

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0030.6
(780 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
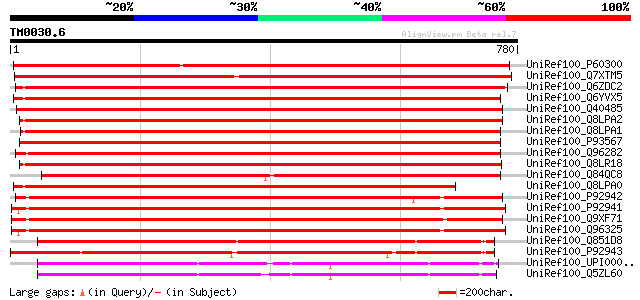
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P60300 Putative chloride channel-like protein CLC-G [A... 1148 0.0
UniRef100_Q7XTM5 OSJNBa0033G05.20 protein [Oryza sativa] 1027 0.0
UniRef100_Q6ZDC2 Putative chloride channel protein [Oryza sativa] 983 0.0
UniRef100_Q6YVX5 Chloride channel [Oryza sativa] 966 0.0
UniRef100_Q40485 C1C-Nt1 protein [Nicotiana tabacum] 964 0.0
UniRef100_Q8LPA2 Chloride channel [Oryza sativa] 962 0.0
UniRef100_Q8LPA1 Chloride channel [Oryza sativa] 959 0.0
UniRef100_P93567 Chloride channel Stclc1 [Solanum tuberosum] 952 0.0
UniRef100_Q96282 Chloride channel protein CLC-c [Arabidopsis tha... 946 0.0
UniRef100_Q8LR18 Putative chloride channel protein [Oryza sativa] 904 0.0
UniRef100_Q84QC8 Chloride channel [Zea mays] 879 0.0
UniRef100_Q8LPA0 Chloride channel [Oryza sativa] 859 0.0
UniRef100_P92942 Chloride channel protein CLC-b [Arabidopsis tha... 822 0.0
UniRef100_P92941 Chloride channel protein CLC-a [Arabidopsis tha... 815 0.0
UniRef100_Q9XF71 CLC-Nt2 protein [Nicotiana tabacum] 811 0.0
UniRef100_Q96325 Voltage-gated chloride channel [Arabidopsis tha... 782 0.0
UniRef100_Q851D8 Putative CLC-d chloride channel protein [Oryza ... 640 0.0
UniRef100_P92943 Chloride channel protein CLC-d [Arabidopsis tha... 640 0.0
UniRef100_UPI00003619FE UPI00003619FE UniRef100 entry 419 e-115
UniRef100_Q5ZL60 Hypothetical protein [Gallus gallus] 405 e-111
>UniRef100_P60300 Putative chloride channel-like protein CLC-G [Arabidopsis thaliana]
Length = 763
Score = 1148 bits (2970), Expect = 0.0
Identities = 562/763 (73%), Positives = 654/763 (85%), Gaps = 2/763 (0%)
Query: 6 LSNGDSEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGS 65
+ N +E + PLL S R NS+SQVAIVGANVCPIESLDYEI EN+FFKQDWR R
Sbjct: 1 MPNSTTEDSVAVPLLPSLRRATNSTSQVAIVGANVCPIESLDYEIAENDFFKQDWRGRSK 60
Query: 66 LQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFF 125
++IFQYV MKWLLCF IG I+ LIGF NNLAVENLAGVKFVVTSNMM+ RF++ FV F
Sbjct: 61 VEIFQYVFMKWLLCFCIGIIVSLIGFANNLAVENLAGVKFVVTSNMMIAGRFAMGFVVFS 120
Query: 126 ASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSS 185
+NL LTLFA++ITA +APAAAGSGI EVKAYLNGVDAP IF+L TLI+KIIG+I+AVS+
Sbjct: 121 VTNLILTLFASVITAFVAPAAAGSGIPEVKAYLNGVDAPEIFSLRTLIIKIIGNISAVSA 180
Query: 186 SLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAA 245
SL IGKAGPM+HTGACVA++LGQGGSKRY LTW+WLR+FKNDRDRRDL+ CG+AAGIAA+
Sbjct: 181 SLLIGKAGPMVHTGACVASILGQGGSKRYRLTWRWLRFFKNDRDRRDLVTCGAAAGIAAS 240
Query: 246 FRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIM 305
FRAPVGGVLFALEEM+SW +ALLWR FF+TA+VAI LRA+IDVCLSGKCGLFGKGGLIM
Sbjct: 241 FRAPVGGVLFALEEMSSW--SALLWRIFFSTAVVAIVLRALIDVCLSGKCGLFGKGGLIM 298
Query: 306 FDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACV 365
FD YS + YH DV V +LGV+GGILGSLYN+L +KVLR YN I EKG KILLAC
Sbjct: 299 FDVYSENASYHLGDVLPVLLLGVVGGILGSLYNFLLDKVLRAYNYIYEKGVTWKILLACA 358
Query: 366 ISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTND 425
IS+FTSCLLFGLP+LASCQPCP D +E CPTIGRSG +KK+QCPPGHYNDLASLIFNTND
Sbjct: 359 ISIFTSCLLFGLPFLASCQPCPVDALEECPTIGRSGNFKKYQCPPGHYNDLASLIFNTND 418
Query: 426 DAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGM 485
DAI+NLFSKNTD EF Y S+ +FF+T FFLSI S G+V PAG+FVP+IVTGASYGRFVGM
Sbjct: 419 DAIKNLFSKNTDFEFHYFSVLVFFVTCFFLSIFSYGIVAPAGLFVPVIVTGASYGRFVGM 478
Query: 486 LFGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVA 545
L G SNLNHGL+AVLGAASFLGG+MR TVS CVI+LELTNNLLLLP++M+VLL+SK+VA
Sbjct: 479 LLGSNSNLNHGLFAVLGAASFLGGTMRMTVSTCVILLELTNNLLLLPMMMVVLLISKTVA 538
Query: 546 DVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTT 605
D FNANIY+LIMK KG PYL +HAEPYMRQL VGDVVTGPLQ+F+G EKV +V VLKTT
Sbjct: 539 DGFNANIYNLIMKLKGFPYLYSHAEPYMRQLLVGDVVTGPLQVFNGIEKVETIVHVLKTT 598
Query: 606 SHNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKR 665
+HNGFPV+D PP + APVL G+ILR H+LTLL+K+VFM +P+A + L QF A++FAK+
Sbjct: 599 NHNGFPVVDGPPLAAAPVLHGLILRAHILTLLKKRVFMPSPVACDSNTLSQFKAEEFAKK 658
Query: 666 GSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIP 725
GS R ++IED+ L++EE++M++DLHPF+NASPYTVVE+MSL KALILFRE+G+RHLLVIP
Sbjct: 659 GSGRSDKIEDVELSEEELNMYLDLHPFSNASPYTVVETMSLAKALILFREVGIRHLLVIP 718
Query: 726 KIPSRSPVVGILTRHDFTAEHILGTHPYLVRSRWKRLRFWQPF 768
K +R PVVGILTRHDF EHILG HP + RS+WKRLR PF
Sbjct: 719 KTSNRPPVVGILTRHDFMPEHILGLHPSVSRSKWKRLRIRLPF 761
>UniRef100_Q7XTM5 OSJNBa0033G05.20 protein [Oryza sativa]
Length = 802
Score = 1027 bits (2655), Expect = 0.0
Identities = 497/765 (64%), Positives = 607/765 (78%), Gaps = 7/765 (0%)
Query: 8 NGDSE-HRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSL 66
+GD E R R+ L + RS N++SQVA+VG VCPIESLDYE+ ENE FKQDWR+RG
Sbjct: 41 DGDEEARRRRRRFLLAGRSQSNTTSQVALVGVGVCPIESLDYELIENEVFKQDWRARGRG 100
Query: 67 QIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFFA 126
I +YV +KW LCFL+G + GF NL VEN+AG KFVVTSN+ML R+ AF F
Sbjct: 101 HILRYVALKWALCFLVGVLSAAAGFVANLGVENVAGAKFVVTSNLMLAGRYGTAFAVFLV 160
Query: 127 SNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSS 186
SN ALT+ AT++T +APAAAGSGI EVKAYLNGVDAP IF+L TL+VKI+G I AVSSS
Sbjct: 161 SNFALTMLATVLTVYVAPAAAGSGIPEVKAYLNGVDAPDIFSLKTLVVKIVGCIAAVSSS 220
Query: 187 LHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAF 246
LH+GKAGP++HTGAC+A++LGQGGS +Y LT KWLRYFKNDRDRRDL+ CG+ AGIAAAF
Sbjct: 221 LHVGKAGPLVHTGACIASILGQGGSSKYHLTCKWLRYFKNDRDRRDLVTCGAGAGIAAAF 280
Query: 247 RAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMF 306
RAPVGGVLFALE ++SWWR+ALLWRAFFTTA+VA+ LRA+ID C S KCGLFGKGGLIMF
Sbjct: 281 RAPVGGVLFALEAVSSWWRSALLWRAFFTTAMVAVVLRALIDFCKSDKCGLFGKGGLIMF 340
Query: 307 DAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACVI 366
D S + YH D+P V LGV+GG+LGSL+N+ +K+ KG K+LLA V+
Sbjct: 341 DVTSDYITYHLVDLPPVITLGVLGGVLGSLHNFFLDKLTF------RKGQKYKLLLAAVV 394
Query: 367 SMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDD 426
++ TSC LFGLPW+ASC+PCP+D E CP+IGRSG +KK+QC YNDLASL FNTNDD
Sbjct: 395 TICTSCCLFGLPWIASCKPCPSDTEEACPSIGRSGNFKKYQCAMNEYNDLASLFFNTNDD 454
Query: 427 AIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGML 486
IRNL+S TD EF SS+ +FF T +FL I S G+ +P+G+FVP+I+TGA+YGR VGML
Sbjct: 455 TIRNLYSAGTDDEFHISSILVFFFTSYFLGIFSYGLALPSGLFVPVILTGATYGRLVGML 514
Query: 487 FGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVAD 546
G +S L+HGL+AVLG+A+ LGGSMR TVS+CV+ILELTNNLL+LPL+M+VLL+SK+VAD
Sbjct: 515 IGSQSTLDHGLFAVLGSAALLGGSMRMTVSVCVVILELTNNLLMLPLVMLVLLISKTVAD 574
Query: 547 VFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTS 606
FNANIYDL++K KG PYLE H EPYMRQL+V DVVTGPLQ F+G EKV ++V VL+TT
Sbjct: 575 AFNANIYDLLVKLKGFPYLEGHVEPYMRQLSVSDVVTGPLQAFNGIEKVGHIVHVLRTTG 634
Query: 607 HNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRG 666
HNGFPV+DEPPFS++PVLFG++LR HLL LLRKK F+ A D +QF DFAK G
Sbjct: 635 HNGFPVVDEPPFSDSPVLFGLVLRAHLLVLLRKKDFIPNCSASALDASKQFLPHDFAKPG 694
Query: 667 SDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPK 726
S + +RIE+I + EE++MF+DLHPFTN SPYTVVE+MSL KA +LFRE+GLRHLLV+PK
Sbjct: 695 SGKHDRIEEIEFSAEELEMFVDLHPFTNTSPYTVVETMSLAKAHVLFREVGLRHLLVLPK 754
Query: 727 IPSRSPVVGILTRHDFTAEHILGTHPYLVRSRWKRLRFWQPFLEN 771
R+PVVGILTRHDF EHILG HP+L ++RWK++RF + N
Sbjct: 755 SSKRAPVVGILTRHDFMPEHILGLHPFLFKTRWKKVRFGKSAFTN 799
>UniRef100_Q6ZDC2 Putative chloride channel protein [Oryza sativa]
