

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0029a.6
(183 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
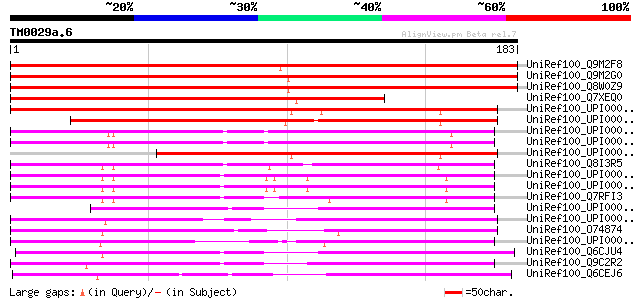
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9M2F8 Hypothetical protein F14P22.170 [Arabidopsis th... 308 4e-83
UniRef100_Q9M2G0 Hypothetical protein F14P22.150 [Arabidopsis th... 296 2e-79
UniRef100_Q8W0Z9 AT3g58560/F14P22_150 [Arabidopsis thaliana] 296 2e-79
UniRef100_Q7XEQ0 Putative chromaffin granule ATPase II homolog [... 228 6e-59
UniRef100_UPI0000282600 UPI0000282600 UniRef100 entry 207 1e-52
UniRef100_UPI000025A86B UPI000025A86B UniRef100 entry 167 1e-40
UniRef100_UPI000045354E UPI000045354E UniRef100 entry 125 5e-28
UniRef100_UPI000042EACA UPI000042EACA UniRef100 entry 125 5e-28
UniRef100_UPI0000325EDE UPI0000325EDE UniRef100 entry 119 5e-26
UniRef100_Q8I3R5 Hypothetical protein PFE0980c [Plasmodium falci... 104 1e-21
UniRef100_UPI000046B111 UPI000046B111 UniRef100 entry 101 8e-21
UniRef100_UPI0000468B91 UPI0000468B91 UniRef100 entry 100 2e-20
UniRef100_Q7RFI3 Arabidopsis thaliana At3g58560/F14P22_150-relat... 96 4e-19
UniRef100_UPI000023456F UPI000023456F UniRef100 entry 86 5e-16
UniRef100_UPI00003AA88A UPI00003AA88A UniRef100 entry 84 2e-15
UniRef100_O74874 SPCC31H12.08c protein [Schizosaccharomyces pombe] 80 2e-14
UniRef100_UPI00003C1BD9 UPI00003C1BD9 UniRef100 entry 80 3e-14
UniRef100_Q6CJU4 Similar to sp|P31384 Saccharomyces cerevisiae Y... 80 3e-14
UniRef100_Q9C2R2 Related to CCR4 protein [Neurospora crassa] 80 3e-14
UniRef100_Q6CEJ6 Yarrowia lipolytica chromosome B of strain CLIB... 78 1e-13
>UniRef100_Q9M2F8 Hypothetical protein F14P22.170 [Arabidopsis thaliana]
Length = 605
Score = 308 bits (789), Expect = 4e-83
Identities = 152/185 (82%), Positives = 163/185 (87%), Gaps = 2/185 (1%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
ANTHVNV QDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFN+ PGSAPH LL MGKV
Sbjct: 420 ANTHVNVQQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNTLPGSAPHTLLVMGKV 479
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFAR--TIGLGYEQHKRRLDSATNEPLF 118
DP HPDLA+DPL+ILRPH+KL HQLPLVSAYSSF R +GLG EQH+RR+D TNEPLF
Sbjct: 480 DPMHPDLAVDPLNILRPHTKLTHQLPLVSAYSSFVRKGIMGLGLEQHRRRIDLNTNEPLF 539
Query: 119 TNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCC 178
TN TRDFIG DYIFYTAD+L+VESLLELLDE+ LRKDTALPSPEWSS+HIALLAEFRC
Sbjct: 540 TNCTRDFIGTHDYIFYTADTLMVESLLELLDEDGLRKDTALPSPEWSSNHIALLAEFRCT 599
Query: 179 KNRSR 183
R
Sbjct: 600 PRTRR 604
>UniRef100_Q9M2G0 Hypothetical protein F14P22.150 [Arabidopsis thaliana]
Length = 597
Score = 296 bits (757), Expect = 2e-79
Identities = 149/184 (80%), Positives = 159/184 (85%), Gaps = 1/184 (0%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
