

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0023.9
(388 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
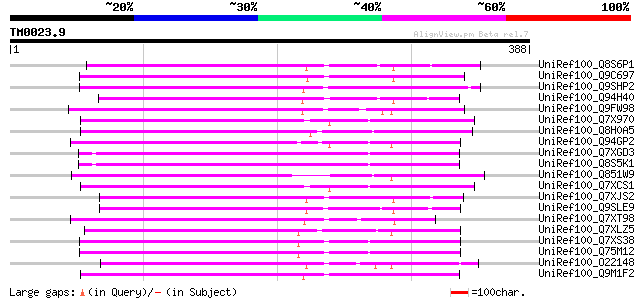
UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa] 206 1e-51
UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [A... 194 4e-48
UniRef100_Q9SHP2 Putative non-LTR retroelement reverse transcrip... 191 4e-47
UniRef100_Q94H40 Putative reverse transcriptase [Oryza sativa] 186 1e-45
UniRef100_Q9FW98 Putative non-LTR retroelement reverse transcrip... 183 6e-45
UniRef100_Q7X970 BZIP-like protein [Oryza sativa] 180 7e-44
UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa] 179 9e-44
UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa] 179 2e-43
UniRef100_Q7XGD3 Putative retroelement [Oryza sativa] 175 2e-42
UniRef100_Q8S5K1 Putative retroelement [Oryza sativa] 175 2e-42
UniRef100_Q851W9 Putative reverse transcriptase [Oryza sativa] 174 4e-42
UniRef100_Q7XCS1 Putative reverse transcriptase [Oryza sativa] 173 6e-42
UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcrip... 171 3e-41
UniRef100_Q9SLE9 Putative non-LTR retroelement reverse transcrip... 170 7e-41
UniRef100_Q7XT98 OSJNBa0052O21.12 protein [Oryza sativa] 169 2e-40
UniRef100_Q7XLZ5 OSJNBa0086O06.14 protein [Oryza sativa] 165 2e-39
UniRef100_Q7XS38 OSJNBa0081G05.2 protein [Oryza sativa] 160 6e-38
UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa] 159 1e-37
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 159 1e-37
UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis tha... 159 2e-37
>UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa]
Length = 1509
Score = 206 bits (523), Expect = 1e-51
Identities = 117/300 (39%), Positives = 166/300 (55%), Gaps = 10/300 (3%)
Query: 58 LEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILIKSIAQ 117
L ++ + +KYLGLP +G+S+T+IF+++KERIW+R++GWKEK LSRAG+EILIK++AQ
Sbjct: 940 LNMQRETTNEKYLGLPVFVGRSRTKIFSYLKERIWQRIQGWKEKLLSRAGKEILIKAVAQ 999
Query: 118 AIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGLGFRFF 177
IP++ M CF L +C I +MIA ++W +HW W L K GGLGFR
Sbjct: 1000 VIPTFAMGCFELTKDLCDQISKMIAKYWWSNQEKDNKMHWLSWNKLTLPKNMGGLGFRDI 1059
Query: 178 RAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMAKKG---YITWSSILKAGGKI 234
F A++AK R+ P+SL RV + Y P K+ TW SI K +
Sbjct: 1060 YIFNLAMLAKQGWRLIQDPDSLCSRVLRAKYFPLGDCFRPKQTSNVSYTWRSIQKGLRVL 1119
Query: 235 EGGGGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDLLD--NGRWNLE 292
+ G+ WRVGDG +IN+ D W+P G S M A L + KV +L+D G W+ +
Sbjct: 1120 Q---NGMIWRVGDGSKINIWADPWIPRGWSRKPMTPRGANL-VTKVEELIDPYTGTWDED 1175
Query: 293 LLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGYKFIRDMMLREEHRPLP 352
LL + F + I +IP+ + D L W+ G + KS YK R+M R P
Sbjct: 1176 LLSQTFWEEDVAAIKSIPVHVE-MEDVLAWHFDARGCFTVKSAYKVQREMERRASRNGCP 1234
>UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [Arabidopsis thaliana]
Length = 1142
Score = 194 bits (493), Expect = 4e-48
Identities = 108/293 (36%), Positives = 159/293 (53%), Gaps = 8/293 (2%)
Query: 53 ELTQLLEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILI 112
++ +L + + YLGLP +G SKT++F+FV++R+ R+ GW K LS+ G+E++I
Sbjct: 518 DIKLILGIHNLGGMGSYLGLPESLGGSKTKVFSFVRDRLQSRINGWSAKFLSKGGKEVMI 577
Query: 113 KSIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGL 172
KS+A +P Y+MSCF LP +I + + +A F+W + RG+HW W LC +K DGGL