Length = 796
Score = 983 bits (2542), Expect = 0.0
Identities = 483/765 (63%), Positives = 598/765 (78%), Gaps = 11/765 (1%)
Query: 10 DSEHR--LRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSLQ 67
+ EHR L + LL RS N++SQVA+VG+N CPIESLDYEI EN+ F Q+WRSRG
Sbjct: 15 EDEHRPPLNRALL--HRSATNNTSQVAMVGSNPCPIESLDYEIIENDLFDQNWRSRGKAD 72
Query: 68 IFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFFAS 127
+YV++KW CF IG I G+ GF NLAVEN+AG+K S +M + AF F +
Sbjct: 73 QVRYVVLKWTFCFAIGIITGIAGFVINLAVENVAGLKHTAVSALMESSSYWTAFWLFAGT 132
Query: 128 NLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSSL 187
NLAL LFA+ ITA ++PAA GSGI EVKAYLNGVDAP IF+L TL VKIIG+I AVSSSL
Sbjct: 133 NLALLLFASSITAFVSPAAGGSGIPEVKAYLNGVDAPNIFSLRTLAVKIIGNIAAVSSSL 192
Query: 188 HIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAFR 247
H+GKAGPM+HTGAC+AA+ GQGGS++YGLT +WLRYFKNDRDRRDL+ G+ AG+ AAFR
Sbjct: 193 HVGKAGPMVHTGACIAAIFGQGGSRKYGLTCRWLRYFKNDRDRRDLVTIGAGAGVTAAFR 252
Query: 248 APVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMFD 307
APVGGVLFALE ++SWWR+AL+WR+FFTTA+VA+ LR I++C SGKCGLFGKGGLIM+D
Sbjct: 253 APVGGVLFALESLSSWWRSALIWRSFFTTAVVAVVLRMFIELCASGKCGLFGKGGLIMYD 312
Query: 308 A---YSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLAC 364
+ + YH KD+P+V ++GVIG ILG+LYN+L KVLR+Y+ INE+G K+LLA
Sbjct: 313 VSTKFDDLMTYHLKDIPIVVLIGVIGAILGALYNFLMMKVLRVYSVINERGNAHKLLLAA 372
Query: 365 VISMFTSCLLFGLPWLASCQPCP--ADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFN 422
V+S+ TSC +FGLPWLA C+PCP P P T +++F CP GHYNDLASL N
Sbjct: 373 VVSILTSCCVFGLPWLAPCRPCPTAGAPSPPNGTCHSLNRFRRFHCPAGHYNDLASLFLN 432
Query: 423 TNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRF 482
NDDAIRNL+S T+ + SM FF+ + L +LS GVV P+G+FVPII+TGA+YGR
Sbjct: 433 INDDAIRNLYSTGTNDVYHPGSMLAFFVASYALGVLSYGVVAPSGLFVPIILTGATYGRL 492
Query: 483 VGMLFGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSK 542
V ML G +S L+HGL A+LG+ASFLGG++R TVS+CVIILELTNNLLLLPL+M+VLL+SK
Sbjct: 493 VAMLLGGRSGLDHGLVAILGSASFLGGTLRMTVSVCVIILELTNNLLLLPLVMLVLLISK 552
Query: 543 SVADVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVL 602
+VAD FN++IYDLI+ KGLP+L+ HAEPYMRQLTVGDVV GPL+ F+G EKV ++V L
Sbjct: 553 TVADSFNSSIYDLILNLKGLPHLDGHAEPYMRQLTVGDVVAGPLRSFNGVEKVGHIVHTL 612
Query: 603 KTTSHNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGDVLR-QFSADD 661
+TT H+ FPV+DEPPFS APVL+G++LR HLL LL+K+ F++AP+ D + +F A D
Sbjct: 613 RTTGHHAFPVVDEPPFSPAPVLYGLVLRAHLLVLLKKREFLTAPVRCPKDYMAGRFEAQD 672
Query: 662 FAKRGSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHL 721
F KRGS + + I D+ L+ EEM+M++DLHPFTN SPYTVVE+MSL KAL+LFRE+GLRHL
Sbjct: 673 FDKRGSGKQDTIADVELSPEEMEMYVDLHPFTNTSPYTVVETMSLAKALVLFREVGLRHL 732
Query: 722 LVIPKIPSRSPVVGILTRHDFTAEHILGTHPYLVRSRWKRLRFWQ 766
LV+PK RSPVVGILTRHDF EHILG HP LV SRWKRLR WQ
Sbjct: 733 LVVPKSCDRSPVVGILTRHDFMPEHILGLHPVLVGSRWKRLR-WQ 776
>UniRef100_Q6YVX5 Chloride channel [Oryza sativa]
Length = 804
Score = 966 bits (2498), Expect = 0.0
Identities = 476/749 (63%), Positives = 573/749 (75%), Gaps = 3/749 (0%)
Query: 7 SNGDSEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSL 66
S G +PLL ++ +N++SQ+AIVGANVCPIESLDYE+ EN+ FKQDWRSR
Sbjct: 46 SGGGRSGSAGEPLL--RKRTMNTTSQIAIVGANVCPIESLDYEVVENDLFKQDWRSRKKK 103
Query: 67 QIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFFA 126
QIFQY+++KW L LIG + GL+GF NNLAVEN+AG K ++T N+ML +R+ AF +
Sbjct: 104 QIFQYIVLKWTLVLLIGLLTGLVGFFNNLAVENIAGFKLLLTGNLMLKERYLTAFFAYGG 163
Query: 127 SNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSS 186
NL L A I A IAPAAAGSGI EVKAYLNGVDA I TL VKI GSI VS+
Sbjct: 164 CNLVLAAAAAAICAYIAPAAAGSGIPEVKAYLNGVDAYSILAPSTLFVKIFGSILGVSAG 223
Query: 187 LHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAF 246
+GK GPM+HTGAC+A LLGQGGS++Y LT WLRYFKNDRDRRDLI CGSAAG+AAAF
Sbjct: 224 FVLGKEGPMVHTGACIANLLGQGGSRKYRLTCNWLRYFKNDRDRRDLITCGSAAGVAAAF 283
Query: 247 RAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMF 306
RAPVGGVLFALEE ASWWR+ALLWRAFFTTA+VA+ LR++I+ C SGKCGLFG+GGLIMF
Sbjct: 284 RAPVGGVLFALEEAASWWRSALLWRAFFTTAVVAVVLRSLIEFCRSGKCGLFGQGGLIMF 343
Query: 307 DAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACVI 366
D S Y D+ + ILG+IGGI G L+N+L +KVLR+Y+ INE+G KILL I
Sbjct: 344 DLSSTVATYSTPDLIAIIILGIIGGIFGGLFNFLLDKVLRVYSIINERGAPFKILLTITI 403
Query: 367 SMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDD 426
S+ TS +GLPWLA+C PCP D VE CPTIGRSG +K FQCPPGHYNDLASL FNTNDD
Sbjct: 404 SIITSMCSYGLPWLAACTPCPVDAVEQCPTIGRSGNFKNFQCPPGHYNDLASLFFNTNDD 463
Query: 427 AIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGML 486
AIRNLFS T+SEF S++FIFF + L IL+ GV VP+G+F+P+I+ GA+YGR VG L
Sbjct: 464 AIRNLFSNGTESEFHMSTLFIFFTAVYCLGILTYGVAVPSGLFIPVILAGATYGRIVGTL 523
Query: 487 FGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVAD 546
G S+L+ GL+A+LGAASFLGG+MR TVS+CVI+LELTN+L +LPL+M+VLL+SK++AD
Sbjct: 524 LGSISDLDPGLFALLGAASFLGGTMRMTVSVCVILLELTNDLAMLPLVMLVLLISKTIAD 583
Query: 547 VFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTS 606
FN +YD I+ KGLPY+E HAEPYMR L GDVV+GPL F G EKV N+V L+ T
Sbjct: 584 NFNKGVYDQIVVMKGLPYMEAHAEPYMRHLVAGDVVSGPLITFSGVEKVGNIVHALRFTG 643
Query: 607 HNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGD-VLRQFSADDFAKR 665
HNGFPV+DEPP +EAP L G++ R HLL LL K+FM + G VL++F A DFAK
Sbjct: 644 HNGFPVVDEPPLTEAPELVGLVTRSHLLVLLNGKMFMKDQLKTSGSFVLQRFGAFDFAKP 703
Query: 666 GSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIP 725
GS +G +I+D+ T EEM+M++DLHP TN SPYTVVE+MSL KA ILFR LGLRHLLV+P
Sbjct: 704 GSGKGLKIQDLDFTDEEMEMYVDLHPVTNTSPYTVVETMSLAKAAILFRALGLRHLLVVP 763
Query: 726 KIPSRSPVVGILTRHDFTAEHILGTHPYL 754
K P R P+VGILTRHDF EHI G P L
Sbjct: 764 KTPDRPPIVGILTRHDFVEEHIHGLFPNL 792
>UniRef100_Q40485 C1C-Nt1 protein [Nicotiana tabacum]
Length = 780
Score = 964 bits (2491), Expect = 0.0
Identities = 468/749 (62%), Positives = 576/749 (76%), Gaps = 2/749 (0%)
Query: 11 SEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSLQIFQ 70
SE +RQPLLSS +S +N++SQ+AI+GANVCPIESLDYEI EN+ FKQDWRSR +QIFQ
Sbjct: 33 SESGVRQPLLSS-KSRVNNTSQIAIIGANVCPIESLDYEIIENDLFKQDWRSRKKVQIFQ 91
Query: 71 YVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFFASNLA 130
Y+ +KW L LIG +GL+GF N+AVEN+AG K ++ S++ML ++ F + NL
Sbjct: 92 YIFLKWTLVLLIGLSVGLVGFFLNIAVENIAGFKLLLISDLMLQDKYFRGFAAYACCNLV 151
Query: 131 LTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSSLHIG 190
L A I+ A IAPAAAGSGI EVKAYLNG+DA I TL VKI GS VS+ +G
Sbjct: 152 LATCAGILCAFIAPAAAGSGIPEVKAYLNGIDAHSILAPSTLFVKIFGSALGVSAGFVVG 211
Query: 191 KAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAFRAPV 250
K GPM+HTGAC+A LLGQGGS++Y LTWKWL+YFKNDRDRRDLI CG+AAG+AAAFRAPV
Sbjct: 212 KEGPMVHTGACIANLLGQGGSRKYHLTWKWLKYFKNDRDRRDLITCGAAAGVAAAFRAPV 271
Query: 251 GGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMFDAYS 310
GGVLFALEE+ASWWR+ALLWR FF+TA+VA+ LR+ I C SGKCGLFG+GGLIM+D S
Sbjct: 272 GGVLFALEEVASWWRSALLWRTFFSTAVVAMVLRSFIVFCRSGKCGLFGQGGLIMYDVNS 331
Query: 311 GSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACVISMFT 370
G+ Y+ DV V ++GV+GG+LGSLYNYL +KVLR Y+ INE+G K+LL IS+ +
Sbjct: 332 GAPNYNTIDVLAVLLIGVLGGLLGSLYNYLVDKVLRTYSIINERGPAFKVLLVMTISILS 391
Query: 371 SCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDAIRN 430
S +GLPW A+C PCP + CPTIGRSG YK FQCP GHYNDLASL NTNDDAIRN
Sbjct: 392 SLCSYGLPWFATCTPCPVGLEDKCPTIGRSGNYKNFQCPAGHYNDLASLFMNTNDDAIRN 451
Query: 431 LFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLFGKK 490
