

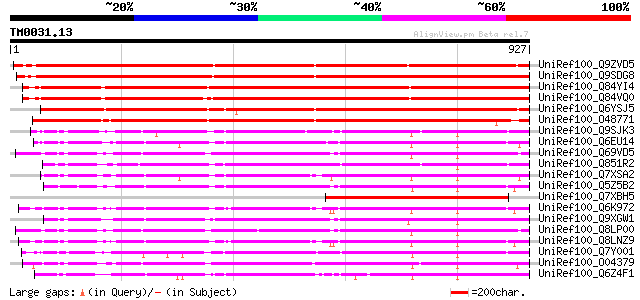
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0031.13
(927 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ZVD5 Putative Argonaute (AGO1) protein [Arabidopsis ... 1357 0.0
UniRef100_Q9SDG8 ESTs AU068544 [Oryza sativa] 1309 0.0
UniRef100_Q84YI4 ARGONAUTE9 protein [Arabidopsis thaliana] 1239 0.0
UniRef100_Q84VQ0 PAZ (Piwi Argonaut and Zwille) family [Arabidop... 1236 0.0
UniRef100_Q6YSJ5 Putative ARGONAUTE9 protein [Oryza sativa] 1079 0.0
UniRef100_O48771 Argonaute (AGO1)-like protein [Arabidopsis thal... 941 0.0
UniRef100_Q9SJK3 Argonaute-like protein At2g27880 [Arabidopsis t... 525 e-147
UniRef100_Q6EU14 Putative argonaute protein [Oryza sativa] 502 e-140
UniRef100_Q69VD5 ZLL/PNH homologous protein [Oryza sativa] 501 e-140
UniRef100_Q851R2 Putative argonaute protein [Oryza sativa] 501 e-140
UniRef100_Q7XSA2 OSJNBa0005N02.3 protein [Oryza sativa] 496 e-138
UniRef100_Q5Z5B2 Putative AGO1 homologous protein [Oryza sativa] 494 e-138
UniRef100_Q7XBH5 Zwille pinhead-like protein [Oryza sativa] 493 e-138
UniRef100_Q6K972 AGO1 homologous protein [Oryza sativa] 493 e-138
UniRef100_Q9XGW1 PINHEAD protein [Arabidopsis thaliana] 493 e-138
UniRef100_Q8LP00 ZLL/PNH homologous protein [Oryza sativa] 493 e-137
UniRef100_Q8LNZ9 AGO1 homologous protein [Oryza sativa] 493 e-137
UniRef100_Q7Y001 Putative leaf development and shoot apical meri... 484 e-135
UniRef100_O04379 Argonaute protein [Arabidopsis thaliana] 480 e-134
UniRef100_Q6Z4F1 Putative leaf development protein Argonaute [Or... 462 e-128
>UniRef100_Q9ZVD5 Putative Argonaute (AGO1) protein [Arabidopsis thaliana]
Length = 924
Score = 1357 bits (3511), Expect = 0.0
Identities = 669/926 (72%), Positives = 792/926 (85%), Gaps = 18/926 (1%)
Query: 7 ENGNG-NGNGNEDLMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLGS 65
E+ NG NG+G + +PPPPP ++P +VEPV+V E +KK P R+P+ARKG G+
Sbjct: 12 ESANGANGSGVTEALPPPPP--VIPPNVEPVRVKT-----ELAEKKGPVRVPMARKGFGT 64
Query: 66 KGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETYGSELN 125
+G K+PLLTNHFKV VAN GHFF YSVAL Y+DGRPVE KGVGRK++DKV +TY S+L+
Sbjct: 65 RGQKIPLLTNHFKVDVANLQGHFFHYSVALFYDDGRPVEQKGVGRKILDKVHQTYHSDLD 124
Query: 126 GKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDG-ASTNDSDKKRMRRP 184
GK+FAYDGEKTLFT G+L NK++F+VVLE+V + R NGN SP+G S +D D+KR+RRP
Sbjct: 125 GKEFAYDGEKTLFTYGALPSNKMDFSVVLEEVSATRANGNGSPNGNESPSDGDRKRLRRP 184
Query: 185 YHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSF 244
SK F+VEIS+AAKIPLQA+ NA+RGQESEN QEAIRVLDIILRQHAA+QGCLLVRQSF
Sbjct: 185 NRSKNFRVEISYAAKIPLQALANAMRGQESENSQEAIRVLDIILRQHAARQGCLLVRQSF 244
Query: 245 FHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNVR 304
FHNDP N VGG +LGCRGFHSSFRTTQ G+SLN+DV+TTMII+PGPVVDFLIANQN R
Sbjct: 245 FHNDPTNCEPVGGNILGCRGFHSSFRTTQGGMSLNMDVTTTMIIKPGPVVDFLIANQNAR 304
Query: 305 DPFSLDWAKAKRTLKNLRIKASPSNQEYKITGLSELPCKEQTFTMKKKGGN-NGEEDATE 363
DP+S+DW+KAKRTLKNLR+K SPS QE+KITGLS+ PC+EQTF +KK+ N NGE + TE
Sbjct: 305 DPYSIDWSKAKRTLKNLRVKVSPSGQEFKITGLSDKPCREQTFELKKRNPNENGEFETTE 364
Query: 364 EEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRS 423
+TV +YF + R IDL+YSADLPCINVGKPKRPTY+P+ELC+LV LQRYTKALTT QRS
Sbjct: 365 --VTVADYFRDTRHIDLQYSADLPCINVGKPKRPTYIPLELCALVPLQRYTKALTTFQRS 422
Query: 424 SLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKFGN 483
+LVEKSRQKP ERM VL++ALK SNY EP+L++CGI+I+S FTQVEGRVL AP+LK G
Sbjct: 423 ALVEKSRQKPQERMTVLSKALKVSNYDAEPLLRSCGISISSNFTQVEGRVLPAPKLKMGC 482
Query: 484 GEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCDVRGLVRDLIKCARLKGIPIDEPY 543
G + PRNGRWN NNK+ V P KI+ W VVNFSARC+VR +V DLIK KGI I P+
Sbjct: 483 GSETFPRNGRWNFNNKEFVEPTKIQRWVVVNFSARCNVRQVVDDLIKIGGSKGIEIASPF 542
Query: 544 EEIFEENGQFRRAPPLVRVEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAE 603
+ +FEE QFRRAPP++RVE MF+ IQ +LPG P F+LC+LP++KNSDLYGPWKKKNL E
Sbjct: 543 Q-VFEEGNQFRRAPPMIRVENMFKDIQSKLPGVPQFILCVLPDKKNSDLYGPWKKKNLTE 601
Query: 604 YGIVTQCISPTRV-NDQYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDV 662
+GIVTQC++PTR NDQYLTN+L+KINAKLGGLNS+L VE P+ ++SKVPTIILGMDV
Sbjct: 602 FGIVTQCMAPTRQPNDQYLTNLLLKINAKLGGLNSMLSVERTPAFTVISKVPTIILGMDV 661
Query: 663 SHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRE 722
SHGSPGQSD+PSIAAVVSSREWPLISKYRA VRTQ K EMI++L K+ + ED+GII+E
Sbjct: 662 SHGSPGQSDVPSIAAVVSSREWPLISKYRASVRTQPSKAEMIESLVKK-NGTEDDGIIKE 720
Query: 723 LLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVA 782
LL+DFY+SS KRKP++IIIFRDGVSESQFNQVLNIEL+QIIEACK LD WNPKFL++VA
Sbjct: 721 LLVDFYTSSNKRKPEHIIIFRDGVSESQFNQVLNIELDQIIEACKLLDANWNPKFLLLVA 780
Query: 783 QKNHHTKFFQPGSPDNVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIG 842
QKNHHTKFFQP SP+NVPPGT+IDNKICHP+NNDFY+CAHAGMIGT+RPTHYHVL D+IG
Sbjct: 781 QKNHHTKFFQPTSPENVPPGTIIDNKICHPKNNDFYLCAHAGMIGTTRPTHYHVLYDEIG 840
Query: 843 FSPDELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLT 902
FS DELQELVHSLSYVYQRST+AISVVAPICYAHLAA Q+G FMKFED+S+TSSSHGG+T
Sbjct: 841 FSADELQELVHSLSYVYQRSTSAISVVAPICYAHLAAAQLGTFMKFEDQSETSSSHGGIT 900
Query: 903 AAGVAPV-VPQLPKLQDSVSSSMFFC 927
A G P+ V QLP+L+D+V++SMFFC
Sbjct: 901 APG--PISVAQLPRLKDNVANSMFFC 924
>UniRef100_Q9SDG8 ESTs AU068544 [Oryza sativa]
Length = 904
Score = 1309 bits (3388), Expect = 0.0
Identities = 640/916 (69%), Positives = 758/916 (81%), Gaps = 15/916 (1%)
Query: 12 NGNGNEDLMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLGSKGTKLP 71
N E+L PPPP PP + EP+K D + K P R +AR G G KG +
Sbjct: 4 NSGEIEELPPPPPLPP----NAEPIKTD------DTKKLSKPKRALMARSGCGKKGQPIQ 53
Query: 72 LLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETYGSELNGKDFAY 131
LLTNHFKV++ +D F Y V L YED RPV+GKG+GRKV+DK+Q+TY SEL KDFAY
Sbjct: 54 LLTNHFKVSLKAADEFFHHYYVNLKYEDDRPVDGKGIGRKVLDKLQQTYASELANKDFAY 113
Query: 132 DGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFK 191
DGEK+LFTIG+L + EFTVVLED + +++ N G + +D+KR+RRPY +KTFK
Sbjct: 114 DGEKSLFTIGALPQVNNEFTVVLEDFNTGKSSANGGSPGNDSPGNDRKRVRRPYQTKTFK 173
Query: 192 VEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKN 251
VE++FAAKIP+ AI ALRGQESEN QEAIRV+DIILRQH+AKQGCLLVRQSFFHN+P N
Sbjct: 174 VELNFAAKIPMSAIAQALRGQESENTQEAIRVIDIILRQHSAKQGCLLVRQSFFHNNPSN 233
Query: 252 YADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNVRDPFSLDW 311
+ D+GGGV+GCRGFHSSFR TQSGLSLNIDVSTTMI++PGPVVDFL+ANQ V P +DW
Sbjct: 234 FVDLGGGVMGCRGFHSSFRATQSGLSLNIDVSTTMIVKPGPVVDFLLANQKVDHPNKIDW 293
Query: 312 AKAKRTLKNLRIKASPSNQEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEY 371
AKAKR LKNLRIK SP+N EYKI GLSE C EQ FT+K++ G+ E E++VYEY
Sbjct: 294 AKAKRALKNLRIKTSPANTEYKIVGLSERNCYEQMFTLKQRNGDGEPEGV---EVSVYEY 350
Query: 372 FVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQ 431
FV R I+LRYS D PCINVGKPKRPTY P+ELCSLV LQRYTKAL+TLQRSSLVEKSRQ
Sbjct: 351 FVKNRGIELRYSGDFPCINVGKPKRPTYFPIELCSLVPLQRYTKALSTLQRSSLVEKSRQ 410
Query: 432 KPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKFGNGEDFNPRN 491
KP ERM+VL+ LK SNY +EPML +CGI+IA GFTQV GRVLQAP+LK GNGED RN
Sbjct: 411 KPEERMSVLSDVLKRSNYDSEPMLNSCGISIARGFTQVAGRVLQAPKLKAGNGEDLFARN 470
Query: 492 GRWNLNNKKVVRPAKIEHWAVVNFSARCDVRGLVRDLIKCARLKGIPIDEPYEEIFEENG 551
GRWN NNK++++ + IE WAVVNFSARC++R LVRD+IKC +KGI +++P++ + EE+
Sbjct: 471 GRWNFNNKRLIKASSIEKWAVVNFSARCNIRDLVRDIIKCGGMKGIKVEDPFD-VIEEDP 529
Query: 552 QFRRAPPLVRVEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCI 611
RRAP RV+ M +++QK+LPG P FLLC+L ERKNSD+YGPWK+K LAE+GI+TQC+
Sbjct: 530 SMRRAPAARRVDGMIDKMQKKLPGQPKFLLCVLAERKNSDIYGPWKRKCLAEFGIITQCV 589
Query: 612 SPTRVNDQYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSD 671
+PTRVNDQY+TNVL+KINAKLGGLNS+L +E +PSIP+VSKVPTIILGMDVSHGSPGQSD
Sbjct: 590 APTRVNDQYITNVLLKINAKLGGLNSLLQIETSPSIPLVSKVPTIILGMDVSHGSPGQSD 649
Query: 672 IPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSS 731
IPSIAAVVSSREWPL+SKYRA VR+QSPK+EMID LFK +ED+G+IRELL+DFY+S+
Sbjct: 650 IPSIAAVVSSREWPLVSKYRASVRSQSPKLEMIDGLFKPQGAQEDDGLIRELLVDFYTST 709
Query: 732 GKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF 791
GKRKPD +IIFRDGVSESQF QVLNIEL+QIIEACKFLDE W+PKF +IVAQKNHHTKFF
Sbjct: 710 GKRKPDQVIIFRDGVSESQFTQVLNIELDQIIEACKFLDENWSPKFTLIVAQKNHHTKFF 769
Query: 792 QPGSPDNVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQEL 851
PGS +NVPPGTV+DN +CHPRNNDFYMCAHAGMIGT+RPTHYH+L D+IGFS D+LQEL
Sbjct: 770 VPGSQNNVPPGTVVDNAVCHPRNNDFYMCAHAGMIGTTRPTHYHILHDEIGFSADDLQEL 829
Query: 852 VHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAPVVP 911
VHSLSYVYQRSTTAISVVAPICYAHLAA Q+ QF+KF++ S+TSSSHGG T+AG AP VP
Sbjct: 830 VHSLSYVYQRSTTAISVVAPICYAHLAAAQVSQFIKFDEMSETSSSHGGHTSAGSAP-VP 888
Query: 912 QLPKLQDSVSSSMFFC 927
+LP+L + V SSMFFC