Sbjct: 578 KSVAATLPRYVMSCFRLPKAITSKLTSAVAKFWWSSNGDSRGMHWMAWDKLCSSKSDGGL 637
Query: 173 GFRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMAKKGY---ITWSSILK 229
GFR F AL+AK R+ P+SL +VFK Y S+ L + K Y W S++
Sbjct: 638 GFRNVDDFNSALLAKQLWRLITAPDSLFAKVFKGRYFRKSNPLDSIKSYSPSYGWRSMIS 697
Query: 230 AGGKIEGGGGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDLLD--NG 287
A + GL RVG G I++ D W+P + KV L+D +
Sbjct: 698 ARSLVY---KGLIKRVGSGASISVWNDPWIPAQFPRPAKYGGSIVDPSLKVKSLIDSRSN 754
Query: 288 RWNLELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGYKFIR 340
WN++LL+E+F + I A+P+ N D L W+ T G Y KSGY R
Sbjct: 755 FWNIDLLKELFDPEDVPLISALPIGNPNMEDTLGWHFTKAGNYTVKSGYHTAR 807
>UniRef100_Q9SHP2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 773
Score = 191 bits (484), Expect = 4e-47
Identities = 114/303 (37%), Positives = 163/303 (53%), Gaps = 7/303 (2%)
Query: 53 ELTQLLEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILI 112
+L+QLL + + + F +YLGLP +G++KT F+F+ + + +++ W K LS AG+E+LI
Sbjct: 189 QLSQLLGIYKTEGFGRYLGLPEFVGRNKTNAFSFIAQTMDQKMDNWYNKLLSPAGKEVLI 248
Query: 113 KSIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGL 172
KSI AIP+Y MSCF+LP + I + F+W + + W W+ L K+ GGL
Sbjct: 249 KSIVTAIPTYSMSCFLLPMRLIHQITSAMRWFWWSNTKVKHKIPWVAWSKLNDPKKMGGL 308
Query: 173 GFRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMAK---KGYITWSSILK 229
R + F AL+AK RI P SLM RVFK Y P +L AK + W SIL
Sbjct: 309 AIRDLKDFNIALLAKQSWRILQQPFSLMARVFKAKYFPKERLLDAKATSQSSYAWKSILH 368
Query: 230 AGGKIEGGGGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDLLDNGRW 289
I GL++ G+G I L +D WLP + + KVSDLL GRW
Sbjct: 369 GTKLI---SRGLKYIAGNGNNIQLWKDNWLPLNPPRPPVGTCDSIYSQLKVSDLLIEGRW 425
Query: 290 NLELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGYKFIRDMMLREEHR 349
N +LL ++ + + I AI S G DA+ W T +G Y KSGY +R + +++H
Sbjct: 426 NEDLLCKLIHQNDIPHIRAIRPSITGANDAITWIYTHDGNYSVKSGYHLLRKLS-QQQHA 484
Query: 350 PLP 352
LP
Sbjct: 485 SLP 487
>UniRef100_Q94H40 Putative reverse transcriptase [Oryza sativa]
Length = 1185
Score = 186 bits (471), Expect = 1e-45
Identities = 108/275 (39%), Positives = 154/275 (55%), Gaps = 11/275 (4%)
Query: 67 DKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILIKSIAQAIPSYIMSC 126
DKYLGLP +IG K+ F + +R+ R+ G KEK LS G+E+LIKSIAQAIP + MS
Sbjct: 566 DKYLGLPAMIGVDKSDCFRHLIDRVNNRINGRKEKMLSTGGKEVLIKSIAQAIPVFAMSV 625
Query: 127 FILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGLGFRFFRAFKEALVA 186
F +P IC I IA F+WG D R +HW W +C K+ GGLGFR F A++A
Sbjct: 626 FRIPKKICDCISEAIARFWWGDDENHRRIHWKAWWRMCIPKQRGGLGFRDLYCFNRAMLA 685
Query: 187 KN**RIYMFPESLMGRVFKVVYSPSSSILMA--KKG-YITWSSILKAGGKIEGGGGGLRW 243
K R P+SL RV + Y P +IL A KKG W SIL + G W
Sbjct: 686 KQIWRFLCDPDSLCARVMRCKYYPDGNILRARPKKGSSYAWQSILSGSECFK----GYIW 741
Query: 244 RVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDLLD--NGRWNLELLEEVFSSS 301
R+GDG Q+N+ +D +P S+ + + + KV +L++ +G W+ +L++ +F
Sbjct: 742 RIGDGSQVNIWEDSLIPSSSNGKVLTPRGRNI-ITKVEELINPVSGGWDEDLIDTLFWPI 800
Query: 302 PMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGY 336
+IL IP+S G D + W+ T +G++ +S Y
Sbjct: 801 DANRILQIPIS-AGREDFVAWHHTRSGIFSVRSAY 834
>UniRef100_Q9FW98 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1382
Score = 183 bits (465), Expect = 6e-45
Identities = 109/305 (35%), Positives = 160/305 (51%), Gaps = 17/305 (5%)
Query: 45 KCVENCFHELTQLLEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLS 104
+C + + L+V D YLG+PT IG + T F F+ ERIWKR+ GW ++ LS
Sbjct: 741 RCPDAVKISVKSCLQVDNEVLQDSYLGMPTEIGLATTNFFKFLPERIWKRVNGWTDRPLS 800
Query: 105 RAGREILIKSIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLC 164
RAG E ++K++AQAIP+Y+MSCF +P SIC + IA +WG + ++ +HW W+ L
Sbjct: 801 RAGMETMLKAVAQAIPNYVMSCFRIPVSICEKMKTCIADHWWGFEDGKKKMHWKSWSWLS 860
Query: 165 KNKRDGGLGFRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMA---KKGY 221
K GG+GFR F F +A++ + R+ P+SL RV K Y P+SS A K
Sbjct: 861 TPKFLGGMGFREFTTFNQAMLGRQCWRLLTDPDSLCSRVLKGRYFPNSSFWEAAQPKSPS 920
Query: 222 ITWSSILKAGGKIEGGGGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRL---- 277
TW S+L + G+RW VGDG I + D W+P + + + L
Sbjct: 921 FTWRSLLFGRELL---AKGVRWGVGDGKTIKIFSDNWIPG-----FRPQLVTTLSPFPTD 972
Query: 278 EKVSDLL--DNGRWNLELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSG 335
VS L+ D W+ +L+ +F ++IL IP+S G+ D W G+Y +S
Sbjct: 973 ATVSCLMNEDARCWDGDLIRSLFPVDIAKEILQIPISRHGDADFASWPHDKLGLYSVRSA 1032
Query: 336 YKFIR 340
Y R
Sbjct: 1033 YNLAR 1037
>UniRef100_Q7X970 BZIP-like protein [Oryza sativa]
Length = 2367
Score = 180 bits (456), Expect = 7e-44
Identities = 104/299 (34%), Positives = 166/299 (54%), Gaps = 10/299 (3%)
Query: 54 LTQLLEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILIK 113
++++L+V+ + D+YLG PT G+ F ++ +IWKR+ W E LS G+E+LIK
Sbjct: 1469 ISEILQVERDRFEDRYLGFPTPEGRMHKGRFQSLQAKIWKRVIQWGENHLSTGGKEVLIK 1528
Query: 114 SIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGLG 173
++ QAIP Y+M F LP+S+ D+ ++ F+W +R HW W +L K K GGLG
Sbjct: 1529 AVIQAIPVYVMGIFKLPESVIDDLTKLTKNFWWDSMNGQRKTHWKAWDSLTKPKSLGGLG 1588
Query: 174 FRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMAKKGYITWSSILKAGGK 233
FR +R F +AL+A+ R+ +P+SL RV K Y P S++ G S+ A
Sbjct: 1589 FRDYRLFNQALLARQAWRLITYPDSLCARVLKAKYFPHGSLIDTSFG----SNSSPAWRS 1644
Query: 234 IEGG----GGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDLL-DNGR 288
IE G G+ WRVG+G I + +D WLP S + A RL+ VSDL+ ++G
Sbjct: 1645 IEYGLDLLKKGIIWRVGNGNSIRIWRDSWLPRDHSRRPI-TGKANCRLKWVSDLITEDGS 1703
Query: 289 WNLELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGYKFIRDMMLREE 347
W++ + + F + + IL I +S++ D + W+ NG++ +S Y+ ++ EE
Sbjct: 1704 WDVPKIHQYFHNLDAEVILNICISSRSEEDFIAWHPDKNGMFSVRSAYRLAAQLVNIEE 1762
>UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa]
Length = 1557
Score = 179 bits (455), Expect = 9e-44
Identities = 102/297 (34%), Positives = 160/297 (53%), Gaps = 8/297 (2%)
Query: 54 LTQLLEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILIK 113
+T L++ + DKYLGLPT G+ F ++ERIWK++ W E LS G+E+LIK
Sbjct: 1104 ITNCLQIASTEFEDKYLGLPTPGGRMHKGRFQSLRERIWKKILQWGENYLSSGGKEVLIK 1163
Query: 114 SIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGLG 173
++ QAIP Y+M F LP+S+C D+ + F+WG + +R HW W +L K+K GGLG
Sbjct: 1164 AVIQAIPVYVMGIFKLPESVCEDLTSLTRNFWWGAEKGKRKTHWKAWKSLTKSKSLGGLG 1223
Query: 174 FRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMAKKGYIT---WSSILKA 230
F+ R F +AL+A+ R+ P+SL RV K Y P+ SI+ G W +I
Sbjct: 1224 FKDIRLFNQALLARQAWRLIDNPDSLCARVLKAKYYPNGSIVDTSFGGNASPGWQAIEHG 1283
Query: 231 GGKIEGGGGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDL-LDNGRW 289
+E G+ WR+G+G + + QD WLP S + R++ V+DL LDNG W