ANTHVNV +LKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFN+ P SAPH LLA+GKV
Sbjct: 410 ANTHVNVPHELKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNTVPASAPHTLLAVGKV 469
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIG-LGYEQHKRRLDSATNEPLFT 119
DP HPDL +DPL ILRPHSKL HQLPLVSAYS FA+ G + EQ +RRLD A++EPLFT
Sbjct: 470 DPLHPDLMVDPLGILRPHSKLTHQLPLVSAYSQFAKMGGNVITEQQRRRLDPASSEPLFT 529
Query: 120 NVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCK 179
N TRDFIG LDYIFYTAD+L VESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRC
Sbjct: 530 NCTRDFIGTLDYIFYTADTLTVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCMP 589
Query: 180 NRSR 183
R
Sbjct: 590 RARR 593
>UniRef100_Q8W0Z9 AT3g58560/F14P22_150 [Arabidopsis thaliana]
Length = 602
Score = 296 bits (757), Expect = 2e-79
Identities = 149/184 (80%), Positives = 159/184 (85%), Gaps = 1/184 (0%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
ANTHVNV +LKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFN+ P SAPH LLA+GKV
Sbjct: 415 ANTHVNVPHELKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNTVPASAPHTLLAVGKV 474
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIG-LGYEQHKRRLDSATNEPLFT 119
DP HPDL +DPL ILRPHSKL HQLPLVSAYS FA+ G + EQ +RRLD A++EPLFT
Sbjct: 475 DPLHPDLMVDPLGILRPHSKLTHQLPLVSAYSQFAKMGGNVITEQQRRRLDPASSEPLFT 534
Query: 120 NVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCK 179
N TRDFIG LDYIFYTAD+L VESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRC
Sbjct: 535 NCTRDFIGTLDYIFYTADTLTVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCMP 594
Query: 180 NRSR 183
R
Sbjct: 595 RARR 598
>UniRef100_Q7XEQ0 Putative chromaffin granule ATPase II homolog [Oryza sativa]
Length = 622
Score = 228 bits (581), Expect = 6e-59
Identities = 112/137 (81%), Positives = 119/137 (86%), Gaps = 2/137 (1%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
ANTHVNVHQDLKDVKLW+V TLLKGLEKIA SADIPMLVCGDFNS PGS+PH LLAMGKV
Sbjct: 421 ANTHVNVHQDLKDVKLWEVQTLLKGLEKIAVSADIPMLVCGDFNSVPGSSPHGLLAMGKV 480
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGY--EQHKRRLDSATNEPLF 118
D HPDLAIDPL ILRP SKL HQLPLVSAYSSFAR +G+GY E +RR+D ATNEPLF
Sbjct: 481 DQLHPDLAIDPLGILRPASKLTHQLPLVSAYSSFARMVGVGYDLEHQRRRMDPATNEPLF 540
Query: 119 TNVTRDFIGALDYIFYT 135
TN TRDF G +DYIFYT
Sbjct: 541 TNCTRDFTGTVDYIFYT 557
>UniRef100_UPI0000282600 UPI0000282600 UniRef100 entry
Length = 210
Score = 207 bits (527), Expect = 1e-52
Identities = 109/181 (60%), Positives = 135/181 (74%), Gaps = 5/181 (2%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKV 60
ANTH++ + +L DVKLWQVHTLLKGLEKIAASA+IPM+VCGDFNS PGSA H LL+ G+V
Sbjct: 23 ANTHIHANTELNDVKLWQVHTLLKGLEKIAASAEIPMVVCGDFNSVPGSAAHNLLSNGRV 82
Query: 61 DPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGL---GYEQHKRRLDS-ATNEP 116
