

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0029a.7
(411 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
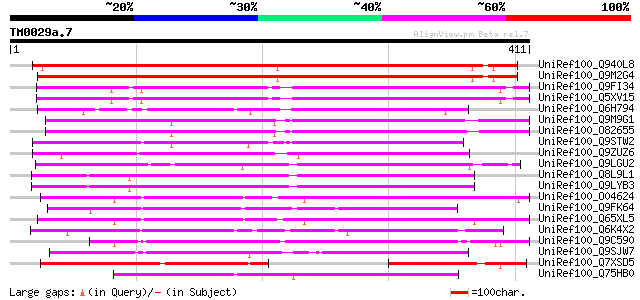
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q940L8 AT3g58520/F14P22_110 [Arabidopsis thaliana] 547 e-154
UniRef100_Q9M2G4 Hypothetical protein F14P22.110 [Arabidopsis th... 542 e-153
UniRef100_Q9FI34 Emb|CAB68190.1 [Arabidopsis thaliana] 266 6e-70
UniRef100_Q5XV15 Hypothetical protein [Arabidopsis thaliana] 265 2e-69
UniRef100_Q6H794 Hypothetical protein P0458B05.12 [Oryza sativa] 213 8e-54
UniRef100_Q9M9G1 F14O23.23 protein [Arabidopsis thaliana] 209 1e-52
UniRef100_O82655 Hypothetical protein ORF2 [Arabidopsis thaliana] 204 5e-51
UniRef100_Q9STW2 Hypothetical protein T22A6.150 [Arabidopsis tha... 196 1e-48
UniRef100_Q9ZUZ6 Hypothetical protein At2g39120 [Arabidopsis tha... 194 4e-48
UniRef100_Q9LGU2 EST AU030398(E51008) corresponds to a region of... 184 5e-45
UniRef100_Q8L9L1 Hypothetical protein [Arabidopsis thaliana] 166 1e-39
UniRef100_Q9LYB3 Hypothetical protein T20O10_190 [Arabidopsis th... 164 3e-39
UniRef100_O04624 A_IG002N01.30 protein [Arabidopsis thaliana] 164 6e-39
UniRef100_Q9FK64 Tyrosine-specific protein phosphatase-like prot... 154 4e-36
UniRef100_Q65XL5 Hypothetical protein OJ1735_C10.3 [Oryza sativa] 153 1e-35
UniRef100_Q6K4X2 Tyrosine-specific protein phosphatase-like [Ory... 151 3e-35
UniRef100_Q9C590 Hypothetical protein At5g21970 [Arabidopsis tha... 150 5e-35
UniRef100_Q9SJW7 Hypothetical protein At2g31290 [Arabidopsis tha... 143 8e-33
UniRef100_Q7XSD5 OSJNBa0079A21.9 protein [Oryza sativa] 132 1e-29
UniRef100_Q75HB0 Expressed protein [Oryza sativa] 131 4e-29
>UniRef100_Q940L8 AT3g58520/F14P22_110 [Arabidopsis thaliana]
Length = 418
Score = 547 bits (1410), Expect = e-154
Identities = 275/396 (69%), Positives = 318/396 (79%), Gaps = 15/396 (3%)
Query: 19 QWRGIA---KVRLKWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKL 75
QWRG+ KVRLKWVKN++LDH+ID ETDLKAA +LKDAI RS TGFLTAKS + WQKL
Sbjct: 21 QWRGMGAMTKVRLKWVKNKNLDHVIDTETDLKAACILKDAIKRSPTGFLTAKSVADWQKL 80
Query: 76 LGLTVPVLRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERL 135
LGLTVPVLRF+RRYPTLFHEFPH R+ SLPCF+LTDTAL+L SQE+ +H HE DTVERL
Sbjct: 81 LGLTVPVLRFLRRYPTLFHEFPHARYASLPCFKLTDTALMLDSQEEIIHQSHEADTVERL 140
Query: 136 SKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWC 195
+ LMM +++TV L SL+ LK+D+GLPD++EKTL+ K+PD F FVKA NG ++L KW
Sbjct: 141 CRVLMMMRSKTVSLRSLHSLKFDLGLPDNYEKTLVMKYPDHFCFVKASNGNPCLKLVKWR 200
Query: 196 EEFAVSALQKSNERD---GGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKL 252
+EFA SALQK NE + G + YR+FKRG++ L FPM FPRGYGAQKKV+ WMDEF KL
Sbjct: 201 DEFAFSALQKRNEDNDVSGEESRYREFKRGQSTLTFPMSFPRGYGAQKKVKAWMDEFQKL 260
Query: 253 PYISPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRI 312
PYISPY D S IDP SDLMEKR V VLHELLSLT+HKKTKRNYLRS+R ELN+PHKFTR+
Sbjct: 261 PYISPYDDPSNIDPESDLMEKRAVAVLHELLSLTIHKKTKRNYLRSMRAELNIPHKFTRL 320
Query: 313 FTRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLY--RGDGS 370
FTRYPGIFYLSLKCKTTTV L+EGY RGKLVDPHPL R RDKFYHVM+TG LY RG G
Sbjct: 321 FTRYPGIFYLSLKCKTTTVILKEGYRRGKLVDPHPLTRLRDKFYHVMRTGFLYRARGLGM 380
Query: 371 IPKPEGGALLL----DEIGDEGSEEEEVETSDEFSE 402
+ K E LLL D++ +EGSEEEE+ E E
Sbjct: 381 VSKEE---LLLDRPEDDLEEEGSEEEEIVEGSELEE 413
>UniRef100_Q9M2G4 Hypothetical protein F14P22.110 [Arabidopsis thaliana]
Length = 394
Score = 542 bits (1397), Expect = e-153
Identities = 271/389 (69%), Positives = 314/389 (80%), Gaps = 12/389 (3%)
Query: 23 IAKVRLKWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLTVPV 82
+ KVRLKWVKN++LDH+ID ETDLKAA +LKDAI RS TGFLTAKS + WQKLLGLTVPV
Sbjct: 4 MTKVRLKWVKNKNLDHVIDTETDLKAACILKDAIKRSPTGFLTAKSVADWQKLLGLTVPV 63
Query: 83 LRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLMMT 142
LRF+RRYPTLFHEFPH R+ SLPCF+LTDTAL+L SQE+ +H HE DTVERL + LMM
Sbjct: 64 LRFLRRYPTLFHEFPHARYASLPCFKLTDTALMLDSQEEIIHQSHEADTVERLCRVLMMM 123