Sbjct: 889 ELPRLHNKVRSSMFFC 904
>UniRef100_Q84YI4 ARGONAUTE9 protein [Arabidopsis thaliana]
Length = 896
Score = 1239 bits (3206), Expect = 0.0
Identities = 607/906 (66%), Positives = 739/906 (80%), Gaps = 22/906 (2%)
Query: 23 PPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIAR-KGLGSKGTKLPLLTNHFKVTV 81
PPPPP VPA++ P EPVKK + LP+AR +G GSKG K+PLLTNHF V
Sbjct: 12 PPPPPFVPANLVP--------EVEPVKKNI--LLPMARPRGSGSKGQKIPLLTNHFGVKF 61
Query: 82 ANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETYGSELNGKDFAYDGEKTLFTIG 141
G+FF YSVA++YEDGRPVE KG+GRK++DKVQETY S+L K FAYDGEKTLFT+G
Sbjct: 62 NKPSGYFFHYSVAINYEDGRPVEAKGIGRKILDKVQETYQSDLGAKYFAYDGEKTLFTVG 121
Query: 142 SLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIP 201
+L NKL+F+VVLE++ S+RN+ G TND+D+KR RRP +K F VEIS+AAKIP
Sbjct: 122 ALPSNKLDFSVVLEEIPSSRNHA-----GNDTNDADRKRSRRPNQTKKFMVEISYAAKIP 176
Query: 202 LQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLG 261
+QAI +AL+G+E+EN Q+A+RVLDIILRQ AA+QGCLLVRQSFFHND KN+ +GGGV G
Sbjct: 177 MQAIASALQGKETENLQDALRVLDIILRQSAARQGCLLVRQSFFHNDVKNFVPIGGGVSG 236
Query: 262 CRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNVRDPFSLDWAKAKRTLKNL 321
CRGFHSSFRTTQ GLSLNID STTMI+QPGP+VDFL+ANQN +DP+ +DW KA+R LKNL
Sbjct: 237 CRGFHSSFRTTQGGLSLNIDTSTTMIVQPGPIVDFLLANQNKKDPYGMDWNKARRVLKNL 296
Query: 322 RIKASPSNQEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLR 381
R++ + SN+EYKI+GLSE CK+Q FT +K N + + E EITV Y+ R I++R
Sbjct: 297 RVQITLSNREYKISGLSEHSCKDQLFTWRKP---NDKGEFEEVEITVLNYYKE-RNIEVR 352
Query: 382 YSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLN 441
YS D PCINVGKPKRPTY P+E C+LVSLQRYTK+LT QR++LVEKSRQKP ERM L
Sbjct: 353 YSGDFPCINVGKPKRPTYFPIEFCNLVSLQRYTKSLTNFQRAALVEKSRQKPPERMASLT 412
Query: 442 QALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKFGNGEDFNPRNGRWNLNNKKV 501
+ LK SNY +P+L++ G++I + FTQVEGR+L P LK G GE+ +P G+WN K +
Sbjct: 413 KGLKDSNYNADPVLQDSGVSIITNFTQVEGRILPTPMLKVGKGENLSPIKGKWNFMRKTL 472
Query: 502 VRPAKIEHWAVVNFSARCDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVR 561
P + WAVVNFSARCD L+RDLIKC R KGI ++ P++++ EN QFR AP VR
Sbjct: 473 AEPTTVTRWAVVNFSARCDTNTLIRDLIKCGREKGINVEPPFKDVINENPQFRNAPATVR 532
Query: 562 VEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRVNDQYL 621
VE MFE+I+ +LP P FLLC+L ERKNSD+YGPWKKK+L + GIVTQCI+PTR+NDQYL
Sbjct: 533 VENMFEQIKSKLPKPPLFLLCILAERKNSDVYGPWKKKDLVDLGIVTQCIAPTRLNDQYL 592
Query: 622 TNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSS 681
TNVL+KINAKLGGLNS+L +E +P++P V++VPTII+GMDVSHGSPGQSDIPSIAAVVSS
Sbjct: 593 TNVLLKINAKLGGLNSLLAMERSPAMPKVTQVPTIIVGMDVSHGSPGQSDIPSIAAVVSS 652
Query: 682 REWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIII 741
R+WPLISKY+ACVRTQS K+EMIDNLFK V+ K DEG+ RELL+DFY SS RKP++III
Sbjct: 653 RQWPLISKYKACVRTQSRKMEMIDNLFKPVNGK-DEGMFRELLLDFYYSSENRKPEHIII 711
Query: 742 FRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPDNVPP 801
FRDGVSESQFNQVLNIEL+Q+++ACKFLD+TW+PKF VIVAQKNHHTKFFQ PDNVPP
Sbjct: 712 FRDGVSESQFNQVLNIELDQMMQACKFLDDTWHPKFTVIVAQKNHHTKFFQSRGPDNVPP 771
Query: 802 GTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQR 861
GT+ID++ICHPRN DFY+CAHAGMIGT+RPTHYHVL D+IGF+ D+LQELVHSLS+VYQR
Sbjct: 772 GTIIDSQICHPRNFDFYLCAHAGMIGTTRPTHYHVLYDEIGFATDDLQELVHSLSHVYQR 831
Query: 862 STTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVS 921
STTAISVVAP+CYAHLAA Q+G MK+E+ S+TSSSHGG+T G P VP +P+L ++VS
Sbjct: 832 STTAISVVAPVCYAHLAAAQMGTVMKYEELSETSSSHGGITTPGTVP-VPPMPQLHNNVS 890
Query: 922 SSMFFC 927
+SMFFC
Sbjct: 891 TSMFFC 896
>UniRef100_Q84VQ0 PAZ (Piwi Argonaut and Zwille) family [Arabidopsis thaliana]
Length = 892
Score = 1236 bits (3197), Expect = 0.0
Identities = 609/906 (67%), Positives = 737/906 (81%), Gaps = 26/906 (2%)
Query: 23 PPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIAR-KGLGSKGTKLPLLTNHFKVTV 81
PPPPP VPA++ P EPVKK + LP+AR +G GSKG K+PLLTNHF V
Sbjct: 12 PPPPPFVPANLVP--------EVEPVKKNI--LLPMARPRGSGSKGQKIPLLTNHFGVKF 61
Query: 82 ANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETYGSELNGKDFAYDGEKTLFTIG 141
G+FF YSVA++YEDGRPVE KG+GRK++DKVQETY S+L K FAYDGEKTLFT+G
Sbjct: 62 NKPSGYFFHYSVAINYEDGRPVEAKGIGRKILDKVQETYQSDLGAKYFAYDGEKTLFTVG 121
Query: 142 SLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIP 201
+L NKL+F+VVLE++ S+RN+ G TND+D+KR RRP +K F VEIS+AAKIP
Sbjct: 122 ALPSNKLDFSVVLEEIPSSRNHA-----GNDTNDADRKRSRRPNQTKKFMVEISYAAKIP 176
Query: 202 LQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLG 261
+QAI +AL+G+E+EN Q+A+RVLDIILRQ AA+QGCLLVRQSFFHND KN+ +GGGV G
Sbjct: 177 MQAIASALQGKETENLQDALRVLDIILRQSAARQGCLLVRQSFFHNDVKNFVPIGGGVSG 236
Query: 262 CRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNVRDPFSLDWAKAKRTLKNL 321
CRGFHSSFRTTQ GLSLNID STTMI+QPGPVVDFL+ANQN +DP+ +DW KA+R LKNL
Sbjct: 237 CRGFHSSFRTTQGGLSLNIDTSTTMIVQPGPVVDFLLANQNKKDPYGMDWNKARRVLKNL 296
Query: 322 RIKASPSNQEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLR 381
R++ + SN+EYKI+GLSE CK+Q K + GE + E EITV Y+ R I++R
Sbjct: 297 RVQITLSNREYKISGLSEHSCKDQL-----KPNDKGEFE--EVEITVLNYYKE-RNIEVR 348
Query: 382 YSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLN 441
YS D PCINVGKPKRPTY P+E C+LVSLQRYTK+LT QR++LVEKSRQKP ERM L
Sbjct: 349 YSGDFPCINVGKPKRPTYFPIEFCNLVSLQRYTKSLTNFQRAALVEKSRQKPPERMASLT 408
Query: 442 QALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKFGNGEDFNPRNGRWNLNNKKV 501
+ LK SNY +P+L++ G++I + FTQVEGR+L P LK G GE+ +P G+WN K +
Sbjct: 409 KGLKDSNYNADPVLQDSGVSIITNFTQVEGRILPTPMLKVGKGENLSPIKGKWNFMRKTL 468
Query: 502 VRPAKIEHWAVVNFSARCDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVR 561
P + WAVVNFSARCD L+RDLIKC R KGI ++ P++++ EN QFR AP VR
Sbjct: 469 AEPTTVTRWAVVNFSARCDTNTLIRDLIKCGREKGINVEPPFKDVINENPQFRNAPATVR 528
Query: 562 VEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRVNDQYL 621
VE MFE+I+ +LP P FLLC+L ERKNSD+YGPWKKKNL + GIVTQCI+PTR+NDQYL
Sbjct: 529 VENMFEQIKSKLPKPPLFLLCILAERKNSDVYGPWKKKNLVDLGIVTQCIAPTRLNDQYL 588
Query: 622 TNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSS 681
TNVL+KINAKLGGLNS+L +E +P++P V++VPTII+GMDVSHGSPGQSDIPSIAAVVSS
Sbjct: 589 TNVLLKINAKLGGLNSLLAMERSPAMPKVTQVPTIIVGMDVSHGSPGQSDIPSIAAVVSS 648
Query: 682 REWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIII 741
R+WPLISKY+ACVRTQS K+EMIDNLFK V+ K DEG+ RELL+DFY SS RKP++III
Sbjct: 649 RQWPLISKYKACVRTQSRKMEMIDNLFKPVNGK-DEGMFRELLLDFYYSSENRKPEHIII 707
Query: 742 FRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPDNVPP 801
FRDGVSESQFNQVLNIEL+Q+++ACKFLD+TW+PKF VIVAQKNHHTKFFQ PDNVPP
Sbjct: 708 FRDGVSESQFNQVLNIELDQMMQACKFLDDTWHPKFTVIVAQKNHHTKFFQSRGPDNVPP 767
Query: 802 GTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQR 861
GT+ID++ICHPRN DFY+CAHAGMIGT+RPTHYHVL D+IGF+ D+LQELVHSLSYVYQR
Sbjct: 768 GTIIDSQICHPRNFDFYLCAHAGMIGTTRPTHYHVLYDEIGFATDDLQELVHSLSYVYQR 827
Query: 862 STTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVS 921
STTAISVVAP+CYAHLAA Q+G MK+E+ S+TSSSHGG+T G P VP +P+L ++VS
Sbjct: 828 STTAISVVAPVCYAHLAAAQMGTVMKYEELSETSSSHGGITTPGAVP-VPPMPQLHNNVS 886
Query: 922 SSMFFC 927
+SMFFC
Sbjct: 887 TSMFFC 892
>UniRef100_Q6YSJ5 Putative ARGONAUTE9 protein [Oryza sativa]
Length = 889
Score = 1079 bits (2791), Expect = 0.0
Identities = 544/880 (61%), Positives = 677/880 (76%), Gaps = 16/880 (1%)
Query: 55 RLPIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVID 114
R+PIAR G +G ++ LL+NHF V ++ D F+QYSV++ ED + ++GKG+GRKV+D
Sbjct: 19 RVPIARPSFGREGKQIKLLSNHFTVKLSGIDAVFYQYSVSIKSEDDKVIDGKGIGRKVMD 78
Query: 115 KVQETYGSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTN 174
KV +TY SEL GK+FAYDGEK LFT+G L +N EFTV+LE+ S G+ S N
Sbjct: 79 KVLQTYSSELAGKEFAYDGEKCLFTVGPLPQNNFEFTVILEETSSRAAGGSLGH--GSPN 136
Query: 175 DSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAK 234
DKKR + + +K V IS+AAKIPL+++ AL+G ES++ Q+A+RVLDI+LRQ AK
Sbjct: 137 QGDKKRSKCTHLAKKIVVGISYAAKIPLKSVALALQGSESDHAQDALRVLDIVLRQQQAK 196
Query: 235 QGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVV 294
+GCLLVRQSFF +D +N D+ GGV GCRG HSSFRTT GLSLN+DVSTTMI+ PGPV
Sbjct: 197 RGCLLVRQSFFSDDFRNLVDLTGGVSGCRGLHSSFRTTIGGLSLNMDVSTTMIVTPGPVF 256
Query: 295 DFLIANQNVRDPFSLDWAKAKRTLKNLRIKASPSNQEYKITGLSELPCKEQTFTMKKKGG 354
DFL+ NQNVRD +DW +AK+ LKNLR+KA +N E+KI GLS+ PC QTF MK +
Sbjct: 257 DFLLTNQNVRDIRDIDWPRAKKMLKNLRVKAIHNNMEFKIIGLSDEPCSRQTFPMKVR-- 314
Query: 355 NNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVE------LCSLV 408
NG + EITV EYF + +++DL LPC++VGKPKRP YVP+E LC +V
Sbjct: 315 -NGSSEGETVEITVQEYFKS-KQVDLTMPY-LPCLDVGKPKRPNYVPIEVAGTNPLCHMV 371
Query: 409 SLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQ 468
SLQRYTKAL++ QR++LVEKSRQKP ERM V+ A+K + Y ++P+L +CGI I T+
