

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0023.16
(463 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
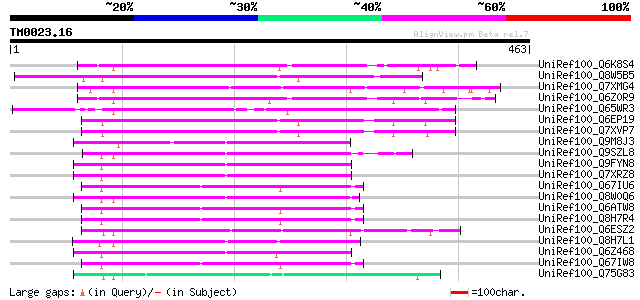
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6K8S4 Putative far-red impaired response protein [Ory... 129 1e-28
UniRef100_Q8W5B5 Putative transposase [Oryza sativa] 124 7e-27
UniRef100_Q7XMG4 OSJNBa0028I23.18 protein [Oryza sativa] 120 8e-26
UniRef100_Q6Z0R9 Putative far-red impaired response protein [Ory... 120 1e-25
UniRef100_Q65WR3 Putative far-red impaired response protein [Ory... 112 2e-23
UniRef100_Q6EP19 Putative far-red impaired response protein [Ory... 110 8e-23
UniRef100_Q7XVP7 OSJNBa0050F15.2 protein [Oryza sativa] 109 2e-22
UniRef100_Q9M8J3 F28L1.19 protein [Arabidopsis thaliana] 102 2e-20
UniRef100_Q9SZL8 Hypothetical protein F20D10.300 [Arabidopsis th... 102 3e-20
UniRef100_Q9FYN8 P0702D12.15 protein [Oryza sativa] 97 1e-18
UniRef100_Q7XRZ8 OSJNBb0002N06.8 protein [Oryza sativa] 97 1e-18
UniRef100_Q67IU6 Far-red impaired response protein-like [Oryza s... 97 1e-18
UniRef100_Q8W0Q6 Putative far-red impaired response protein [Sor... 97 1e-18
UniRef100_Q6ATW8 Hypothetical protein P0015C02.8 [Oryza sativa] 96 2e-18
UniRef100_Q8H7R4 Hypothetical protein OSJNBa0081P02.17 [Oryza sa... 96 2e-18
UniRef100_Q6ESZ2 Putative far-red impaired response protein [Ory... 95 5e-18
UniRef100_Q8H7L1 Putative transposase [Oryza sativa] 94 8e-18
UniRef100_Q6Z468 Far-red impaired response protein-like protein ... 94 1e-17
UniRef100_Q67IW8 Far-red impaired response protein-like [Oryza s... 93 1e-17
UniRef100_Q75G83 Putative transposase [Oryza sativa] 92 2e-17
>UniRef100_Q6K8S4 Putative far-red impaired response protein [Oryza sativa]
Length = 880
Score = 129 bits (325), Expect = 1e-28
Identities = 103/389 (26%), Positives = 174/389 (44%), Gaps = 49/389 (12%)
Query: 61 TEKYLCAWHLLRNANQNLGNPLLTKGFKNCM--------FRKKWDEL-VLNLGLQDNCWV 111
+E +LC WH+ +N ++L P + F+ + F +KW E V + G +DN W+
Sbjct: 366 SEHWLCTWHIEQNMARHL-RPDMLSDFRTLVHAPYDHEEFERKWVEFKVKHKGCEDNQWL 424
Query: 112 KETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHM 171
Y R W A+ +G FF G ++ R E S+L +++ + +L ++ ++C+ M
Sbjct: 425 VRMYNLRKKWATAYTKGLFFLGMKSNQRSESLNSKLHRYLDRKMSLVLLVEHYEHCLSRM 484
Query: 172 RFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAV 231
R+RE E DC + +P + +E A + FT VF + V+ + +V+DC +
Sbjct: 485 RYREAELDCKASQSIPFTSNDASFIEKDAARIFTPAVFKKLKLVIAKSMDWEVIDCIEED 544
Query: 232 TCLNYTVA------KLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRI 285
+ Y ++ L L + V S K +C KM+ L CEH+V ++ HL++
Sbjct: 545 NLVKYVISMKGDSEMLQILNCTY---VESTMKSINCTCRKMDRECLPCEHMVAIMHHLKL 601
Query: 286 EELSECFIFKRWSKGAKDGVYSGKNIENDFWDFQKSSRCSALMDLYRALGYLNSNTAADF 345
+ + E I +RW+ AK S + E W Q M+ Y L + T F
Sbjct: 602 DAIPEACIVRRWTIKAKHMFPSDRYGEVYTWSNQ--------MERYHRL--RSMGTEVFF 651
Query: 346 NDATQKASELIEQSKAKK------SCQREGS-LSLS--------QTNILN---LKDPIRL 387
+ K + L+ +K C+ + S LSL +T+I + + DP+ +
Sbjct: 652 KCSVSKETTLMVMEMLEKLDLETDECETKNSDLSLCGHVLAQSLRTDIESHDKVHDPMEI 711
Query: 388 RRKGRGTKNKSSLGLTVKHVINCLIFKTP 416
KG TK G K V C I + P
Sbjct: 712 VPKGAPTKRLR--GFMEKRVRRCGIAEAP 738
Score = 42.7 bits (99), Expect = 0.022
Identities = 19/61 (31%), Positives = 31/61 (50%)
Query: 1 MLKNDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDV 60
+L D+ +++ D +F E+ D+ L+ LFW D R+ Y +GDV+ FD
Sbjct: 230 ILGGDAERVIKYFQWRQKHDMEFFFEYETDEAGRLKRLFWSDPQSRIDYDAFGDVVVFDS 289
Query: 61 T 61
T
Sbjct: 290 T 290
>UniRef100_Q8W5B5 Putative transposase [Oryza sativa]
Length = 682