LFS + SEF SS+F+FF + L +++ G+ +P+G+F+P+I+ GASYGRFVG + G
Sbjct: 452 LFSSDNSSEFHLSSLFVFFAGVYCLGVVTYGIAIPSGLFIPVILAGASYGRFVGTVLGSI 511
Query: 491 SNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNA 550
SNLN+GL+A+LGAASFLGG+MR TVS+CVI+LELT++LL+LPL+M+VLL+SK+VAD FN
Sbjct: 512 SNLNNGLFALLGAASFLGGTMRMTVSICVILLELTDDLLMLPLVMLVLLISKTVADCFNH 571
Query: 551 NIYDLIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTSHNGF 610
+YD I+K KGLPYLE HAEPYMRQL GDV +GPL F G EKV N++ LK T HNGF
Sbjct: 572 GVYDQIVKMKGLPYLEAHAEPYMRQLVAGDVCSGPLITFSGVEKVGNIIHALKFTRHNGF 631
Query: 611 PVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRG 670
PVID PPFS+AP G+ LR HLL LL+ K F + G +LR F A DFAK GS +G
Sbjct: 632 PVIDAPPFSDAPEFCGLALRSHLLVLLKAKKFTKLSVLSGSSILRSFHAFDFAKPGSGKG 691
Query: 671 ERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIP-KIPS 729
++ED+ T EEM+M++DLHP TN SPYTVVE+MSL KA ILFR+LGLRHL V+P K
Sbjct: 692 PKLEDLSFTDEEMEMYVDLHPVTNTSPYTVVETMSLAKAAILFRQLGLRHLCVVPKKTTG 751
Query: 730 RSPVVGILTRHDFTAEHILGTHPYLVRSR 758
R P+VGILTRHDF EHI G +P+LV +
Sbjct: 752 RDPIVGILTRHDFMPEHIKGLYPHLVHHK 780
>UniRef100_Q8LPA2 Chloride channel [Oryza sativa]
Length = 801
Score = 962 bits (2487), Expect = 0.0
Identities = 470/745 (63%), Positives = 577/745 (77%), Gaps = 5/745 (0%)
Query: 16 RQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSLQIFQYVLMK 75
RQPLL ++ +N++SQ+AIVGANVCPIESLDYEI EN+ FKQDWRSR QIFQY+++K
Sbjct: 58 RQPLL--RKRTMNTTSQIAIVGANVCPIESLDYEIVENDLFKQDWRSRKKKQIFQYIVLK 115
Query: 76 WLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHK--RFSLAFVTFFASNLALTL 133
W L LIG + G++GF NNLAVEN+AG+K ++TS++ML + R+ AF+ + NL L
Sbjct: 116 WALVLLIGMLTGIVGFFNNLAVENIAGLKLLLTSDLMLKQKCRYFTAFLAYGGCNLVLAT 175
Query: 134 FATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSSLHIGKAG 193
A I A IAPAAAGSGI EVKAYLNGVDA I TL VKI GSI VS+ +GK G
Sbjct: 176 TAAAICAYIAPAAAGSGIPEVKAYLNGVDAYSILAPSTLFVKIFGSILGVSAGFVLGKEG 235
Query: 194 PMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAFRAPVGGV 253
PM+HTGAC+A LLGQGGS++Y LTW WLRYFKNDRDRRDLI CGSAAG+AAAFRAPVGGV
Sbjct: 236 PMVHTGACIANLLGQGGSRKYHLTWNWLRYFKNDRDRRDLITCGSAAGVAAAFRAPVGGV 295
Query: 254 LFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMFDAYSGSL 313
LFALEE ASWWR+ALLWR FFTTA+VA+ LR +I+ C SGKCGLFG+GGLIMFD S
Sbjct: 296 LFALEEAASWWRSALLWRTFFTTAVVAVVLRGLIEFCRSGKCGLFGQGGLIMFDLSSTIP 355
Query: 314 MYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACVISMFTSCL 373
Y A+DV + +LG+IGG+ G L+N+L +++LR Y+ INE+G KILL +IS+ TS
Sbjct: 356 TYTAQDVVAIIVLGIIGGVFGGLFNFLLDRILRAYSIINERGPPFKILLTMIISIITSAC 415
Query: 374 LFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDAIRNLFS 433
+GLPWLA C PCPAD E CPTIGRSG +K FQCPPGHYN LASL FNTNDDAIRNLFS
Sbjct: 416 SYGLPWLAPCTPCPADAAEECPTIGRSGNFKNFQCPPGHYNGLASLFFNTNDDAIRNLFS 475
Query: 434 KNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLFGKKSNL 493
T+ EF S++F+FF + L +++ G+ VP+G+F+P+I+ GA+YGR VG L G S+L
Sbjct: 476 SGTEKEFHMSTLFVFFTAIYCLGLVTYGIAVPSGLFIPVILAGATYGRIVGTLLGPISDL 535
Query: 494 NHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNANIY 553
+ GL+A+LGAASFLGG+MR TVS+CVI+LELTN+L +LPL+M+VLL+SK++AD FN +Y
Sbjct: 536 DPGLFALLGAASFLGGTMRMTVSVCVILLELTNDLHMLPLVMLVLLISKTIADSFNKGVY 595
Query: 554 DLIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTSHNGFPVI 613
D I+ KGLP++E HAEP+MR L GDVV+GPL F G EKV N+V L+ T HNGFPV+
Sbjct: 596 DQIVVMKGLPFMEAHAEPFMRNLVAGDVVSGPLITFSGVEKVGNIVHALRITGHNGFPVV 655
Query: 614 DEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGD-VLRQFSADDFAKRGSDRGER 672
DEPP SEAP L G++LR HLL LL+ + FM + G VLR+F A DFAK GS +G +
Sbjct: 656 DEPPVSEAPELVGLVLRSHLLVLLKGRSFMKEKVKTSGSFVLRRFGAFDFAKPGSGKGLK 715
Query: 673 IEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSP 732
IED+ LT EE+DM++DLHP TN SPYTVVE+MSL KA +LFR LGLRHLLV+PK P R P
Sbjct: 716 IEDLDLTDEELDMYVDLHPITNTSPYTVVETMSLAKAAVLFRALGLRHLLVVPKTPGRPP 775
Query: 733 VVGILTRHDFTAEHILGTHPYLVRS 757
+VGILTRHDF EHI G P L +S
Sbjct: 776 IVGILTRHDFMHEHIHGLFPNLGKS 800
>UniRef100_Q8LPA1 Chloride channel [Oryza sativa]
Length = 756
Score = 959 bits (2480), Expect = 0.0
Identities = 474/741 (63%), Positives = 570/741 (75%), Gaps = 5/741 (0%)
Query: 17 QPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSLQIFQYVLMKW 76
+PLL ++ +N++SQ+AIVGANVCPIESLDYE+ EN+ FKQDWRSR QIFQY+++KW
Sbjct: 6 EPLL--RKRTMNTTSQIAIVGANVCPIESLDYEVVENDLFKQDWRSRKKKQIFQYIVLKW 63
Query: 77 LLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHK--RFSLAFVTFFASNLALTLF 134
L LIG + GL+GF NNLAVEN+AG K ++T N+ML R+ AF + NL L
Sbjct: 64 TLVLLIGLLTGLVGFFNNLAVENIAGFKLLLTGNLMLKGKCRYLTAFFAYGGCNLVLAAA 123
Query: 135 ATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSSLHIGKAGP 194
A I A IAPAAAGSGI EVKAYLNGVDA I TL VKI GSI VS+ +GK GP
Sbjct: 124 AAAICAYIAPAAAGSGIPEVKAYLNGVDAYSILAPSTLFVKIFGSILGVSAGFVLGKEGP 183
Query: 195 MLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAFRAPVGGVL 254
M+HTGAC+A LLGQGGS++Y LT WLRYFKNDRDRRDLI CGSAAG+AAAFRAPVGGVL
Sbjct: 184 MVHTGACIANLLGQGGSRKYRLTCNWLRYFKNDRDRRDLITCGSAAGVAAAFRAPVGGVL 243
Query: 255 FALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMFDAYSGSLM 314
FALEE ASWWR+ALLWRAFFTTA+VA+ LR++I+ C SGKCGLFG+GGLIMFD S
Sbjct: 244 FALEEAASWWRSALLWRAFFTTAVVAVVLRSLIEFCRSGKCGLFGQGGLIMFDLSSTVAT 303
Query: 315 YHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACVISMFTSCLL 374
Y D+ + ILG+IGGI G L+N+L +KVLR+Y+ INE+G KILL IS+ TS
Sbjct: 304 YSTPDLIAIIILGIIGGIFGGLFNFLLDKVLRVYSIINERGAPFKILLTITISIITSMCS 363
Query: 375 FGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDAIRNLFSK 434
+GLPWLA+C PCP D VE CPTIGRSG +K FQCPPGHYNDLASL FNTNDDAIRNLFS
Sbjct: 364 YGLPWLAACTPCPVDAVEQCPTIGRSGNFKNFQCPPGHYNDLASLFFNTNDDAIRNLFSN 423
Query: 435 NTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLFGKKSNLN 494
T+SEF S++FIFF + L IL+ GV VP+G+F+P+I+ GA+YGR VG L G S+L+
Sbjct: 424 GTESEFHMSTLFIFFTAVYCLGILTYGVAVPSGLFIPVILAGATYGRIVGTLLGSISDLD 483
Query: 495 HGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNANIYD 554
GL+A+LGAASFLGG+MR TVS+CVI+LELTN+L +LPL+M+VLL+SK++AD FN +YD
Sbjct: 484 PGLFALLGAASFLGGTMRMTVSVCVILLELTNDLAMLPLVMLVLLISKTIADNFNKGVYD 543
Query: 555 LIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTSHNGFPVID 614
I+ KGLPY+E HAEPYMR L GDVV+GPL F G EKV N+V L+ T HNGFPV+D
Sbjct: 544 QIVVMKGLPYMEAHAEPYMRHLVAGDVVSGPLITFSGVEKVGNIVHALRFTGHNGFPVVD 603
Query: 615 EPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGD-VLRQFSADDFAKRGSDRGERI 673
EPP +EAP L G++ R HLL LL K+FM + G VL++F A DFAK GS +G +I
Sbjct: 604 EPPLTEAPELVGLVTRSHLLVLLNGKMFMKDQLKTSGSFVLQRFGAFDFAKPGSGKGLKI 663
Query: 674 EDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPV 733
+D+ T EEM+M++DLHP TN SPYTVVE+MSL KA ILFR LGLRHLLV+PK P R P+
Sbjct: 664 QDLDFTDEEMEMYVDLHPVTNTSPYTVVETMSLAKAAILFRALGLRHLLVVPKTPDRPPI 723
Query: 734 VGILTRHDFTAEHILGTHPYL 754
VGILTRHDF EHI G P L
Sbjct: 724 VGILTRHDFVEEHIHGLFPNL 744
>UniRef100_P93567 Chloride channel Stclc1 [Solanum tuberosum]
Length = 764
Score = 952 bits (2461), Expect = 0.0
Identities = 464/741 (62%), Positives = 564/741 (75%), Gaps = 1/741 (0%)
Query: 15 LRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSLQIFQYVLM 74
+R PLL S +S +N++SQ+AIVGANV PIESLDY+I EN+ FKQDWRSR ++IFQY+ +
Sbjct: 22 IRVPLLKS-KSRVNNTSQIAIVGANVYPIESLDYDIVENDLFKQDWRSRKKVEIFQYIFL 80
Query: 75 KWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFFASNLALTLF 134
KW L LIG GL+GF NN+ VEN+AG K ++TSN+ML ++ AF F N+