Sbjct: 372 SLQRYTKALSSQQRATLVEKSRQKPQERMRVVTDAVKNNRYDDDPILSSCGIKIEKQLTR 431
Query: 469 VEGRVLQAPRLKFGNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCDVRGLVRDL 528
V+GRVL AP L GN ED P GRWN NNK++ P KIE WA+VNFSARCD+ + RDL
Sbjct: 432 VDGRVLSAPTLVVGNSEDCIPNRGRWNYNNKRLFEPVKIERWAIVNFSARCDMSRISRDL 491
Query: 529 IKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKMFERIQKELPGAPSFLLCLLPERK 588
I C R KGI I+ P+ + +E+ Q RR P+VRVE MFE+++ LPG P FLLC+LPERK
Sbjct: 492 INCGRTKGIIIERPFT-LVDEDSQSRRCTPVVRVESMFEKVKANLPGPPEFLLCVLPERK 550
Query: 589 NSDLYGPWKKKNLAEYGIVTQCISPT-RVNDQYLTNVLMKINAKLGGLNSVLGVEMNPSI 647
N DLYGPWKKKNL E GI+TQCI P+ ++NDQY TNVL+KINAKLGG+NS L +E I
Sbjct: 551 NCDLYGPWKKKNLHEMGIITQCIVPSVKMNDQYYTNVLLKINAKLGGMNSKLSLEHRHMI 610
Query: 648 PIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNL 707
PIV++ PT+ILGMDVSHGSPG++D+PSIAAVV SR WPLIS+YRA VRTQSPKVEMID+L
Sbjct: 611 PIVNQTPTLILGMDVSHGSPGRADVPSIAAVVGSRCWPLISRYRASVRTQSPKVEMIDSL 670
Query: 708 FKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACK 767
FK + + +D+GIIRELL+DFY +S +RKP IIIFRDGVSESQF+QVLN+ELNQII+A +
Sbjct: 671 FKPLDDGKDDGIIRELLLDFYKTSQQRKPKQIIIFRDGVSESQFSQVLNVELNQIIKAYQ 730
Query: 768 FLDETWNPKFLVIVAQKNHHTKFFQPGSPDNVPPGTVIDNKICHPRNNDFYMCAHAGMIG 827
++D+ PKF VI+AQKNHHTK FQ +PDNVPPGTV+D+ I HPR DFYM AHAG IG
Sbjct: 731 YMDQGPIPKFTVIIAQKNHHTKLFQENTPDNVPPGTVVDSGIVHPRQYDFYMYAHAGPIG 790
Query: 828 TSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMK 887
TSRPTHYHVLLD+IGF PD++Q+LV SLSYVYQRSTTAISVVAPICYAHLAA Q+GQFMK
Sbjct: 791 TSRPTHYHVLLDEIGFLPDDVQKLVLSLSYVYQRSTTAISVVAPICYAHLAAAQMGQFMK 850
Query: 888 FEDKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVSSSMFFC 927
FE+ ++TSS GG+ ++ A VVP+LP+L V SSMFFC
Sbjct: 851 FEEFAETSSGSGGVPSSSGA-VVPELPRLHADVCSSMFFC 889
>UniRef100_O48771 Argonaute (AGO1)-like protein [Arabidopsis thaliana]
Length = 887
Score = 941 bits (2431), Expect = 0.0
Identities = 485/903 (53%), Positives = 641/903 (70%), Gaps = 38/903 (4%)
Query: 41 LDLPPEPVKKKLPTRLPI---ARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSY 97
L L P ++ + P+ R+G+G+ G + L TNHF V+V D F+QY+V+++
Sbjct: 7 LPLSPISIEPEQPSHRDYDITTRRGVGTTGNPIELCTNHFNVSVRQPDVVFYQYTVSITT 66
Query: 98 EDGRPVEGKGVGRKVIDKVQETYGSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDV 157
E+G V+G G+ RK++D++ +TY S+L+GK AYDGEKTL+T+G L +N+ +F V++E
Sbjct: 67 ENGDAVDGTGISRKLMDQLFKTYSSDLDGKRLAYDGEKTLYTVGPLPQNEFDFLVIVEGS 126
Query: 158 ISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQES--E 215
S R+ G DG S++ + K R +R + +++KV+I +AA+IPL+ ++ RG + +
Sbjct: 127 FSKRDCG--VSDGGSSSGTCK-RSKRSFLPRSYKVQIHYAAEIPLKTVLGTQRGAYTPDK 183
Query: 216 NYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSG 275
+ Q+A+RVLDI+LRQ AA++GCLLVRQ+FFH+D + VGGGV+G RG HSSFR T G
Sbjct: 184 SAQDALRVLDIVLRQQAAERGCLLVRQAFFHSDG-HPMKVGGGVIGIRGLHSSFRPTHGG 242
Query: 276 LSLNIDVSTTMIIQPGPVVDFLIANQNVRDPFSLDWAK-AKRTLKNLRIKASPSNQEYKI 334
LSLNIDVSTTMI++PGPV++FL ANQ+V P +DW K A + LK++R+KA+ N E+KI
Sbjct: 243 LSLNIDVSTTMILEPGPVIEFLKANQSVETPRQIDWIKVAAKMLKHMRVKATHRNMEFKI 302
Query: 335 TGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKP 394
GLS PC +Q F+MK K +GE + EITVY+YF + SA PC++VGKP
Sbjct: 303 IGLSSKPCNQQLFSMKIK---DGEREVPIREITVYDYFKQTYTEPIS-SAYFPCLDVGKP 358
Query: 395 KRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPM 454
RP Y+P+E C+LVSLQRYTK L+ QR LVE SRQKPLER+ LN A+ T Y +P
Sbjct: 359 DRPNYLPLEFCNLVSLQRYTKPLSGRQRVLLVESSRQKPLERIKTLNDAMHTYCYDKDPF 418
Query: 455 LKNCGITIASGFTQVEGRVLQAPRLKFGNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVN 514
L CGI+I TQVEGRVL+ P LKFG EDF P NGRWN NNK ++ P I+ WA+VN
Sbjct: 419 LAGCGISIEKEMTQVEGRVLKPPMLKFGKNEDFQPCNGRWNFNNKMLLEPRAIKSWAIVN 478
Query: 515 FSARCDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKMFERIQKELP 574
FS CD + R+LI C KGI ID P+ + EE+ Q+++A P+ RVEKM ++ + P
Sbjct: 479 FSFPCDSSHISRELISCGMRKGIEIDRPFA-LVEEDPQYKKAGPVERVEKMIATMKLKFP 537
Query: 575 GAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRVNDQYLTNVLMKINAKLGG 634
P F+LC+LPERK SD+YGPWKK L E GI TQCI P +++DQYLTNVL+KIN+KLGG
Sbjct: 538 DPPHFILCILPERKTSDIYGPWKKICLTEEGIHTQCICPIKISDQYLTNVLLKINSKLGG 597
Query: 635 LNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACV 694
+NS+LG+E + +IP+++K+PT+ILGMDVSHG PG++D+PS+AAVV S+ WPLIS+YRA V
Sbjct: 598 INSLLGIEYSYNIPLINKIPTLILGMDVSHGPPGRADVPSVAAVVGSKCWPLISRYRAAV 657
Query: 695 RTQSPKVEMIDNLFKQV--SEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFN 752
RTQSP++EMID+LF+ + +EK D GI+ EL ++FY +S RKP IIIFRDGVSESQF
Sbjct: 658 RTQSPRLEMIDSLFQPIENTEKGDNGIMNELFVEFYRTSRARKPKQIIIFRDGVSESQFE 717
Query: 753 QVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPDNVPPGTVIDNKICHP 812
QVL IE++QII+A + L E+ PKF VIVAQKNHHTK FQ P+NVP GTV+D KI HP
Sbjct: 718 QVLKIEVDQIIKAYQRLGESDVPKFTVIVAQKNHHTKLFQAKGPENVPAGTVVDTKIVHP 777
Query: 813 RNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAIS----- 867
N DFYMCAHAG IGTSRP HYHVLLD+IGFSPD+LQ L+HSLSY S +S
Sbjct: 778 TNYDFYMCAHAGKIGTSRPAHYHVLLDEIGFSPDDLQNLIHSLSYKLLNSIFNVSSLLCV 837
Query: 868 ---VVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVSSSM 924
VAP+ YAHLAA Q+ QF KFE S+ VP+LP+L ++V +M
Sbjct: 838 FVLSVAPVRYAHLAAAQVAQFTKFEGISEDGK-------------VPELPRLHENVEGNM 884
Query: 925 FFC 927
FFC
Sbjct: 885 FFC 887
>UniRef100_Q9SJK3 Argonaute-like protein At2g27880 [Arabidopsis thaliana]
Length = 997
Score = 525 bits (1351), Expect = e-147
Identities = 350/921 (38%), Positives = 499/921 (54%), Gaps = 84/921 (9%)
Query: 37 KVDLLDLPPEPVKKKLPTRLPIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALS 96
K ++ LPP K P+ R G G+ G K+ + NHF V VA+ D + Y V+++
Sbjct: 131 KPEMTSLPPASSKA---VTFPV-RPGRGTLGKKVMVRANHFLVQVADRD--LYHYDVSIN 184
Query: 97 YEDGRPVEGKGVGRKVIDKVQETY-GSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLE 155
E V K V R V+ + + Y S L GK AYDG K+L+T G L + EF V L
Sbjct: 185 PE----VISKTVNRNVMKLLVKNYKDSHLGGKSPAYDGRKSLYTAGPLPFDSKEFVVNLA 240
Query: 156 DVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESE 215
+ + DG+S D + FKV + L + L ++ E
Sbjct: 241 EKRA---------DGSSGKD------------RPFKVAVKNVTSTDLYQLQQFLDRKQRE 279
Query: 216 NYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLG-----CRGFHSSFR 270
+ I+VLD++LR + + V +SFFH A G G LG RG+ S R
Sbjct: 280 APYDTIQVLDVVLRDKPSND-YVSVGRSFFHTSLGKDARDGRGELGDGIEYWRGYFQSLR 338
Query: 271 TTQSGLSLNIDVSTTMIIQPGPVVDFLIANQNVRD---PF-SLDWAKAKRTLKNLRIKAS 326
TQ GLSLNIDVS +P V DF+ N+RD P D K K+ L+ L++K
Sbjct: 339 LTQMGLSLNIDVSARSFYEPIVVTDFISKFLNIRDLNRPLRDSDRLKVKKVLRTLKVKLL 398
Query: 327 PSN--QEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSA 384
N + KI+G+S LP +E FT++ K E TV +YF ++Y A
Sbjct: 399 HWNCTKSAKISGISSLPIRELRFTLEDKS-----------EKTVVQYFAEKYNYRVKYQA 447
Query: 385 DLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQAL 444
LP I G RP Y+P+ELC + QRYTK L Q ++L++ + Q+P +R N + +
Sbjct: 448 -LPAIQTGSDTRPVYLPMELCQIDEGQRYTKRLNEKQVTALLKATCQRPPDRENSIKNLV 506
Query: 445 KTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF---GNGEDFNPRNGRWNLNNKKV 501
+NY N+ + K G+++ + +E RVL P LK+ G + NPR G+WN+ +KK+
Sbjct: 507 VKNNY-NDDLSKEFGMSVTTQLASIEARVLPPPMLKYHDSGKEKMVNPRLGQWNMIDKKM 565
Query: 502 VRPAKIEHWAVVNFSARCDVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVR 561
V AK+ W V+FS R D RGL ++ C +L G+ + + E + F PP
Sbjct: 566 VNGAKVTSWTCVSFSTRID-RGLPQEF--CKQLIGMCVSKGMEFKPQPAIPFISCPP-EH 621
Query: 562 VEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRVND--- 618
+E+ I K PG L+ +LP+ S YG K+ E GIV+QC P +VN
Sbjct: 622 IEEALLDIHKRAPGL-QLLIVILPDVTGS--YGKIKRICETELGIVSQCCQPRQVNKLNK 678
Query: 619 QYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAV 678
QY+ NV +KIN K GG N+VL + +IP+++ PTII+G DV+H PG+ PSIAAV
Sbjct: 679 QYMENVALKINVKTGGRNTVLNDAIRRNIPLITDRPTIIMGADVTHPQPGEDSSPSIAAV 738
Query: 679 VSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKE----DEGIIRELLIDFYSSSGKR 734
V+S +WP I+KYR V Q+ + E+I +L+K V + + G+IRE I F ++G+
Sbjct: 739 VASMDWPEINKYRGLVSAQAHREEIIQDLYKLVQDPQRGLVHSGLIREHFIAFRRATGQI 798
Query: 735 KPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFF--Q 792
P II +RDGVSE QF+QVL E+ I +AC L E + P+ ++ QK HHT+ F Q
Sbjct: 799 -PQRIIFYRDGVSEGQFSQVLLHEMTAIRKACNSLQENYVPRVTFVIVQKRHHTRLFPEQ 857
Query: 793 PGSPD------NVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPD 846
G+ D N+ PGTV+D KICHP DFY+ +HAG+ GTSRP HYHVLLD+ GF+ D
Sbjct: 858 HGNRDMTDKSGNIQPGTVVDTKICHPNEFDFYLNSHAGIQGTSRPAHYHVLLDENGFTAD 917
Query: 847 ELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGV 906
+LQ L ++L Y Y R T ++S+V P YAHLAA + +M+ E SD SS + GV
Sbjct: 918 QLQMLTNNLCYTYARCTKSVSIVPPAYYAHLAAFRARYYMESE-MSDGGSSRSRSSTTGV 976
Query: 907 APVVPQLPKLQDSVSSSMFFC 927
