sbjct: 568 -EKAAHAEAKKASEADKAALADAKKALDAEKAAAADAQKALEAEKAAAADAKKALEAEKA 626
Query: 516 YERAKTDAET-----ANKKLKSAEEKCAKLTEDLAASDLLLQKTKSLKEAINDKHTAVQA 570
+ + ET A K + A+ K K ED A + + + + ++ K TA ++
Sbjct: 627 SAKKALETETAATKEAKKAAEDAQSKLKKAEEDTKALNKRVADAEKESKELSTKATAAES 686
Query: 571 KYQKLEKKYDRLN 583
+ ++LE K L+
Sbjct: 687 RTKELEAKVKELS 699
Score = 34.7 bits (78), Expect = 9.7
Identities = 45/206 (21%), Positives = 79/206 (37%), Gaps = 20/206 (9%)
Query: 489 AGMAQCVKELIAAKNRFEKKAADYKTAYERAKTDAETANKKLKSAEEKCAKLTEDL-AAS 547
A + Q + E K +K+ + A E+AK + +KLKS + + L
Sbjct: 108 AAVDQQLAEAQKGKEGAQKETLEKIDALEKAKAELNAQVEKLKSEVADVSSKNDSLRQEQ 167
Query: 548 DLLLQKTKSLKEAINDKHTAVQAKYQKLEKKYDRLNASILGRASLQYAQGFLAAKEQINV 607
LL++T S K+ + + AK L D A L RA+ + A +EQ+
Sbjct: 168 SKLLEETNSAKDTLK---AELDAKIVALTSDLDAAKAD-LSRANEEAATTKTKLEEQVKT 223
Query: 608 VEPGFDLSRIGWLKEIKDGQVVGDDDISLDLLPQFDDESEPEEDGEDGNEQHRNEDQE-- 665
++ D ++ K+ + G ++ ++ ++ EED E N+ E
Sbjct: 224 LQAELDATK----KDAQAAASKGTEEAKSEVTSLNTKIAKLEEDLEAANKSTETAQAEAA 279
Query: 666 ---------KEDPQAGTSQGNNANNE 682
+ED A SQ + E
Sbjct: 280 TLKTKISKLEEDLAAAKSQSDKLTQE 305
>UniRef100_Q94GJ8 Putative retrotransposon protein [Oryza sativa]
Length = 577
Score = 43.9 bits (102), Expect = 0.016
Identities = 128/571 (22%), Positives = 209/571 (36%), Gaps = 94/571 (16%)
Query: 141 VYGYFFLDLNIKLPFSPFICHVLSFLNVAPCQLQPNAWGFLRCFEILCEHLSFTPTYPLF 200
V+ +FF LP S F +L F ++ L PN+ + F CE +F P F
Sbjct: 57 VFSHFFYG-GFSLPTSKFFRGILEFYGISLHHLNPNSIVHIANFVHACE--AFLGIKPHF 113
Query: 201 FLFYKSVTTKPTSVKWVPLLA--------RKNMSR---FTALKSHYKVWQKKYFKVMESP 249
LF++ + KP K P + R +S+ K+ K W +F V ++P
Sbjct: 114 ALFHRILFLKPQPNKNKPCVVGGGVGFQLRGTLSQKYFSMPFKTSNKGWHANWFYV-QNP 172
Query: 250 EIKNLFRDSDNNPLFPFYWTKNP-----RRKISVSYDALDESEQGVADYLRTLPVISGHD 304
E L S P + W P + +++ L EQG+ T I
Sbjct: 173 E-PALSEYSCLPPTYQDTWNSLPMGDEAAQALTLFNCMLKLKEQGLQGEQITRHYIKSRM 231
Query: 305 L-IEASKSGTLNQFFTTMGKGKVDANPIKMKEYLAQSAAAAKKRAAETEQKKKNEGTSGS 363
I+ K ++ F GK + + K ++ +A AA+ E K+N + GS
Sbjct: 232 APIKERKPSDIDPF------GKDSDSTVNSKTAEKEAGSAT---AADAEPSKENR-SIGS 281
Query: 364 DNVRDPKRQKTSGAAGGKPLHQSTLDSKSRPAEKKKG---HDNVPPPQ-QDSSTLINRPS 419
+V + G + T ++ P + G +D++P + Q SS +N+
Sbjct: 282 QSVTSDVKAGNEPPTGNQSADTETSANQEPPTRNQPGDEQNDDIPETEGQASSPPLNKGR 341
Query: 420 TPFGQAGPSSA----IGGEAPPP---------LLNLSDPHFNGLEFMTRTFDNQIHKDIS 466
AGP ++ + G A PP L PH LE TR ++H+
Sbjct: 342 --ITDAGPEASTFDKVEGLARPPPKIITAKTATLEKLVPHLGTLE-ETRA---KLHE--- 392
Query: 467 GQGPPNIASMAIHHALSAASTVAGMAQCVKELIAAKNRFEKKAADYKTAYERAKTDAETA 526
+A H VA + EL ++ EK D + A D E
Sbjct: 393 ---TRELARKTEH---DLTDRVAELQNANFELSSSLKALEKDKDDLTKERDLAIKDFE-- 444
Query: 527 NKKLKSAEEKCAKLTEDLAASDLLLQKTKSLKEAINDKHTAVQAKYQKLEKKYDRLNASI 586
++K+KS A D+L+ + K L+ A N+ A Q + Y+ S
Sbjct: 445 DRKIKSQ-----------AQFDILVNRIKKLEGARNEVANAATPLIQAM--FYNNNGPST 491
Query: 587 LGRASLQYAQGFLAAKEQINVVEPGFDLSRIGW----------LKEIKDGQVVGDDDISL 636
+ + G A + P D+ I L+ I D Q + I+
Sbjct: 492 IDVVEEAGSIGASMALAMTKSLYPNIDIDAIDGFTDGTSEEDALELINDAQKAA-NKIAA 550
Query: 637 DLLPQFDDESEPEEDGEDGNEQHRNEDQEKE 667
D++ +F D + G G+ +E E E
Sbjct: 551 DVVERFQDNNL----GPTGSHDSDDEKTESE 577
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,229,907,926
Number of Sequences: 2790947
Number of extensions: 56634027
Number of successful extensions: 242745
Number of sequences better than 10.0: 1255
Number of HSP's better than 10.0 without gapping: 310
Number of HSP's successfully gapped in prelim test: 978
Number of HSP's that attempted gapping in prelim test: 236352
Number of HSP's gapped (non-prelim): 5050
length of query: 685
length of database: 848,049,833
effective HSP length: 134
effective length of query: 551
effective length of database: 474,062,935
effective search space: 261208677185
effective search space used: 261208677185
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0037a.11