Query: 143 KTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCEEFAVSA 202
+++TV L SL+ LK+D+GLPD++EKTL+ K+PD F FVKA NG ++L KW +EFA SA
Sbjct: 124 RSKTVSLRSLHSLKFDLGLPDNYEKTLVMKYPDHFCFVKASNGNPCLKLVKWRDEFAFSA 183
Query: 203 LQKSNERD---GGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYISPYA 259
LQK NE + G + YR+FKRG++ L FPM FPRGYGAQKKV+ WMDEF KLPYISPY
Sbjct: 184 LQKRNEDNDVSGEESRYREFKRGQSTLTFPMSFPRGYGAQKKVKAWMDEFQKLPYISPYD 243
Query: 260 DSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYPGI 319
D S IDP SDLMEKR V VLHELLSLT+HKKTKRNYLRS+R ELN+PHKFTR+FTRYPGI
Sbjct: 244 DPSNIDPESDLMEKRAVAVLHELLSLTIHKKTKRNYLRSMRAELNIPHKFTRLFTRYPGI 303
Query: 320 FYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLY--RGDGSIPKPEGG 377
FYLSLKCKTTTV L+EGY RGKLVDPHPL R RDKFYHVM+TG LY RG G + K E
Sbjct: 304 FYLSLKCKTTTVILKEGYRRGKLVDPHPLTRLRDKFYHVMRTGFLYRARGLGMVSKEE-- 361
Query: 378 ALLL----DEIGDEGSEEEEVETSDEFSE 402
LLL D++ +EGSEEEE+ E E
Sbjct: 362 -LLLDRPEDDLEEEGSEEEEIVEGSELEE 389
>UniRef100_Q9FI34 Emb|CAB68190.1 [Arabidopsis thaliana]
Length = 422
Score = 266 bits (681), Expect = 6e-70
Identities = 161/399 (40%), Positives = 224/399 (55%), Gaps = 27/399 (6%)
Query: 22 GIAKVRLKWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLT-- 79
G+ V+LKWVK+R LD ++ +E L+A L I+ S L + LGL
Sbjct: 31 GLVNVKLKWVKDRELDAVVVREKHLRAVCNLVSVISASPDLRLPIFKLLPHRGQLGLPQE 90
Query: 80 VPVLRFIRRYPTLFHEFPHPRWPS----LPCFRLTDTALLLHSQEQSLHNHHENDTVERL 135
+ + FIRRYP +F E H W S +PCF LT + L+ +E + +E D + RL
Sbjct: 91 LKLSAFIRRYPNIFVE--HCYWDSAGTSVPCFGLTRETIDLYYEEVDVSRVNERDVLVRL 148
Query: 136 SKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWC 195
K LM+T RT+ L S+ L+WD+GLP + +LI K PD F VK + + ++L W
Sbjct: 149 CKLLMLTCERTLSLHSIDHLRWDLGLPYDYRDSLITKHPDLFSLVKLSSDLDGLKLIHWD 208
Query: 196 EEFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYI 255
E AVS +Q D G+D + FP++F RG+G ++K W+ E+ +LPY
Sbjct: 209 EHLAVSQMQL--REDVGND---------ERMAFPVKFTRGFGLKRKSIEWLQEWQRLPYT 257
Query: 256 SPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTR 315
SPY D+S +DP +DL EKR VGV HELL LT+ KKT+R + +LR+ LP KFT++F R
Sbjct: 258 SPYVDASHLDPRTDLSEKRNVGVFHELLHLTIGKKTERKNVSNLRKPFALPQKFTKVFER 317
Query: 316 YPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYRGDGSIPKPE 375
+PGIFY+S+KC T TV LRE Y R L++ HPLV R+KF ++M G L R G K
Sbjct: 318 HPGIFYISMKCDTQTVILREAYDRRHLIEKHPLVEVREKFANMMNEGFLDRSRGLYQKSV 377
Query: 376 GGALLLDEIGDE---GSEEEEVETSDEFSEGEVSGSDSE 411
L + I S+EEE E +SG DS+
Sbjct: 378 EADLEKNNIKTSYPVWSDEEE-----ELDNNLISGYDSD 411
>UniRef100_Q5XV15 Hypothetical protein [Arabidopsis thaliana]
Length = 422
Score = 265 bits (676), Expect = 2e-69
Identities = 160/399 (40%), Positives = 223/399 (55%), Gaps = 27/399 (6%)
Query: 22 GIAKVRLKWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLT-- 79
G+ V+LKWVK+R LD ++ +E L+A L I+ S L + LGL
Sbjct: 31 GLVNVKLKWVKDRELDAVVVREKHLRAVCNLVSVISASPDLRLPIFKLLPHRGQLGLPQE 90
Query: 80 VPVLRFIRRYPTLFHEFPHPRWPS----LPCFRLTDTALLLHSQEQSLHNHHENDTVERL 135
+ + FIRRYP +F E H W S +PCF LT + L+ +E + +E D + RL
Sbjct: 91 LKLSAFIRRYPNIFVE--HCYWDSAGTSVPCFGLTRETIDLYYEEVDVSRVNERDVLVRL 148
Query: 136 SKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWC 195
K LM+T RT+ L S+ L+WD+G P + +LI K PD F VK + + ++L W
Sbjct: 149 CKLLMLTCERTLSLHSIDHLRWDLGXPYDYRDSLITKHPDLFSLVKLSSDLDGLKLIHWD 208
Query: 196 EEFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYI 255
E AVS +Q D G+D + FP++F RG+G ++K W+ E+ +LPY
Sbjct: 209 EHLAVSQMQL--REDVGND---------ERMAFPVKFTRGFGLKRKSIEWLQEWQRLPYT 257
Query: 256 SPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTR 315
SPY D+S +DP +DL EKR VGV HELL LT+ KKT+R + +LR+ LP KFT++F R
Sbjct: 258 SPYVDASHLDPRTDLSEKRNVGVFHELLHLTIGKKTERKNVSNLRKPFALPQKFTKVFER 317
Query: 316 YPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYRGDGSIPKPE 375
+PGIFY+S+KC T TV LRE Y R L++ HPLV R+KF ++M G L R G K
Sbjct: 318 HPGIFYISMKCDTQTVILREAYDRRHLIEKHPLVEVREKFANMMNEGFLDRSRGLYQKSV 377