V+ QLP ++D+V MF+C
Sbjct: 977 GQVISQLPAIKDNVKEVMFYC 997
>UniRef100_Q6EU14 Putative argonaute protein [Oryza sativa]
Length = 1082
Score = 502 bits (1292), Expect = e-140
Identities = 338/929 (36%), Positives = 492/929 (52%), Gaps = 85/929 (9%)
Query: 43 LPPEPVKKKLPTRLPIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRP 102
+ P P K R P+ R G G+ G + + NHF + + D H QY V+++ E
Sbjct: 195 IQPAPPSSK-SVRFPM-RPGKGTFGDRCIVKANHFFAELPDKDLH--QYDVSITPE---- 246
Query: 103 VEGKGVGRKVIDKVQETYG-SELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNR 161
V +GV R VI ++ Y S L G+ YDG K+L+T G L F V+L+D +
Sbjct: 247 VPSRGVNRAVIGEIVTQYRQSHLGGRLPVYDGRKSLYTAGPLPFTSRTFDVILQDEEESL 306
Query: 162 NNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAI 221
G GA +R RP FKV I FAA+ L + L G++++ QEA+
Sbjct: 307 AVGQ----GA-------QRRERP-----FKVVIKFAARADLHHLAMFLAGRQADAPQEAL 350
Query: 222 RVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNID 281
+VLDI+LR+ + + R SF+ + +G G+ RGF+ S R TQ GLSLNID
Sbjct: 351 QVLDIVLRELPTARYSPVAR-SFYSPNLGRRQQLGEGLESWRGFYQSIRPTQMGLSLNID 409
Query: 282 VSTTMIIQPGPVVDFLIANQN----VRDPFSLDWAKAKRTLKNLRIKASPSN---QEYKI 334
+S+T I+P PV+DF+ N VR D K K+ L+ ++++ + ++Y+I
Sbjct: 410 MSSTAFIEPLPVIDFVAQLLNRDISVRPLSDADRVKIKKALRGVKVEVTHRGNMRRKYRI 469
Query: 335 TGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKP 394
+GL+ +E +F + G TV +YF +++++ LPC+ VG
Sbjct: 470 SGLTSQATRELSFPIDNHGTVK----------TVVQYFQETYGFNIKHTT-LPCLQVGNQ 518
Query: 395 KRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPM 454
+RP Y+P+E+C +V QRY+K L Q ++L++ + Q+P ER + Q + + Y +P
Sbjct: 519 QRPNYLPMEVCKIVEGQRYSKRLNEKQITALLKVTCQRPQERELDILQTVHHNAYHQDPY 578
Query: 455 LKNCGITIASGFTQVEGRVLQAPRLKF---GNGEDFNPRNGRWNLNNKKVVRPAKIEHWA 511
+ GI I VE RVL P LK+ G +D PR G+WN+ NKK+V ++ +W
Sbjct: 579 AQEFGIRIDERLASVEARVLPPPWLKYHDSGREKDVLPRIGQWNMMNKKMVNGGRVNNWT 638
Query: 512 VVNFSARCD---VRGLVRDLIKCARLKGIP--IDEPYEEIFEENGQFRRAPPLVRVEKMF 566
+NFS R R+L ++ G+ ID + RA E M
Sbjct: 639 CINFSRHVQDNAARSFCRELAIMCQISGMDFSIDPVVPLVTARPEHVERALKARYQEAM- 697
Query: 567 ERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRV---NDQYLTN 623
I K G L+ +LP+ N LYG K+ + G+V+QC V + QYL N
Sbjct: 698 -NILKPQGGELDLLIAILPDN-NGSLYGDLKRICETDLGLVSQCCLTKHVFKMSKQYLAN 755
Query: 624 VLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSRE 683
V +KIN K+GG N+VL + IP+VS PTII G DV+H PG+ PSIAAVV+S++
Sbjct: 756 VALKINVKVGGRNTVLVDALTRRIPLVSDRPTIIFGADVTHPHPGEDSSPSIAAVVASQD 815
Query: 684 WPLISKYRACVRTQSPKVEMIDNLFKQVSEKE----DEGIIRELLIDFYSSSGKRKPDNI 739
WP ++KY V Q+ + E+I +LFK + + G+IRELLI F ++G+ KP I
Sbjct: 816 WPEVTKYAGLVSAQAHRQELIQDLFKVWKDPQRGTVSGGMIRELLISFKRATGQ-KPQRI 874
Query: 740 IIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPD-- 797
I +RDGVSE QF QVL EL+ I +AC L+ + P +V QK HHT+ F D
Sbjct: 875 IFYRDGVSEGQFYQVLFYELDAIRKACASLEADYQPPVTFVVVQKRHHTRLFANNHKDQR 934
Query: 798 ------NVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQEL 851
N+ PGTV+D+KICHP DFY+C+HAG+ GTSRP HYHVL D+ F+ D LQ L
Sbjct: 935 TVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIQGTSRPAHYHVLWDENKFTADGLQTL 994
Query: 852 VHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAPV-- 909
++L Y Y R T ++S+V P YAHLAA + +M+ D SD+ S G G P+
Sbjct: 995 TNNLCYTYARCTRSVSIVPPAYYAHLAAFRARFYME-PDTSDSGSMASGAHTRGGGPLPG 1053
Query: 910 -----------VPQLPKLQDSVSSSMFFC 927
V LP L+++V MF+C
Sbjct: 1054 ARSTKPAGNVAVRPLPDLKENVKRVMFYC 1082
>UniRef100_Q69VD5 ZLL/PNH homologous protein [Oryza sativa]
Length = 979
Score = 501 bits (1290), Expect = e-140
Identities = 334/950 (35%), Positives = 503/950 (52%), Gaps = 85/950 (8%)
Query: 11 GNGNGNEDLMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLGSKGTKL 70
G G G P P PA V + PP K R P G G+ G +
Sbjct: 82 GRGRGGRAGAGPGPGLAAAPAVVVAPAARAVIGPPVASKGLSFCRRP----GFGTVGARC 137
Query: 71 PLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETY-GSELNGKDF 129
+ NHF + + D QY V ++ E V + V R ++ ++ Y S+L G+
Sbjct: 138 VVKANHFLAELPDKD--LTQYDVKITPE----VSSRSVNRAIMSELVRLYHDSDLGGRLP 191
Query: 130 AYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKT 189
AYDG K L+T G+L + EF V L D DG P +
Sbjct: 192 AYDGRKNLYTAGTLPFDAREFVVRLTD----------DDDGTGV----------PPRERE 231
Query: 190 FKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDP 249
++V I FAA+ L + + G++++ QEA++VLDI+LR+ A ++ + + +SF+ D
Sbjct: 232 YRVAIKFAARADLHHLRQFIAGRQADAPQEALQVLDIVLRELANRR-YVSIGRSFYSPDI 290
Query: 250 KNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDF---LIANQNVRDP 306
+ +G G+ GF+ S R TQ GLSLNID+S+T I+P PV++F ++ + P
Sbjct: 291 RKPQRLGDGLQSWCGFYQSIRPTQMGLSLNIDMSSTAFIEPLPVIEFVAQILGKDVISRP 350
Query: 307 FS-LDWAKAKRTLKNLRIKASPSN---QEYKITGLSELPCKEQTFTMKKKGGNNGEEDAT 362
S + K K+ L+ ++++ + ++Y+I+GL+ P E F + D
Sbjct: 351 LSDANRIKIKKALRGVKVEVTHRGNVRRKYRISGLTTQPTHELIFPI----------DDQ 400
Query: 363 EEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQR 422
+V EYF +++ LPC+ VG K+ Y+P+E C +V QRYTK L Q
Sbjct: 401 MNMKSVVEYFKEMYGFTIQHP-HLPCLQVGNQKKANYLPMEACKIVEGQRYTKRLNEKQI 459
Query: 423 SSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF- 481
+SL++ + ++P E+ + Q ++ + Y +P K GI I+ T VE RVL AP LK+
Sbjct: 460 TSLLKVTCRRPREQEMDILQTVQQNGYEQDPYAKEFGINISEKLTSVEARVLPAPWLKYH 519
Query: 482 --GNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD---VRGLVRDLIKCARLKG 536
G ++ P+ G+WN+ NKKV+ K+ HWA +NFS RG ++L + ++ G
Sbjct: 520 DTGKEKECLPQVGQWNMVNKKVINGCKVNHWACINFSRSVQETTARGFCQELAQMCQISG 579
Query: 537 IPID-EPYEEIFEEN-GQFRRAPPLVRVEKMFERIQKELPGAPSFLLCLLPERKNSDLYG 594
+ + EP I+ Q +A V + + KEL LL +LP+ N LYG
Sbjct: 580 MEFNSEPVIPIYSARPDQVEKALKHVYNMSLNKLKGKEL----ELLLAILPDN-NGSLYG 634
Query: 595 PWKKKNLAEYGIVTQCISPT---RVNDQYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVS 651
K+ + G+++QC +++ QYL NV +KIN K+GG N+VL ++ IP+VS
Sbjct: 635 DIKRICETDLGLISQCCLTKHVFKISKQYLANVSLKINVKMGGRNTVLLDAISWRIPLVS 694
Query: 652 KVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQV 711
+PTII G DV+H G+ PSIAAVV+S++WP ++KY V Q+ + E+I +L+K
Sbjct: 695 DIPTIIFGADVTHPETGEDSSPSIAAVVASQDWPEVTKYAGLVCAQAHRQELIQDLYKTW 754
Query: 712 SEKE----DEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACK 767
+ + G+IRELLI F ++G+ KP II +RDGVSE QF QVL EL+ I +AC
Sbjct: 755 HDPQRGTVTGGMIRELLISFRKATGQ-KPLRIIFYRDGVSEGQFYQVLLYELDAIRKACA 813
Query: 768 FLDETWNPKFLVIVAQKNHHTKFFQPGSPD--------NVPPGTVIDNKICHPRNNDFYM 819
L+ + P +V QK HHT+ F D N+ PGTV+D+KICHP DFY+
Sbjct: 814 SLEPNYQPPVTFVVVQKRHHTRLFANNHKDRSSTDKSGNILPGTVVDSKICHPSEFDFYL 873
Query: 820 CAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVAPICYAHLAA 879
C+HAG+ GTSRP HYHVL D+ F+ DE+Q L ++L Y Y R T ++SVV P YAHLAA
Sbjct: 874 CSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAYYAHLAA 933
Query: 880 TQIGQFMKFE--DKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVSSSMFFC 927
+ +M+ E + TS S G V P LP +++ V MF+C
Sbjct: 934 FRARFYMEPEMSENQTTSKSSTGTNGTSVKP----LPAVKEKVKRVMFYC 979
>UniRef100_Q851R2 Putative argonaute protein [Oryza sativa]
Length = 1058
Score = 501 bits (1289), Expect = e-140
Identities = 330/901 (36%), Positives = 486/901 (53%), Gaps = 75/901 (8%)
Query: 59 ARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQE 118
AR G G+ G K+ + NHF V VA D + F Y V+++ E + + R+V++++ +
Sbjct: 201 ARPGFGAAGKKVMIRANHFLVNVA--DNNLFHYDVSINPES----KSRATNREVLNELIK 254
Query: 119 TYG-SELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSD 177
+G + L GK AYDG K+L+T GSL EF V L D D
Sbjct: 255 LHGKTSLGGKLPAYDGRKSLYTAGSLPFESEEFVVKLID----------------PEKKD 298
Query: 178 KKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGC 237
K+R R Y K+ I A + L + L G++ + QE I+VLD++LR+ +
Sbjct: 299 KERAEREY-----KITIRIAGRTDLYHLQQFLLGRQRDMPQETIQVLDVVLRE-SPSWNY 352
Query: 238 LLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFL 297
+ V +SFF + D+G G+ RG++ S R TQ GLSLNID+S T +P V+ F+
Sbjct: 353 VTVSRSFFSTQFGHRGDIGEGLECWRGYYQSLRPTQMGLSLNIDISATSFFKPVTVIQFV 412
Query: 298 IANQNVRD---PFS-LDWAKAKRTLKNLRIKASPSNQE---YKITGLSELPCKEQTFTMK 350
N+RD P S D K K+ L+ +RI+ + + YKITG++ +P + F +
Sbjct: 413 EEFLNIRDTSRPLSDRDRVKIKKALRGVRIETNHQEDQIRRYKITGITPIPMSQLIFPVD 472
Query: 351 KKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSL 410
G TV +YF + L+Y A PC+ G RP Y+P+E+C +V
Sbjct: 473 DNGTRK----------TVVQYFWDRYNYRLKY-ASWPCLQSGSDSRPVYLPMEVCKIVEG 521
Query: 411 QRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVE 470
QRY+K L Q ++++ + Q+P +R +++ + + Y + + GI + + V
Sbjct: 522 QRYSKKLNDKQVTNILRATCQRPQQREQSIHEMVLHNKYTEDRFAQEFGIKVCNDLVSVP 581
Query: 471 GRVLQAPRLKF---GNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARC--DVRGLV 525
RVL P LK+ G + P G+WN+ NKK++ +++W ++FS +V+