D +HP+LA DP ILRP SKL H LPLVSAY++ + L E+ + R+D+ T EP
Sbjct: 83 DGAHPELATDPFGILRPPSKLQHPLPLVSAYTALTKQPCLESEAAERQRTRMDAQGTGEP 142
Query: 117 LFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLR-KDTALPSPEWSSDHIALLAEF 175
+FTN T+DF G LDYIFYT D+L SLLEL E+ R K LP+ + SSDH+AL+AEF
Sbjct: 143 IFTNCTKDFFGTLDYIFYTDDTLAPLSLLELPSEKECRNKYGGLPNTQCSSDHVALMAEF 202
Query: 176 R 176
+
Sbjct: 203 Q 203
>UniRef100_UPI000025A86B UPI000025A86B UniRef100 entry
Length = 171
Score = 167 bits (423), Expect = 1e-40
Identities = 88/157 (56%), Positives = 111/157 (70%), Gaps = 4/157 (2%)
Query: 23 LKGLEKIAASADIPMLVCGDFNSNPGSAPHALLAMGKVDPSHPDLAIDPLSILRPHSKLV 82
LKGLEKIA SADIPM+VCGDFNS PGSA H+LL+ G+V HP+L IDP IL+P +KL
Sbjct: 1 LKGLEKIATSADIPMVVCGDFNSVPGSAAHSLLSTGRVPNDHPELGIDPFGILQPSTKLS 60
Query: 83 HQLPLVSAYSSFARTI--GLGYEQHKRRLDSATNEPLFTNVTRDFIGALDYIFYTADSLV 140
H LPLVSAY++ + E+ + R+D EPLFTN T+DF GALDY+FYT D+L
Sbjct: 61 HPLPLVSAYTNLHKPCLDSDALERQRDRVD-VIGEPLFTNCTKDFNGALDYVFYTEDTLA 119
Query: 141 VESLLELLDEESLR-KDTALPSPEWSSDHIALLAEFR 176
S+LEL E +R K LP+ +WSSDH+ L+ EF+
Sbjct: 120 PVSVLELPGEREVRAKYGGLPNTQWSSDHVCLMTEFQ 156
>UniRef100_UPI000045354E UPI000045354E UniRef100 entry
Length = 783
Score = 125 bits (314), Expect = 5e-28
Identities = 75/189 (39%), Positives = 112/189 (58%), Gaps = 16/189 (8%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASAD-----IP-MLVCGDFNSNPGSAPHAL 54
ANTH+ + + DVK+WQ TL+ LE+ +P +++CGDFNS P SA + L
Sbjct: 593 ANTHIVANPEANDVKIWQAQTLVSVLEEYLHDCYRRQPVLPGLIICGDFNSTPDSALYRL 652
Query: 55 LAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATN 114
LA G + +H DLA+D +L +L H + L SAYS A+ + G+ + + +
Sbjct: 653 LATGTCEKTHKDLAMDRYGLLSD-LQLGHSMRLRSAYS-MAKAMVEGHNPNMLVSSTESL 710
Query: 115 EPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTA--------LPSPEWSS 166
EP+FTN T +++G LDY+FYT + L + +LELLDEE+L ++ A LP+P+ S
Sbjct: 711 EPVFTNYTPNYLGCLDYVFYTDERLRLGGVLELLDEEALIREAAALQLPDWSLPNPQRPS 770
Query: 167 DHIALLAEF 175
DH+ LL EF
Sbjct: 771 DHLPLLTEF 779
>UniRef100_UPI000042EACA UPI000042EACA UniRef100 entry
Length = 782
Score = 125 bits (314), Expect = 5e-28
Identities = 75/189 (39%), Positives = 112/189 (58%), Gaps = 16/189 (8%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASAD-----IP-MLVCGDFNSNPGSAPHAL 54
ANTH+ + + DVK+WQ TL+ LE+ +P +++CGDFNS P SA + L
Sbjct: 592 ANTHIVANPEANDVKIWQAQTLVSVLEEYLHDCYRRQPVLPGLIICGDFNSTPDSALYRL 651
Query: 55 LAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATN 114
LA G + +H DLA+D +L +L H + L SAYS A+ + G+ + + +
Sbjct: 652 LATGTCEKTHKDLAMDRYGLLSD-LQLGHSMRLRSAYS-MAKAMVEGHNPNMLVSSTESL 709
Query: 115 EPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTA--------LPSPEWSS 166
EP+FTN T +++G LDY+FYT + L + +LELLDEE+L ++ A LP+P+ S