Sbjct: 1284 ---LELVKKGIIWRIGNGRSVRVWQDPWLPRDLSRRPITPK-NNCRIKWVADLMLDNGMW 1339
Query: 290 NLELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGYKFIRDMMLRE 346
+ + ++F ++ IL + S++ D + W+ G + ++ Y+ + E
Sbjct: 1340 DANKINQIFLPVDVEIILKLRTSSRDEEDFIAWHPDKLGNFSVRTAYRLAENWAKEE 1396
>UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa]
Length = 1833
Score = 179 bits (453), Expect = 2e-43
Identities = 105/298 (35%), Positives = 157/298 (52%), Gaps = 11/298 (3%)
Query: 46 CVENCFHELTQLLEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSR 105
C + E+ L+V +KYLGLPT G+ K + F + +++ KR+ W EK +S
Sbjct: 639 CPQERQDEIKSALQVHSSTFEEKYLGLPTPDGRMKAEQFQPINDKLSKRMSDWNEKYMSS 698
Query: 106 AGREILIKSIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCK 165
+E+LIKS+AQA+P+YIM F + D C D RM+ F+WG D R +HW W +
Sbjct: 699 GAKEVLIKSVAQALPTYIMGVFKMTDKFCDDYTRMVRNFWWGHDNGERKVHWIAWEKILY 758
Query: 166 NKRDGGLGFRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMAKKGYITWS 225
K GGLGFR + F +AL+A+ R+ P+SL RVFK Y P+ +++ + T S
Sbjct: 759 PKSHGGLGFRDMKCFNQALLARQAWRLLTSPDSLCARVFKAKYYPNGNLV--DTAFPTVS 816
Query: 226 SILKAGGKIEGG----GGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVS 281
S + G I+ G G WRVG+G I + + KW+P G + + E RL V+
Sbjct: 817 SSVWKG--IQHGLDLLKQGTIWRVGNGHNIKIWRHKWVPHGEHINVL-EKKGRNRLIYVN 873
Query: 282 DLL--DNGRWNLELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGYK 337
+L+ +N WN L+ V ++L I L N D W+ +G++ KS YK
Sbjct: 874 ELINEENRCWNETLVRHVLKEEDANEVLKIRLPNHQMDDFPAWHHEKSGLFTVKSAYK 931
>UniRef100_Q7XGD3 Putative retroelement [Oryza sativa]
Length = 1694
Score = 175 bits (444), Expect = 2e-42
Identities = 101/287 (35%), Positives = 157/287 (54%), Gaps = 5/287 (1%)
Query: 52 HELTQLLEVKEVKSF-DKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREI 110
+++ Q L++ SF DKYLG PT G+ F ++ER+WKR+ W EK LS G+EI
Sbjct: 1347 NQIKQTLQISN--SFEDKYLGFPTPEGRMTKGKFQSLQERLWKRIMLWGEKFLSTGGKEI 1404
Query: 111 LIKSIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDG 170
LIK++ QA+P Y+M F LP+S+C ++ +++ F+WG + R HW W L K G
Sbjct: 1405 LIKAVLQAMPVYVMGLFKLPESVCEELTKLVRNFWWGAEKGMRRTHWKSWDCLIAQKLKG 1464
Query: 171 GLGFRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMAKKGYITWSSILKA 230
G+GFR FR F +AL+A+ R+ P+SL R+ K +Y P+ S++ G
Sbjct: 1465 GMGFRDFRLFNQALLARQAWRLLDRPDSLCARILKAIYYPNGSLIDTSFGGNASPGWRAI 1524
Query: 231 GGKIEGGGGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDLLD-NGRW 289
+E G+ WR+G+G + L +D W+P S + A RL VSDL+D +G W
Sbjct: 1525 EYGLELLKKGIIWRIGNGKSVRLWRDPWIPRSYSRRPI-SAKRNCRLRWVSDLIDQSGSW 1583
Query: 290 NLELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGY 336
+ + + + F + IL I LS++ D + W+ G + +S Y
Sbjct: 1584 DTDKISQHFLPMDAEAILNIRLSSRLEDDFIAWHPDKLGRFSVRSAY 1630
>UniRef100_Q8S5K1 Putative retroelement [Oryza sativa]
Length = 1888
Score = 175 bits (444), Expect = 2e-42
Identities = 101/287 (35%), Positives = 157/287 (54%), Gaps = 5/287 (1%)
Query: 52 HELTQLLEVKEVKSF-DKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREI 110
+++ Q L++ SF DKYLG PT G+ F ++ER+WKR+ W EK LS G+EI
Sbjct: 1304 NQIKQTLQISN--SFEDKYLGFPTPEGRMTKGKFQSLQERLWKRIMLWGEKFLSTGGKEI 1361
Query: 111 LIKSIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDG 170
LIK++ QA+P Y+M F LP+S+C ++ +++ F+WG + R HW W L K G
Sbjct: 1362 LIKAVLQAMPVYVMGLFKLPESVCEELTKLVRNFWWGAEKGMRRTHWKSWDCLIAQKLKG 1421