Sbjct: 81 KWTLVLLIGLSTGLVGFFNNIGVENIAGFKLLLTSNLMLDGKYFQAFAAFAGCNVFFATC 140
Query: 135 ATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSSLHIGKAGP 194
A + A IAPAAAGSGI EVKAYLNG+DA I TL+VKI GSI VS+ +GK GP
Sbjct: 141 AAALCAFIAPAAAGSGIPEVKAYLNGIDAHSILAPSTLLVKIFGSILGVSAGFVVGKEGP 200
Query: 195 MLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAFRAPVGGVL 254
M+HTGAC+A LLGQGGS++Y LTWKWL+YFKNDRDRRDLI CG+AAG+AAAFRAPVGGVL
Sbjct: 201 MVHTGACIANLLGQGGSRKYHLTWKWLKYFKNDRDRRDLITCGAAAGVAAAFRAPVGGVL 260
Query: 255 FALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMFDAYSGSLM 314
FALEE+ASWWR+ALLWR FFTTAIVA+ LR++I C G CGLFG+GGLIMFD SG
Sbjct: 261 FALEEIASWWRSALLWRTFFTTAIVAMVLRSLIQFCRGGNCGLFGQGGLIMFDVNSGVSN 320
Query: 315 YHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACVISMFTSCLL 374
Y+ DV + +GV+GG+LGSLYNYL +KVLR Y INE+G KILL +S+ TSC
Sbjct: 321 YNTIDVLALIFIGVLGGLLGSLYNYLVDKVLRTYAVINERGPAFKILLVMSVSILTSCCS 380
Query: 375 FGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDAIRNLFSK 434
+GLPW A C PCP E CPTIGRSG YK FQCP GHYNDLASL NTNDDAIRNLFS
Sbjct: 381 YGLPWFAGCIPCPVGLEEKCPTIGRSGNYKNFQCPAGHYNDLASLFLNTNDDAIRNLFSS 440
Query: 435 NTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLFGKKSNLN 494
N +EF S++ IFF + L I++ G+ +P+G+F+P+I+ GASYGR G G SNLN
Sbjct: 441 NNSNEFHISTLLIFFAGVYCLGIITYGIAIPSGLFIPVILAGASYGRIFGRALGSLSNLN 500
Query: 495 HGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNANIYD 554
GL+++LGAASFLGG+MR TVS+CVI+LELTNNLL+LPL+M+VLL+SK+VAD+FN +YD
Sbjct: 501 VGLFSLLGAASFLGGTMRMTVSICVILLELTNNLLMLPLVMLVLLISKTVADIFNKGVYD 560
Query: 555 LIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTSHNGFPVID 614
I+K KGLP+LE HAEP+MR L GDV +GPL F G EKV N+V LK T HNGFPVID
Sbjct: 561 QIVKMKGLPFLEAHAEPFMRNLVAGDVCSGPLLSFSGVEKVGNIVHALKYTRHNGFPVID 620
Query: 615 EPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIE 674
EPPFSE P L G++LR HLL LL K F + ++L +F A DFAK GS +G + E
Sbjct: 621 EPPFSETPELCGLVLRSHLLVLLNGKKFTKQRVLSASNILSRFHAFDFAKPGSGKGLKFE 680
Query: 675 DIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVV 734
D+ +T+EEM+M+IDLHP TN SPYTVVE+MSL KA ILFR+LGLRHL V+PK R+P+V
Sbjct: 681 DLVITEEEMEMYIDLHPITNTSPYTVVETMSLAKAAILFRQLGLRHLCVVPKKTGRAPIV 740
Query: 735 GILTRHDFTAEHILGTHPYLV 755
GILTRHDF EHI +P+LV
Sbjct: 741 GILTRHDFMHEHISNLYPHLV 761
>UniRef100_Q96282 Chloride channel protein CLC-c [Arabidopsis thaliana]
Length = 779
Score = 946 bits (2446), Expect = 0.0
Identities = 460/746 (61%), Positives = 563/746 (74%), Gaps = 3/746 (0%)
Query: 10 DSEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSLQIF 69
D RQPLL+ R N++SQ+AIVGAN CPIESLDYEIFEN+FFKQDWRSR ++I
Sbjct: 32 DGSVGFRQPLLARNRK--NTTSQIAIVGANTCPIESLDYEIFENDFFKQDWRSRKKIEIL 89
Query: 70 QYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFFASNL 129
QY +KW L FLIG GL+GF NNL VEN+AG K ++ N+ML +++ AF F NL
Sbjct: 90 QYTFLKWALAFLIGLATGLVGFLNNLGVENIAGFKLLLIGNLMLKEKYFQAFFAFAGCNL 149
Query: 130 ALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSSLHI 189
L A + A IAPAAAGSGI EVKAYLNG+DA I TL VKI GSI V++ +
Sbjct: 150 ILATAAASLCAFIAPAAAGSGIPEVKAYLNGIDAYSILAPSTLFVKIFGSIFGVAAGFVV 209
Query: 190 GKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAFRAP 249
GK GPM+HTGAC+A LLGQGGSK+Y LTWKWLR+FKNDRDRRDLI CG+AAG+AAAFRAP
Sbjct: 210 GKEGPMVHTGACIANLLGQGGSKKYRLTWKWLRFFKNDRDRRDLITCGAAAGVAAAFRAP 269
Query: 250 VGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMFDAY 309
VGGVLFALEE ASWWR ALLWR FFTTA+VA+ LR++I+ C SG+CGLFGKGGLIMFD
Sbjct: 270 VGGVLFALEEAASWWRNALLWRTFFTTAVVAVVLRSLIEFCRSGRCGLFGKGGLIMFDVN 329
Query: 310 SGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACVISMF 369
SG ++Y D+ + LGVIGG+LGSLYNYL +KVLR Y+ INEKG KI+L +S+
Sbjct: 330 SGPVLYSTPDLLAIVFLGVIGGVLGSLYNYLVDKVLRTYSIINEKGPRFKIMLVMAVSIL 389
Query: 370 TSCLLFGLPWLASCQPCPADPVE-PCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDAI 428
+SC FGLPWL+ C PCP E CP++GRS IYK FQCPP HYNDL+SL+ NTNDDAI
Sbjct: 390 SSCCAFGLPWLSQCTPCPIGIEEGKCPSVGRSSIYKSFQCPPNHYNDLSSLLLNTNDDAI 449
Query: 429 RNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLFG 488
RNLF+ +++EF S++ IFF+ + L I++ G+ +P+G+F+P+I+ GASYGR VG L G
Sbjct: 450 RNLFTSRSENEFHISTLAIFFVAVYCLGIITYGIAIPSGLFIPVILAGASYGRLVGRLLG 509
Query: 489 KKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVF 548
S L+ GL+++LGAASFLGG+MR TVSLCVI+LELTNNLL+LPL+M+VLL+SK+VAD F
Sbjct: 510 PVSQLDVGLFSLLGAASFLGGTMRMTVSLCVILLELTNNLLMLPLVMLVLLISKTVADCF 569
Query: 549 NANIYDLIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTSHN 608
N +YD I+ KGLPY+E HAEPYMR L DVV+G L F EKV + LK T HN
Sbjct: 570 NRGVYDQIVTMKGLPYMEDHAEPYMRNLVAKDVVSGALISFSRVEKVGVIWQALKMTRHN 629
Query: 609 GFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSD 668
GFPVIDEPPF+EA L GI LR HLL LL+ K F G +LR A DF K G
Sbjct: 630 GFPVIDEPPFTEASELCGIALRSHLLVLLQGKKFSKQRTTFGSQILRSCKARDFGKAGLG 689
Query: 669 RGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIP 728
+G +IED+ L++EEM+M++DLHP TN SPYTV+E++SL KA ILFR+LGLRHL V+PK P
Sbjct: 690 KGLKIEDLDLSEEEMEMYVDLHPITNTSPYTVLETLSLAKAAILFRQLGLRHLCVVPKTP 749
Query: 729 SRSPVVGILTRHDFTAEHILGTHPYL 754
R P+VGILTRHDF EH+LG +P++
Sbjct: 750 GRPPIVGILTRHDFMPEHVLGLYPHI 775
>UniRef100_Q8LR18 Putative chloride channel protein [Oryza sativa]
Length = 773
Score = 904 bits (2336), Expect = 0.0
Identities = 443/745 (59%), Positives = 556/745 (74%), Gaps = 5/745 (0%)
Query: 15 LRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSLQIFQYVLM 74
L +PLL +R N++SQ+AIVGANVCPIESLDYE+ ENE +KQDWRSRG LQIF Y ++
Sbjct: 30 LERPLL--RRRGTNTTSQMAIVGANVCPIESLDYELVENEVYKQDWRSRGKLQIFHYQIL 87
Query: 75 KWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFFASNLALTLF 134
KW+L L+G I+GLIGF NN+AVEN+AG K ++T+N+ML R+ AF+ F + N L
Sbjct: 88 KWVLALLVGLIVGLIGFFNNIAVENIAGFKLLLTTNLMLQNRYKAAFLWFISCNAMLAAA 147
Query: 135 ATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSSLHIGKAGP 194
A + A PAAAGSGI EVKAYLNGVDAP I TL VKI+GSI VS+ +GK GP
Sbjct: 148 AAALCAYFGPAAAGSGIPEVKAYLNGVDAPSILAPSTLFVKIVGSIFGVSAGFVLGKEGP 207
Query: 195 MLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAFRAPVGGVL 254
M+HTGACVA+ LGQGGS++YG TW WLRYFKND DRRDLI CG+AAG+ AAFRAPVGGVL
Sbjct: 208 MVHTGACVASFLGQGGSRKYGFTWNWLRYFKNDLDRRDLITCGAAAGVTAAFRAPVGGVL 267
Query: 255 FALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMFDAYSGSLM 314
FALEE SWWR+ALLWR F TTA+ A+ LR++I+ C SG CGLFGKGGLIMFD S
Sbjct: 268 FALEEATSWWRSALLWRTFSTTAVAAMVLRSLIEYCRSGNCGLFGKGGLIMFDVSSQVTS 327
Query: 315 YHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACVISMFTSCLL 374
Y D+ V +L ++GG+LG+L+N+L N++LR+Y+ INEKG KI+L VIS+ TSC
Sbjct: 328 YTTMDLAAVVLLAIVGGLLGALFNFLLNRILRVYSYINEKGAPYKIILTVVISLVTSCCS 387
Query: 375 FGLPWLASCQPCPADPVEP--CPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDAIRNLF 432
FGLPWL +C PCP + CPTIGRSG +K F+CPPG YN +ASL NTNDDAIRNLF
Sbjct: 388 FGLPWLTACTPCPPELAASGHCPTIGRSGNFKNFRCPPGQYNAMASLFLNTNDDAIRNLF 447
Query: 433 SKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLFGKKSN 492
S T+SEF + FF + L +++ GV VP+G+F+P+I++GAS+GR +G L G +
Sbjct: 448 SGGTESEFGVPMLLAFFTAVYSLGLVTYGVAVPSGLFIPVILSGASFGRLLGKLLGVLTG 507
Query: 493 LNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNANI 552
L+ GL+A+LGAASFLGG+MR TVS+CVI+LELTN+LLLLPLIM+VLLVSK+VAD FN +
Sbjct: 508 LDTGLFALLGAASFLGGTMRMTVSVCVILLELTNDLLLLPLIMLVLLVSKTVADCFNKGV 567
Query: 553 YDLIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTSHNGFPV 612
Y+ +++ KGLPYLE HAEP MR L GDVV+ PL F E V VV L+ T HNGFPV