Score = 124 bits (310), Expect = 7e-27
Identities = 92/377 (24%), Positives = 163/377 (42%), Gaps = 19/377 (5%)
Query: 5 DSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVTEKY 64
D+ +Q++ ++ +F ++ D+ L ++FW D RL Y +G + FD T +
Sbjct: 294 DAEDVLQYLTRKQEENPQFFFKYTTDEEGRLSNVFWADAESRLDYAAFGGGVIFDSTYRV 353
Query: 65 --LCAWHLLRNANQNLGNP-------LLTKGFKNCMFRKKWDELVLNLGLQDNCWVKETY 115
LC W++ N +++L P L+ + F ++W + N G + W+ Y
Sbjct: 354 NKLCTWYIEENMSRHLRKPKLDELRKLIYESMDEEEFERRWADFKENGGTGNGQWIALMY 413
Query: 116 EKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHMRFRE 175
R W AA+ GK+ G R+ E S+L + +L ++ ++ C++ +R +E
Sbjct: 414 RLREKWAAAYTDGKYLLGMRSNQCSESLNSKLHTLMKRNMSLMCLVKHVKLCIQRLRKKE 473
Query: 176 IEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAVTCLN 235
+ D S + VP + + + LE A + +T +F R ++ ++V+ T+
Sbjct: 474 AQLDIKSTNSVPFCRIDTDPLEKDAARIYTTVMFKKVRAQIRLIAGLEVISGTNQDGSSL 533
Query: 236 YTVAKLSFLAKEW-HVSVSSDN---KDFKYSCMKMESRGLACEHIVDVLAHLRIEELSEC 291
Y V L + W V V+ K + C KME + C HI VL L + +
Sbjct: 534 YVVG-LKDDNEVWDEVRVTFKGQALKGVECHCRKMECEDIPCSHIFVVLKFLGFDTIPHF 592
Query: 292 FIFKRWSKGAKDGVYSGKNIENDFWDFQKSSRCSALMDLYRALGYLNSNTAADFNDATQK 351
+ RW+ GAK S +N + + W CS R LG AA + T+K
Sbjct: 593 CVVDRWTMGAKATFRSDRNTDPNVWSEHMVRYCS-----LRNLGNDAFFEAARNPEQTEK 647
Query: 352 ASELIEQSKAKKSCQRE 368
+ ++ K S E
Sbjct: 648 VMDFLKGFLDKGSSSHE 664
>UniRef100_Q7XMG4 OSJNBa0028I23.18 protein [Oryza sativa]
Length = 1002
Score = 120 bits (301), Expect = 8e-26
Identities = 110/417 (26%), Positives = 177/417 (42%), Gaps = 45/417 (10%)
Query: 61 TEKYLCAWHL----LRNANQNLGNPLLTKGFKNCM--------FRKKWDELVLNLGLQDN 108
T +CAWHL RN NQ + K FK C+ F WD ++ L DN
Sbjct: 508 TTHVICAWHLKHAATRNINQLKSDSDFMKEFKACINLYEEETEFLTSWDAMINKHNLHDN 567
Query: 109 CWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCV 168
W+K+ +E++ W ++RG F AG + T + FQS++ H+ N+ L+ L+ +
Sbjct: 568 VWLKKVFEEKEKWARPYMRGVFSAGMKGTRLNDRFQSDVRDHLRPEVNILLLLRHLETVI 627
Query: 169 KHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCT 228
R +E+E + S + K L A + +TN +F LF+ + +++
Sbjct: 628 NDRRRKELEVEYSSRLKLLYFKIKAPIL-TQAFEAYTNTIFPLFQEEYEEFQSAYIVNRD 686
Query: 229 DAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEEL 288
++ C Y V+ + K + V + + SC K E G C H + +L + I+ L
Sbjct: 687 ESGPCREYVVSVVE-KEKRYTVYGNPTEQTVSCSCKKFERNGFLCSHALKILDAMDIKYL 745
Query: 289 SECFIFKRWSKGAK---DGVYSGKNIENDFWDFQKSSRCSALMDLYRALGYLNSNTAADF 345
+ +I KRW+K A+ G G+ I+ D + SSR + Y L S +
Sbjct: 746 PDRYIMKRWTKYARTLMSGDVQGQAIQADKLS-ESSSRYQYMCPKYVRLVARASECEESY 804
Query: 346 NDATQ---KASELIEQSKAKKSCQREGSLSLSQTNILNLKDPI----------RLRRKGR 392
Q S +E+ K++C +L+QT++ NLK + + K
Sbjct: 805 RVLDQCWVDLSNKVEEILQKQTCV---DATLTQTDVQNLKVSLPSITNGTQAENIMDKSS 861
Query: 393 GTKNKSSLGLTVKHVI---NC----LIFKTPGHNKLTGTYC----SQQGHTMSDFEG 438
GT K S K+ I NC L K H++ Y SQ G+ FEG
Sbjct: 862 GTTAKESKKKGQKNKIQSRNCIEKGLRKKQKVHSEQPSEYALLGGSQSGNMFQAFEG 918
>UniRef100_Q6Z0R9 Putative far-red impaired response protein [Oryza sativa]
Length = 690
Score = 120 bits (300), Expect = 1e-25
Identities = 97/405 (23%), Positives = 172/405 (41%), Gaps = 60/405 (14%)
Query: 61 TEKYLCAWHLLRNANQNLGNPLLTKGFKNCM--------FRKKWDELVLNLGL-QDNCWV 111
+E LC WH+ +N ++L P + F+ + F +KW E L + +DN W+
Sbjct: 300 SEHRLCTWHIEQNMARHL-RPKMISDFRVLVHATYSAEEFEEKWVEFKLKRKVAEDNQWL 358
Query: 112 KETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHM 171
Y R W AA+ +G +F G ++ R E S+L +H+ + +L ++ ++C+ M
Sbjct: 359 GRMYNLRKKWAAAYTKGMYFLGMKSNQRSESLNSKLHRHLDRKMSLVLLVEHYEHCLSRM 418
Query: 172 RFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDA- 230