Sbjct: 582 ARVLPPPMLKYHDSGREKTCAPSVGQWNMINKKMINGGTVDNWTCLSFSRMRPEEVQRFC 641
Query: 526 RDLIKCARLKGIPID-EPYEEIFEENGQFRRAPPLVRVEKMFERIQKELPGAPSFLLCLL 584
DLI+ G+ + P ++ N + E + +E G L+ +L
Sbjct: 642 GDLIQMCNATGMSFNPRPVVDVRSTNPNNIENALRDVHRRTSELLAREGKGGLQLLIVIL 701
Query: 585 PERKNSDLYGPWKKKNLAEYGIVTQCISP---TRVNDQYLTNVLMKINAKLGGLNSVLG- 640
PE S YG K+ + GIV+QC P +R N QYL NV +KIN K+GG N+VL
Sbjct: 702 PEVSGS--YGKIKRVCETDLGIVSQCCLPRHASRPNKQYLENVALKINVKVGGRNTVLER 759
Query: 641 VEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPK 700
+ IP VS+VPTII G DV+H PG+ SIAAVV+S +WP I+KYR V Q +
Sbjct: 760 AFIRNGIPFVSEVPTIIFGADVTHPPPGEDSASSIAAVVASMDWPEITKYRGLVSAQPHR 819
Query: 701 VEMIDNLF---KQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNI 757
E+I++LF K + + G+IRELLI F +G+R P+ II +RDGVSE QF+ VL
Sbjct: 820 QEIIEDLFSVGKDPVKVVNGGMIRELLIAFRKKTGRR-PERIIFYRDGVSEGQFSHVLLH 878
Query: 758 ELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQP--GSPD------NVPPGTVIDNKI 809
E++ I +AC L+E + P +V QK HHT+ F G D N+ PGTV+D +I
Sbjct: 879 EMDAIRKACASLEEGYLPPVTFVVVQKRHHTRLFPEVHGRRDMTDKSGNILPGTVVDRQI 938
Query: 810 CHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVV 869
CHP DFY+C+HAG+ GTSRPTHYHVL D+ F+ D LQ L ++L Y Y R T A+SVV
Sbjct: 939 CHPTEFDFYLCSHAGIQGTSRPTHYHVLYDENHFTADALQSLTNNLCYTYARCTRAVSVV 998
Query: 870 APICYAHLAATQIGQFMKFEDKSDTSSSHG--GLTAAGVAPV-VPQLPKLQDSVSSSMFF 926
P YAHLAA + +++ E SD S+ G G A PV V QLPK++++V MF+
Sbjct: 999 PPAYYAHLAAFRARYYVEGE-SSDGGSTPGSSGQAVAREGPVEVRQLPKIKENVKDVMFY 1057
Query: 927 C 927
C
Sbjct: 1058 C 1058
>UniRef100_Q7XSA2 OSJNBa0005N02.3 protein [Oryza sativa]
Length = 1101
Score = 496 bits (1276), Expect = e-138
Identities = 328/923 (35%), Positives = 486/923 (52%), Gaps = 97/923 (10%)
Query: 55 RLPIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVID 114
R P+ R G G+ G + + NHF + + D H QY V+++ E V +GV R V+
Sbjct: 226 RFPL-RPGKGTYGDRCIVKANHFFAELPDKDLH--QYDVSITPE----VTSRGVNRAVMF 278
Query: 115 KVQETYG-SELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGAST 173
++ Y S L G+ AYDG K+L+T G L F + L+D D
Sbjct: 279 ELVTLYRYSHLGGRLPAYDGRKSLYTAGPLPFASRTFEITLQD----------EEDSLGG 328
Query: 174 NDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAA 233
++R R F+V I FAA+ L + L G++++ QEA++VLDI+LR+
Sbjct: 329 GQGTQRRER------LFRVVIKFAARADLHHLAMFLAGRQADAPQEALQVLDIVLRELPT 382
Query: 234 KQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPV 293
+ + R SF+ + +G G+ RGF+ S R TQ GLSLNID+S+T I+P PV
Sbjct: 383 TRYSPVGR-SFYSPNLGRRQQLGEGLESWRGFYQSIRPTQMGLSLNIDMSSTAFIEPLPV 441
Query: 294 VDFLIANQN----VRDPFSLDWAKAKRTLKNLRIKASPSN---QEYKITGLSELPCKEQT 346
+DF+ N VR D K K+ L+ ++++ + ++Y+I+GL+ +E +
Sbjct: 442 IDFVAQLLNRDISVRPLSDSDRVKIKKALRGVKVEVTHRGNMRRKYRISGLTSQATRELS 501
Query: 347 FTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCS 406
F + +G TV +YF+ ++++ LPC+ VG +RP Y+P+E+C
Sbjct: 502 FPVDDRGTVK----------TVVQYFLETYGFSIQHTT-LPCLQVGNQQRPNYLPMEVCK 550
Query: 407 LVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGF 466
+V QRY+K L Q ++L++ + Q+P ER + + + + Y + + GI I
Sbjct: 551 IVEGQRYSKRLNEKQITALLKVTCQRPQERELDILRTVSHNAYHEDQYAQEFGIKIDERL 610
Query: 467 TQVEGRVLQAPRLKF---GNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD--- 520
VE RVL PRLK+ G +D PR G+WN+ NKK+V ++ +WA +NFS
Sbjct: 611 ASVEARVLPPPRLKYHDSGREKDVLPRVGQWNMMNKKMVNGGRVNNWACINFSRNVQDSA 670
Query: 521 VRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKMFERIQKEL------- 573
RG +L ++ G+ D E + PPL + ER K
Sbjct: 671 ARGFCHELAIMCQISGM--DFALEPVL---------PPLTARPEHVERALKARYQDAMNM 719
Query: 574 ---PGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRV---NDQYLTNVLMK 627
G LL ++ N LYG K+ + G+V+QC V + QYL NV +K
Sbjct: 720 LRPQGRELDLLIVILPDNNGSLYGDLKRICETDLGLVSQCCLTKHVFKMSKQYLANVALK 779
Query: 628 INAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLI 687
IN K+GG N+VL + IP+VS PTII G DV+H PG+ PSIAAVV+S++WP +
Sbjct: 780 INVKVGGRNTVLVDALTRRIPLVSDRPTIIFGADVTHPHPGEDSSPSIAAVVASQDWPEV 839
Query: 688 SKYRACVRTQSPKVEMIDNLFKQVSEKE----DEGIIRELLIDFYSSSGKRKPDNIIIFR 743
+KY V Q+ + E+I +LFK + G+I+ELLI F ++G+ KP II +R
Sbjct: 840 TKYAGLVSAQAHRQELIQDLFKVWQDPHRGTVTGGMIKELLISFKRATGQ-KPQRIIFYR 898
Query: 744 DGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPD------ 797
DGVSE QF QVL EL+ I +AC L+ + P +V QK HHT+ F D
Sbjct: 899 DGVSEGQFYQVLLYELDAIRKACASLEPNYQPPVTFVVVQKRHHTRLFANNHNDQRTVDR 958
Query: 798 --NVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSL 855
N+ PGTV+D+KICHP DFY+C+HAG+ GTSRP HYHVL D+ F+ DELQ L ++L
Sbjct: 959 SGNILPGTVVDSKICHPTEFDFYLCSHAGIQGTSRPAHYHVLWDENKFTADELQTLTNNL 1018
Query: 856 SYVYQRSTTAISVVAPICYAHLAATQIGQFMKFEDKSDTSSSHGGLTAAGVAP------- 908
Y Y R T ++S+V P YAHLAA + +M+ E S + G T+ G+ P
Sbjct: 1019 CYTYARCTRSVSIVPPAYYAHLAAFRARFYMEPETSDSGSMASGAATSRGLPPGVRSARV 1078
Query: 909 ----VVPQLPKLQDSVSSSMFFC 927
V LP L+++V MF+C
Sbjct: 1079 AGNVAVRPLPALKENVKRVMFYC 1101
>UniRef100_Q5Z5B2 Putative AGO1 homologous protein [Oryza sativa]
Length = 1038
Score = 494 bits (1272), Expect = e-138
Identities = 333/910 (36%), Positives = 491/910 (53%), Gaps = 87/910 (9%)
Query: 60 RKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQET 119
R G GS GT+ + NHF + + D H QY V+++ E + + V +++ + +
Sbjct: 174 RPGSGSIGTRCLVKANHFFAQLPDKDLH--QYDVSITPELTSRIRSRAVMEELVRLHKMS 231
Query: 120 YGSELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKK 179
Y L G+ AYDG K+L+T G L EF + L + DG+ +
Sbjct: 232 Y---LGGRLPAYDGRKSLYTAGPLPFTSKEFRISLLE----------EDDGSGSE----- 273
Query: 180 RMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLL 239
RR KT+ V I FAA+ L + L G+++E QEA++VLDI+LR+ +
Sbjct: 274 --RR---QKTYNVVIKFAARADLHRLEQFLAGRQAEAPQEALQVLDIVLRELPTARYAPF 328
Query: 240 VRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFLIA 299
R SFF D +G G+ RGF+ S R TQ GLSLNID+S T +P PV+DF+I
Sbjct: 329 GR-SFFSPDLGRRRSLGEGLETWRGFYQSIRPTQMGLSLNIDMSATAFFEPLPVIDFVIQ 387
Query: 300 --NQNVRD-PFS-LDWAKAKRTLKNLRIKASPSN---QEYKITGLSELPCKEQTFTMKKK 352
N ++R P S + K K+ L+ +++ + ++Y+I+GL+ +E TF + +
Sbjct: 388 LLNTDIRSRPLSDAERVKIKKALRGVKVGVTHRGNMRRKYRISGLTSQATRELTFPVDQG 447
Query: 353 GGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQR 412
G +V +YF ++++ LPC+ VG +RP Y+P+E+C +V QR
Sbjct: 448 GTVK----------SVVQYFQETYGFAIQHTY-LPCLQVGNQQRPNYLPMEVCKIVEGQR 496
Query: 413 YTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGR 472
Y+K L Q +L+E++ Q+P +R + Q + ++Y +P K GI I+ VE R
Sbjct: 497 YSKRLNQNQIRALLEETCQRPHDRERDIIQMVNHNSYHEDPYAKEFGIKISERLALVEAR 556
Query: 473 VLQAPRLKF---GNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD---VRGLVR 526
+L APRLK+ G +D PR G+WN+ NKK+V ++ W VNF+ G R
Sbjct: 557 ILPAPRLKYNETGREKDCLPRVGQWNMMNKKMVNGGRVRSWICVNFARNVQESVASGFCR 616
Query: 527 DLIKCARLKGIP--IDEPYEEIFEENGQFRRAPPLVRVEKM--FERIQKELPGAPSFLLC 582
+L + + G+ ++ ++ Q RA + M KEL L+
Sbjct: 617 ELARMCQASGMDFALEPVLPSMYARPDQVERALKARFHDAMNILGPQHKEL----DLLIG 672
Query: 583 LLPERKNSDLYGPWKKKNLAEYGIVTQCISPTRV---NDQYLTNVLMKINAKLGGLNSVL 639
LLP+ N LYG K+ + G+V+QC +V N Q L N+ +KIN K+GG N+VL
Sbjct: 673 LLPDN-NGSLYGDLKRICEIDLGLVSQCCCTKQVFKMNKQILANLALKINVKVGGRNTVL 731
Query: 640 GVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSP 699
++ IP+V+ PTII G DV+H PG+ PSIAAVV+S++WP ++KY V QS
Sbjct: 732 VDAVSRRIPLVTDRPTIIFGADVTHPHPGEDSSPSIAAVVASQDWPEVTKYAGLVSAQSH 791
Query: 700 KVEMIDNLFKQVSEKEDE----GIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVL 755
+ E+ID+L+ + G++RELLI F S+G+ KP II +RDGVSE QF QVL
Sbjct: 792 RQELIDDLYNITHDPHRGPICGGMVRELLISFKRSTGQ-KPQRIIFYRDGVSEGQFYQVL 850
Query: 756 NIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPD--------NVPPGTVIDN 807
EL+ I +AC L+ + P+ IV QK HHT+ F D N+ PGTV+D+
Sbjct: 851 LHELDAIRKACASLEANYQPQVTFIVVQKRHHTRLFAHNHNDQNSVDRSGNILPGTVVDS 910
Query: 808 KICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAIS 867
KICHP DF++C+HAG+ GTSRP HYHVL D+ F+ D LQ L ++L Y Y R T ++S
Sbjct: 911 KICHPTEFDFFLCSHAGIKGTSRPAHYHVLWDENNFTADALQTLTNNLCYTYARCTRSVS 970
Query: 868 VVAPICYAHLAATQIGQFMKFEDKSDTSSSHGG----------LTAAGVAPVVPQLPKLQ 917
+V P YAHLAA + +M+ D SD+ S G AAG V P LP L+
Sbjct: 971 IVPPAYYAHLAAFRARFYME-SDSSDSGSMASGRGGGSSTSRSTRAAGGGAVRP-LPALK 1028
Query: 918 DSVSSSMFFC 927
DSV + MF+C
Sbjct: 1029 DSVKNVMFYC 1038
>UniRef100_Q7XBH5 Zwille pinhead-like protein [Oryza sativa]
Length = 361
Score = 493 bits (1270), Expect = e-138
Identities = 239/327 (73%), Positives = 282/327 (86%), Gaps = 1/327 (0%)
Query: 565 MFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPT-RVNDQYLTN 623
MFE+++ LPG P FLLC+LPERKN DLYGPWKKKNL E GI+TQCI P+ ++NDQY TN
Sbjct: 1 MFEKVKANLPGPPEFLLCVLPERKNCDLYGPWKKKNLHEMGIITQCIVPSVKMNDQYYTN 60