Query: 376 GGALLLDEIGDE---GSEEEEVETSDEFSEGEVSGSDSE 411
L + I S+EEE E +SG DS+
Sbjct: 378 EADLEKNNIKTSYPVWSDEEE-----ELDNNLISGYDSD 411
>UniRef100_Q6H794 Hypothetical protein P0458B05.12 [Oryza sativa]
Length = 386
Score = 213 bits (542), Expect = 8e-54
Identities = 138/351 (39%), Positives = 194/351 (54%), Gaps = 27/351 (7%)
Query: 23 IAKVRLKWVKNRSLDHIIDKETDLKAASLLKDAIN-----RSSTGFLTAKSFSGWQKLLG 77
+ V+LKWVK+R+LD + +E DL+AA L D ++ R S L A S ++ G
Sbjct: 23 LVNVKLKWVKDRALDGAVSRERDLRAAHHLLDVVSARPGHRVSRPELLADS--SVRRAFG 80
Query: 78 LTVPVLRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSK 137
V F+ RY TLF R + LTD AL L +E E D V RL +
Sbjct: 81 GVDGVDAFLARYHTLF---ALRRGGGVS---LTDAALDLRRREVDCLVESEPDLVSRLRR 134
Query: 138 FLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAV--RLSKWC 195
LM+T R++PL ++ L+WD+GLP + +++ ++PD F + P G V RL W
Sbjct: 135 LLMLTLPRSLPLHTVDLLRWDLGLPRDYRASILRRYPDHFA-LDQPEGDERVWLRLLWWD 193
Query: 196 EEFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYI 255
+ AVS L+KS GG D L FP+ F +G+G + K W+ E+ LPY
Sbjct: 194 DGLAVSELEKSTAGGGGGD--------TTCLPFPVSFTKGFGLRSKCINWLKEWQALPYT 245
Query: 256 SPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTR 315
SPYAD S +D +D+ EKR VGV HELL LT+ K+T+R + ++R+ L +P KFT++F R
Sbjct: 246 SPYADPSGLDRRTDVSEKRNVGVFHELLHLTVAKRTERRNVSNMRKLLGMPQKFTKVFER 305
Query: 316 YPGIFYLSLKCKTTTVTLREGYARGKLV---DPHPLVRHRDKFYHVMKTGL 363
+PGIFYLS T TV LRE Y G L+ HPL R+++ VM+ L
Sbjct: 306 HPGIFYLSRVLGTQTVVLREAYGGGSLLLAKHAHPLATIREEYSAVMRAAL 356
>UniRef100_Q9M9G1 F14O23.23 protein [Arabidopsis thaliana]
Length = 470
Score = 209 bits (531), Expect = 1e-52
Identities = 129/390 (33%), Positives = 211/390 (54%), Gaps = 23/390 (5%)
Query: 29 KWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLTVPVLRFIRR 88
K+V++R LDH +++E +L+ +KD I + + + L + + + FIR
Sbjct: 30 KFVRDRGLDHAVEREKNLRPLLSIKDLIRSEPAKSVPISVITSQKDSLRVPLRPIEFIRS 89
Query: 89 YPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNH--HENDTVERLSKFLMMTKTRT 146
+P++F EF P LT L + EQ ++ ++ +RL K LM+ +
Sbjct: 90 FPSVFQEFLPGGIGIHPHISLTPEILNHDADEQLVYGSETYKQGLADRLLKLLMINRINK 149
Query: 147 VPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAP-NGVSA-VRLSKWCEEFAVSALQ 204
+PL L LKWD+GLP + +T++P+FPD F+ +K+ G S + L W E AVS L+
Sbjct: 150 IPLEILDLLKWDLGLPKDYVETMVPEFPDYFRVIKSKLRGCSGELELVCWSNEHAVSVLE 209
Query: 205 KSNE--RDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYISPYADSS 262
K R G ++ +G +A+ FPM+F G+ KK++ W+D++ KLPYISPY ++
Sbjct: 210 KKARTLRKG------EYTKG-SAIAFPMKFSNGFVVDKKMKKWIDDWQKLPYISPYENAL 262
Query: 263 KIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYPGIFYL 322
+ SD +K VLHE+++L + KK +++ + L E + L +F R+ +PGIFYL
Sbjct: 263 HLSATSDESDKWAAAVLHEIMNLFVSKKVEKDAILHLGEFMGLRSRFKRVLHNHPGIFYL 322
Query: 323 SLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYRGDGSIPKPEGGALLLD 382
S K +T TV LR+GY RG L++ + LV R+++ +M T + K
Sbjct: 323 SSKLRTHTVVLRDGYKRGMLIESNELVTSRNRYMKLMNT---------VKKDNKAVSSSS 373
Query: 383 EIGDEGSEEEEVETSDEFSEG-EVSGSDSE 411
+ D+G E EV +D +E ++SGSD E
Sbjct: 374 KKEDKGKVEGEVCDTDAKAENDDISGSDVE 403
>UniRef100_O82655 Hypothetical protein ORF2 [Arabidopsis thaliana]
Length = 470
Score = 204 bits (518), Expect = 5e-51
Identities = 127/390 (32%), Positives = 209/390 (53%), Gaps = 23/390 (5%)
Query: 29 KWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLTVPVLRFIRR 88
K+V++R LDH +++E +L+ +KD I + + + L + + + FIR
Sbjct: 30 KFVRDRGLDHAVEREKNLRPLLSIKDLIRSEPAKSVPISVITSQKDSLRVPLRPIEFIRS 89
Query: 89 YPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNH--HENDTVERLSKFLMMTKTRT 146
+P++F EF P LT L + EQ ++ ++ +RL K LM+ +
Sbjct: 90 FPSVFQEFLPGGIGIHPHISLTPEILNHDADEQLVYGSETYKQGLADRLLKLLMINRINK 149
Query: 147 VPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAP-NGVSA-VRLSKWCEEFAVSALQ 204
+PL L LKWD+GLP + +T++P+FPD F+ +K+ G S + L W E AVS L+
Sbjct: 150 IPLEILDLLKWDLGLPKDYVETMVPEFPDYFRVIKSKLRGCSGELELVCWSNEHAVSVLE 209