R RE E D + VP + +E A + FT +VF + + + +V+DC D
Sbjct: 419 RHREAELDSKASQSVPFTANDASLIEKDAARIFTPSVFKKLKMDIIKSKDCEVIDCIDEE 478
Query: 231 -----VTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRI 285
+ C+ + K++ L + V S K+ +C KME L C+H+ ++ +L +
Sbjct: 479 NIIKYIICMKGKIDKITILTCSY---VESSLKNMSCTCKKMECESLPCQHMFTIMDYLNL 535
Query: 286 EELSECFIFKRWSKGAKDGVYSGKNIENDFWDFQKSSRCSALMDLYRALGYLNSNT---A 342
+ + + + RW+ AK S + E W Q M+ YR + + S
Sbjct: 536 QAIPDVCVVPRWTVKAKIAFPSDRYGEVYTWSDQ--------MERYRRMRAIGSEVLLKC 587
Query: 343 ADFNDATQKASELIEQSKAKKSCQREG--------------SLSLSQTNILNLKDPIRLR 388
+ + T K E+ E+ + + EG SL ++ + DP+ +
Sbjct: 588 SMSEEYTLKVMEMFEKLDLETNA-IEGDEFDVTLCGHVVAQSLRSDIASVQKVHDPMEVV 646
Query: 389 RKGRGTKNKSSLGLTVKHVINCLIFKTPGHNKLTGTYCSQQGHTM 433
KG TK F G+ + YC +GHT+
Sbjct: 647 PKGAPTKRLRG-------------FLEKGNRRC--GYCRVRGHTV 676
Score = 42.7 bits (99), Expect = 0.022
Identities = 19/49 (38%), Positives = 28/49 (56%), Gaps = 1/49 (2%)
Query: 14 VELASKDD-NYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
++ KDD +F E+ D+ L+ LFW D R+ Y +GDV+ FD T
Sbjct: 176 MQARQKDDMEFFYEYETDEAGRLKRLFWADPQSRIDYDAFGDVVVFDST 224
>UniRef100_Q65WR3 Putative far-red impaired response protein [Oryza sativa]
Length = 710
Score = 112 bits (280), Expect = 2e-23
Identities = 97/408 (23%), Positives = 180/408 (43%), Gaps = 34/408 (8%)
Query: 3 KNDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVTE 62
+ D+ +++M + AS+D +F D+ + + ++FW D Y+ +GDV A
Sbjct: 214 QGDTGGVLEYMEKKASEDVKFFYSIQVDEDDLITNIFWADSKMVADYEDFGDVNAC---- 269
Query: 63 KYLCAWHLLRNANQNLGNPLLTKGFKNCM--------FRKKWDELVLNLGLQDNCWVKET 114
K+L +++ + F+NC+ F + W +L+ LQ N W++
Sbjct: 270 KHLAG--CVKDYKK------FNADFQNCIYDQEEEEEFLRAWGQLLEKYKLQQNTWLQRI 321
Query: 115 YEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHMRFR 174
++KR W + R F A TT R E +EL ++ +Y++ F + V R+
Sbjct: 322 FKKREQWALVYGRNTFSADMSTTQRNESLNNELKGYISVKYDMLTFFEHFDRLVGDKRYE 381
Query: 175 EIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAVTCL 234
E++ D + P +K L L A + +T V+ +F +M+ L+C D C
Sbjct: 382 EVKCDFRATQSTPKLKAELRILRYVA-EVYTPAVYKIF-----EEEVMQTLNC-DIFYCG 434
Query: 235 NYTVAKLSFLAK-----EWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEELS 289
V K+ + E V S + K SC K E G+ C H + +L I+ +
Sbjct: 435 EVDVEKVYKIKANGKHLEHVVRYSPLESNVKCSCKKFEFAGILCSHALKILDVNNIKSVP 494
Query: 290 ECFIFKRWSKGAKDGVYSGKNIENDFWDFQKSSRCSALMDLYRALGYLNSNTAADFNDAT 349
+ +I KRW+ AK SG + +D + S R S+L ++ + + + ++ +T
Sbjct: 495 QQYILKRWTIDAKVLHISGNSNMHDDPRIKISDRYSSLCRMFVRIASTAAKSEETYSMST 554
Query: 350 QKASELIEQSKAKKSCQREGSLSLSQTNILNLKDPIRLRRKGRGTKNK 397
A +L E + + + L S + + ++ ++ +K RG K K
Sbjct: 555 NCAEKLAEDIEKFLRIRSDPDLDKSPSPQVGKEN--QMPKKPRGIKLK 600
>UniRef100_Q6EP19 Putative far-red impaired response protein [Oryza sativa]
Length = 688
Score = 110 bits (275), Expect = 8e-23
Identities = 92/359 (25%), Positives = 154/359 (42%), Gaps = 35/359 (9%)
Query: 65 LCAWHLLRNANQNLGNP-------LLTKGFKNCMFRKKWDELVLNLGLQDNCWVKETYEK 117
LC WH+ N +++L P L+ + F ++W + N G + W+ Y
Sbjct: 327 LCTWHIEENMSRHLRKPKLDELRKLIYESMDEEEFERRWADFKENGGTGNEQWIALMYRL 386
Query: 118 RSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHMRFREIE 177
R W AA+ GK+ G R+ R E S+L + +L ++ ++ C++ +R +E +
Sbjct: 387 REKWAAAYTDGKYLLGMRSNQRSESLNSKLHTLLKRNMSLMCLVKHVKLCIQGLRKKEAQ 446
Query: 178 DDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAVTCLNYT 237
D S + VP + + + LE A + +T VF R ++ ++V+ T+ Y
Sbjct: 447 LDAKSTNSVPFCRIDADPLEKDAARIYTAVVFKKVRAQIRLIAGLEVISGTNQDGSSLYV 506