Sbjct: 710 EPVFTNYTPNYLGCLDYVFYTDERLRLGGVLELLDEEALIREAAALQLPDWSLPNPQRPS 769
Query: 167 DHIALLAEF 175
DH+ LL EF
Sbjct: 770 DHLPLLTEF 778
>UniRef100_UPI0000325EDE UPI0000325EDE UniRef100 entry
Length = 144
Score = 119 bits (297), Expect = 5e-26
Identities = 67/129 (51%), Positives = 85/129 (64%), Gaps = 6/129 (4%)
Query: 54 LLAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGL-----GYEQHKRR 108
LL G VD +HP+L IDPL IL P SKL H LPLVSAYSS R E+ + R
Sbjct: 7 LLTRGMVDNNHPELQIDPLGILHPTSKLSHPLPLVSAYSSALRRDNRLLESEALERLRDR 66
Query: 109 LDSATNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLR-KDTALPSPEWSSD 167
+D EP+FTN T+DF G+LDY+FYT D+L +LLEL E+ +R K LP+ + SSD
Sbjct: 67 VDPRMAEPIFTNCTKDFFGSLDYVFYTEDTLCPVALLELPGEKDVRAKYGGLPNTQLSSD 126
Query: 168 HIALLAEFR 176
HI+L+AEF+
Sbjct: 127 HISLMAEFQ 135
>UniRef100_Q8I3R5 Hypothetical protein PFE0980c [Plasmodium falciparum]
Length = 2488
Score = 104 bits (259), Expect = 1e-21
Identities = 72/190 (37%), Positives = 101/190 (52%), Gaps = 19/190 (10%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAAS-----ADIP-MLVCGDFNSNPGSAPHAL 54
ANTH+ + + VK+WQ L+K +E + + IP +++CGDFNS P SA + L
Sbjct: 2298 ANTHIVANPEANYVKIWQTQILVKVIEYLKINFIKKYETIPSLIICGDFNSTPSSAVYQL 2357
Query: 55 LAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYS-SFARTIGLGYEQHKRRLDSAT 113
+ +H D D S+L KL H L L SAY+ S + L E++ +
Sbjct: 2358 IYKKTCSRTHEDFNSDKYSLLTD-LKLGHNLNLKSAYAISKLLSQKLNPEEYN---NLEL 2413
Query: 114 NEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESL--------RKDTALPSPEWS 165
EPLFTN T +FIG LDYIFY ++L + S + + DE L D ALPSP
Sbjct: 2414 YEPLFTNYTSNFIGCLDYIFYNDENLNIISTVNVADENQLIQEAQMYQLSDCALPSPIRP 2473
Query: 166 SDHIALLAEF 175
SDH+ L+A+F
Sbjct: 2474 SDHLPLIAQF 2483
>UniRef100_UPI000046B111 UPI000046B111 UniRef100 entry
Length = 958
Score = 101 bits (252), Expect = 8e-21
Identities = 73/196 (37%), Positives = 105/196 (53%), Gaps = 22/196 (11%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAAS-----ADIP-MLVCGDFNSNPGSAPHAL 54
ANTH+ + + VK+WQ L+K +E + + IP +++CGDFNS P SA + L
Sbjct: 759 ANTHIIANPEATYVKIWQTQILVKVIEYLKINFIQKYEIIPSIIICGDFNSTPNSAVYQL 818
Query: 55 LAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAY--SSF-ARTIGLGYEQHK----R 107
L K P+H D+ D +L + + H L L SAY S+F ++TI +
Sbjct: 819 LYKKKCFPTHHDINSDEHGLLE-YLPMSHNLNLKSAYAISNFLSQTINTEESINNIIINN 877
Query: 108 RLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKD--------TAL 159
+D EP FTN T +FIG LDYIFY + L + S + + DE L ++ +AL
Sbjct: 878 TIDLDKFEPPFTNYTSNFIGCLDYIFYNDEDLNIISTVNIPDENQLIQESQIYHLSSSAL 937
Query: 160 PSPEWSSDHIALLAEF 175
PSP SSDH L+A+F
Sbjct: 938 PSPIRSSDHFPLIAKF 953
>UniRef100_UPI0000468B91 UPI0000468B91 UniRef100 entry
Length = 752
Score = 100 bits (249), Expect = 2e-20