Query: 205 KSNE--RDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYISPYADSS 262
K R G ++ +G +A+ FPM+F G+ KK++ W+D++ KLPYISPY ++
Sbjct: 210 KKARTLRKG------EYTKG-SAIAFPMKFSNGFVVDKKMKKWIDDWQKLPYISPYENAL 262
Query: 263 KIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYPGIFYL 322
+ SD +K VLHE+++L + KK +++ + L E + L +F R+ +PGIFYL
Sbjct: 263 HLSATSDESDKWAAAVLHEIMNLFVSKKVEKDAILHLGEFMGLRSRFKRVLHNHPGIFYL 322
Query: 323 SLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYRGDGSIPKPEGGALLLD 382
S K +T TV LR+GY RG L++ + LV R++++ + K
Sbjct: 323 SSKLRTHTVVLRDGYKRGMLIESNELVTSRNRYHETHEYS---------KKDNKAVSSSS 373
Query: 383 EIGDEGSEEEEVETSDEFSEG-EVSGSDSE 411
+ D+G E EV +D +E ++SGSD E
Sbjct: 374 KKEDKGKVEGEVCDTDAKAENDDISGSDVE 403
>UniRef100_Q9STW2 Hypothetical protein T22A6.150 [Arabidopsis thaliana]
Length = 395
Score = 196 bits (497), Expect = 1e-48
Identities = 123/349 (35%), Positives = 193/349 (55%), Gaps = 15/349 (4%)
Query: 19 QWRGIAKVRLKWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGL 78
Q R R+KWV + LD + +E +LK LKD I S + L S S + L+ L
Sbjct: 14 QRRTFVNARVKWVSDHYLDEAVQREKNLKQVISLKDRIVSSPSKSLPLSSLSLLKPLVNL 73
Query: 79 TVPVLRFIRRYPTLFHEF-PHPRWPSLPCFRLTDTALLLHSQEQSLHNH--HENDTVERL 135
+ F ++YP++F F P P P RLT AL LH +E+++H N TV+RL
Sbjct: 74 HITAAAFFQKYPSVFTTFQPSPSHPLH--VRLTPQALTLHKEEETIHLSPPQRNVTVQRL 131
Query: 136 SKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVS-----AVR 190
+KFLM+T ++PL L ++D+GLP + +LI +P+ F+ + + ++ A+
Sbjct: 132 TKFLMLTGAGSLPLYVLDRFRFDLGLPRDYITSLIGDYPEYFEVTEIKDRLTGEKTLALT 191
Query: 191 LSKWCEEFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFH 250
+S VS +++ R+ DG R K+G + + M FP+GY K+V+ W++++
Sbjct: 192 ISSRRNNLPVSEMER---REATIDGSR-VKKG-LRIRYSMNFPKGYELHKRVKNWVEQWQ 246
Query: 251 KLPYISPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFT 310
LPYISPY ++ + SD EK V VLHELL L + KKT+ + + L E L +F
Sbjct: 247 NLPYISPYENAFHLGSYSDQAEKWAVAVLHELLFLLVSKKTETDNVICLGEYLGFGMRFK 306
Query: 311 RIFTRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVM 359
+ +PGIFY+S K +T TV LRE Y + L++ HPL+ R ++ ++M
Sbjct: 307 KALVHHPGIFYMSHKIRTQTVVLREAYHKVFLLEKHPLMGIRHQYIYLM 355
>UniRef100_Q9ZUZ6 Hypothetical protein At2g39120 [Arabidopsis thaliana]
Length = 387
Score = 194 bits (493), Expect = 4e-48
Identities = 119/351 (33%), Positives = 187/351 (52%), Gaps = 15/351 (4%)
Query: 19 QWRGIAKVRLKWVKNRSLDHI--IDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLL 76
Q R V +KW ++ D+I I + + LK+ LK+ I + + + S +
Sbjct: 20 QKRTYVDVYMKWKRDPYFDNIEHILRSSQLKSVVSLKNCIVQEPNRCIPISAISKKTRQF 79
Query: 77 GLTVPVLRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLS 136
++ + F+R++P++F EF P + +LP FRLT A L QE+ ++ +D +RL
Sbjct: 80 DVSTKIAHFLRKFPSIFEEFVGPEY-NLPWFRLTPEATELDRQERVVYQTSADDLRDRLK 138
Query: 137 KFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCE 196
K ++M+K +PL + +KW +GLPD + + F+FV +GV + +
Sbjct: 139 KLILMSKDNVLPLSIVQGMKWYLGLPDDYLQFPDMNLDSSFRFVDMEDGVKGLAVDYNGG 198
Query: 197 EFAVSALQKSNERDGGDDGYRDFKRGKAALV---FPMRFPRGYGAQKKVRFWMDEFHKLP 253
+ +S LQK+ + +RG+ +L FP+ +G + K+ W+ EF KLP
Sbjct: 199 DKVLSVLQKNAMKK---------RRGEVSLEEIEFPLFPSKGCRLRVKIEDWLMEFQKLP 249
Query: 254 YISPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIF 313
Y+SPY D S +DP+SD+ EKRVVG LHELL L + +R L L++ LP K + F
Sbjct: 250 YVSPYDDYSCLDPSSDIAEKRVVGFLHELLCLFVEHSAERKKLLCLKKHFGLPQKVHKAF 309
Query: 314 TRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLL 364
R+P IFYLS+K KT T LRE Y V+ HP++ R K+ +MK L
Sbjct: 310 ERHPQIFYLSMKNKTCTAILREPYRDKASVETHPVLGVRKKYIQLMKNSEL 360
>UniRef100_Q9LGU2 EST AU030398(E51008) corresponds to a region of the predicted gene
[Oryza sativa]
Length = 498
Score = 184 bits (466), Expect = 5e-45
Identities = 127/410 (30%), Positives = 210/410 (50%), Gaps = 44/410 (10%)
Query: 21 RGIAKVRLKWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLTV 80
R I+ +++ W ++ +LD I ++ + AS L + S L + S ++ + L V
Sbjct: 105 RRISSLKVPWRRDAALDAAILRDRRYRLASRLVREVLLSPGRRLLLRYLSKRRQRIRLPV 164