Query: 238 VAKLSFLAKEW-HVSVSSDN---KDFKYSCMKMESRGLACEHIVDVLAHLRIEELSECFI 293
V L + W V VS + + C KME + C HI V+ L + + C +
Sbjct: 507 VG-LKDDNEVWDEVRVSFKGQALEGVECHCRKMECEDIPCSHIFVVVKFLGFDTIPRCCV 565
Query: 294 FKRWSKGAKDGVYSGKNIENDFWDFQKSSRCSALMDLYRALGYLNSNT---AADFNDATQ 350
RW+ GAK S +N + + W S M YR+L L S+ AA + T+
Sbjct: 566 VDRWTMGAKAAFRSDRNADPNVW--------SEHMVRYRSLRNLGSDAFFEAARNPEQTE 617
Query: 351 KASELIEQSKAKKSCQREG------------SLSLSQTNILNLKDPIRLRRKGRGTKNK 397
KA + ++ K S E LS +Q + DP +R KG +K +
Sbjct: 618 KAMDFLKGILDKGSSSDENIVAGDFGPMPTHFLSSNQPLEKRVLDPEEIRAKGAPSKRR 676
Score = 42.7 bits (99), Expect = 0.022
Identities = 25/115 (21%), Positives = 49/115 (41%), Gaps = 15/115 (13%)
Query: 4 NDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT-- 61
+D+ ++++ +D +F ++ D+ L+++FW D RL Y +G V+ FD T
Sbjct: 190 HDAEDVLKYLTRKQEEDAEFFFKYTTDEEGRLRNVFWADAESRLDYAAFGGVVIFDSTYR 249
Query: 62 -EKYLCAWHLLRNANQNLGNPLLTKGFKNCMFRKKWDELVLNLGLQDNCWVKETY 115
KY + N + + G ++ N + CW+ ET+
Sbjct: 250 VNKYNLPFIPFIGVNHHRSTTIFGCG------------ILSNESVNSYCWLLETF 292
>UniRef100_Q7XVP7 OSJNBa0050F15.2 protein [Oryza sativa]
Length = 688
Score = 109 bits (272), Expect = 2e-22
Identities = 91/359 (25%), Positives = 154/359 (42%), Gaps = 35/359 (9%)
Query: 65 LCAWHLLRNANQNLGNP-------LLTKGFKNCMFRKKWDELVLNLGLQDNCWVKETYEK 117
LC WH+ N +++L P L+ + F ++W + N G + W+ Y
Sbjct: 327 LCTWHIEENMSRHLRKPKLDELRKLIYESMDEEEFERRWADFKENGGTGNGQWIALMYRL 386
Query: 118 RSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHMRFREIE 177
R W AA+ GK+ G R+ R E S+L + +L ++ ++ C++ +R +E +
Sbjct: 387 REKWAAAYTDGKYLLGMRSNQRSESLNSKLHTLLKRNMSLMCLVKHVKLCIQGLRKKEAQ 446
Query: 178 DDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAVTCLNYT 237
D S + VP + + + LE A + +T VF R ++ ++V+ T+ Y
Sbjct: 447 LDAKSTNSVPFCRIDADPLEKDAARIYTAVVFKKVRAQIRLIAGLEVISGTNQDGSSLYV 506
Query: 238 VAKLSFLAKEW-HVSVSSDN---KDFKYSCMKMESRGLACEHIVDVLAHLRIEELSECFI 293
V L + W V VS + + C KME + C HI V+ L + + C +
Sbjct: 507 VG-LKDDNEVWDEVRVSFKGQALEGVECHCRKMECEDIPCSHIFVVVKFLGFDTIPRCCV 565
Query: 294 FKRWSKGAKDGVYSGKNIENDFWDFQKSSRCSALMDLYRALGYLNSNT---AADFNDATQ 350
RW+ GAK S +N + + W S M YR+L L S+ AA + T+
Sbjct: 566 VDRWTMGAKAAFRSDRNADPNVW--------SEHMVRYRSLRNLGSDAFFEAARNPEQTE 617
Query: 351 KASELIEQSKAKKSCQREGSL------------SLSQTNILNLKDPIRLRRKGRGTKNK 397
KA + ++ K S E + S +Q + DP +R KG +K +
Sbjct: 618 KAMDFLKGILDKGSSSDENIVAGDFGPMPTHFSSSNQPLEKRVLDPEEIRAKGAPSKRR 676
Score = 42.7 bits (99), Expect = 0.022
Identities = 25/115 (21%), Positives = 49/115 (41%), Gaps = 15/115 (13%)
Query: 4 NDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT-- 61
+D+ ++++ +D +F ++ D+ L+++FW D RL Y +G V+ FD T
Sbjct: 190 HDAEDVLKYLTRKQEEDAEFFFKYTTDEEGRLRNVFWADAESRLDYAAFGGVVIFDSTYR 249
Query: 62 -EKYLCAWHLLRNANQNLGNPLLTKGFKNCMFRKKWDELVLNLGLQDNCWVKETY 115
KY + N + + G ++ N + CW+ ET+
Sbjct: 250 VNKYNLPFIPFIGVNHHRSTTIFGCG------------ILSNESVNSYCWLLETF 292
>UniRef100_Q9M8J3 F28L1.19 protein [Arabidopsis thaliana]
Length = 764
Score = 102 bits (255), Expect = 2e-20
Identities = 72/257 (28%), Positives = 119/257 (46%), Gaps = 14/257 (5%)
Query: 58 FDVTEKYLCAWHLLRNANQNLGN-PLLTK-GFKNCMFRKK--------WDELVLNLGLQD 107
F T AW + +NL + P K ++ C+++ + W LV GL+D
Sbjct: 454 FPGTHHRFSAWQIRSKERENLRSFPNEFKYEYEKCLYQSQTTVEFDTMWSSLVNKYGLRD 513
Query: 108 NCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNC 167
N W++E YEKR WV A+LR FF G + F G ++S +L +F+ + +
Sbjct: 514 NMWLREIYEKREKWVPAYLRASFFGGIHVDGTFDPF---YGTSLNSLTSLREFISRYEQG 570
Query: 168 VKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDC 227
++ R E ++D +S + P ++T E +E + +T +F +F+ L + L