Identities = 74/196 (37%), Positives = 104/196 (52%), Gaps = 22/196 (11%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAAS-----ADIP-MLVCGDFNSNPGSAPHAL 54
ANTH+ + + VK+WQ L+K +E + + IP M++CGDFNS P SA + L
Sbjct: 553 ANTHIIANPEATYVKIWQTQILVKVIEYLKINFIQKYEIIPSMIICGDFNSTPNSAVYQL 612
Query: 55 LAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAY--SSF-ARTIGLGYEQHK----R 107
L K +H DL D +L + + H L L SAY S+F ++TI +
Sbjct: 613 LYKKKCCRTHNDLNSDEHGLLE-YLPMSHNLNLKSAYAISNFLSQTISSEESINNIIINN 671
Query: 108 RLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKD--------TAL 159
+D EP FTN T +FIG LDYIFY + L + S + + DE L ++ +AL
Sbjct: 672 TIDLDRFEPAFTNYTSNFIGCLDYIFYNDEDLNIISTVNIPDENQLIQESQVYHLPTSAL 731
Query: 160 PSPEWSSDHIALLAEF 175
PSP SSDH L+A+F
Sbjct: 732 PSPIRSSDHFPLVAKF 747
>UniRef100_Q7RFI3 Arabidopsis thaliana At3g58560/F14P22_150-related [Plasmodium yoelii
yoelii]
Length = 1534
Score = 95.9 bits (237), Expect = 4e-19
Identities = 68/201 (33%), Positives = 102/201 (49%), Gaps = 32/201 (15%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAAS-----ADIP-MLVCGDFNSNPGSAPHAL 54
+NTH+ + + VK+WQ L+K +E + + IP +++CGDFNS P SA + L
Sbjct: 1335 SNTHIIANPEATYVKIWQTQILVKVIEYLKINFIQKYEIIPSIIICGDFNSTPNSAVYQL 1394
Query: 55 LAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATN 114
L K P+H D+ D +L+ + + H L + SAY+ I Q + +S N
Sbjct: 1395 LYKKKCFPTHNDIHSDEHGLLK-YLPMSHNLNIKSAYA-----ISNFLSQRINKEESINN 1448
Query: 115 ------------EPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKD------ 156
EP FTN T +FIG LDYIFY + L + S + + +E L ++
Sbjct: 1449 IIINNTIELDKFEPAFTNYTSNFIGCLDYIFYNDEDLNIISTVNIPNETQLIQESQIYHL 1508
Query: 157 --TALPSPEWSSDHIALLAEF 175
+ALPSP SDH L+A+F
Sbjct: 1509 STSALPSPVRPSDHFPLVAKF 1529
>UniRef100_UPI000023456F UPI000023456F UniRef100 entry
Length = 675
Score = 85.9 bits (211), Expect = 5e-16
Identities = 55/146 (37%), Positives = 76/146 (51%), Gaps = 20/146 (13%)
Query: 30 AASADIPMLVCGDFNSNPGSAPHALLAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVS 89
A+ IP+ +CGDFNS+PGSA + L+A G + HPDL L + H L S
Sbjct: 531 ASGDQIPLFMCGDFNSSPGSAAYNLIANGGLIEEHPDLEKRMYGNLSKVG-MTHPFKLKS 589
Query: 90 AYSSFARTIGLGYEQHKRRLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLD 149
AY A E FTN T DF LDYI+Y+++++ V LL +D
Sbjct: 590 AYG-------------------AIGELSFTNYTPDFKDILDYIWYSSNTVHVSGLLGEVD 630
Query: 150 EESLRKDTALPSPEWSSDHIALLAEF 175
++ L++ P+ + SDHIALLAEF
Sbjct: 631 KDYLQRVPGFPNYHFPSDHIALLAEF 656
>UniRef100_UPI00003AA88A UPI00003AA88A UniRef100 entry
Length = 561
Score = 83.6 bits (205), Expect = 2e-15
Identities = 54/178 (30%), Positives = 82/178 (45%), Gaps = 25/178 (14%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAASA--DIPMLVCGDFNSNPGSAPHALLAMG 58
ANTH+ H +++L Q+ L ++ +A +IP++ CGDFNS P S + + G
Sbjct: 405 ANTHLYWHPKGGNIRLIQIAVALSHIKYVACDLYPNIPLIFCGDFNSTPSSGTYGFINTG 464