Query: 81 PVLRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLM 140
V F+RRYPTL P P + P L + L S+ +LH+ RL+K LM
Sbjct: 165 LVPTFLRRYPTLLSVSPPPNPVASPSPHLL-SFLEFASRHHALHSPL---LASRLAKLLM 220
Query: 141 MTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAP----NGVSAVRLSKWCE 196
++ TR +P+ + K D GLPD F +L+P++P F+ V P +G + + L W +
Sbjct: 221 ISSTRALPVPKIAAAKRDFGLPDDFLTSLVPRYPHLFRLVGDPGPDASGNAFLELVSWDD 280
Query: 197 EFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYIS 256
+ A S ++ +++ G R F ++ PRG+ +K++R W+ ++ +LPY+S
Sbjct: 281 QLAKSVIELRADKEADVVGIRPRPN------FTVKLPRGFYLKKEMREWVRDWLELPYVS 334
Query: 257 PYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRY 316
PYAD+S + P S EKR++GVLHE+LSL++ ++ + +E L + F FTR+
Sbjct: 335 PYADTSGLHPASPEAEKRLIGVLHEVLSLSVERRMAVPIIGKFCDEFRLSNAFANAFTRH 394
Query: 317 PGIFYLSLKCKTTTVTLREGY-ARGKLVDPHPLVRHRDKFYHVMKTG------------- 362
PGIFY+SLK T LRE Y G+LVD P++ +++F +M G
Sbjct: 395 PGIFYVSLKGGIKTAVLREAYDENGELVDKDPMIELKERFVAIMDEGHREYLEELRKKRE 454
Query: 363 --------LLYRGDGSIPKPEGGALLLDEIGDEGSEEEEVETSDEFSEGE 404
YRG + G + DE+ +EG++E + + D+ EGE
Sbjct: 455 ELEKQRLQEAYRG------AKVGTGIEDEMEEEGTDESDED--DDSEEGE 496
>UniRef100_Q8L9L1 Hypothetical protein [Arabidopsis thaliana]
Length = 404
Score = 166 bits (420), Expect = 1e-39
Identities = 111/356 (31%), Positives = 179/356 (50%), Gaps = 12/356 (3%)
Query: 18 LQWRGIAKVRLKWVKNRSLDHIIDKETDLK-AASLLKDAINRSSTGFLTAKSFSGWQKLL 76
+Q R I+ +++ W K+ LD I+++ K A ++K+ +N + + ++ L
Sbjct: 26 IQIRWISSLKVVWKKDTRLDEAIEQDKRYKLCARVVKEVLNEPGQ-VIPLRYLEKRRERL 84
Query: 77 GLTVPVLRFIRRYPTLF---HEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVE 133
LT F+ P+LF ++ P+ + R T +EQ +++ +E V
Sbjct: 85 RLTFKAKSFVEMNPSLFEIYYDRIKPKSDPVQFLRPTPRLRAFLDEEQRIYSENEPLLVS 144
Query: 134 RLSKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPN-GVSAVRLS 192
+L K LMM K + + L +K D G P+ F L+ K+P+ F+ P G S + L
Sbjct: 145 KLCKLLMMAKDKVISADKLVHVKRDFGFPNDFLMKLVQKYPNYFRLTGLPEEGKSFLELV 204
Query: 193 KWCEEFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKL 252
W +FA S ++ E + G R + ++ P G+ +K++R W ++ +
Sbjct: 205 SWNPDFAKSKIELRAEDETLKTGVRIRPN------YNVKLPSGFFLRKEMREWTRDWLEQ 258
Query: 253 PYISPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRI 312
YISPY D S +D S MEKR VGV+HELLSL++ K+ L +E + F+ +
Sbjct: 259 DYISPYEDVSHLDQASKEMEKRTVGVVHELLSLSLLKRVPVPTLGKFCDEFRFSNAFSSV 318
Query: 313 FTRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYRGD 368
FTR+ GIFYLSLK T LRE Y +LVD PL+ +DKF +++ G R D
Sbjct: 319 FTRHSGIFYLSLKGGIKTAVLREAYKDDELVDRDPLLAIKDKFLRLLEEGWQERKD 374
>UniRef100_Q9LYB3 Hypothetical protein T20O10_190 [Arabidopsis thaliana]
Length = 404
Score = 164 bits (416), Expect = 3e-39
Identities = 110/356 (30%), Positives = 179/356 (49%), Gaps = 12/356 (3%)
Query: 18 LQWRGIAKVRLKWVKNRSLDHIIDKETDLK-AASLLKDAINRSSTGFLTAKSFSGWQKLL 76
+Q R I+ +++ W K+ LD I+++ K A ++K+ +N + + ++ L
Sbjct: 26 IQIRWISSLKVVWKKDTRLDEAIEQDKRYKLCARVVKEVLNEPGQ-VIPLRYLEKRRERL 84
Query: 77 GLTVPVLRFIRRYPTLF---HEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVE 133
LT F+ P+LF ++ P+ + R T +EQ +++ +E V
Sbjct: 85 RLTFKAKSFVEMNPSLFEIYYDRIKPKSDPVQFLRPTPRLRAFLDEEQRIYSENEPLLVS 144
Query: 134 RLSKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPN-GVSAVRLS 192
+L + LMM K + + L +K D G P+ F L+ K+P+ F+ P G S + L
Sbjct: 145 KLCQLLMMAKDKVISADKLVHVKRDFGFPNDFLMKLVQKYPNYFRLTGLPEEGKSFLELV 204
Query: 193 KWCEEFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKL 252
W +FA S ++ E + G R + ++ P G+ +K++R W ++ +
Sbjct: 205 SWNPDFAKSKIELRAEDETLKTGVRIRPN------YNVKLPSGFFLRKEMREWTRDWLEQ 258
Query: 253 PYISPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRI 312
YISPY D S +D S MEKR VGV+HELLSL++ K+ L +E + F+ +
Sbjct: 259 DYISPYEDVSHLDQASKEMEKRTVGVVHELLSLSLLKRVPVPTLGKFCDEFRFSNAFSSV 318
Query: 313 FTRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYRGD 368
FTR+ GIFYLSLK T LRE Y +LVD PL+ +DKF +++ G R D