Query: 624 VLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSRE 683
VL+KINAKLGG+NS L +E IPIV++ PT+ILGMDVSHGSPG++D+PSIAAV SR
Sbjct: 61 VLLKINAKLGGMNSKLSLEHRHMIPIVNQTPTLILGMDVSHGSPGRADVPSIAAVAGSRC 120
Query: 684 WPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFR 743
WPLIS+YRA VRTQSPKVEMID+LFK + + +D+GIIRELL+DFY +S +RKP IIIFR
Sbjct: 121 WPLISRYRASVRTQSPKVEMIDSLFKPLDDGKDDGIIRELLLDFYKTSQQRKPKQIIIFR 180
Query: 744 DGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPDNVPPGT 803
DGVSESQF+QVLN+ELNQII+A +++D+ PKF VI+AQKNHHTK FQ +PDNVPPGT
Sbjct: 181 DGVSESQFSQVLNVELNQIIKAYQYMDQGPIPKFTVIIAQKNHHTKLFQENTPDNVPPGT 240
Query: 804 VIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRST 863
V+D+ I HPR DFYM AHAG IGTSRPTHYHVLLD+IGF PD++Q+LV SLSYVYQRST
Sbjct: 241 VVDSGIVHPRQYDFYMYAHAGPIGTSRPTHYHVLLDEIGFLPDDVQKLVLSLSYVYQRST 300
Query: 864 TAISVVAPICYAHLAATQIGQFMKFED 890
TAISVVAPICYAHLAA Q+GQFMKFE+
Sbjct: 301 TAISVVAPICYAHLAAAQMGQFMKFEE 327
>UniRef100_Q6K972 AGO1 homologous protein [Oryza sativa]
Length = 1011
Score = 493 bits (1270), Expect = e-138
Identities = 354/966 (36%), Positives = 505/966 (51%), Gaps = 117/966 (12%)
Query: 16 NEDLMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLGSKGTKLPLLTN 75
+E P P PP P D P R P+ R G G+ GT+ + N
Sbjct: 109 SELAQPSPTPPQEQPVDAATTT------PHHIPSSSKSIRFPL-RPGKGTIGTRCMVKAN 161
Query: 76 HFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETY-GSELNGKDFAYDGE 134
HF + N D H Y V+++ E V + V R VI ++ Y S L G+ AYDG
Sbjct: 162 HFFAHLPNKDLH--HYDVSITPE----VTSRIVNRAVIKELVNLYKASYLGGRLPAYDGR 215
Query: 135 KTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEI 194
K+L+T G L EF + L D DG+ + ++R R TF+V I
Sbjct: 216 KSLYTAGPLPFTSQEFQITLLD----------DDDGSGS----ERRQR------TFRVVI 255
Query: 195 SFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYAD 254
FAA+ L + L G+ +E QEA++VLDI+LR+ + + R SFF
Sbjct: 256 KFAARADLHRLELFLAGRHAEAPQEALQVLDIVLRELPSARYAPFGR-SFFSPYLGRRQP 314
Query: 255 VGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFL--IANQNVRD-PFS-LD 310
+G G+ RGF+ S R TQ GLSLNID+S T I+P PV+DF+ + N ++ P S +
Sbjct: 315 LGEGLESWRGFYQSIRPTQMGLSLNIDMSATAFIEPLPVIDFVAQLLNSDIHSRPLSDAE 374
Query: 311 WAKAKRTLKNLRIKASPSN---QEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEIT 367
K K+ L+ ++++ + ++Y+I+GL+ P +E TF + + G +
Sbjct: 375 RVKIKKALRGVKVEVTHRGNMRRKYRISGLTIQPTRELTFPVDEGGTVK----------S 424
Query: 368 VYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVE 427
V +YF ++++ LPC+ V +R Y+P+E+C +V QRY+K L Q +L+E
Sbjct: 425 VVQYFQETYGFAIQHTY-LPCLTV---QRLNYLPMEVCKIVEGQRYSKRLNQNQIRALLE 480
Query: 428 KSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF---GNG 484
++ Q P +R + + +K + Y ++P K GI I+ VE R+L APRLK+ G
Sbjct: 481 ETCQHPRDRERDIIKMVKHNAYQDDPYAKEFGIKISDRLASVEARILPAPRLKYNETGRE 540
Query: 485 EDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD---VRGLVRDLIKCARLKGIPIDE 541
+D PR G+WN+ NKK+V K+ W VNF+ VRG +L + G+ D
Sbjct: 541 KDCLPRVGQWNMMNKKMVNGGKVRSWMCVNFARNVQESVVRGFCHELALMCQASGM--DF 598
Query: 542 PYEEIFEENGQFRRAPPLVRVEKMFERIQK-------ELPGAP----SFLLCLLPERKNS 590
E I PPL ER K + G L+ +LP+ N
Sbjct: 599 APEPIL---------PPLNAHPDQVERALKARYHDAMNVLGPQRRELDLLIGILPDN-NG 648
Query: 591 DLYGPWKKKNLAEYGIVTQCISPTRV---NDQYLTNVLMKINAKLGGLNSVLGVEMNPSI 647
LYG K+ + GIV+QC +V N Q L N+ +KIN K+GG N+VL ++ I
Sbjct: 649 SLYGDLKRVCEIDLGIVSQCCCTKQVFKMNKQILANLALKINVKVGGRNTVLVDAVSRRI 708
Query: 648 PIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNL 707
P+V+ PTII G DV+H PG+ PSIAAVV+S++WP ++KY V Q+ + E+I++L
Sbjct: 709 PLVTDRPTIIFGADVTHPHPGEDSSPSIAAVVASQDWPEVTKYAGLVSAQAHRQELIEDL 768
Query: 708 FKQVSEKE----DEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQII 763
+K + + G+IRELLI F S+G+ KP II +RDGVSE QF QVL ELN I
Sbjct: 769 YKIWQDPQRGTVSGGMIRELLISFKRSTGE-KPQRIIFYRDGVSEGQFYQVLLYELNAIR 827
Query: 764 EACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPD--------NVPPGTVIDNKICHPRNN 815
+AC L+ + PK IV QK HHT+ F D N+ PGTV+D+KICHP
Sbjct: 828 KACASLETNYQPKVTFIVVQKRHHTRLFAHNHNDQNSVDRSGNILPGTVVDSKICHPTEF 887
Query: 816 DFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVAPICYA 875
DFY+C+HAG+ GTSRP HYHVL D+ F+ D LQ L ++L Y Y R T ++S+V P YA
Sbjct: 888 DFYLCSHAGIKGTSRPAHYHVLWDENNFTADALQILTNNLCYTYARCTRSVSIVPPAYYA 947
Query: 876 HLAATQIGQFMKFEDKSDTSSSHGG--------------LTAAGVAPVVPQLPKLQDSVS 921
HLAA + +M+ D SD+SS G A G A V P LP L+DSV
Sbjct: 948 HLAAFRARFYME-PDTSDSSSVVSGPGVRGPLSGSSTSRTRAPGGAAVKP-LPALKDSVK 1005
Query: 922 SSMFFC 927
MF+C
Sbjct: 1006 RVMFYC 1011
>UniRef100_Q9XGW1 PINHEAD protein [Arabidopsis thaliana]
Length = 988
Score = 493 bits (1270), Expect = e-138
Identities = 322/902 (35%), Positives = 487/902 (53%), Gaps = 78/902 (8%)
Query: 60 RKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQET 119
R G G+ GTK + NHF + D + QY V ++ E V K V R +I ++
Sbjct: 131 RPGFGTLGTKCIVKANHFLADLPTKDLN--QYDVTITPE----VSSKSVNRAIIAELVRL 184
Query: 120 YG-SELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDK 178
Y S+L + AYDG K+L+T G L EF+V + D NG
Sbjct: 185 YKESDLGRRLPAYDGRKSLYTAGELPFTWKEFSVKIVDEDDGIING-------------- 230
Query: 179 KRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCL 238
P +++KV I F A+ + + L G+ ++ QEA+++LDI+LR+ + K+ C
Sbjct: 231 -----PKRERSYKVAIKFVARANMHHLGEFLAGKRADCPQEAVQILDIVLRELSVKRFCP 285
Query: 239 LVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDF-- 296
+ R SFF D K +G G+ GF+ S R TQ GLSLNID+++ I+P PV++F
Sbjct: 286 VGR-SFFSPDIKTPQRLGEGLESWCGFYQSIRPTQMGLSLNIDMASAAFIEPLPVIEFVA 344
Query: 297 -LIANQNVRDPFS-LDWAKAKRTLKNLRIKASPS---NQEYKITGLSELPCKEQTFTMKK 351
L+ + P S D K K+ L+ ++++ + ++Y++ GL+ P +E F +
Sbjct: 345 QLLGKDVLSKPLSDSDRVKIKKGLRGVKVEVTHRANVRRKYRVAGLTTQPTRELMFPV-- 402
Query: 352 KGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQ 411
+E+ T + +V EYF ++++ LPC+ VG K+ +Y+P+E C +V Q
Sbjct: 403 ------DENCTMK--SVIEYFQEMYGFTIQHT-HLPCLQVGNQKKASYLPMEACKIVEGQ 453
Query: 412 RYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEG 471
RYTK L Q ++L++ + Q+P +R N + + ++ + Y +P K G+ I+ VE
Sbjct: 454 RYTKRLNEKQITALLKVTCQRPRDRENDILRTVQHNAYDQDPYAKEFGMNISEKLASVEA 513
Query: 472 RVLQAPRLKF---GNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD---VRGLV 525
R+L AP LK+ G +D P+ G+WN+ NKK++ + WA VNFS RG
Sbjct: 514 RILPAPWLKYHENGKEKDCLPQVGQWNMMNKKMINGMTVSRWACVNFSRSVQENVARGFC 573
Query: 526 RDLIKCARLKGIPID-EPYEEIFEEN-GQFRRAPPLVRVEKMFERIQKELPGAPSFLLCL 583
+L + + G+ + EP I+ Q +A V M + KEL LL +
Sbjct: 574 NELGQMCEVSGMEFNPEPVIPIYSARPDQVEKALKHVYHTSMNKTKGKEL----ELLLAI 629
Query: 584 LPERKNSDLYGPWKKKNLAEYGIVTQCISPT---RVNDQYLTNVLMKINAKLGGLNSVLG 640
LP+ N LYG K+ E G+++QC +++ QYL NV +KIN K+GG N+VL
Sbjct: 630 LPDN-NGSLYGDLKRICETELGLISQCCLTKHVFKISKQYLANVSLKINVKMGGRNTVLV 688
Query: 641 VEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPK 700
++ IP+VS +PTII G DV+H G+ PSIAAVV+S++WP ++KY V Q+ +
Sbjct: 689 DAISCRIPLVSDIPTIIFGADVTHPENGEESSPSIAAVVASQDWPEVTKYAGLVCAQAHR 748
Query: 701 VEMIDNLFKQ----VSEKEDEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLN 756
E+I +L+K V G+IR+LLI F ++G+ KP II +RDGVSE QF QVL
Sbjct: 749 QELIQDLYKTWQDPVRGTVSGGMIRDLLISFRKATGQ-KPLRIIFYRDGVSEGQFYQVLL 807
Query: 757 IELNQIIEACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPD--------NVPPGTVIDNK 808
EL+ I +AC L+ + P IV QK HHT+ F D N+ PGTV+D K
Sbjct: 808 YELDAIRKACASLEPNYQPPVTFIVVQKRHHTRLFANNHRDKNSTDRSGNILPGTVVDTK 867
Query: 809 ICHPRNNDFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISV 868
ICHP DFY+C+HAG+ GTSRP HYHVL D+ F+ D +Q L ++L Y Y R T ++S+
Sbjct: 868 ICHPTEFDFYLCSHAGIQGTSRPAHYHVLWDENNFTADGIQSLTNNLCYTYARCTRSVSI 927
Query: 869 VAPICYAHLAATQIGQFMK---FEDKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVSSSMF 925
V P YAHLAA + +++ +D + T G V P LP L+++V MF
Sbjct: 928 VPPAYYAHLAAFRARFYLEPEIMQDNGSPGKKNTKTTTVGDVGVKP-LPALKENVKRVMF 986
Query: 926 FC 927
+C
Sbjct: 987 YC 988
>UniRef100_Q8LP00 ZLL/PNH homologous protein [Oryza sativa]
Length = 978
Score = 493 bits (1269), Expect = e-137
Identities = 333/951 (35%), Positives = 504/951 (52%), Gaps = 88/951 (9%)
Query: 11 GNGNGNEDLMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLGSKGTKL 70
G G G P P PA V + PP K R P G G+ G +
Sbjct: 82 GRGRGGRAGAGPGPGLAAAPAVVVAPAARAVIGPPVASKGLSFCRRP----GFGTVGARC 137
Query: 71 PLLTNHFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETY-GSELNGKDF 129
+ NHF + + D QY V ++ E V + V R ++ ++ Y S+L G+
Sbjct: 138 VVKANHFLAELPDKD--LTQYDVKITPE----VSSRSVNRAIMSELVRLYHDSDLGGRLP 191
Query: 130 AYDGEKTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKT 189
AYDG K L+T G+L + EF V L D DG P +
Sbjct: 192 AYDGRKNLYTAGTLPFDAREFVVRLTD----------DDDGTGV----------PPRERE 231