Sbjct: 571 LEQRREEERKEDFNSYNLQPFLQTK-EPVEEQCRRLYTLTIFRIFQSELAQSYNYLGLKT 629
Query: 228 TDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEE 287
+ + V K ++ V+ S+ N + SC E GL C HI+ V L I E
Sbjct: 630 YEEGAISRFLVRKCGNENEKHAVTFSASNLNASCSCQMFEYEGLLCRHILKVFNLLDIRE 689
Query: 288 LSECFIFKRWSKGAKDG 304
L +I RW+K A+ G
Sbjct: 690 LPSRYILHRWTKNAEFG 706
>UniRef100_Q9SZL8 Hypothetical protein F20D10.300 [Arabidopsis thaliana]
Length = 788
Score = 102 bits (253), Expect = 3e-20
Identities = 74/307 (24%), Positives = 128/307 (41%), Gaps = 33/307 (10%)
Query: 66 CAWHLLRNANQNLGN-----PLLTKGFKNCM--------FRKKWDELVLNLGLQDNCWVK 112
C WH+L+ + L + P F C+ F + W L+ L+D+ W++
Sbjct: 386 CKWHILKKCQEKLSHVFLKHPSFESDFHKCVNLTESVEDFERCWFSLLDKYELRDHEWLQ 445
Query: 113 ETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHMR 172
Y R WV +LR FFA T R + S ++++ NL+ F + + ++
Sbjct: 446 AIYSDRRQWVPVYLRDTFFADMSLTHRSDSINSYFDGYINASTNLSQFFKLYEKALESRL 505
Query: 173 FREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAVT 232
+E++ D +++ P +KT +E A++ +T +F+ F+ L D
Sbjct: 506 EKEVKADYDTMNSPPVLKTP-SPMEKQASELYTRKLFMRFQEELVGTLTFMASKADDDGD 564
Query: 233 CLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEELSECF 292
+ Y VAK K V + SC E G+ C HI+ V + L +
Sbjct: 565 LVTYQVAKYGEAHKAHFVKFNVLEMRANCSCQMFEFSGIICRHILAVFRVTNLLTLPPYY 624
Query: 293 IFKRWSKGAKDGVYSGKNIENDFWDFQKSSRCSALMDLYRALGYLNSNTAADFNDATQKA 352
I KRW++ AK V I +D+ +L+ YL S+T +N KA
Sbjct: 625 ILKRWTRNAKSSV-----IFDDY-------------NLHAYANYLESHTVR-YNTLRHKA 665
Query: 353 SELIEQS 359
S ++++
Sbjct: 666 SNFVQEA 672
>UniRef100_Q9FYN8 P0702D12.15 protein [Oryza sativa]
Length = 832
Score = 97.1 bits (240), Expect = 1e-18
Identities = 65/262 (24%), Positives = 117/262 (43%), Gaps = 15/262 (5%)
Query: 58 FDVTEKYLCAWHLLRNANQNLGN-------------PLLTKGFKNCMFRKKWDELVLNLG 104
F T +C WH+L+ A + +GN +LT+ F W +L+ +
Sbjct: 366 FPNTIHRVCKWHVLKKAKEFMGNIYSKRHTFKKAFHKVLTQTLTEEEFVAAWHKLIRDYN 425
Query: 105 LQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQL 164
L+ + +++ ++ R W + +FFAG TT R E V ++ F+++
Sbjct: 426 LEKSVYLRHIWDIRRKWAFVYFSHRFFAGMTTTQRSESANHVFKMFVSPSSSMNGFVKRY 485
Query: 165 QNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKV 224
+E ++ + + +KT +E+ A++ +T VF LF L + V
Sbjct: 486 DRFFNEKLQKEDSEEFQTSNDKVEIKTR-SPIEIHASQVYTRAVFQLFSEELTDSLSYMV 544
Query: 225 LDCTDAVTCLNYTV-AKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHL 283
D T + ++ SFL KE+ VS + ++F C E +G+ C HI+ VL
Sbjct: 545 KPGEDESTVQVVRMNSQESFLRKEYQVSCDLEREEFSCVCKMFEHKGILCSHILRVLVQY 604
Query: 284 RIEELSECFIFKRWSKGAKDGV 305
+ + E +I KRW+K A+D +
Sbjct: 605 GLSRIPERYILKRWTKDARDTI 626
Score = 43.9 bits (102), Expect = 0.010
Identities = 18/60 (30%), Positives = 34/60 (56%)
Query: 4 NDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVTEK 63
+D T+ + +L + N+F D+ ++++FW V RL+++ +GDV+ FD T K
Sbjct: 236 DDMQKTLDVLKDLQKRSKNFFYSIQVDEACRVKNIFWSHAVSRLNFEHFGDVITFDTTYK 295
>UniRef100_Q7XRZ8 OSJNBb0002N06.8 protein [Oryza sativa]
Length = 885
Score = 97.1 bits (240), Expect = 1e-18
Identities = 65/262 (24%), Positives = 117/262 (43%), Gaps = 15/262 (5%)
Query: 58 FDVTEKYLCAWHLLRNANQNLGN-------------PLLTKGFKNCMFRKKWDELVLNLG 104
F T +C WH+L+ A + +GN +LT+ F W +L+ +
Sbjct: 419 FPNTIHRVCKWHVLKKAKEFMGNIYSKRHTFKKAFHKVLTQTLTEEEFVAAWHKLIRDYN 478
Query: 105 LQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQL 164
L+ + +++ ++ R W + +FFAG TT R E V ++ F+++
Sbjct: 479 LEKSVYLRHIWDIRRKWAFVYFSHRFFAGMTTTQRSESANHVFKMFVSPSSSMNGFVKRY 538
Query: 165 QNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKV 224