Sbjct: 319 FTRHSGIFYLSLKGGIKTAVLREAYKDDELVDRDPLLAIKDKFLRLLEEGWQERKD 374
>UniRef100_O04624 A_IG002N01.30 protein [Arabidopsis thaliana]
Length = 878
Score = 164 bits (414), Expect = 6e-39
Identities = 114/400 (28%), Positives = 190/400 (47%), Gaps = 20/400 (5%)
Query: 25 KVRLKWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLTVP--V 82
+ +K K + D ++ ++ LK ++ + ++ + +++ LGL
Sbjct: 127 RAAVKRRKELTFDSVVQRDKKLKLVLNIRKILVSQPDRMMSLRGLGKYRRDLGLKKRRRF 186
Query: 83 LRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLMMT 142
+ +R+YP +F + F++T A L+ E + N E+ V +L K +MM+
Sbjct: 187 IALLRKYPGVFEIVEEGAYSLR--FKMTSEAERLYLDEMRIRNELEDVLVVKLRKLVMMS 244
Query: 143 KTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCEEFAVSA 202
+ + L + LK D+GLP F T+ ++P F+ V P G A+ L+ W E AVSA
Sbjct: 245 IDKRILLEKISHLKTDLGLPLEFRDTICQRYPQYFRVVPTPRG-PALELTHWDPELAVSA 303
Query: 203 LQKSNERDGGDDGYRDFKRGKAALVFPMRF-----PRGYGAQKKVRFWMDEFHKLPYISP 257
+ S + D+ R+ + + P +F PRG K + +F + YISP
Sbjct: 304 AELSED----DNRTRESEERNLIIDRPPKFNRVKLPRGLNLSKSETRKISQFRDMQYISP 359
Query: 258 YADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYP 317
Y D S + + EK GV+HELLSLT K+T ++L REE + + R+P
Sbjct: 360 YKDFSHLRSGTLEKEKHACGVIHELLSLTTEKRTLVDHLTHFREEFRFSQQLRGMLIRHP 419
Query: 318 GIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYRGDGSIPKPEGG 377
+FY+SLK + +V LRE Y +L+D PL ++K ++ R G EG
Sbjct: 420 DLFYVSLKGERDSVFLREAYRNSELIDKDPLTLVKEKMRALVSVPRFPRRGGPRKDEEGR 479
Query: 378 ALLLDEIGDEGSEEEEVETSDEFS------EGEVSGSDSE 411
+ +D +G EEEE ++E+S EGE G+D +
Sbjct: 480 EVEIDGSDADGEEEEEESDAEEWSDVDGYLEGEDGGNDDD 519
>UniRef100_Q9FK64 Tyrosine-specific protein phosphatase-like protein [Arabidopsis
thaliana]
Length = 423
Score = 154 bits (389), Expect = 4e-36
Identities = 95/329 (28%), Positives = 178/329 (53%), Gaps = 13/329 (3%)
Query: 31 VKNRSLDHIIDKET-DLKAASLLKDAINRSSTG----FLTAKSFSGWQKLLGLTVPVLRF 85
++NR+ DH +DK ++ +++ + S+ F++ + S W+ L+GL V V F
Sbjct: 49 LENRTRDHQLDKIIPQIRKLNIILEISKLMSSKKRGPFVSLQLMSRWKNLVGLNVNVGAF 108
Query: 86 IRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLMMTKTR 145
I +YP F F HP +L C ++T+ +L +E+++ E D V+R+ K L+++K
Sbjct: 109 IGKYPHAFEIFTHPFSKNL-CCKITEKFKVLIDEEENVVRECEVDAVKRVKKLLLLSKHG 167
Query: 146 TVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCEEFAVSALQK 205
+ + +L ++ ++GLP+ F +++ K+ +F+ V + + + E V+ +++
Sbjct: 168 VLRVHALRLIRKELGLPEDFRDSILAKYSSEFRLVDLE---TLELVDRDDESLCVAKVEE 224
Query: 206 SNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYISPYADSSKID 265
E + + F+ A FP+ P G+ +K R + + ++PY+ PY D +I
Sbjct: 225 WREVEYREKWLSKFETNYA---FPIHLPTGFKIEKGFREELKNWQRVPYVKPY-DRKEIS 280
Query: 266 PNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYPGIFYLSLK 325
+ EKRVV V+HELLSLT+ K + L R++L + + ++PGIFY+S K
Sbjct: 281 RGLERFEKRVVAVIHELLSLTVEKMVEVERLAHFRKDLGIEVNVREVILKHPGIFYVSTK 340
Query: 326 CKTTTVTLREGYARGKLVDPHPLVRHRDK 354
+ T+ LRE Y++G L++P+P+ R K
Sbjct: 341 GSSQTLFLREAYSKGCLIEPNPIYNVRRK 369
>UniRef100_Q65XL5 Hypothetical protein OJ1735_C10.3 [Oryza sativa]
Length = 509
Score = 153 bits (386), Expect = 1e-35
Identities = 112/401 (27%), Positives = 192/401 (46%), Gaps = 19/401 (4%)
Query: 23 IAKVRLKWVKNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLTVP- 81
+A+ +K K D++I ++ LK L++ + + ++ + +++ LGLT
Sbjct: 56 VAQAAVKRRKEIPFDNVIQRDKKLKLVLKLRNILVSNPDRVMSLRDLGRFRRDLGLTRKR 115
Query: 82 -VLRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLM 140
++ ++R+P +F + FRLT A L+ E L N E V +L K LM
Sbjct: 116 RLIALLKRFPGVFEVVEEGVYSLR--FRLTPAAERLYLDELHLKNESEGLAVTKLRKLLM 173
Query: 141 MTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCEEFAV 200
M++ + + + + LK D+GLP F T+ ++P F+ V+ G + L+ W E AV
Sbjct: 174 MSQDKRILIEKIAHLKNDLGLPPEFRDTICLRYPQYFRVVQMDRG-PGLELTHWDPELAV 232
Query: 201 SALQKSNERDGGDDGYRDFKRGKAALVFPMRF-----PRGYGAQKKVRFWMDEFHKLPYI 255
SA + + E + R+ + + P++F P+G + + +F ++PYI
Sbjct: 233 SAAEVAEEENRA----REEQERNLIIDRPLKFNRVKLPQGLKLSRGEARRVAQFKEMPYI 288