Query: 171 GLGFRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMAKKGYITWSSILKA 230
G+GFR FR F +AL+A+ R+ P+SL R+ K +Y P+ S++ G
Sbjct: 1422 GMGFRDFRLFNQALLARQAWRLLDRPDSLCARILKAIYYPNGSLIDTSFGGNASPGWRAI 1481
Query: 231 GGKIEGGGGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDLLD-NGRW 289
+E G+ WR+G+G + L +D W+P S + A RL VSDL+D +G W
Sbjct: 1482 EYGLELLKKGIIWRIGNGKSVRLWRDPWIPRSYSRRPI-SAKRNCRLRWVSDLIDQSGSW 1540
Query: 290 NLELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGY 336
+ + + + F + IL I LS++ D + W+ G + +S Y
Sbjct: 1541 DTDKISQHFLPMDAEAILNIRLSSRLEDDFIAWHPDKLGRFSVRSAY 1587
>UniRef100_Q851W9 Putative reverse transcriptase [Oryza sativa]
Length = 435
Score = 174 bits (441), Expect = 4e-42
Identities = 99/311 (31%), Positives = 163/311 (51%), Gaps = 31/311 (9%)
Query: 47 VENCFHELTQLLEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRA 106
++N + LL+VK+ +KYLGLPT G+ K+ F +KE+ KRL W EK+LS
Sbjct: 8 IDNTQQSIKTLLKVKQTVFEEKYLGLPTPSGRIKSGKFQSLKEKFEKRLSDWCEKNLSMG 67
Query: 107 GREILIKSIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKN 166
+E+LIKS+ QA+P Y+MS +P S+C + ++I F+WG D +R +HW W L K
Sbjct: 68 RKEVLIKSVLQALPMYMMSVSKIPVSLCEEYMQLIRKFWWGEDRHKRKVHWISWHQLVKP 127
Query: 167 KRDGGLGFRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMAKKGYITWSS 226
K GG+ FR+ + F +AL+A+ R+ +P+SL +V K Y P+
Sbjct: 128 KSQGGISFRYLKLFNQALLARQAWRLIQYPDSLCAKVLKAKYYPN--------------- 172
Query: 227 ILKAGGKIEGGGGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDLL-- 284
G+ WR+G +I + +D W+P +L + + + RL+ V+DL+
Sbjct: 173 -------------GIIWRIGSESKIIIWRDNWIPREHNLKVLGKR-SRARLKWVADLMLP 218
Query: 285 DNGRWNLELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGYKFIRDMML 344
+ W+ L+ ++F + IL I LS++ D + W+ G++ +S YK D+
Sbjct: 219 NKQEWDERLIRQLFYPADADIILNIQLSHRMTEDFIAWHYDRVGMFTVRSAYKLALDIQT 278
Query: 345 REEHRPLPCRS 355
R + + C S
Sbjct: 279 RSQGNVVGCSS 289
>UniRef100_Q7XCS1 Putative reverse transcriptase [Oryza sativa]
Length = 791
Score = 173 bits (439), Expect = 6e-42
Identities = 102/299 (34%), Positives = 164/299 (54%), Gaps = 10/299 (3%)
Query: 54 LTQLLEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILIK 113
++++L+V+ + D+YL PT G+ F ++ +IWKR+ W E LS G+E+LIK
Sbjct: 145 ISEILQVERDRFEDRYLEFPTPEGRMHKGRFQSLQAKIWKRVIQWGENHLSTGGKEVLIK 204
Query: 114 SIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGLG 173
++ QAIP Y+M F LP+S+ D+ ++ F+W +R HW W +L K K GGLG
Sbjct: 205 AVIQAIPVYVMGIFKLPESVIDDLTKLTKNFWWDSMNGQRKTHWKAWDSLTKPKSLGGLG 264
Query: 174 FRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMAKKGYITWSSILKAGGK 233
FR + F +AL+A+ R+ +P+SL RV K Y P S++ G S+ A
Sbjct: 265 FRDYWLFNQALLARQAWRLITYPDSLCARVLKAKYFPHGSLIDTSFG----SNSSPAWRS 320
Query: 234 IEGG----GGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDLL-DNGR 288
IE G G+ WRVG+G I + +D WLP S + A RL+ VSDL+ ++G
Sbjct: 321 IEYGLDLLKKGIIWRVGNGNSIRIWRDPWLPRDHSRRPI-TGKANCRLKWVSDLITEDGS 379
Query: 289 WNLELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGYKFIRDMMLREE 347
W++ + + F + + IL I +S++ D + W+ NG++ +S Y+ ++ EE
Sbjct: 380 WDVPKIHQYFHNLDAEVILNICISSRSEEDFIAWHPDKNGMFSVRSAYRLAAQLVNIEE 438
>UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1344
Score = 171 bits (433), Expect = 3e-41
Identities = 103/278 (37%), Positives = 153/278 (54%), Gaps = 11/278 (3%)
Query: 68 KYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILIKSIAQAIPSYIMSCF 127
KYLGLP + SK +F F+KE++ RL GW K+LS+ G+E+L+KSIA A+P Y+MSCF
Sbjct: 727 KYLGLPECLSGSKRDLFGFIKEKLQSRLTGWYAKTLSQGGKEVLLKSIALALPVYVMSCF 786
Query: 128 ILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGLGFRFFRAFKEALVAK 187
LP ++C + ++ F+W +R +HW W L K GG GF+ + F +AL+AK
Sbjct: 787 KLPKNLCQKLTTVMMDFWWNSMQQKRKIHWLSWQRLTLPKDQGGFGFKDLQCFNQALLAK 846
Query: 188 N**RIYMFPESLMGRVFKVVYSPSSSILMAKKG---YITWSSILKAGGKIEGGGGGLRWR 244
R+ SL RVF+ Y +S L A +G W SIL + GLR
Sbjct: 847 QAWRVLQEKGSLFSRVFQSRYFSNSDFLSATRGSRPSYAWRSILFGRELLM---QGLRTV 903
Query: 245 VGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDLLD--NGRWNLELLEEVFSSSP 302
+G+G + + DKWL DGS+ ++ KVS L+D + WNL +L ++F
Sbjct: 904 IGNGQKTFVWTDKWLHDGSNRRPLNRRRFINVDLKVSQLIDPTSRNWNLNMLRDLFPWKD 963
Query: 303 MQKIL-AIPLSNQGNTDALYWYDTFNGVYVCKSGYKFI 339
++ IL PL + D+ W + NG+Y K+GY+F+
Sbjct: 964 VEIILKQRPLFFK--EDSFCWLHSHNGLYSVKTGYEFL 999
>UniRef100_Q9SLE9 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1319
Score = 170 bits (430), Expect = 7e-41
Identities = 103/277 (37%), Positives = 157/277 (56%), Gaps = 13/277 (4%)
Query: 68 KYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILIKSIAQAIPSYIMSCF 127
KYLGLP + SK +F ++KE++ L GW +K+LS+ G+EIL+KSIA A+P YIM+CF
Sbjct: 701 KYLGLPENLSGSKQDLFGYIKEKLQSHLSGWYDKTLSQGGKEILLKSIALALPVYIMTCF 760
Query: 128 ILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGLGFRFFRAFKEALVAK 187
LP +C + ++ F+W +HW L K GG GF+ + F +AL+AK
Sbjct: 761 RLPKGLCTKLTSVMMDFWWNSMEFSNKIHWIGGKKLTLPKSLGGFGFKDLQCFNQALLAK 820
Query: 188 N**RIYMFPESLMGRVFKVVYSPSSSILMAKKG---YITWSSILKAGGKIEGGGGGLRWR 244
R++ +S++ ++FK Y ++ L A++G TW SIL + GGL+
Sbjct: 821 QAWRLFSDSKSIVSQIFKSRYFMNTDFLNARQGTRPSYTWRSILYGRELL---NGGLKRL 877
Query: 245 VGDGFQINLLQDKWLPDGSSLVYMH-EAIAELRLEKVSDLLD--NGRWNLELLEEVFSSS 301
+G+G Q N+ DKWL DG S M+ ++ + + KVS L+D WNL+ L E+F
Sbjct: 878 IGNGEQTNVWIDKWLFDGHSRRPMNLHSLMNIHM-KVSHLIDPLTRNWNLKKLTELFHEK 936
Query: 302 PMQKIL-AIPLSNQGNTDALYWYDTFNGVYVCKSGYK 337
+Q I+ PL + + D+ W T NG+Y KSGY+
Sbjct: 937 DVQLIMHQRPLIS--SEDSYCWAGTNNGLYTVKSGYE 971
>UniRef100_Q7XT98 OSJNBa0052O21.12 protein [Oryza sativa]
Length = 427
Score = 169 bits (427), Expect = 2e-40
Identities = 97/279 (34%), Positives = 151/279 (53%), Gaps = 11/279 (3%)
Query: 46 CVENCFHELTQLLEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSR 105
C+E+ + + + L+V D YLG+PT IG++ T F+F+ ER+W+R+ GW ++ +SR
Sbjct: 151 CLEDIKNSVKETLQVSVEILQDTYLGMPTEIGRASTGSFHFLPERVWRRVNGWNDRPMSR 210
Query: 106 AGREILIKSIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCK 165
A +E ++K++AQAIP+Y+MSCF LP S C + I +WG + ++ +HW W +
Sbjct: 211 ARKETMLKAVAQAIPTYVMSCFKLPVSTCEKMKSCILDHWWGFEDGKKKMHWRSWEWMTT 270
Query: 166 NKRDGGLGFRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMAKK---GYI 222
K GG+GFR F +A++A+ RI SL RV K Y P+S + A K
Sbjct: 271 PKSLGGMGFRDLGLFNQAMLARQGWRIVTDLVSLCARVLKGRYFPNSDLWNAPKPTATSF 330
Query: 223 TWSSILKAGGKIEGGGGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSD 282
TW SIL + G+RW +G+G + +L+D W+P M + L + D
Sbjct: 331 TWPSILFGRDLLR---KGVRWGIGNGSSVKILKDHWIP--GIKPSMVRPLLPLPEDVTVD 385
Query: 283 LLDN---GRWNLELLEEVFSSSPMQKILAIPLSNQGNTD 318
L N G W+ + + F + Q+IL IP+S G D
Sbjct: 386 FLVNDAIGEWDEDKVFSFFDETTAQQILQIPVSAHGGED 424
>UniRef100_Q7XLZ5 OSJNBa0086O06.14 protein [Oryza sativa]
Length = 1205