Query: 59 KVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSATNEPLF 118
+ H D A + + +PL H +L SA EP +
Sbjct: 465 GIAEDHEDWASN-------GEEERCNMPL----------------SHPFKLQSACGEPAY 501
Query: 119 TNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFR 176
TN F G LDY+F ++L VE ++ L E + ALPS SDHIAL+ + +
Sbjct: 502 TNYVGGFYGCLDYVFIDQNALEVEQVIPLPSHEEVTTHQALPSVSHPSDHIALVCDLK 559
>UniRef100_O74874 SPCC31H12.08c protein [Schizosaccharomyces pombe]
Length = 690
Score = 80.5 bits (197), Expect = 2e-14
Identities = 60/200 (30%), Positives = 89/200 (44%), Gaps = 45/200 (22%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAAS-----------------------ADIPM 37
AN H++ +DVK+ QV L+ + ++A IP+
Sbjct: 494 ANCHIHWDPQFRDVKVIQVAMLMDEIAQVATKFRNMPSKIPSDQLKDERPTYPEYLKIPI 553
Query: 38 LVCGDFNSNPGSAPHALLAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFART 97
L+CGDFNS GS + L+ G + +H D + + + H L SAY
Sbjct: 554 LICGDFNSVQGSGVYDFLSSGSISQNHEDFMNNDYGEYTVNGRS-HAFNLKSAYGE---- 608
Query: 98 IGLGYEQHKRRLDSATNEPL-FTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKD 156
+E L FTN T F GA+D+I+YT +SL V LL+ +D++ L
Sbjct: 609 ----------------SEALSFTNYTPGFKGAIDHIWYTGNSLEVTGLLKGVDKDYLSGV 652
Query: 157 TALPSPEWSSDHIALLAEFR 176
P+ + SDHI LLAEF+
Sbjct: 653 VGFPNAHFPSDHICLLAEFK 672
>UniRef100_UPI00003C1BD9 UPI00003C1BD9 UniRef100 entry
Length = 670
Score = 79.7 bits (195), Expect = 3e-14
Identities = 61/201 (30%), Positives = 89/201 (43%), Gaps = 49/201 (24%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGLEKIAA-----------------------SADIPM 37
AN H + +DVKL QV L+ +EK A + IP
Sbjct: 461 ANVHTHWDPAFRDVKLVQVAMLMDEVEKAGARFAKLPPKPSVAEGYPPPPKYTHANQIPT 520
Query: 38 LVCGDFNSNPGSAPHALLAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFART 97
++CGDFNS P + + LA G V H D + Y ++
Sbjct: 521 IICGDFNSVPETGVYDFLANGAVPGDHEDF-------------------MDHVYGNYTAQ 561
Query: 98 IGLGYEQHKRRLDSA---TNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLR 154
GL QH +L+S+ E FTN T + GA+DYIFYT ++L V +L +D++ L
Sbjct: 562 -GL---QHSYKLESSYVPIGELPFTNYTPGYEGAIDYIFYTKNTLSVTGVLGEIDKQYLS 617
Query: 155 KDTALPSPEWSSDHIALLAEF 175
K P+ + SDHI +++EF
Sbjct: 618 KVVGFPNAHFPSDHICIMSEF 638
>UniRef100_Q6CJU4 Similar to sp|P31384 Saccharomyces cerevisiae YAL021c CCR4
transcriptional regulator [Kluyveromyces lactis]
Length = 790
Score = 79.7 bits (195), Expect = 3e-14
Identities = 58/190 (30%), Positives = 81/190 (42%), Gaps = 30/190 (15%)
Query: 3 THVNVHQDLKDVKLWQVHTLLKGLEKIAAS----------ADIPMLVCGDFNSNPGSAPH 52
TH++ DVK +QV +L LE + IPM++CGDFNS SA
Sbjct: 613 THLHWDPQFNDVKTFQVGVMLDYLETLIKQHHHVNNNNDIKKIPMVICGDFNSQLDSAVV 672
Query: 53 ALLAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQHKRRLDSA 112
L G V +H D+ + H L L S+Y A