Query: 256 SPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTR 315
SPY+D S + S EK GV+HE+LSLT+ K+T ++L REE + R
Sbjct: 289 SPYSDFSHLRSGSAEKEKHACGVVHEILSLTLEKRTLVDHLTHFREEFRFSQSLRGMLIR 348
Query: 316 YPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYR-----GDGS 370
+P +FY+SLK +V LRE Y +LV+ LV ++K ++ R
Sbjct: 349 HPDMFYVSLKGDRDSVFLREAYKNSQLVEKSKLVLLKEKMRALVAVPRFPRRGVPATSEE 408
Query: 371 IPKPEGGALLLDEIGDEGSEEEEVETSDEFSEGEVSGSDSE 411
+ G A +L E D +E+E + E E+SG S+
Sbjct: 409 ADRTNGAAQMLSEGSDVEDDEDEGLSDMEDLISEISGGKSD 449
>UniRef100_Q6K4X2 Tyrosine-specific protein phosphatase-like [Oryza sativa]
Length = 402
Score = 151 bits (382), Expect = 3e-35
Identities = 101/381 (26%), Positives = 190/381 (49%), Gaps = 20/381 (5%)
Query: 17 KLQWRGIAKVRLKWVKNRSLDHIIDKET----DLKAASLLKDAINRSSTGFLTAKSFSGW 72
K +W+ ++ R+ DH +DK +L+ A L + I++ + + + S W
Sbjct: 35 KRRWKKPVDSARTRLEGRTRDHKLDKLMIQLKNLRLALDLHELISQQRNKYASLQLLSRW 94
Query: 73 QKLLGLTVPVLRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTV 132
+ +GL + + F+++YP +F + HP + C ++T L +QE + +E
Sbjct: 95 RHEVGLNIEIGAFLKKYPHIFDIYVHPIKRN-ECCKVTPKMSELIAQEDAAIRENEPAIA 153
Query: 133 ERLSKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLS 192
+RL K L ++K T+ + +L+ ++ ++GLPD + +++P F + +P+ ++ +
Sbjct: 154 KRLKKLLTLSKDGTLNMHALWLVRRELGLPDDYRCSILPNHQSDFS-LSSPDTLTLITRD 212
Query: 193 KWCEEFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKL 252
E AV+ +++ ++ + + + FP+ FP G+ +K R + + +L
Sbjct: 213 ---ENLAVADVEEWRAKEYTEKWLAE---SETKYAFPINFPTGFKIEKGFRGKLGNWQRL 266
Query: 253 PYISPYADSSKIDP--NSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFT 310
PY Y + +++ P N + +EKR+VG+LHELLSLT+ K L R+ +
Sbjct: 267 PYTKAY-EKNELHPIRNIERLEKRIVGILHELLSLTIEKMIPLERLSHFRKPFEMEVNLR 325
Query: 311 RIFTRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGLLYRGDGS 370
+ ++PGIFY+S K T TV LRE Y++G LV P+P+ R K GL+ G
Sbjct: 326 ELILKHPGIFYISTKGSTQTVLLRESYSKGCLVHPNPVYNVRRKM-----LGLILSGCCG 380
Query: 371 IPKPEGGALLLDEIGDEGSEE 391
+ + + +E E S E
Sbjct: 381 VDEMDSPLWFSEEYNQESSNE 401
>UniRef100_Q9C590 Hypothetical protein At5g21970 [Arabidopsis thaliana]
Length = 449
Score = 150 bits (380), Expect = 5e-35
Identities = 111/366 (30%), Positives = 177/366 (48%), Gaps = 27/366 (7%)
Query: 64 LTAKSFSGWQKLLGLTVP--VLRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQ 121
+T +SF +++ + L P + FIR+ P LF + R L C LT+ L +
Sbjct: 89 MTVRSFEQYRRQINLPKPHKISDFIRKSPKLFELYKDQRGV-LWC-GLTEGGEDLLDEHD 146
Query: 122 SLHNHHENDTVERLSKFLMMTKTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVK 181
L + + E +++ LMM+ + +PL + + D GLP F + FP F+ VK
Sbjct: 147 KLLEENGDKAAEHVTRCLMMSVDKKLPLDKIVHFRRDFGLPLDFRINWVYNFPQHFKVVK 206
Query: 182 APNGVSAVRLSKWCEEFAVSALQKSNERDGGDDGYRDFKRGKAALVFPMRFPRGYGAQKK 241
+G + L W +A++ L+K D + K G +L FPM+FP Y +
Sbjct: 207 LGDGEEYLELVSWNPAWAITELEKKTLGITEDC---EHKPGMLSLAFPMKFPPSYKKMYR 263
Query: 242 VRFWMDEFHKLPYISPYADSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLRE 301
R ++ F K Y+SPYAD+ ++ S +KR + V+HELLS T+ K+ ++L R
Sbjct: 264 YRGKIEHFQKRSYLSPYADARGLEAGSKEFDKRAIAVMHELLSFTLEKRLVTDHLTHFRR 323
Query: 302 ELNLPHKFTRIFTRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMKT 361
E +P K RIF ++ GIFY+S + K +V L EGY +L++ PL+ ++K + T
Sbjct: 324 EFVMPQKLMRIFLKHCGIFYVSERGKRFSVFLTEGYEGPELIEKCPLILWKEKL--LKFT 381
Query: 362 GLLYRGDGSIPKPEGGALLLDE-------IGDE---------GSEEEEVETSDEFSEGEV 405
G YRG + L ++E GDE G ++ V DE GEV
Sbjct: 382 G--YRGRKRDIQTYSDTLDMEERELLESGSGDEDLSVGFEKDGDYDDVVTDDDEMDIGEV 439
Query: 406 SGSDSE 411
+ + E
Sbjct: 440 NDAYEE 445
>UniRef100_Q9SJW7 Hypothetical protein At2g31290 [Arabidopsis thaliana]
Length = 415
Score = 143 bits (361), Expect = 8e-33
Identities = 103/334 (30%), Positives = 160/334 (47%), Gaps = 20/334 (5%)
Query: 32 KNRSLDHIIDKETDLKAASLLKDAINRSSTGFLTAKSFSGWQKLLGLTVPVLRFIRRYPT 91
K+ L+ + + S LK+ I R + K K L L L ++++YP
Sbjct: 30 KDPDLESALSRNKRWIVNSRLKNIILRCPNQVASIKFLQKKFKTLDLQGKALNWLKKYPC 89