Sbjct: 568 YEQMVRMKGLPYLEAHAEPCMRSLVAGDVVSAPLIAFSSVESVGTVVDTLRRTGHNGFPV 627
Query: 613 IDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMG-GDVLRQFSADDFAKRGSDRGE 671
I++ PF+ P L G++LR HLL LLR K F + + G +V R+ + DFAK GS +G
Sbjct: 628 IEDAPFAPEPELCGLVLRSHLLVLLRAKTFTADRVKTGAAEVFRKLAPFDFAKPGSGKGL 687
Query: 672 RIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRS 731
++D+ LT+EEM M++DLHP N SPYTVVE+MSL KA +LFR+LGLRH+ V+P+ P R
Sbjct: 688 TVDDLDLTEEEMAMYVDLHPIANRSPYTVVENMSLAKAAVLFRQLGLRHMCVVPRTPGRP 747
Query: 732 PVVGILTRHDFTAEHILGTHPYLVR 756
PVVGILTRHDF +I G P ++R
Sbjct: 748 PVVGILTRHDFMPGYIRGLFPNVLR 772
>UniRef100_Q84QC8 Chloride channel [Zea mays]
Length = 719
Score = 879 bits (2271), Expect = 0.0
Identities = 439/712 (61%), Positives = 529/712 (73%), Gaps = 12/712 (1%)
Query: 50 IFENEFFKQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTS 109
+ EN+ FKQDWRSR QIFQY+++KW L LIG + G +GF NNLAVEN+AG K ++TS
Sbjct: 1 VVENDLFKQDWRSRKKKQIFQYIVLKWSLVLLIGLLTGFVGFFNNLAVENIAGFKLLLTS 60
Query: 110 NMMLHKRFSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTL 169
++ML R+ AF + NL L A I A IAPAAAGSGI EVKAYLNGVDA I
Sbjct: 61 DLMLKGRYIRAFFVYGGCNLVLAAAAAAICAYIAPAAAGSGIPEVKAYLNGVDAYSILAP 120
Query: 170 PTLIVKIIGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRD 229
TL VKI GSI VS+ +GK GPM+HTGA +A LLGQGGS++Y LT WLRYFKNDRD
Sbjct: 121 STLFVKIFGSILGVSAGFVLGKEGPMVHTGAFIANLLGQGGSRKYHLTCNWLRYFKNDRD 180
Query: 230 RRDLIVCGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDV 289
RRDLI CGSAAG+AAAFRAPVGGVLFALEE ASWWR+ALLWR FFTTA+VA+ LR +I+
Sbjct: 181 RRDLITCGSAAGVAAAFRAPVGGVLFALEEAASWWRSALLWRTFFTTAVVAVVLRGLIEF 240
Query: 290 CLSGKCGLFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYN 349
C SGKCGLFG+GGLIMFD S +Y D+ + +LG+IGGI G L+NYL +K+LR+Y+
Sbjct: 241 CRSGKCGLFGQGGLIMFDLSSTVAVYSTPDLIAIIVLGIIGGIFGGLFNYLLDKILRVYS 300
Query: 350 AINEKGTICKILLACVISMFTSCLLFGLPWLASCQPCPADPVE------PCPTIGRSGIY 403
INE+G KILL IS+ TS +GLPWLA+C PCP D V P P I R+
Sbjct: 301 IINERGAPFKILLTIPISIITSMGSYGLPWLAACTPCPVDAVGAVSHCWPLPEIIRTS-- 358
Query: 404 KKFQCPPGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVV 463
+CPPGHYN LASL FNTNDDAIRNLFS T +EFQ SS+FIFF + L +++ G+
Sbjct: 359 ---KCPPGHYNGLASLFFNTNDDAIRNLFSNGTSTEFQMSSLFIFFTAIYCLGLVTYGIA 415
Query: 464 VPAGVFVPIIVTGASYGRFVGMLFGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILE 523
VP+G+F+P+I+ GA+YGR VG L G S+L+ GL+A+LGAASFLGG+MR TVS+CVI+LE
Sbjct: 416 VPSGLFIPVILAGATYGRIVGTLLGSISDLDPGLFALLGAASFLGGTMRMTVSVCVILLE 475
Query: 524 LTNNLLLLPLIMMVLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDVVT 583
LTN+L +LPL+M+VLL+SK++AD FN +YD I+ KGLPY+E HAEPYMR L GDVV+
Sbjct: 476 LTNDLPMLPLVMLVLLISKTIADSFNKGVYDQIVVMKGLPYMEAHAEPYMRHLVAGDVVS 535
Query: 584 GPLQIFHGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFM 643
GPL F G EKV N+V L+ T HNGFPV+DEPP +E P L G++ R HLL LL K FM
Sbjct: 536 GPLITFSGVEKVGNIVHALRLTGHNGFPVLDEPPITETPELVGLVTRSHLLVLLNSKNFM 595
Query: 644 SAPMAMGGD-VLRQFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVE 702
+ G VLR+F A DFAK GS +G +IED+ T EEMDM++DLHP TN SPYTVVE
Sbjct: 596 KGRVKTSGSFVLRRFGAFDFAKPGSGKGLKIEDLDFTDEEMDMYVDLHPITNTSPYTVVE 655
Query: 703 SMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHDFTAEHILGTHPYL 754
+MSL KA ILFRELGLRHLLV+PK P R P+VGILTRHDF EHI P L
Sbjct: 656 TMSLAKAAILFRELGLRHLLVVPKTPDRPPIVGILTRHDFMPEHIHSLFPNL 707
>UniRef100_Q8LPA0 Chloride channel [Oryza sativa]
Length = 726
Score = 859 bits (2220), Expect = 0.0
Identities = 426/683 (62%), Positives = 518/683 (75%), Gaps = 5/683 (0%)
Query: 7 SNGDSEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSL 66
S G +PLL ++ +N++SQ+AIVGANVCPIESLDYE+ EN+ FKQDWRSR
Sbjct: 46 SGGGRSGSAGEPLL--RKRTMNTTSQIAIVGANVCPIESLDYEVVENDLFKQDWRSRKKK 103
Query: 67 QIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHK--RFSLAFVTF 124
QIFQY+++KW L LIG + GL+GF NNLAVEN+AG K ++T N+ML R+ AF +
Sbjct: 104 QIFQYIVLKWTLVLLIGLLTGLVGFFNNLAVENIAGFKLLLTGNLMLKGKCRYLTAFFAY 163
Query: 125 FASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVS 184
NL L A I A IAPAAAGSGI EVKAYLNGVDA I TL VKI GSI VS
Sbjct: 164 GGCNLVLAAAAAAICAYIAPAAAGSGIPEVKAYLNGVDAYSILAPSTLFVKIFGSILGVS 223
Query: 185 SSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAA 244
+ +GK GPM+HTGAC+A LLGQGGS++Y LT WLRYFKNDRDRRDLI CGSAAG+AA
Sbjct: 224 AGFVLGKEGPMVHTGACIANLLGQGGSRKYRLTCNWLRYFKNDRDRRDLITCGSAAGVAA 283
Query: 245 AFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLI 304
AFRAPVGGVLFALEE ASWWR+ALLWRAFFTTA+VA+ LR++I+ C SGKCGLFG+GGLI
Sbjct: 284 AFRAPVGGVLFALEEAASWWRSALLWRAFFTTAVVAVVLRSLIEFCRSGKCGLFGQGGLI 343
Query: 305 MFDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLAC 364
MFD S Y D+ + ILG+IGGI G L+N+L +KVLR+Y+ INE+G KILL
Sbjct: 344 MFDLSSTVATYSTPDLIAIIILGIIGGIFGGLFNFLLDKVLRVYSIINERGAPFKILLTI 403
Query: 365 VISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTN 424
IS+ TS +GLPWLA+C PCP D VE CPTIGRSG +K FQCPPGHYNDLASL FNTN
Sbjct: 404 TISIITSMCSYGLPWLAACTPCPVDAVEQCPTIGRSGNFKNFQCPPGHYNDLASLFFNTN 463
Query: 425 DDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVG 484
DDAIRNLFS T+SEF S++FIFF + L IL+ GV VP+G+F+P+I+ GA+YGR VG
Sbjct: 464 DDAIRNLFSNGTESEFHMSTLFIFFTAVYCLGILTYGVAVPSGLFIPVILAGATYGRIVG 523
Query: 485 MLFGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSV 544
L G S+L+ GL+A+LGAASFLGG+MR TVS+CVI+LELTN+L +LPL+M+VLL+SK++
Sbjct: 524 TLLGSISDLDPGLFALLGAASFLGGTMRMTVSVCVILLELTNDLAMLPLVMLVLLISKTI 583
Query: 545 ADVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKT 604
AD FN +YD I+ KGLPY+E HAEPYMR L GDVV+GPL F G EKV N+V L+
Sbjct: 584 ADNFNKGVYDQIVVMKGLPYMEAHAEPYMRHLVAGDVVSGPLITFSGVEKVGNIVHALRF 643
Query: 605 TSHNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAPMAMGGD-VLRQFSADDFA 663
T HNGFPV+DEPP +EAP L G++ R HLL LL K+FM + G VL++F A DFA
Sbjct: 644 TGHNGFPVVDEPPLTEAPELVGLVTRSHLLVLLNGKMFMKDQLKTSGSFVLQRFGAFDFA 703
Query: 664 KRGSDRGERIEDIHLTQEEMDMF 686
K GS +G +I+D+ T EEM+M+
Sbjct: 704 KPGSGKGLKIQDLDFTDEEMEMY 726
>UniRef100_P92942 Chloride channel protein CLC-b [Arabidopsis thaliana]
Length = 780
Score = 822 bits (2122), Expect = 0.0
Identities = 409/762 (53%), Positives = 546/762 (70%), Gaps = 18/762 (2%)
Query: 9 GDSE-HRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGSLQ 67
GD E + L QPL+ + R++ SS+ +A+VGA V IESLDYEI EN+ FK DWR R Q
Sbjct: 20 GDPESNTLNQPLVKANRTL--SSTPLALVGAKVSHIESLDYEINENDLFKHDWRKRSKAQ 77
Query: 68 IFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVTFFAS 127
+ QYV +KW L L+G GLI NLAVEN+AG K + + + +R+ + +
Sbjct: 78 VLQYVFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGHFLTQERYVTGLMVLVGA 137
Query: 128 NLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAVSSSL 187
NL LTL A+++ AP AAG GI E+KAYLNGVD P +F T+IVKI+GSI AV++ L
Sbjct: 138 NLGLTLVASVLCVCFAPTAAGPGIPEIKAYLNGVDTPNMFGATTMIVKIVGSIGAVAAGL 197
Query: 188 HIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIAAAFR 247
+GK GP++H G+C+A+LLGQGG+ + + W+WLRYF NDRDRRDLI CGSAAG+ AAFR
Sbjct: 198 DLGKEGPLVHIGSCIASLLGQGGTDNHRIKWRWLRYFNNDRDRRDLITCGSAAGVCAAFR 257
Query: 248 APVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGLIMFD 307
+PVGGVLFALEE+A+WWR+ALLWR FF+TA+V + LR I++C SGKCGLFGKGGLIMFD
Sbjct: 258 SPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLREFIEICNSGKCGLFGKGGLIMFD 317
Query: 308 AYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLACVIS 367
+ YH D+ V ++GVIGGILGSLYN+L +KVLR+YN INEKG I K+LL+ +S