+E ++ + + +KT +E+ A++ +T VF LF L + V
Sbjct: 539 DRFFNEKLQKEDSEEFQTSNDKVEIKTR-SPIEIHASQVYTRAVFQLFSEELTDSLSYMV 597
Query: 225 LDCTDAVTCLNYTV-AKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHL 283
D T + ++ SFL KE+ VS + ++F C E +G+ C HI+ VL
Sbjct: 598 KPGEDESTVQVVRMNSQESFLRKEYQVSCDLEREEFSCVCKMFEHKGILCSHILRVLVQY 657
Query: 284 RIEELSECFIFKRWSKGAKDGV 305
+ + E +I KRW+K A+D +
Sbjct: 658 GLSRIPERYILKRWTKDARDTI 679
Score = 43.9 bits (102), Expect = 0.010
Identities = 18/60 (30%), Positives = 34/60 (56%)
Query: 4 NDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVTEK 63
+D T+ + +L + N+F D+ ++++FW V RL+++ +GDV+ FD T K
Sbjct: 289 DDMQKTLDVLKDLQKRSKNFFYSIQVDEACRVKNIFWSHAVSRLNFEHFGDVITFDTTYK 348
>UniRef100_Q67IU6 Far-red impaired response protein-like [Oryza sativa]
Length = 823
Score = 96.7 bits (239), Expect = 1e-18
Identities = 64/267 (23%), Positives = 114/267 (41%), Gaps = 19/267 (7%)
Query: 65 LCAWHLLRNANQNLGN-------------PLLTKGFKNCMFRKKWDELVLNLGLQDNCWV 111
+C WH+L+NA +NLGN +L + F K W +L+ L+ + ++
Sbjct: 352 VCKWHVLKNAKENLGNIYSKRSSFKQEFHRVLNEPQTEAEFEKAWSDLMEQYNLESSVYL 411
Query: 112 KETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHM 171
+ ++ + W A+ R FFA TT R E L K+V +L F ++ +N
Sbjct: 412 RRMWDMKKKWAPAYFREFFFARMSTTQRSESMNHVLKKYVKPSSSLHGFAKRYENFYND- 470
Query: 172 RFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAV 231
R + + H + +E A++ +T F F+ + + V +D
Sbjct: 471 RIEAEDAEEHDTYNEKVSTLTSSPIEKHASRVYTRGAFSRFKEQFKLSFSFMVYHTSDQH 530
Query: 232 TCLNYTVAK---LSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEEL 288
+ S+ +KE+ V V +D C E G+ C HI+ V+ E+
Sbjct: 531 VLQLVHIGDDTLQSWGSKEFKVQVDLTEQDLSCGCKLFEHLGIICSHIIRVMVQYGFTEI 590
Query: 289 SECFIFKRWSKGAKDGVYSGKNIENDF 315
+ +I KRW+K A+D + K++E +
Sbjct: 591 PKKYILKRWTKDARDSI--PKHLEESY 615
Score = 42.4 bits (98), Expect = 0.028
Identities = 18/58 (31%), Positives = 31/58 (53%)
Query: 4 NDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
ND T+ F E+ + N+F D + ++++FW + RLS+ +GD + FD T
Sbjct: 215 NDVQKTLNFFREMQCRSKNFFYTIQIDGASRIKNIFWCHSLSRLSFDHFGDAITFDTT 272
>UniRef100_Q8W0Q6 Putative far-red impaired response protein [Sorghum bicolor]
Length = 1142
Score = 96.7 bits (239), Expect = 1e-18
Identities = 66/268 (24%), Positives = 109/268 (40%), Gaps = 15/268 (5%)
Query: 58 FDVTEKYLCAWHLLRNANQNLGN-----PLLTKGFKNCM--------FRKKWDELVLNLG 104
F +T C WH++ A L P + F NC+ F W L+ +
Sbjct: 385 FPLTRLQFCKWHIMNEAQDKLSYLLDAFPSFHEDFINCVNMSDTINEFETNWKALISKVS 444
Query: 105 LQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQL 164
Q W+ Y R WV +LR FF +C S ++ ++ + F+QQ
Sbjct: 445 SQKTEWLDLVYNCRHQWVPVYLRDTFFGDVSLKLQCSSRSSLFEGYISAKTDSQSFIQQY 504
Query: 165 QNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKV 224
+ + +E++++ + + +P +KT +E + +T ++FL F+ L A +
Sbjct: 505 EKALDCCYEKEVKEEFETKYSLPDIKTP-SPIEKQGAELYTRSMFLKFQQELIDASVYTA 563
Query: 225 LDCTDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLR 284
+ YTV + K V SS SC E G+ C HI+ V
Sbjct: 564 EMVKEVGNASVYTVTRSEGSEKSVTVEFSSSGSSATCSCRMFEYFGIVCRHILTVFGVRG 623
Query: 285 IEELSECFIFKRWSKGAKDGVYSGKNIE 312
+ L + KRW+K A D SGKN++
Sbjct: 624 VSALPSHYFVKRWTKNALDR-SSGKNVD 650
>UniRef100_Q6ATW8 Hypothetical protein P0015C02.8 [Oryza sativa]
Length = 909
Score = 95.9 bits (237), Expect = 2e-18
Identities = 64/267 (23%), Positives = 114/267 (41%), Gaps = 19/267 (7%)
Query: 65 LCAWHLLRNANQNLGN-------------PLLTKGFKNCMFRKKWDELVLNLGLQDNCWV 111
+C WH+L+NA +NLGN +L + F K W +L+ L+ + ++
Sbjct: 438 VCKWHVLKNAKENLGNIYSKRSSFKQEFHRVLNEPQTEAEFEKAWSDLMEQYNLESSVYL 497