Sbjct: 673 ELFNSGHVTANHKDIDQRDFGYMS-QKNFSHNLSLRSSYG-------------------A 712
Query: 113 TNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALL 172
E FTN+T F +DYI+Y++ SL V LL +DEE K P+ ++ SDHI L+
Sbjct: 713 IGELPFTNMTPSFTDVIDYIWYSSQSLRVRGLLGKIDEEYASKFIGFPNDKFPSDHIPLV 772
Query: 173 AEFRCCKNRS 182
F + +
Sbjct: 773 TRFEISRGNA 782
>UniRef100_Q9C2R2 Related to CCR4 protein [Neurospora crassa]
Length = 766
Score = 79.7 bits (195), Expect = 3e-14
Identities = 55/190 (28%), Positives = 87/190 (44%), Gaps = 31/190 (16%)
Query: 1 ANTHVNVHQDLKDVKLWQVHTLLKGL---------------EKIAASADIPMLVCGDFNS 45
ANTH+ L DVKL Q L++ + ++ ++ DIP++VCGD+NS
Sbjct: 552 ANTHLAWEPTLADVKLVQTAILMENITNDIPKPEMPEPGPSQEYRSNTDIPLIVCGDYNS 611
Query: 46 NPGSAPHALLAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTIGLGYEQH 105
S+ + LL+MG+V P D + H + SAY
Sbjct: 612 TQESSVYELLSMGRVTPEQSDFGGHQYGNFT-RDGVAHPFSMRSAYV------------- 657
Query: 106 KRRLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWS 165
L+ +E FTN F +DYI+Y+ ++L V LL D+ L++ P+ +
Sbjct: 658 --HLNGTPDELSFTNYVPGFQEVIDYIWYSTNTLEVVELLGPPDQNHLKRVPGFPNYHFP 715
Query: 166 SDHIALLAEF 175
+DHI ++AEF
Sbjct: 716 ADHIQIMAEF 725
>UniRef100_Q6CEJ6 Yarrowia lipolytica chromosome B of strain CLIB99 of Yarrowia
lipolytica [Yarrowia lipolytica]
Length = 705
Score = 78.2 bits (191), Expect = 1e-13
Identities = 59/203 (29%), Positives = 89/203 (43%), Gaps = 44/203 (21%)
Query: 2 NTHVNVHQDLKDVKLWQVHTLLKGLEKIA-----------------------ASADIPML 38
NTH++ DVKL QV LL +EK A + +P++
Sbjct: 500 NTHLHWDPAFNDVKLIQVALLLDEVEKFAERVAKDSNRVSARNQDGNNVKYESGKKLPLV 559
Query: 39 VCGDFNSNPGSAPHALLAMGKVDPSHPDLAIDPLSILRPHSKLVHQLPLVSAYSSFARTI 98
+CGDFNS S ++L + G V +H D++ + H L SAYS+
Sbjct: 560 ICGDFNSTTDSGVYSLFSQGTVT-NHKDMSGRAYGKFTDEG-MNHGFTLKSAYSNIG--- 614
Query: 99 GLGYEQHKRRLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTA 158
E FTN T +F+ +DY++Y++++L V LL +D +
Sbjct: 615 ----------------ELAFTNYTPNFVDVIDYVWYSSNALSVRGLLGGIDPDYTSNMVG 658
Query: 159 LPSPEWSSDHIALLAEFRCCKNR 181
PS + SDHI+LLAEF K +
Sbjct: 659 FPSVHYPSDHISLLAEFSFKKQK 681
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 305,632,861
Number of Sequences: 2790947
Number of extensions: 11812922
Number of successful extensions: 26931
Number of sequences better than 10.0: 181
Number of HSP's better than 10.0 without gapping: 116
Number of HSP's successfully gapped in prelim test: 65
Number of HSP's that attempted gapping in prelim test: 26535
Number of HSP's gapped (non-prelim): 253
length of query: 183
length of database: 848,049,833
effective HSP length: 119
effective length of query: 64
effective length of database: 515,927,140
effective search space: 33019336960
effective search space used: 33019336960
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0029a.6