Sbjct: 318 VSHVTYTYHVTDIIPVMLIGVIGGILGSLYNHLLHKVLRLYNLINEKGKIHKVLLSLTVS 377
Query: 368 MFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNTNDDA 427
+FTS L+GLP+LA C+PC E CPT GRSG +K+F CP G+YNDLA+L+ TNDDA
Sbjct: 378 LFTSVCLYGLPFLAKCKPCDPSIDEICPTNGRSGNFKQFHCPKGYYNDLATLLLTTNDDA 437
Query: 428 IRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFVGMLF 487
+RNLFS NT +EF S++IFF+ L + + G+ P+G+F+PII+ GA+YGR +G
Sbjct: 438 VRNLFSSNTPNEFGMGSLWIFFVLYCILGLFTFGIATPSGLFLPIILMGAAYGRMLGAAM 497
Query: 488 GKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADV 547
G ++++ GLYAVLGAA+ + GSMR TVSLCVI LELTNNLLLLP+ M+VLL++K+V D
Sbjct: 498 GSYTSIDQGLYAVLGAAALMAGSMRMTVSLCVIFLELTNNLLLLPITMIVLLIAKTVGDS 557
Query: 548 FNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDV--VTGPLQIFHGYEKVRNVVFVLKTT 605
FN +IYD+I+ KGLP+LE + EP+MR LTVG++ P+ G EKV N+V VLK T
Sbjct: 558 FNPSIYDIILHLKGLPFLEANPEPWMRNLTVGELGDAKPPVVTLQGVEKVSNIVDVLKNT 617
Query: 606 SHNGFPVIDEPPFSE------APVLFGIILRHHLLTLLRKKVFMSAP-MAMGGDVLRQFS 658
+HN FPV+DE + A L G+ILR HL+ +L+K+ F++ +V +F
Sbjct: 618 THNAFPVLDEAEVPQVGLATGATELHGLILRAHLVKVLKKRWFLTEKRRTEEWEVREKFP 677
Query: 659 ADDFAKRGSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFRELGL 718
D+ A +R + +D+ +T EM+M++DLHP TN +PYTV+E+MS+ KAL+LFR++GL
Sbjct: 678 WDELA----EREDNFDDVAITSAEMEMYVDLHPLTNTTPYTVMENMSVAKALVLFRQVGL 733
Query: 719 RHLLVIPKIPSRS--PVVGILTRHDFTAEHILGTHPYLVRSR 758
RHLL++PKI + PVVGILTR D A +IL P L +S+
Sbjct: 734 RHLLIVPKIQASGMCPVVGILTRQDLRAYNILQAFPLLEKSK 775
>UniRef100_P92941 Chloride channel protein CLC-a [Arabidopsis thaliana]
Length = 775
Score = 815 bits (2106), Expect = 0.0
Identities = 407/772 (52%), Positives = 547/772 (70%), Gaps = 17/772 (2%)
Query: 3 TNHLSNGDSE------HRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFF 56
+N NG+ E + L QPLL R++ SS+ +A+VGA V IESLDYEI EN+ F
Sbjct: 10 SNSNYNGEEEGEDPENNTLNQPLLKRHRTL--SSTPLALVGAKVSHIESLDYEINENDLF 67
Query: 57 KQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKR 116
K DWRSR Q+FQY+ +KW L L+G GLI NLAVEN+AG K + + R
Sbjct: 68 KHDWRSRSKAQVFQYIFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGYYIAQDR 127
Query: 117 FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKI 176
F + F +NL LTL AT++ AP AAG GI E+KAYLNG+D P +F T++VKI
Sbjct: 128 FWTGLMVFTGANLGLTLVATVLVVYFAPTAAGPGIPEIKAYLNGIDTPNMFGFTTMMVKI 187
Query: 177 IGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVC 236
+GSI AV++ L +GK GP++H G+C+A+LLGQGG + + W+WLRYF NDRDRRDLI C
Sbjct: 188 VGSIGAVAAGLDLGKEGPLVHIGSCIASLLGQGGPDNHRIKWRWLRYFNNDRDRRDLITC 247
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCG 296
GSA+G+ AAFR+PVGGVLFALEE+A+WWR+ALLWR FF+TA+V + LRA I++C SGKCG
Sbjct: 248 GSASGVCAAFRSPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLRAFIEICNSGKCG 307
Query: 297 LFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGT 356
LFG GGLIMFD + YHA D+ V ++GV GGILGSLYN+L +KVLR+YN IN+KG
Sbjct: 308 LFGSGGLIMFDVSHVEVRYHAADIIPVTLIGVFGGILGSLYNHLLHKVLRLYNLINQKGK 367
Query: 357 ICKILLACVISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDL 416
I K+LL+ +S+FTS LFGLP+LA C+PC E CPT GRSG +K+F CP G+YNDL
Sbjct: 368 IHKVLLSLGVSLFTSVCLFGLPFLAECKPCDPSIDEICPTNGRSGNFKQFNCPNGYYNDL 427
Query: 417 ASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTG 476
++L+ TNDDA+RN+FS NT +EF S++IFF L +++ G+ P+G+F+PII+ G
Sbjct: 428 STLLLTTNDDAVRNIFSSNTPNEFGMVSLWIFFGLYCILGLITFGIATPSGLFLPIILMG 487
Query: 477 ASYGRFVGMLFGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMM 536
++YGR +G G +N++ GLYAVLGAAS + GSMR TVSLCVI LELTNNLLLLP+ M
Sbjct: 488 SAYGRMLGTAMGSYTNIDQGLYAVLGAASLMAGSMRMTVSLCVIFLELTNNLLLLPITMF 547
Query: 537 VLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDV--VTGPLQIFHGYEK 594
VLL++K+V D FN +IY++I+ KGLP+LE + EP+MR LTVG++ P+ +G EK
Sbjct: 548 VLLIAKTVGDSFNLSIYEIILHLKGLPFLEANPEPWMRNLTVGELNDAKPPVVTLNGVEK 607
Query: 595 VRNVVFVLKTTSHNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAP-MAMGGDV 653
V N+V VL+ T+HN FPV+D + L G+ILR HL+ +L+K+ F++ +V
Sbjct: 608 VANIVDVLRNTTHNAFPVLDGADQNTGTELHGLILRAHLVKVLKKRWFLNEKRRTEEWEV 667
Query: 654 LRQFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILF 713
+F+ + A +R + +D+ +T EM +++DLHP TN +PYTVV+SMS+ KAL+LF
Sbjct: 668 REKFTPVELA----EREDNFDDVAITSSEMQLYVDLHPLTNTTPYTVVQSMSVAKALVLF 723
Query: 714 RELGLRHLLVIPKIPS--RSPVVGILTRHDFTAEHILGTHPYLVRSRWKRLR 763
R +GLRHLLV+PKI + SPV+GILTR D A +IL P+L + + + R
Sbjct: 724 RSVGLRHLLVVPKIQASGMSPVIGILTRQDLRAYNILQAFPHLDKHKSGKAR 775
>UniRef100_Q9XF71 CLC-Nt2 protein [Nicotiana tabacum]
Length = 786
Score = 811 bits (2094), Expect = 0.0
Identities = 402/766 (52%), Positives = 542/766 (70%), Gaps = 17/766 (2%)
Query: 4 NHLSNGDSEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQDWRSR 63
N + L QPLL R++ SSS A+VGA V IESLDYEI EN+ FK DWR R
Sbjct: 21 NEEERDPESNSLHQPLLKRNRTL--SSSPFALVGAKVSHIESLDYEINENDLFKHDWRRR 78
Query: 64 GSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSLAFVT 123
+Q+ QYV +KW L FL+G + G+ NLA+EN+AG K N + +R+ + F
Sbjct: 79 SRVQVLQYVFLKWTLAFLVGLLTGVTATLINLAIENMAGYKLRAVVNYIEDRRYLMGFAY 138
Query: 124 FFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGSITAV 183
F +N LTL A ++ AP AAG GI E+KAYLNGVD P ++ TL VKIIGSI AV
Sbjct: 139 FAGANFVLTLIAALLCVCFAPTAAGPGIPEIKAYLNGVDTPNMYGATTLFVKIIGSIAAV 198
Query: 184 SSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSAAGIA 243
S+SL +GK GP++H GAC A+LLGQGG Y L W+WLRYF NDRDRRDLI CGS++G+
Sbjct: 199 SASLDLGKEGPLVHIGACFASLLGQGGPDNYRLRWRWLRYFNNDRDRRDLITCGSSSGVC 258
Query: 244 AAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFGKGGL 303
AAFR+PVGGVLFALEE+A+WWR+ALLWR FF+TA+V + LRA I+ C SG CGLFG+GGL
Sbjct: 259 AAFRSPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVILRAFIEYCKSGNCGLFGRGGL 318
Query: 304 IMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGTICKILLA 363
IMFD S+ YH D+ V ++G+IGG+LGSLYN++ +K+LR+YN INEKG + K+LLA
Sbjct: 319 IMFDVSGVSVSYHVVDIIPVVVIGIIGGLLGSLYNHVLHKILRLYNLINEKGKLHKVLLA 378
Query: 364 CVISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDLASLIFNT 423
+S+FTS ++GLP+LA C+PC CP G +G +K+F CP G+YNDLA+L+ T
Sbjct: 379 LSVSLFTSICMYGLPFLAKCKPCDPSLPGSCPGTGGTGNFKQFNCPDGYYNDLATLLLTT 438
Query: 424 NDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTGASYGRFV 483
NDDA+RN+FS NT EFQ S+ +F+ L +++ G+ VP+G+F+PII+ G++YGR +
Sbjct: 439 NDDAVRNIFSINTPGEFQVMSLISYFVLYCILGLITFGIAVPSGLFLPIILMGSAYGRLL 498
Query: 484 GMLFGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMMVLLVSKS 543
+ G + ++ GLYAVLGAAS + GSMR TVSLCVI LELTNNLLLLP+ M+VLL++KS
Sbjct: 499 AIAMGSYTKIDPGLYAVLGAASLMAGSMRMTVSLCVIFLELTNNLLLLPITMLVLLIAKS 558
Query: 544 VADVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDV--VTGPLQIFHGYEKVRNVVFV 601
V D FN +IY++I++ KGLP+L+ + EP+MR +T G++ V P+ G EKV +V
Sbjct: 559 VGDCFNLSIYEIILELKGLPFLDANPEPWMRNITAGELADVKPPVVTLCGVEKVGRIVEA 618
Query: 602 LKTTSHNGFPVIDE---PPFS---EAPVLFGIILRHHLLTLLRKKVFM-SAPMAMGGDVL 654
LK T++NGFPV+DE PP A L G++LR HLL +L+KK F+ +V
Sbjct: 619 LKNTTYNGFPVVDEGVVPPVGLPVGATELHGLVLRTHLLLVLKKKWFLHERRRTEEWEVR 678
Query: 655 RQFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILFR 714
+F+ D A+RG +IED+ +T++EM+M++DLHP TN +PYTVVES+S+ KA++LFR
Sbjct: 679 EKFTWIDLAERGG----KIEDVLVTKDEMEMYVDLHPLTNTTPYTVVESLSVAKAMVLFR 734
Query: 715 ELGLRHLLVIPKIPSR--SPVVGILTRHDFTAEHILGTHPYLVRSR 758
++GLRH+L++PK + SPVVGILTR D A +IL P+L +S+
Sbjct: 735 QVGLRHMLIVPKYQAAGVSPVVGILTRQDLRAHNILSVFPHLEKSK 780
>UniRef100_Q96325 Voltage-gated chloride channel [Arabidopsis thaliana]