Query: 112 KETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHM 171
+ ++ + W A+ R FFA TT R E L K+V +L F ++ +N
Sbjct: 498 RRMWDMKKKWAPAYFREFFFARMSTTQRSESMNHVLKKYVKPSSSLHGFAKRYENFYND- 556
Query: 172 RFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAV 231
R + + H + +E A++ +T F F+ + + V +D
Sbjct: 557 RIEAEDAEEHDTYNEKVSTLTSSPIEKHASRVYTRGEFSRFKEQFKLSFSFMVYHTSDQH 616
Query: 232 TCLNYTVAK---LSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEEL 288
+ S+ +KE+ V V +D C E G+ C HI+ V+ E+
Sbjct: 617 VLQLVHIGDDTLQSWGSKEFKVQVDLTEQDLSCGCKLFEHLGIICSHIIRVMVQYGFTEI 676
Query: 289 SECFIFKRWSKGAKDGVYSGKNIENDF 315
+ +I KRW+K A+D + K++E +
Sbjct: 677 PKKYILKRWTKDARDSI--PKHLEESY 701
Score = 42.4 bits (98), Expect = 0.028
Identities = 18/58 (31%), Positives = 31/58 (53%)
Query: 4 NDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
ND T+ F E+ + N+F D + ++++FW + RLS+ +GD + FD T
Sbjct: 301 NDVQKTLNFFREMQCRSKNFFYTIQIDGASRIKNIFWCHSLSRLSFDHFGDAITFDTT 358
>UniRef100_Q8H7R4 Hypothetical protein OSJNBa0081P02.17 [Oryza sativa]
Length = 909
Score = 95.9 bits (237), Expect = 2e-18
Identities = 64/267 (23%), Positives = 114/267 (41%), Gaps = 19/267 (7%)
Query: 65 LCAWHLLRNANQNLGN-------------PLLTKGFKNCMFRKKWDELVLNLGLQDNCWV 111
+C WH+L+NA +NLGN +L + F K W +L+ L+ + ++
Sbjct: 438 VCKWHVLKNAKENLGNIYSKRSSFKQEFHRVLNEPQTEAEFDKAWSDLMEQYNLESSVYL 497
Query: 112 KETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHM 171
+ ++ + W A+ R FFA TT R E L K+V +L F ++ +N
Sbjct: 498 RRMWDMKKKWAPAYFREFFFARMSTTQRSESMNHVLKKYVKPSSSLHGFAKRYENFYND- 556
Query: 172 RFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAV 231
R + + H + +E A++ +T F F+ + + V +D
Sbjct: 557 RIEAEDAEEHDTYNEKVSTLTSSPIEKHASRVYTRGAFSRFKEQFKLSFSFMVYHTSDQH 616
Query: 232 TCLNYTVAK---LSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEEL 288
+ S+ +KE+ V V +D C E G+ C HI+ V+ E+
Sbjct: 617 VLQLVHIGDDTLQSWGSKEFKVQVDLTEQDLSCGCKLFEHLGIICSHIIRVMVQYGFTEI 676
Query: 289 SECFIFKRWSKGAKDGVYSGKNIENDF 315
+ +I KRW+K A+D + K++E +
Sbjct: 677 PKKYILKRWTKDARDSI--PKHLEESY 701
Score = 42.4 bits (98), Expect = 0.028
Identities = 18/58 (31%), Positives = 31/58 (53%)
Query: 4 NDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
ND T+ F E+ + N+F D + ++++FW + RLS+ +GD + FD T
Sbjct: 301 NDVQKTLNFFREMQCRSKNFFYTIQIDGASRIKNIFWCHSLSRLSFDHFGDAITFDTT 358
>UniRef100_Q6ESZ2 Putative far-red impaired response protein [Oryza sativa]
Length = 815
Score = 94.7 bits (234), Expect = 5e-18
Identities = 83/356 (23%), Positives = 156/356 (43%), Gaps = 27/356 (7%)
Query: 65 LCAWHLLRNANQNLGNPL-----LTKGFKNCM--------FRKKWDELVLNLGLQDNCWV 111
+C W+L ++A NL + + K FK C+ F W+ ++ L +N W+
Sbjct: 355 ICTWNLKQSAKSNLNHLIRGDCGFIKEFKACINDYEEEVEFFTAWEAMISKYNLHNNVWL 414
Query: 112 KETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHM 171
++ +E++ W + + F AG + T E S++ ++ S ++ FL+ L+ V
Sbjct: 415 QKVFEEKEKWARPYTKWIFSAGMKNTQLNERLHSDVRDYLKSDVDIISFLKHLKKLVNDR 474
Query: 172 RFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAV 231
R+ E+E + S +P K L A++ +T +F LF+ + ++ ++
Sbjct: 475 RYIELEVEFSSRLKLPDFKIRAPILR-QASEAYTGMIFQLFQEEYEEFQSAYIVTRDESG 533
Query: 232 TCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEELSEC 291
Y VA L + + V + + SC K E+ G C H + VL + I+ + +
Sbjct: 534 PSREYIVAILE-KERRYKVHGNPCEQTVTCSCRKFETLGFLCSHALKVLDTMDIKYMPDR 592
Query: 292 FIFKRWSKGAK---DGVYSGKNIENDFWDFQKSSRCSALMDLYRALGYLNSNTAADFNDA 348
+I KRW+K + G+ ++ D + SSR L +Y L S +
Sbjct: 593 YILKRWTKYGRCLTAPQVEGRKVQAD-TTLEFSSRYEYLCPVYVRLVARASECEESYRVL 651
Query: 349 TQKASELIEQSKAKKSCQREGSLSLS--QTNILNLKDPIRLRRKGRGTKNKSSLGL 402