Query: 92 LFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLMMTKTRTVPLVS 151
FH + C RLT + L +E+ + + E +RL+K LM++ + + +V
Sbjct: 90 CFHVYLEN--DEYYC-RLTKPMMTLVEEEELVKDTQEPVLADRLAKLLMLSVNQRLNVVK 146
Query: 152 LYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSA--VRLSKWCEEFAVSALQKSNER 209
L K G PD + ++PK+ D F+ V S+ + L W E AVSA++ + ++
Sbjct: 147 LNEFKRSFGFPDDYVIRIVPKYSDVFRLVNYSGRKSSMEIELLLWKPELAVSAVEAAAKK 206
Query: 210 DGGDDGYRDFKRGKAALVFPMRFPRGYGAQKKVRFWMDEFHKLPYISPYADSSKIDPNSD 269
G + + + P + + + RF EF+ PYISPY D + S
Sbjct: 207 CGSEPSF--------SCSLPSTWTKPWE-----RFM--EFNAFPYISPYEDHGDLVEGSK 251
Query: 270 LMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYPGIFYLSLKCKTT 329
EKR VG++HELLSLT+ KK L + E LP K + ++PGIFY+ K +
Sbjct: 252 ESEKRSVGLVHELLSLTLWKKLSIVKLSHFKREFGLPEKLNGMLLKHPGIFYVGNKYQVH 311
Query: 330 TVTLREGYARGKLVDPHPLVRHRDKFYHVMKTGL 363
TV LREGY +L+ PLV +DKF +M+ GL
Sbjct: 312 TVILREGYNGSELIHKDPLVVVKDKFGELMQQGL 345
>UniRef100_Q7XSD5 OSJNBa0079A21.9 protein [Oryza sativa]
Length = 328
Score = 132 bits (333), Expect = 1e-29
Identities = 80/182 (43%), Positives = 115/182 (62%), Gaps = 5/182 (2%)
Query: 25 KVRLKWVKNRSLDHIIDKETDLKAASLLKDAINR-SSTGFLTAKSFSGWQKLLGLTVPVL 83
KVRLKWVKNR LDHII + T ++A+ LL D + R S+ + A+S + QK LGLTVPVL
Sbjct: 3 KVRLKWVKNRGLDHIIARTTSIRASCLLLDHLARLPSSSPVPARSLARLQKPLGLTVPVL 62
Query: 84 RFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLMMTK 143
RF+RR+PTLF E HPR+P+LP F LT + +L + L D+ RL++ L++T+
Sbjct: 63 RFLRRHPTLFAETLHPRFPTLPSFSLTPASDILLGR---LARASALDSHLRLARLLLLTR 119
Query: 144 TRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCEEFAVSAL 203
++++PL S+ PL++D+GLP +F PD F +SA S E A+S+L
Sbjct: 120 SKSLPLASVLPLRFDLGLPYNFAAAFPVAHPDLFAVSNNHISLSAT-ASGLPEGIAISSL 178
Query: 204 QK 205
Q+
Sbjct: 179 QR 180
Score = 128 bits (321), Expect = 3e-28
Identities = 68/114 (59%), Positives = 79/114 (68%), Gaps = 11/114 (9%)
Query: 301 EELNLPHKFTRIFTRYPGIFYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKFYHVMK 360
EEL LPHKFTR+FTRYPG+FYLSLKCKTTTV LREGY RGKLV+ HPL RDK ++VM+
Sbjct: 202 EELGLPHKFTRLFTRYPGVFYLSLKCKTTTVVLREGYERGKLVEQHPLAAVRDKVFYVMR 261
Query: 361 TGLLYRGDGSIPKPEGGALLLDEIGDE-----GSEEEEVETSDEFSEGEVSGSD 409
TG+L+RG G L+LDE GDE G EE E DE ++ E G D
Sbjct: 262 TGVLFRGKGL------SKLVLDEDGDEEVVMDGDEEFHGEGMDEDADVECFGMD 309
>UniRef100_Q75HB0 Expressed protein [Oryza sativa]
Length = 360
Score = 131 bits (329), Expect = 4e-29
Identities = 82/276 (29%), Positives = 136/276 (48%), Gaps = 4/276 (1%)
Query: 83 LRFIRRYPTLFHEFPHPRWPSLPCFRLTDTALLLHSQEQSLHNHHENDTVERLSKFLMMT 142
L I R+P +FH LT+ A + S+E E V+ L K LMM+
Sbjct: 69 LSLIERHPNIFHVSGGSASREPISVTLTEKARKISSEEIQARELMEPILVKNLRKLLMMS 128
Query: 143 KTRTVPLVSLYPLKWDIGLPDSFEKTLIPKFPDQFQFVKAPNGVSAVRLSKWCEEFAVSA 202
+PL + ++ ++GLP++F+ LIPK+P+ F V+ G+ + L W AV+A
Sbjct: 129 LDCQIPLEKIELIQSELGLPNNFKNNLIPKYPELFS-VRDVKGLDHLCLESWDSSLAVTA 187
Query: 203 LQKSNERDGGDDGYRDFKRGK---AALVFPMRFPRGYGAQKKVRFWMDEFHKLPYISPYA 259
++ +G YR + F +++P G+ + + + KL + SPY
Sbjct: 188 REEKLNFEGFQMDYRGIPKDGNIVGPFAFRLKYPAGFRPNRNYLEEVVRWQKLAFPSPYL 247
Query: 260 DSSKIDPNSDLMEKRVVGVLHELLSLTMHKKTKRNYLRSLREELNLPHKFTRIFTRYPGI 319
++ +++P + KR V VLHE+LSLTM+++ + L E LP K + GI
Sbjct: 248 NARRVEPATPQARKRAVAVLHEILSLTMNRRLTSDKLEIFHNEYRLPCKLLLCLIKNHGI 307
Query: 320 FYLSLKCKTTTVTLREGYARGKLVDPHPLVRHRDKF 355
FY++ K +TV L+E Y LV+ PL++ D+F
Sbjct: 308 FYITNKGARSTVFLKEAYDDSNLVEKCPLLKFHDRF 343
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 715,320,813
Number of Sequences: 2790947
Number of extensions: 31204684
Number of successful extensions: 81787
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 81646
Number of HSP's gapped (non-prelim): 70
length of query: 411
length of database: 848,049,833
effective HSP length: 130
effective length of query: 281
effective length of database: 485,226,723
effective search space: 136348709163
effective search space used: 136348709163
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0029a.7