Score = 165 bits (417), Expect = 2e-39
Identities = 96/286 (33%), Positives = 149/286 (51%), Gaps = 9/286 (3%)
Query: 57 LLEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILIKSIA 116
+L+V+ DK LGLPT G+ K + F +KER KRL W E+ LS AG+E LIKS+A
Sbjct: 726 VLQVERTCFDDKCLGLPTPDGRMKAEQFQPIKERFEKRLTDWSERFLSLAGKEALIKSVA 785
Query: 117 QAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGLGFRF 176
QA+P+Y M F +P+ C + ++++ F+WG + + +HW W L K GGLGFR
Sbjct: 786 QALPTYTMGVFKMPERFCEEYEQLVRNFWWGHEKGEKKVHWIAWEKLTSPKLLGGLGFRD 845
Query: 177 FRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSI---LMAKKGYITWSSILKAGGK 233
R F +AL+A+ R+ P+SL RV K Y P+ +I TW I+
Sbjct: 846 IRCFNQALLARQAWRLIESPDSLCARVLKAKYYPNGTITDTAFPSVSSPTWKGIVHG--- 902
Query: 234 IEGGGGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDLL--DNGRWNL 291
+E GL WR+GDG + + ++ W+ G +L + + R+ V +L+ D WN
Sbjct: 903 LELLKKGLIWRIGDGSKTKIWRNHWVAHGENLKILEKKTWN-RVIYVRELIVTDTKTWNE 961
Query: 292 ELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGYK 337
L+ + +IL I + + D W+ G++ +S Y+
Sbjct: 962 PLIRHIIREEDADEILKIRIPQREEEDFPAWHYEKTGIFSVRSVYR 1007
>UniRef100_Q7XS38 OSJNBa0081G05.2 protein [Oryza sativa]
Length = 1711
Score = 160 bits (405), Expect = 6e-38
Identities = 97/289 (33%), Positives = 150/289 (51%), Gaps = 8/289 (2%)
Query: 53 ELTQLLEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILI 112
++ L+V+ D+YLG PT G+ F ++ +I KR+ W E LS G+EILI
Sbjct: 1218 DIRNTLQVERDNFEDRYLGFPTPEGRMHKGRFQSLEAKIAKRVILWGENFLSSGGKEILI 1277
Query: 113 KSIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGL 172
K++ QAIP Y+M F P+S+C D+ +M F+W D RR HW W +L K K +GGL
Sbjct: 1278 KAVIQAIPVYVMGLFKFPESVCDDLTKMTRNFWWVADNGRRRTHWRAWDSLTKAKINGGL 1337
Query: 173 GFRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSI---LMAKKGYITWSSILK 229
GFR ++ F +AL+A R+ FP SL +V K Y P S+ + TW I
Sbjct: 1338 GFRDYKLFNQALLAPQAWRLIEFPNSLCAQVLKAKYFPHGSLTDTTFSANASPTWHGIEY 1397
Query: 230 AGGKIEGGGGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDLL-DNGR 288
++ G+ WR+G+G + + +D W+P S + + A RL+ VSDL+ ++G
Sbjct: 1398 GLDLLK---KGIIWRIGNGNSVRIWRDPWIPKDLSRRPV-SSKANCRLKWVSDLIAEDGT 1453
Query: 289 WNLELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGYK 337
W+ + + F I I +S + D + W+ G + +S YK
Sbjct: 1454 WDSAKINQYFLKIDADIIQKICISARLEEDFIAWHPDKTGRFSVRSAYK 1502
>UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa]
Length = 1936
Score = 159 bits (403), Expect = 1e-37
Identities = 96/289 (33%), Positives = 150/289 (51%), Gaps = 8/289 (2%)
Query: 53 ELTQLLEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILI 112
++ L+V+ D+YLG PT G+ F ++ +I KR+ W E LS G+EILI
Sbjct: 1374 DIRNTLQVERDNFEDRYLGFPTPEGRMHKGRFQSLQAKIAKRVIQWGENFLSSGGKEILI 1433
Query: 113 KSIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGL 172
K++ QAIP Y+M F PDS+ ++ +M F+WG D RR HW W +L K K +GGL
Sbjct: 1434 KAVIQAIPVYVMGLFKFPDSVYDELTKMTRNFWWGADNGRRRTHWRAWDSLTKAKINGGL 1493
Query: 173 GFRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSI---LMAKKGYITWSSILK 229
GFR ++ F +AL+ + R+ FP SL +V K Y P S+ + TW I
Sbjct: 1494 GFRDYKLFNQALLTRQAWRLIEFPNSLCAQVLKAKYFPHGSLTDTTFSANASPTWHGIEY 1553
Query: 230 AGGKIEGGGGGLRWRVGDGFQINLLQDKWLPDGSSLVYMHEAIAELRLEKVSDLL-DNGR 288
++ G+ WR+G+G + + +D W+P S + + A RL+ VSDL+ ++G
Sbjct: 1554 GLDLLK---KGIIWRIGNGNSVRIWRDPWIPRDLSRRPV-SSKANCRLKWVSDLIAEDGT 1609