Length = 773
Score = 782 bits (2020), Expect = 0.0
Identities = 399/772 (51%), Positives = 538/772 (69%), Gaps = 19/772 (2%)
Query: 3 TNHLSNGDSE------HRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFF 56
+N NG+ E + L QPLL R++ SS+ +A+VGA V IESLDYEI EN+ F
Sbjct: 10 SNSNYNGEEEGEDPENNTLNQPLLKRHRTL--SSTPLALVGAKVSHIESLDYEINENDLF 67
Query: 57 KQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKR 116
K DWRSR Q+FQY+ +KW L L+G GLI NLAVEN+AG K + + R
Sbjct: 68 KHDWRSRSKAQVFQYIFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGYYIAQDR 127
Query: 117 FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKI 176
F + F +NL LTL AT++ AP AAG GI E+KAYLNG+D P +F T++VKI
Sbjct: 128 FWTGLMVFTGANLGLTLVATVLVVYFAPTAAGPGIPEIKAYLNGIDTPNMFGFTTMMVKI 187
Query: 177 IGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVC 236
+GSI AV++ L +GK GP++H G+C+A+LLGQGG + + W+WLRYF NDRDRRDLI C
Sbjct: 188 VGSIGAVAAGLDLGKEGPLVHIGSCIASLLGQGGPDNHRIKWRWLRYFNNDRDRRDLITC 247
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCG 296
GSA+G+ AAFR+PVGGVLFALEE+A+WWR+ALLWR FF+TA+V + LRA I++C SGKCG
Sbjct: 248 GSASGVCAAFRSPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLRAFIEICNSGKCG 307
Query: 297 LFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLWNKVLRIYNAINEKGT 356
LFG GGLIMFD + YHA D+ V ++GV GGILGSLYN+L +KVLR+YN IN+KG
Sbjct: 308 LFGSGGLIMFDVSHVEVRYHAADIIPVTLIGVFGGILGSLYNHLLHKVLRLYNLINQKGK 367
Query: 357 ICKILLACVISMFTSCLLFGLPWLASCQPCPADPVEPCPTIGRSGIYKKFQCPPGHYNDL 416
I K+LL+ +S+ LLFGLP+LA+ + C E CPT GRSG +K+F CP G+YNDL
Sbjct: 368 IHKVLLSLGVSLLHQ-LLFGLPFLAN-EACDPTIDEICPTNGRSGNFKQFNCPNGYYNDL 425
Query: 417 ASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGVFVPIIVTG 476
++L+ TNDDA+R F EF S++IFF L +++ G+ P+G+F+PII+ G
Sbjct: 426 STLLLTTNDDAVRKHFLFKHSYEFGMVSLWIFFGLYCILGLITFGIATPSGLFLPIILMG 485
Query: 477 ASYGRFVGMLFGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELTNNLLLLPLIMM 536
++YGR +G G +N++ GLYAVLGAAS + GSMR TVSLCVI LELTNNLLLLP+ M
Sbjct: 486 SAYGRMLGTAMGSYTNIDQGLYAVLGAASLMAGSMRMTVSLCVIFLELTNNLLLLPITMF 545
Query: 537 VLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGDV--VTGPLQIFHGYEK 594
VLL++K+V D FN +IY++I+ KGLP+LE + EP+MR LTVG++ P+ +G EK
Sbjct: 546 VLLIAKTVGDSFNLSIYEIILHLKGLPFLEANPEPWMRNLTVGELNDAKPPVVTLNGVEK 605
Query: 595 VRNVVFVLKTTSHNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRKKVFMSAP-MAMGGDV 653
V N+V VL+ T+HN FPV+D + L G+ILR HL+ +L+K+ F++ +V
Sbjct: 606 VANIVDVLRNTTHNAFPVLDGADQNTGTELHGLILRAHLVKVLKKRWFLNEKRRTEEWEV 665
Query: 654 LRQFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDLHPFTNASPYTVVESMSLGKALILF 713
+F+ + A +R + +D+ +T EM +++DLHP TN +PYTVV+SMS+ KAL+LF
Sbjct: 666 REKFTPVELA----EREDNFDDVAITSSEMQLYVDLHPLTNTTPYTVVQSMSVAKALVLF 721
Query: 714 RELGLRHLLVIPKIPS--RSPVVGILTRHDFTAEHILGTHPYLVRSRWKRLR 763
R +GLRHLLV+PKI + SPV+GILTR D A +IL P+L + + + R
Sbjct: 722 RSVGLRHLLVVPKIQASGMSPVIGILTRQDLRAYNILQAFPHLDKHKSGKAR 773
>UniRef100_Q851D8 Putative CLC-d chloride channel protein [Oryza sativa]
Length = 782
Score = 640 bits (1651), Expect = 0.0
Identities = 342/716 (47%), Positives = 468/716 (64%), Gaps = 20/716 (2%)
Query: 43 IESLDYEIFENEFFKQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAG 102
+ESLDYE+ EN ++++ R + YV++KWL LIG GL NLAVEN +G
Sbjct: 41 VESLDYEVIENYAYREEQAQRSKFWVPYYVMLKWLFSLLIGVGTGLAAIFINLAVENFSG 100
Query: 103 VKFVVTSNMMLHKRFSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVD 162
K+ T ++ H F + F + NLAL + I APAAAGSGI E+K YLNGVD
Sbjct: 101 WKYAATFAIIQHSYF-VGFFVYIVFNLALVFSSVYIVTNFAPAAAGSGIPEIKGYLNGVD 159
Query: 163 APGIFTLPTLIVKIIGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLR 222
GI TL+ KI GSI +V L +GK GP++HTGAC+A+LLGQGGS +Y L+ +W+R
Sbjct: 160 THGILLFRTLVGKIFGSIGSVGGGLALGKEGPLVHTGACIASLLGQGGSAKYHLSSRWVR 219
Query: 223 YFKNDRDRRDLIVCGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIF 282
F++DRDRRDL+ CG AAG+AAAFRAPVGGVLFALEE+ SWWR+ L+WR FFT+A+VA+
Sbjct: 220 IFESDRDRRDLVTCGCAAGVAAAFRAPVGGVLFALEEVTSWWRSHLMWRVFFTSAVVAVV 279
Query: 283 LRAMIDVCLSGKCGLFGKGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYLW- 341
+R+ ++ C SGKCG FG GG I++D G Y +++ + I+GVIGG+LG+L+N L
Sbjct: 280 VRSAMNWCKSGKCGHFGSGGFIIWDISGGQEDYSYQELLPMAIIGVIGGLLGALFNQLTL 339
Query: 342 --NKVLRIYNAINEKGTICKILLACVISMFTSCLLFGLPWLASCQPCP---ADPVEPCP- 395
K R Y +++KG KI AC+IS+ TS + F LP + C CP + CP
Sbjct: 340 YITKWRRTY--LHKKGKRVKIFEACLISLVTSTISFVLPLMRKCSSCPQLETNSGIECPR 397
Query: 396 TIGRSGIYKKFQC-PPGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFF 454
G G + F C YNDLA++ FNT DDAIRNLFS T E+ S+ F + +
Sbjct: 398 PPGTDGNFVNFYCSKDNEYNDLATIFFNTQDDAIRNLFSAKTFHEYSAQSLITFLVMFYS 457
Query: 455 LSILSCGVVVPAGVFVPIIVTGASYGRFVGML---FGKKSNLNHGLYAVLGAASFLGGSM 511
L++++ G VPAG FVP I+ G++YGR VGM F KK N+ G YA+LGAASFLGGSM
Sbjct: 458 LAVVTFGTAVPAGQFVPGIMIGSTYGRLVGMFVVKFYKKLNVEEGTYALLGAASFLGGSM 517
Query: 512 RTTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEP 571
R TVSLCVI++E+TNNL LLPLIM+VLL+SK+V D FN +Y++ + +G+P L++ +
Sbjct: 518 RMTVSLCVIMVEITNNLKLLPLIMLVLLISKAVGDFFNEGLYEVQAQLRGIPLLDSRPKQ 577
Query: 572 YMRQLTVGDVVTGPLQI-FHGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEAPVLFGIILR 630
MR ++ D + ++ +++ VL++ HNGFPV+D E+ V+ G+ILR
Sbjct: 578 VMRNMSAKDACKNQKVVSLPRVSRIVDIISVLRSNKHNGFPVVDRGQNGESLVI-GLILR 636
Query: 631 HHLLTLLRKKV-FMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDL 689
HLL LL+ KV F ++P G +L + + DF K S +G+ I+DIHLT++E+ +++DL
Sbjct: 637 SHLLVLLQSKVDFQNSPFPCGPGILNRHNTSDFVKPASSKGKSIDDIHLTEDELGLYLDL 696
Query: 690 HPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHDFTAE 745
PF N SPY V E MSL K LFR+LGLRH+ V+P+ PSR VVG++TR D E
Sbjct: 697 APFLNPSPYIVPEDMSLAKVYNLFRQLGLRHIFVVPR-PSR--VVGLITRQDLLLE 749
>UniRef100_P92943 Chloride channel protein CLC-d [Arabidopsis thaliana]
Length = 792
Score = 640 bits (1650), Expect = 0.0
Identities = 362/768 (47%), Positives = 482/768 (62%), Gaps = 38/768 (4%)
Query: 1 MSTNHLSNG-DSEHRLRQPLLSSQRSIINSSSQVAIVGANVCPIESLDYEIFENEFFKQD 59
M +NHL NG +S++ L + S + + + + + SLDYE+ EN ++++
Sbjct: 1 MLSNHLQNGIESDNLLWSRVPESDDTSTDDITLLNSHRDGDGGVNSLDYEVIENYAYREE 60
Query: 60 WRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGVKFVVTSNMMLHKRFSL 119
RG L + YV +KW LIG GL NL+VEN AG KF +T ++ K +
Sbjct: 61 QAHRGKLYVGYYVAVKWFFSLLIGIGTGLAAVFINLSVENFAGWKFALTF-AIIQKSYFA 119
Query: 120 AFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLNGVDAPGIFTLPTLIVKIIGS 179
F+ + NL L + I APAAAGSGI E+K YLNG+D PG TLI KI GS
Sbjct: 120 GFIVYLLINLVLVFSSAYIITQFAPAAAGSGIPEIKGYLNGIDIPGTLLFRTLIGKIFGS 179
Query: 180 ITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWKWLRYFKNDRDRRDLIVCGSA 239
I +V L +GK GP++HTGAC+A+LLGQGGS +Y L +W + FK+DRDRRDL+ CG A
Sbjct: 180 IGSVGGGLALGKEGPLVHTGACIASLLGQGGSTKYHLNSRWPQLFKSDRDRRDLVTCGCA 239
Query: 240 AGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIVAIFLRAMIDVCLSGKCGLFG 299
AG+AAAFRAPVGGVLFALEE+ SWWR+ L+WR FFT+AIVA+ +R + C SG CG FG
Sbjct: 240 AGVAAAFRAPVGGVLFALEEVTSWWRSQLMWRVFFTSAIVAVVVRTAMGWCKSGICGHFG 299
Query: 300 KGGLIMFDAYSGSLMYHAKDVPLVFILGVIGGILGSLYNYL------WNKVLRIYNAINE 353
GG I++D G Y+ K++ + ++GVIGG+LG+L+N L W + N++++
Sbjct: 300 GGGFIIWDVSDGQDDYYFKELLPMAVIGVIGGLLGALFNQLTLYMTSWRR-----NSLHK 354
Query: 354 KGTICKILLACVISMFTSCLLFGLPWLASCQPCP---ADPVEPCP-TIGRSGIYKKFQC- 408
KG KI+ AC+IS TS + FGLP L C PCP D CP G G Y F C
Sbjct: 355 KGNRVKIIEACIISCITSAISFGLPLLRKCSPCPESVPDSGIECPRPPGMYGNYVNFFCK 414