Query: 190 FKVEISFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDP 249
++V I FAA+ L + + G++++ QEA++VLDI+LR+ A ++ + + +SF+ D
Sbjct: 232 YRVAIKFAARADLHHLRQFIAGRQADAPQEALQVLDIVLRELANRR-YVSIGRSFYSPDI 290
Query: 250 KNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDF---LIANQNVRDP 306
+ +G G+ GF+ S R TQ GLSLNID+S+T I+P PV++F ++ + P
Sbjct: 291 RKPQRLGDGLQSWCGFYQSIRPTQMGLSLNIDMSSTAFIEPLPVIEFVAQILGKDVISRP 350
Query: 307 FS-LDWAKAKRTLKNLRIKASPSN---QEYKITGLSELPCKEQTFTMKKKGGNNGEEDAT 362
S + K K+ L+ ++++ + ++Y+I+GL+ P E F + D
Sbjct: 351 LSDANRIKIKKALRGVKVEVTHRGNVRRKYRISGLTTQPTHELIFPI----------DDQ 400
Query: 363 EEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQR 422
+V +YF +++ LPC+ VG K+ Y+P+E C +V QRYTK L Q
Sbjct: 401 MNMKSVVQYFKEMYGFTIQHP-HLPCLQVGNQKKANYLPMEACKIVEGQRYTKRLNEKQI 459
Query: 423 SSLVEKSRQKPLER-MNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF 481
+SL++ + ++P E+ M++L + + Y +P K GI I+ T VE RVL AP LK+
Sbjct: 460 TSLLKVTCRRPREQEMDILQS--QQNGYEQDPYAKEFGINISEKLTSVEARVLPAPWLKY 517
Query: 482 GNG---EDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD---VRGLVRDLIKCARLK 535
+ ++ P+ G+WN+ NKKV+ K+ HWA +NFS RG ++L + ++
Sbjct: 518 HDTVKEKECLPQVGQWNMVNKKVINGCKVNHWACINFSRSVQETTARGFCQELAQMCQIS 577
Query: 536 GIPID-EPYEEIFEEN-GQFRRAPPLVRVEKMFERIQKELPGAPSFLLCLLPERKNSDLY 593
G+ + EP I+ Q +A V + + KEL LL +LP+ N LY
Sbjct: 578 GMEFNSEPVIPIYSARPDQVEKALKHVYNMSLNKLKGKEL----ELLLAILPDN-NGSLY 632
Query: 594 GPWKKKNLAEYGIVTQCISPT---RVNDQYLTNVLMKINAKLGGLNSVLGVEMNPSIPIV 650
G K+ + G+++QC +++ QYL NV +KIN K+GG N+VL ++ IP+V
Sbjct: 633 GDIKRICETDLGLISQCCLTKHVFKISKQYLANVSLKINVKMGGRNTVLLDAISWRIPLV 692
Query: 651 SKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQ 710
S +PTII G DV+H G+ PSIAAVV+S++WP ++KY V Q+ + E+I +L+K
Sbjct: 693 SDIPTIIFGADVTHPETGEDSSPSIAAVVASQDWPEVTKYAGLVCAQAHRQELIQDLYKT 752
Query: 711 VSEKE----DEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEAC 766
+ + G+IRELLI F ++G+ KP II +RDGVSE QF QVL EL+ I +AC
Sbjct: 753 WHDPQRGTVTGGMIRELLISFRKATGQ-KPLRIIFYRDGVSEGQFYQVLLYELDAIRKAC 811
Query: 767 KFLDETWNPKFLVIVAQKNHHTKFFQPGSPD--------NVPPGTVIDNKICHPRNNDFY 818
L+ + P +V QK HHT+ F D N+ PGTV+D+KICHP DFY
Sbjct: 812 ASLEPIYQPPVTFVVVQKRHHTRLFANNHKDRSSTDKSGNILPGTVVDSKICHPSEFDFY 871
Query: 819 MCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVAPICYAHLA 878
+C+HAG+ GTSRP HYHVL D+ F+ DE+Q L ++L Y Y R T ++SVV P YAHLA
Sbjct: 872 LCSHAGIQGTSRPAHYHVLWDENNFTADEMQTLTNNLCYTYARCTRSVSVVPPAYYAHLA 931
Query: 879 ATQIGQFMKFE--DKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVSSSMFFC 927
A + +M+ E + TS S G V P LP +++ V MF+C
Sbjct: 932 AFRARFYMEPEMSENQTTSKSSTGTNGTSVKP----LPAVKEKVKRVMFYC 978
>UniRef100_Q8LNZ9 AGO1 homologous protein [Oryza sativa]
Length = 909
Score = 493 bits (1269), Expect = e-137
Identities = 353/966 (36%), Positives = 504/966 (51%), Gaps = 117/966 (12%)
Query: 16 NEDLMPPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLGSKGTKLPLLTN 75
+E P P PP P D P R P+ R G G+ GT+ + N
Sbjct: 7 SELAQPSPTPPQEQPVDAATTT------PHHIPSSSKSIRFPL-RPGKGTIGTRCMVKAN 59
Query: 76 HFKVTVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETY-GSELNGKDFAYDGE 134
HF + N D H Y V+++ E V + V R VI ++ Y S L G+ AYDG
Sbjct: 60 HFFAHLPNKDLH--HYDVSITPE----VTSRIVNRAVIKELVNLYKASYLGGRLPAYDGR 113
Query: 135 KTLFTIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEI 194
K+L+T G L EF + L D DG+ + ++R R TF+V I
Sbjct: 114 KSLYTAGPLPFTSQEFQITLLD----------DDDGSGS----ERRQR------TFRVVI 153
Query: 195 SFAAKIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYAD 254
FAA+ L + L G+ +E QEA++VLDI+LR+ + + R SFF
Sbjct: 154 KFAARADLHRLELFLAGRHAEAPQEALQVLDIVLRELPSARYAPFGR-SFFSPYLGRRQP 212
Query: 255 VGGGVLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFL--IANQNV--RDPFSLD 310
+G G+ RGF+ S R TQ GLSLNID+S T I+P PV+DF+ + N ++ R F +
Sbjct: 213 LGEGLESWRGFYQSIRPTQMGLSLNIDMSATAFIEPLPVIDFVAQLLNSDIHSRSLFDAE 272
Query: 311 WAKAKRTLKNLRIKASPSN---QEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEIT 367
K K+ L+ ++++ + ++Y+I+GL+ P +E TF + + G +
Sbjct: 273 RVKIKKALRGVKVEVTHRGNMRRKYRISGLTIQPTRELTFPVDEGGTVK----------S 322
Query: 368 VYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVE 427
V +YF ++++ LPC+ V +R Y+P+E+C +V QRY+K L Q +L+E
Sbjct: 323 VVQYFQETYGFAIQHTY-LPCLTV---QRLNYLPMEVCKIVEGQRYSKRLNQNQIRALLE 378
Query: 428 KSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF---GNG 484
++ Q P +R + + +K + Y ++P K GI I+ VE R+L APRLK+ G
Sbjct: 379 ETCQHPRDRERDIIKMVKHNAYQDDPYAKEFGIKISDRLASVEARILPAPRLKYNETGRE 438
Query: 485 EDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD---VRGLVRDLIKCARLKGIPIDE 541
+D PR G+WN+ NKK+V K+ W VNF+ VRG +L + G+ D
Sbjct: 439 KDCLPRVGQWNMMNKKMVNGGKVRSWMCVNFARNVQESVVRGFCHELALMCQASGM--DF 496
Query: 542 PYEEIFEENGQFRRAPPLVRVEKMFERIQK-------ELPGAP----SFLLCLLPERKNS 590
E I PPL ER K + G L+ LP+ N
Sbjct: 497 APEPIL---------PPLNAHPDQVERALKARYHDAMNVLGPQRRELDLLIGKLPDN-NG 546
Query: 591 DLYGPWKKKNLAEYGIVTQCISPTRV---NDQYLTNVLMKINAKLGGLNSVLGVEMNPSI 647
LYG K+ + GIV+QC +V N Q L N+ +KIN K+GG N+VL ++ I
Sbjct: 547 SLYGDLKRVCEIDLGIVSQCCCTKQVFKMNKQILANLALKINVKVGGRNTVLVDAVSRRI 606
Query: 648 PIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNL 707
P+V+ PTII G DV+H PG+ PSIAAVV+S++WP ++KY V Q+ + E+I++L
Sbjct: 607 PLVTDRPTIIFGADVTHPHPGEDSSPSIAAVVASQDWPEVTKYAGLVSAQAHRQELIEDL 666
Query: 708 FKQVSEKE----DEGIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQII 763
+K + + G+IRELLI F S+G+ KP II +RDGVSE QF QVL ELN I
Sbjct: 667 YKIWQDPQRGTVSGGMIRELLISFKRSTGE-KPQRIIFYRDGVSEGQFYQVLLYELNAIR 725
Query: 764 EACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPD--------NVPPGTVIDNKICHPRNN 815
+AC L+ + PK IV QK HHT+ F D N+ PGTV+D+KICHP
Sbjct: 726 KACASLETNYQPKVTFIVVQKRHHTRLFAHNHNDQNSVDRSGNILPGTVVDSKICHPTEF 785
Query: 816 DFYMCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVAPICYA 875
DFY+C+HAG+ GTSRP HYHVL ++ F+ D LQ L ++L Y Y R T ++S+V P YA
Sbjct: 786 DFYLCSHAGIKGTSRPAHYHVLWNENNFTADALQILTNNLCYTYARCTRSVSIVPPAYYA 845
Query: 876 HLAATQIGQFMKFEDKSDTSSSHGG--------------LTAAGVAPVVPQLPKLQDSVS 921
HLAA + +M+ D SD+SS G A G A V P LP L+DSV
Sbjct: 846 HLAAFRARFYME-PDTSDSSSVVSGPGVRGPLSGSSTSRTRAPGGAAVKP-LPALKDSVK 903
Query: 922 SSMFFC 927
MF+C
Sbjct: 904 RVMFYC 909
>UniRef100_Q7Y001 Putative leaf development and shoot apical meristem regulating
protein [Oryza sativa]
Length = 1055
Score = 484 bits (1245), Expect = e-135
Identities = 325/949 (34%), Positives = 493/949 (51%), Gaps = 109/949 (11%)
Query: 21 PPPPPPPIVPADVEPVKVDLLDLPPEPVKKKLPTRLPIARKGLGSKGTKLPLLTNHFKVT 80
P P PPP LPP K P AR +G+ G + + NHF V
Sbjct: 174 PAPAPPPAT-------------LPPSSSKA---VTFP-ARPDVGTIGRRCRVRANHFLVQ 216
Query: 81 VANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETYGSELNGKDFAYDGEKTLFTI 140
VA+ D + Y V ++ E + R +I+K+ + L+G+ YDG K+++T
Sbjct: 217 VADKD--IYHYDVVITPESTY----RERNRSIINKLVALHKQFLDGRLPVYDGRKSIYTA 270
Query: 141 GSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKI 200
G L +F V + +P +R + +KV I A+K
Sbjct: 271 GPLPFKTKDFVVK-----------HINP------------LRGNQREEEYKVTIKQASKT 307
Query: 201 PLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGC-----LLVRQSFFHNDPKNYADV 255
L ++ L G++ E Q+ I+ LDI LR+ + + +SFF + ++
Sbjct: 308 DLYSLKQFLVGRQRELPQDTIQALDIALRECPTSVNFTCDRYVSISRSFFSQSFGHGGEI 367
Query: 256 GGGVLGCRGFHSSFRTTQSGLSLNI------DVSTTMIIQPGPVVDFLIANQNVRDPF-- 307
G G RG++ S R TQ GLSLNI ++S T + PV+DF + N+RD
Sbjct: 368 GSGTECWRGYYQSLRPTQMGLSLNIGMDLPQNISATAFYKAQPVMDFAVQYLNIRDVSRR 427
Query: 308 --SLDWAKAKRTLKNLRIKASPSNQE---YKITGLSELPCKEQTFTMKKKGGNNGEEDAT 362
D K K+ LK ++I A+ ++ YKITG+ P E F D
Sbjct: 428 LSDQDRIKLKKALKGVQIVATHWKEKSIRYKITGIPSAPMNELMF------------DLD 475
Query: 363 EEEITVYEYFVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQR 422
I+V +YF L++ + PC+ G RP Y+P+E+CS++ QRY+K L Q
Sbjct: 476 GNRISVVQYFKKQYNYSLKH-VNWPCLQAGSDSRPKYLPMEVCSILEGQRYSKKLNEHQV 534
Query: 423 SSLVEKSRQKPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF- 481
++++ + ++P +R + + + + T++YGN+ K GI +A+ V+ RVL PRLK+
Sbjct: 535 TNILRMTCERPAQRESSIIEIVNTNSYGNDDCAKEFGIKVANQLAVVDARVLPTPRLKYH 594
Query: 482 --GNGEDFNPRNGRWNLNNKKVVRPAKIEHWAVVNFSARC---DVRGLVRDLIKCARLKG 536
G + NP G+WN+ NK++V I HW ++F++R D+R DL+ G
Sbjct: 595 DSGREKVCNPSVGQWNMINKRMVNGGCINHWTCLSFASRMHVNDIRMFCEDLVGMCNNIG 654
Query: 537 IPID-EPYEEIFEENGQFRRAPPLVRV--EKMFERI-QKELPGAP-SFLLCLLPERKNSD 591
+ ++ P +I + GQ R +R + E++ Q++L G L+ +L E S
Sbjct: 655 MQMNTRPCVDIIQ--GQQRNIEGAIRNIHRQSSEKLDQQDLTGQQLQLLIVILTEISGS- 711
Query: 592 LYGPWKKKNLAEYGIVTQCISPTRVND---QYLTNVLMKINAKLGGLNSVLGVEMNPSIP 648
YG K+ E G++TQC +P + QYL N+ +K+N K+GG N+VL ++ IP
Sbjct: 712 -YGRIKRICETEVGVITQCCAPKSLQKGGKQYLENLALKMNVKVGGRNTVLEDALHKKIP 770