Query: 289 WNLELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGYK 337
W+ + + F I I +S + D + W+ G + +S YK
Sbjct: 1610 WDSAKINQYFLKIDADIIQKICISARLEEDFIAWHPDKTGRFSVRSAYK 1658
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 159 bits (402), Expect = 1e-37
Identities = 101/296 (34%), Positives = 153/296 (51%), Gaps = 20/296 (6%)
Query: 69 YLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILIKSIAQAIPSYIMSCFI 128
YLGLP SK +++K+R+ K++ GW+ LS G+EIL+K++A A+P+Y MSCF
Sbjct: 750 YLGLPESFQGSKVATLSYLKDRLGKKVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFK 809
Query: 129 LPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGLGFRFFRAFKEALVAKN 188
+P +IC I+ ++A F+W RGLHW W +L + K GGLGF+ AF AL+ K
Sbjct: 810 IPKTICQQIESVMAEFWWKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQ 869
Query: 189 **RIYMFPESLMGRVFKVVYSPSSSILMAKKG---YITWSSILKAGGKIEGGGGGLRWRV 245
R+ +SLM +VFK Y S L A G W SI +A I+ G+R +
Sbjct: 870 LWRMITEKDSLMAKVFKSRYFSKSDPLNAPLGSRPSFAWKSIYEAQVLIK---QGIRAVI 926
Query: 246 GDGFQINLLQDKWLPDGSSLVYMHEAI---------AELRLEKVSDLL--DNGRWNLELL 294
G+G IN+ D W+ G+ +A+ A + V DLL D WN L+
Sbjct: 927 GNGETINVWTDPWI--GAKPAKAAQAVKRSHLVSQYAANSIHVVKDLLLPDGRDWNWNLV 984
Query: 295 EEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGYKFIRDMMLREEHRP 350
+F + + ILA+ + D W + +G Y KSGY ++ ++ + + P
Sbjct: 985 SLLFPDNTQENILALRPGGKETRDRFTWEYSRSGHYSVKSGY-WVMTEIINQRNNP 1039
>UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis thaliana]
Length = 851
Score = 159 bits (401), Expect = 2e-37
Identities = 102/288 (35%), Positives = 145/288 (49%), Gaps = 8/288 (2%)
Query: 54 LTQLLEVKEVKSFDKYLGLPTIIGKSKTQIFNFVKERIWKRLKGWKEKSLSRAGREILIK 113
L LL + KYLGLP G+ K ++FN++ +R+ +R W K LS AG+EIL+K
Sbjct: 356 LKTLLNIPNQGGGGKYLGLPEQFGRKKKEMFNYIIDRVKERTASWSAKFLSPAGKEILLK 415
Query: 114 SIAQAIPSYIMSCFILPDSICADIDRMIACFFWGGDVTRRGLHWTRWANLCKNKRDGGLG 173
S+A A+P Y MSCF LP I ++I+ ++ F+W +RG+ W W L +K++GGLG
Sbjct: 416 SVALAMPVYAMSCFKLPQGIVSEIESLLMNFWWEKASNKRGIPWVAWKRLQYSKKEGGLG 475
Query: 174 FRFFRAFKEALVAKN**RIYMFPESLMGRVFKVVYSPSSSILMAK---KGYITWSSILKA 230
FR F +AL+AK RI +P SL RV K Y +SI+ AK + WSS+L
Sbjct: 476 FRDLAKFNDALLAKQAWRIIQYPNSLFARVMKARYFKDNSIIDAKTRSQQSYGWSSLLSG 535
Query: 231 GGKIEGGGGGLRWRVGDGFQINLLQDKWLPDGSSL-VYMHEAIAELRLEKVSDLLDNGR- 288
+ G R+ +GDG I L D + + E L L+ + + R
Sbjct: 536 IALLR---KGTRYVIGDGKTIRLGIDNVVDSHPPRPLLTDEQHNGLSLDNLFQHRGHSRC 592
Query: 289 WNLELLEEVFSSSPMQKILAIPLSNQGNTDALYWYDTFNGVYVCKSGY 336
W+ L+ S I I LS + TD L W G Y +SGY
Sbjct: 593 WDNAKLQTFVDQSDHDYIKRIYLSTRSKTDRLIWSYNSTGDYTVRSGY 640
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.334 0.147 0.477
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 641,108,650
Number of Sequences: 2790947
Number of extensions: 27020533
Number of successful extensions: 92910
Number of sequences better than 10.0: 235
Number of HSP's better than 10.0 without gapping: 169
Number of HSP's successfully gapped in prelim test: 66
Number of HSP's that attempted gapping in prelim test: 92488
Number of HSP's gapped (non-prelim): 314
length of query: 388
length of database: 848,049,833
effective HSP length: 129
effective length of query: 259
effective length of database: 488,017,670
effective search space: 126396576530
effective search space used: 126396576530
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0023.9