Query: 409 PPGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSILSCGVVVPAGV 468
YNDLA++ FNT DDAIRNLFS T EF S+ F + L++++ G VPAG
Sbjct: 415 TDNEYNDLATIFFNTQDDAIRNLFSAKTMREFSAQSLLTFLAMFYTLAVVTFGTAVPAGQ 474
Query: 469 FVPIIVTGASYGRFVGML---FGKKSNLNHGLYAVLGAASFLGGSMRTTVSLCVIILELT 525
FVP I+ G++YGR VGM F KK N+ G YA+LGAASFLGGSMR TVSLCVI++E+T
Sbjct: 475 FVPGIMIGSTYGRLVGMFVVRFYKKLNIEEGTYALLGAASFLGGSMRMTVSLCVIMVEIT 534
Query: 526 NNLLLLPLIMMVLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEPYMRQLTVGD----- 580
NNL LLPLIM+VLL+SK+V D FN +Y++ + KG+P LE+ + +MRQ+ +
Sbjct: 535 NNLKLLPLIMLVLLISKAVGDAFNEGLYEVQARLKGIPLLESRPKYHMRQMIAKEACQSQ 594
Query: 581 -VVTGPLQIFHGYEKVRNVVFVLKTTSHNGFPVIDEPPFSEAPVLFGIILRHHLLTLLRK 639
V++ P I +V +V +L + HNGFPVID E V+ G++LR HLL LL+
Sbjct: 595 KVISLPRVI-----RVADVASILGSNKHNGFPVIDHTRSGETLVI-GLVLRSHLLVLLQS 648
Query: 640 KV-FMSAPMAMGGDVLR-QFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDLHPFTNASP 697
KV F +P+ + S +FAK S +G IEDIHLT ++++M+IDL PF N SP
Sbjct: 649 KVDFQHSPLPCDPSARNIRHSFSEFAKPVSSKGLCIEDIHLTSDDLEMYIDLAPFLNPSP 708
Query: 698 YTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHDFTAE 745
Y V E MSL K LFR+LGLRHL V+P+ PSR V+G++TR D E
Sbjct: 709 YVVPEDMSLTKVYNLFRQLGLRHLFVVPR-PSR--VIGLITRKDLLIE 753
>UniRef100_UPI00003619FE UPI00003619FE UniRef100 entry
Length = 729
Score = 419 bits (1076), Expect = e-115
Identities = 263/722 (36%), Positives = 402/722 (55%), Gaps = 32/722 (4%)
Query: 44 ESLDYEIFENEFFKQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGV 103
ESLDY+ EN+ F ++ R + + +W++C LIG + GLI ++ VE LAG+
Sbjct: 16 ESLDYDNIENQLFLEEERRMSYMSFRCLEISRWVVCGLIGFLTGLIACFIDIVVEELAGI 75
Query: 104 KF-VVTSNMMLHKR---FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLN 159
K+ VV N+ S++ + + N + +II A P AAGSGI ++K YLN
Sbjct: 76 KYQVVKENIDKFTEVGGLSISLILWAVLNSVFVMLGSIIVAFFEPIAAGSGIPQIKCYLN 135
Query: 160 GVDAPGIFTLPTLIVKIIGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWK 219
GV P + L TL+VK+ G I +V L +GK GPM+H+GA VAA + QG S +K
Sbjct: 136 GVKIPRVVRLKTLLVKVCGVICSVVGGLAVGKEGPMIHSGAVVAAGVSQGRSTSLKRDFK 195
Query: 220 WLRYFKNDRDRRDLIVCGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIV 279
YF+ D ++RD + G+AAG++AAF APVGGVLF+LEE AS+W L WR FF + I
Sbjct: 196 MFEYFRRDTEKRDFVSAGAAAGVSAAFGAPVGGVLFSLEEGASFWNQMLTWRIFFASMIS 255
Query: 280 AIFLRAMIDVCLSGKCGLFGKGGLIMFDAY-SGSLMYHAKDVPLVFILGVIGGILGSLYN 338
L + + + G GLI F + S S+ Y+ ++PL +G IGG+LG+L+N
Sbjct: 256 TFTLNFFLSI-YNNNPGDLSNPGLINFGRFESESVAYNLYEIPLFIAMGAIGGLLGALFN 314
Query: 339 YLWNKVLRIYNAINEKGTICKILLACVISMFTSCLLFGLPWLAS-CQPCPADPVEPCPTI 397
L N L I+ +++ A +++ T+ + F + + ++ CQP ++ E P
Sbjct: 315 IL-NYWLTIFRIRYVHRPCLQVMEAMLVAAVTATVSFTMIYFSNDCQPLGSEHSEEYPL- 372
Query: 398 GRSGIYKKFQCPPGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLSI 457
+ C G YN +A+ FNT + ++R+LF N + ++ +F +T FFL+
Sbjct: 373 -------QLFCADGEYNSMATAFFNTPERSVRSLF-HNQPRTYNPLTLGLFTLTYFFLAC 424
Query: 458 LSCGVVVPAGVFVPIIVTGASYGRFVGMLFGKKSN-----LNHGLYAVLGAASFLGGSMR 512
+ G+ V AGVF+P ++ GA++GR G+L ++ + G YA++GAA+ LGG +R
Sbjct: 425 WTYGLAVSAGVFIPSLLIGAAWGRLCGILLASITSTGSIWADPGKYALIGAAAQLGGIVR 484
Query: 513 TTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEPY 572
T+SL VI++E T N+ IM+VL+ +K V D F +YD+ +K + +P+L A
Sbjct: 485 MTLSLTVIMVEATGNVTYGLPIMLVLMTAKIVGDYFVEGLYDIHIKLQSVPFLHGEAPGT 544
Query: 573 MRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTS--HNGFPVIDEPPFSEAPV-LFGIIL 629
LT +V++ P+ + EKV +V L TS HNGFPV+ + P L G+IL
Sbjct: 545 SHWLTAREVMSSPVTCLNRIEKVGTIVDTLSNTSTNHNGFPVVVQADVLFQPAKLCGLIL 604
Query: 630 RHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDL 689
R L+ LL+ KVF+ +A R+ DF + R I+ IH++Q+E + +DL
Sbjct: 605 RSQLIVLLKHKVFVE--LARSRLTQRKLQLKDF-RDAYPRFPPIQSIHVSQDERECMMDL 661
Query: 690 HPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHDFTAEHILG 749
F N +PYTV + SL + LFR LGLRHL+V + + VVG++TR D H LG
Sbjct: 662 TEFMNPTPYTVPQETSLPRVFKLFRALGLRHLVV---VDDENRVVGLVTRKDLARYH-LG 717
Query: 750 TH 751
H
Sbjct: 718 KH 719
>UniRef100_Q5ZL60 Hypothetical protein [Gallus gallus]
Length = 802
Score = 405 bits (1041), Expect = e-111
Identities = 259/720 (35%), Positives = 400/720 (54%), Gaps = 34/720 (4%)
Query: 44 ESLDYEIFENEFFKQDWRSRGSLQIFQYVLMKWLLCFLIGTILGLIGFCNNLAVENLAGV 103
ESLDY+ EN+ F ++ R + +W++C +IG + GL+ ++ VENLAG+
Sbjct: 91 ESLDYDNSENQLFLEEERRINHAAFRTVEIKRWVICAMIGILTGLVACFIDIVVENLAGL 150
Query: 104 KF-VVTSNMMLHKR---FSLAFVTFFASNLALTLFATIITAMIAPAAAGSGISEVKAYLN 159
K+ VV N+ S + + + N ++ + ++I A I P AAGSGI ++K YLN
Sbjct: 151 KYRVVKDNIDKFTEKGGLSFSLLLWATLNASVVMVGSVIVAFIEPVAAGSGIPQIKCYLN 210
Query: 160 GVDAPGIFTLPTLIVKIIGSITAVSSSLHIGKAGPMLHTGACVAALLGQGGSKRYGLTWK 219
GV P + L TL++K+ G I +V L +GK GPM+H+GA +AA + QG S +K
Sbjct: 211 GVKIPHVVRLKTLVIKVCGVILSVVGGLAVGKEGPMIHSGAVIAAGISQGRSTSLKRDFK 270
Query: 220 WLRYFKNDRDRRDLIVCGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTAIV 279
YF+ D ++RD + G+AAG++AAF APVGGVLF+LEE AS+W L WR FF + I
Sbjct: 271 IFEYFRRDTEKRDFVSAGAAAGVSAAFGAPVGGVLFSLEEGASFWNQFLTWRIFFASMIS 330
Query: 280 AIFLRAMIDVCLSGKCGLFGKGGLIMFDAYSGSLM-YHAKDVPLVFILGVIGGILGSLYN 338
L +++ V G GLI F + M Y +++P+ +GV+GGILG+L+N
Sbjct: 331 TFTLNSVLSV-YHGNAWDLSSPGLINFGRFDSEKMGYTIQEIPIFIFMGVVGGILGALFN 389
Query: 339 YLWNKVLRIYNAINEKGTICKILLACVISMFTSCLLFGLPWLA-SCQPCPADPVEPCPTI 397
L N L ++ +++ A +++ T+ + F + + + CQP
Sbjct: 390 AL-NYWLTMFRIRYIHRPCLQVIEAMLVAAVTAAVGFVMIYCSRDCQPIQ---------- 438
Query: 398 GRSGIYK-KFQCPPGHYNDLASLIFNTNDDAIRNLFSKNTDSEFQYSSMFIFFITGFFLS 456
G S Y + C G YN +A+ FNT + ++ NLF + + ++ +F + FFL+
Sbjct: 439 GSSVAYPLQLFCADGEYNSMATAFFNTPEKSVVNLF-HDPPGSYNPMTLGMFTLMYFFLA 497
Query: 457 ILSCGVVVPAGVFVPIIVTGASYGRFVGMLFGKKSN----LNHGLYAVLGAASFLGGSMR 512
+ G+ V AGVF+P ++ GA++GR G+ S + G YA++GAA+ LGG +R
Sbjct: 498 CWTYGLTVSAGVFIPSLLIGAAWGRLFGISLSYLSKGSIWADPGKYALMGAAAQLGGIVR 557
Query: 513 TTVSLCVIILELTNNLLLLPLIMMVLLVSKSVADVFNANIYDLIMKSKGLPYLETHAEPY 572
T+SL VI++E T N+ IM+VL+ +K V D F +YD+ ++ + +P+L A
Sbjct: 558 MTLSLTVIMMEATGNVTYGFPIMLVLMTAKIVGDYFVEGLYDMHIQLQSVPFLHWEAPVT 617
Query: 573 MRQLTVGDVVTGPLQIFHGYEKVRNVVFVLKTTS--HNGFPVID-EPPFSEAPVLFGIIL 629
LT +V++ P+ E+V VV +L TS HNGFPV++ P ++ L G+IL
Sbjct: 618 SHSLTAREVMSTPVTCLRRIERVGTVVDILSDTSSNHNGFPVVESNPNTTQVAGLRGLIL 677
Query: 630 RHHLLTLLRKKVFMSAPMAMGGDVLRQFSADDFAKRGSDRGERIEDIHLTQEEMDMFIDL 689
R L+ LL+ KVF+ A V R+ DF + R I+ IH++Q+E + IDL
Sbjct: 678 RSQLIVLLKHKVFVE--RANLNLVQRRLKLKDF-RDAYPRFPPIQSIHVSQDERECMIDL 734
Query: 690 HPFTNASPYTVVESMSLGKALILFRELGLRHLLVIPKIPSRSPVVGILTRHDFTAEHILG 749
F N SPYTV SL + LFR LGLRHL+V + + + VVG++TR D A + LG
Sbjct: 735 SEFMNPSPYTVPREASLPRVFKLFRALGLRHLVV---VNNHNEVVGMVTRKDL-ARYRLG 790
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.327 0.142 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,320,672,779
Number of Sequences: 2790947
Number of extensions: 57904318
Number of successful extensions: 179656
Number of sequences better than 10.0: 614
Number of HSP's better than 10.0 without gapping: 477
Number of HSP's successfully gapped in prelim test: 138
Number of HSP's that attempted gapping in prelim test: 176717
Number of HSP's gapped (non-prelim): 1385
length of query: 780
length of database: 848,049,833
effective HSP length: 135
effective length of query: 645
effective length of database: 471,271,988
effective search space: 303970432260
effective search space used: 303970432260
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 79 (35.0 bits)
Lotus: description of TM0030.6