Query: 649 IVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLF 708
I++ PTI+ G DV+H SPG+ PSIAAVV+S +WP ++KY+ V TQS + E+I NL+
Sbjct: 771 ILTDRPTIVFGADVTHPSPGEDASPSIAAVVASMDWPEVTKYKCLVSTQSHREEIISNLY 830
Query: 709 KQVSEKEDE----GIIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIE 764
+V + G+IRELL FY +G+ KP II +RDG+SE QF+QVL E++ I +
Sbjct: 831 TEVKDPLKGIIRGGMIRELLRSFYQETGQ-KPSRIIFYRDGISEGQFSQVLLYEMDAIRK 889
Query: 765 ACKFLDETWNPKFLVIVAQKNHHTKFFQPGSPD------NVPPGTVIDNKICHPRNNDFY 818
AC L E + P +V QK HHT+ F D N+ PGTV+D ICHP DFY
Sbjct: 890 ACASLQEGYLPPVTFVVVQKRHHTRLFPENRRDMMDRSGNILPGTVVDTMICHPSEFDFY 949
Query: 819 MCAHAGMIGTSRPTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVAPICYAHLA 878
+C+H+G+ GTSRPTHYHVLLD+ GF D LQ L ++LSY Y R T A+S+V P YAHL
Sbjct: 950 LCSHSGIKGTSRPTHYHVLLDENGFKADTLQTLTYNLSYTYARCTRAVSIVPPAYYAHLG 1009
Query: 879 ATQIGQFMKFEDKSDTSSSHGGLTAAGVAPVVPQLPKLQDSVSSSMFFC 927
A + +M ED+ S +T P LP+++++V MF+C
Sbjct: 1010 AFRARYYM--EDEHSDQGSSSSVTTRTDRSTKP-LPEIKENVKRFMFYC 1055
>UniRef100_O04379 Argonaute protein [Arabidopsis thaliana]
Length = 1048
Score = 480 bits (1235), Expect = e-134
Identities = 322/948 (33%), Positives = 492/948 (50%), Gaps = 87/948 (9%)
Query: 24 PPPPIVPADVEPVKVDL----LDLPPEPVKKKLPTRLPIARKGLGSKGTKLPLLTNHFKV 79
P P ++ E + V+ + P P K + P+ R G G G + + NHF
Sbjct: 144 PEPTVLAQQFEQLSVEQGAPSQAIQPIPSSSKA-FKFPM-RPGKGQSGKRCIVKANHFFA 201
Query: 80 TVANSDGHFFQYSVALSYEDGRPVEGKGVGRKVIDKVQETY-GSELNGKDFAYDGEKTLF 138
+ + D H Y V ++ E V +GV R V+ ++ + Y S L + AYDG K+L+
Sbjct: 202 ELPDKDLH--HYDVTITPE----VTSRGVNRAVMKQLVDNYRDSHLGSRLPAYDGRKSLY 255
Query: 139 TIGSLARNKLEFTVVLEDVISNRNNGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAA 198
T G L N EF + L D ++R R FKV I A
Sbjct: 256 TAGPLPFNSKEFRINLLD--------------EEVGAGGQRRERE------FKVVIKLVA 295
Query: 199 KIPLQAIVNALRGQESENYQEAIRVLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGG 258
+ L + L G++S+ QEA++VLDI+LR+ + + V +SF+ D +G G
Sbjct: 296 RADLHHLGMFLEGKQSDAPQEALQVLDIVLRELPTSR-YIPVGRSFYSPDIGKKQSLGDG 354
Query: 259 VLGCRGFHSSFRTTQSGLSLNIDVSTTMIIQPGPVVDFL--IANQNVRD-PFS-LDWAKA 314
+ RGF+ S R TQ GLSLNID+S+T I+ PV+ F+ + N+++ P S D K
Sbjct: 355 LESWRGFYQSIRPTQMGLSLNIDMSSTAFIEANPVIQFVCDLLNRDISSRPLSDADRVKI 414
Query: 315 KRTLKNLRIKASPSN---QEYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEY 371
K+ L+ ++++ + ++Y+I+GL+ + +E TF + D + +V EY
Sbjct: 415 KKALRGVKVEVTHRGNMRRKYRISGLTAVATRELTFPV----------DERNTQKSVVEY 464
Query: 372 FVNYRKIDLRYSADLPCINVGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQ 431
F ++++ LPC+ VG RP Y+P+E+C +V QRY+K L Q ++L++ + Q
Sbjct: 465 FHETYGFRIQHT-QLPCLQVGNSNRPNYLPMEVCKIVEGQRYSKRLNERQITALLKVTCQ 523
Query: 432 KPLERMNVLNQALKTSNYGNEPMLKNCGITIASGFTQVEGRVLQAPRLKF---GNGEDFN 488
+P++R + Q ++ ++Y + + GI I++ VE R+L P LK+ G
Sbjct: 524 RPIDREKDILQTVQLNDYAKDNYAQEFGIKISTSLASVEARILPPPWLKYHESGREGTCL 583
Query: 489 PRNGRWNLNNKKVVRPAKIEHWAVVNFSARCD---VRGLVRDLIKCARLKGIPIDEPYEE 545
P+ G+WN+ NKK++ + +W +NFS + R ++L + + G+ + E
Sbjct: 584 PQVGQWNMMNKKMINGGTVNNWICINFSRQVQDNLARTFCQELAQMCYVSGMAFNP--EP 641
Query: 546 IFEENGQFRRAPPLVRVEKMFERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYG 605
+ V + + K G LL ++ N LYG K+ E G
Sbjct: 642 VLPPVSARPEQVEKVLKTRYHDATSKLSQGKEIDLLIVILPDNNGSLYGDLKRICETELG 701
Query: 606 IVTQCISPTRV---NDQYLTNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDV 662
IV+QC V + QY+ NV +KIN K+GG N+VL ++ IP+VS PTII G DV
Sbjct: 702 IVSQCCLTKHVFKMSKQYMANVALKINVKVGGRNTVLVDALSRRIPLVSDRPTIIFGADV 761
Query: 663 SHGSPGQSDIPSIAAVVSSREWPLISKYRACVRTQSPKVEMIDNLFKQVSEKEDE----G 718
+H PG+ PSIAAVV+S++WP I+KY V Q+ + E+I +LFK+ + + G
Sbjct: 762 THPHPGEDSSPSIAAVVASQDWPEITKYAGLVCAQAHRQELIQDLFKEWKDPQKGVVTGG 821
Query: 719 IIRELLIDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFL 778
+I+ELLI F S+G KP II +RDGVSE QF QVL EL+ I +AC L+ + P
Sbjct: 822 MIKELLIAFRRSTG-HKPLRIIFYRDGVSEGQFYQVLLYELDAIRKACASLEAGYQPPVT 880
Query: 779 VIVAQKNHHTKFFQPGSPD--------NVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSR 830
+V QK HHT+ F D N+ PGTV+D+KICHP DFY+C+HAG+ GTSR
Sbjct: 881 FVVVQKRHHTRLFAQNHNDRHSVDRSGNILPGTVVDSKICHPTEFDFYLCSHAGIQGTSR 940
Query: 831 PTHYHVLLDDIGFSPDELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMKFED 890
P HYHVL D+ F+ D LQ L ++L Y Y R T ++S+V P YAHLAA + +M+ E
Sbjct: 941 PAHYHVLWDENNFTADGLQSLTNNLCYTYARCTRSVSIVPPAYYAHLAAFRARFYMEPET 1000
Query: 891 KSDTSSSHGGLTAAG-----------VAPVVPQLPKLQDSVSSSMFFC 927
S + G + G V V LP L+++V MF+C
Sbjct: 1001 SDSGSMASGSMARGGGMAGRSTRGPNVNAAVRPLPALKENVKRVMFYC 1048
>UniRef100_Q6Z4F1 Putative leaf development protein Argonaute [Oryza sativa]
Length = 1052
Score = 462 bits (1190), Expect = e-128
Identities = 313/934 (33%), Positives = 480/934 (50%), Gaps = 110/934 (11%)
Query: 44 PPEPVKKKLPTRLPIARKGLGSKGTKLPLLTNHFKVTVANSDGHFFQYSVALSYEDGRPV 103
PP P+ P +R G G+ G ++ + NHF V V+++D + Y V+LS P
Sbjct: 179 PPAPIPVSSKGVAPPSRPGFGTVGERIVVRANHFLVRVSDND-MIYLYDVSLS----PPP 233
Query: 104 EGKGVGRKVIDKVQETYG-SELNGKDFAYDGEKTLFTIGSLARNKLEFTVVLEDVISNRN 162
+ + + R V+ ++ + S L G FAYDG K L+T G L + ++F +
Sbjct: 234 KTRRINRVVMSELARLHRESHLGGISFAYDGSKALYTAGKLPFDSMDFKI---------- 283
Query: 163 NGNCSPDGASTNDSDKKRMRRPYHSKTFKVEISFAAKIPLQAIVNALRGQESENYQEAIR 222
++ + +KV I A + L + + G++ ++ Q+ I+
Sbjct: 284 -----------------KLGKELREIEYKVTIRRAGQADLHHLHEFIAGRQRDSQQQTIQ 326
Query: 223 VLDIILRQHAAKQGCLLVRQSFFHNDPKNYADVGGGVLGCRGFHSSFRTTQSGLSLNIDV 282
LD++LR+ + ++ R F++ D+G G+ +G++ S R TQ GLSLNID+
Sbjct: 327 ALDVVLRESPSLNYVIVSRS--FYSTMFGRQDIGDGLECWKGYYQSLRPTQMGLSLNIDI 384
Query: 283 STTMIIQPGPVVDFLI------ANQNVRDP----FSLDWAKAKRTLKNLRIKASPSNQ-- 330
S+T +P VV+++ N N DP +D K K+ L+ +R++ + +
Sbjct: 385 SSTPFFKPISVVEYVKNCLGTPTNANGPDPRRPLSDIDRLKVKKALRGVRVETTHQGKSS 444
Query: 331 EYKITGLSELPCKEQTFTMKKKGGNNGEEDATEEEITVYEYFVNYRKIDLRYSADLPCIN 390
+YKIT ++ P + F+M D T + TV +YF K L+Y++ PC+
Sbjct: 445 KYKITTITSEPLSQLNFSM----------DGTTQ--TVIQYFSQRYKYRLQYTS-WPCLQ 491
Query: 391 VGKPKRPTYVPVELCSLVSLQRYTKALTTLQRSSLVEKSRQKPLERMNVLNQALKTSNYG 450
G P P Y+P+E+C++V QRY+K L Q + L+ + Q P +R + + ++ +NY
Sbjct: 492 SGNPSNPIYLPMEVCTIVEGQRYSKKLNDKQVTGLLRATCQPPQKREQKIIEMVQHNNYP 551
Query: 451 NEPMLKNCGITIASGFTQVEGRVLQAPRLKF---GNGEDFNPRNGRWNLNNKKVVRPAKI 507
+ ++ + I I++ + RVL AP L++ G + NPR G+WN+ NKK+V A +
Sbjct: 552 ADKVVSDFRINISNQMATMPARVLPAPTLRYHDSGKEKTCNPRVGQWNMINKKMVGGAVV 611
Query: 508 EHWAVVNFSARC--DVRGLVRDLIKCARLKGIPIDEPYEEIFEENGQFRRAPPLVRVEKM 565
+ W VNFS V L +L+ G+ +E E + A P +E
Sbjct: 612 QKWTCVNFSRMHIDAVHRLCGELVYTCNAIGMVFNEMPEI------EVGSAAPN-NIEAA 664
Query: 566 FERIQKELPGAPSFLLCLLPERKNSDLYGPWKKKNLAEYGIVTQCISPTR----VNDQYL 621
I P L+ +LP+ YG K+ E GIV+QC+ P R ++ Q+L
Sbjct: 665 LSNIHTRAPQL-QLLIVILPDVNG--YYGRIKRVCETELGIVSQCLKPGRKLLSLDRQFL 721
Query: 622 TNVLMKINAKLGGLNSVLGVEMNPSIPIVSKVPTIILGMDVSHGSPGQSDIPSIAAVVSS 681
NV +KIN K GG NSVL P +P + TII G DV+H + G+ SIAAVV+S
Sbjct: 722 ENVSLKINVKAGGRNSVL---QRPLVPGGLENTTIIFGADVTHPASGEDSSASIAAVVAS 778
Query: 682 REWPLISKYRACVRTQSPKVEMIDNLFK--QVSEKEDE---------------GIIRELL 724
+WP I+KY+A V Q P+ E+I +LF +V++ D G+ RELL
Sbjct: 779 MDWPEITKYKALVSAQPPRQEIIQDLFTMTEVAQNADAPAQKAEGSKKNFICGGMFRELL 838
Query: 725 IDFYSSSGKRKPDNIIIFRDGVSESQFNQVLNIELNQIIEACKFLDETWNPKFLVIVAQK 784
+ FYS + KRKP II +RDGVS+ QF VL E++ I +A LD + P +V QK
Sbjct: 839 MSFYSKNAKRKPQRIIFYRDGVSDGQFLHVLLYEMDAIKKAIASLDPAYRPLVTFVVVQK 898
Query: 785 NHHTKFFQP--GSPD------NVPPGTVIDNKICHPRNNDFYMCAHAGMIGTSRPTHYHV 836
HHT+ F G D NV PGTV+D ICHP DFY+C+HAG+ GTSRPTHYHV
Sbjct: 899 RHHTRLFPEVHGRQDLTDRSGNVRPGTVVDTNICHPSEFDFYLCSHAGIQGTSRPTHYHV 958
Query: 837 LLDDIGFSPDELQELVHSLSYVYQRSTTAISVVAPICYAHLAATQIGQFMK---FEDKSD 893
L D+ FS D+LQ L ++L Y Y R T ++SVV P YAHLAA + + + + S
Sbjct: 959 LHDENRFSADQLQMLTYNLCYTYARCTRSVSVVPPAYYAHLAAFRARYYDEPPAMDGASS 1018
Query: 894 TSSSHGGLTAAGVAPVVPQLPKLQDSVSSSMFFC 927
S A G P V +LP+++++V MF+C
Sbjct: 1019 VGSGGNQAAAGGQPPAVRRLPQIKENVKDVMFYC 1052
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,659,446,707
Number of Sequences: 2790947
Number of extensions: 76605714
Number of successful extensions: 297578
Number of sequences better than 10.0: 454
Number of HSP's better than 10.0 without gapping: 322
Number of HSP's successfully gapped in prelim test: 143
Number of HSP's that attempted gapping in prelim test: 293954
Number of HSP's gapped (non-prelim): 1138
length of query: 927
length of database: 848,049,833
effective HSP length: 137
effective length of query: 790
effective length of database: 465,690,094
effective search space: 367895174260
effective search space used: 367895174260
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0031.13