Q + EL K ++ Q++ S+ S Q++I + + + GT N+S L
Sbjct: 652 DQCSVEL--GKKIEEILQKQTSIDASAPQSDI----EDVTISLSANGTDNESERAL 701
>UniRef100_Q8H7L1 Putative transposase [Oryza sativa]
Length = 778
Score = 94.0 bits (232), Expect = 8e-18
Identities = 63/270 (23%), Positives = 119/270 (43%), Gaps = 16/270 (5%)
Query: 57 AFDVTEKYLCAWHLLRNANQNL-----GNPLLTKGFKNCM--------FRKKWDELVLNL 103
A+ T LC W + +N+ ++L G+ K F C+ F W+ ++
Sbjct: 389 AWPGTVHRLCPWQVYQNSVKHLNHIFQGSKTFAKDFGKCVYDYDDEENFLLGWNTMLEKY 448
Query: 104 GLQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQ 163
L++N W+K+ ++ R W + R F A +++ + E ++ L K + +++L F +
Sbjct: 449 DLRNNEWIKKIFDDRDKWSPVYNRHVFTADIKSSLQSESVRNALKKSLSPQFDLLSFFKH 508
Query: 164 LQNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMK 223
+ + R+ E++ D H+ P + + + A +T VF +FR +
Sbjct: 509 YERMLDEFRYAELQADFHASQSFPRIPPS--KMLRQAANMYTPVVFEIFRREFEMFVDSV 566
Query: 224 VLDCTDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHL 283
+ C + Y VA ++ E +V S + SC K E+ G+ C H++ VL
Sbjct: 567 IYSCGEDGNAFEYRVA-VTDRPGEHYVRFDSGDLSVVCSCKKFEAMGIQCCHVLKVLDFR 625
Query: 284 RIEELSECFIFKRWSKGAKDGVYSGKNIEN 313
I+EL + + KRW K K + + N
Sbjct: 626 NIKELPQKYFMKRWKKDVKSASTGNQELLN 655
Score = 37.7 bits (86), Expect = 0.70
Identities = 16/63 (25%), Positives = 32/63 (50%)
Query: 1 MLKNDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDV 60
M D+ +T++++ + + ++F D+ + L ++FW D R + Y DV+ D
Sbjct: 255 MQPGDAGVTVKYLQSMQLSNPSFFYAVQLDEDDKLTNIFWADSKSRTDFSYYSDVVCLDT 314
Query: 61 TEK 63
T K
Sbjct: 315 TYK 317
>UniRef100_Q6Z468 Far-red impaired response protein-like protein [Oryza sativa]
Length = 749
Score = 93.6 bits (231), Expect = 1e-17
Identities = 63/264 (23%), Positives = 116/264 (43%), Gaps = 17/264 (6%)
Query: 58 FDVTEKYLCAWHLLRNANQNLGN-------------PLLTKGFKNCMFRKKWDELVLNLG 104
F T +C WH+L+ A + +GN +LT+ F W +L+ +
Sbjct: 270 FPNTIHRVCKWHVLKKAKEFMGNIYSKRRSFKKAFHKVLTQTLTEEDFEAAWHKLISDYQ 329
Query: 105 LQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQL 164
L+++ +++ ++ R W + +FFAG TT R E V ++ F+++
Sbjct: 330 LENSVYLRHIWDIRRKWAFVYFAHRFFAGMTTTQRSESANHVFKMFVKPSSSMNRFVKRY 389
Query: 165 QNCVKHMRFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKV 224
++ ++ + + +KT+ +E+ A++ +T VF LF L + V
Sbjct: 390 DRFFNEKLQKDDSEEFQTSNDKVEIKTS-SPIEIHASQVYTRAVFQLFSEELTDSLSYMV 448
Query: 225 LDCTDAVTCLNY---TVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLA 281
D T T SFL KE+ V + ++F C E +G+ C H+ VL
Sbjct: 449 KPREDESTVQVVRMCTEESSSFLRKEYQVYYDVEREEFSCVCKMFEHKGILCSHVFRVLV 508
Query: 282 HLRIEELSECFIFKRWSKGAKDGV 305
+ ++ + +I KRW+K A+D V
Sbjct: 509 QYGLSKIPDRYILKRWTKNARDSV 532
>UniRef100_Q67IW8 Far-red impaired response protein-like [Oryza sativa]
Length = 845
Score = 93.2 bits (230), Expect = 1e-17
Identities = 62/267 (23%), Positives = 113/267 (42%), Gaps = 19/267 (7%)
Query: 65 LCAWHLLRNANQNLGN-------------PLLTKGFKNCMFRKKWDELVLNLGLQDNCWV 111
+C WH+L+NA +NLGN +L + F K W +L+ L+ + ++
Sbjct: 352 VCKWHVLKNAKENLGNIYSKRSSFKQEFHRVLNEPQTEAEFEKAWSDLMEQYNLESSVYL 411
Query: 112 KETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQLQNCVKHM 171
+ ++ + W + R F+A TT R E L K+V +L F ++ +N
Sbjct: 412 RRMWDMKKKWAPDYFREFFYARMSTTQRSESMNHVLKKYVKPSSSLHGFAKRYENFYND- 470
Query: 172 RFREIEDDCHSIHGVPAMKTNLESLEMSATKHFTNNVFLLFRPVLQHAPLMKVLDCTDAV 231
R + + H + +E A++ +T F F+ + + V +D
Sbjct: 471 RIEAEDAEEHDTYNEKVSTLTSSPIEKHASRVYTRGAFSRFKEQFKLSFSFMVYHTSDQH 530
Query: 232 TCLNYTVAK---LSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAHLRIEEL 288
+ S+ +KE+ V V +D C E G+ C HI+ V+ E+
Sbjct: 531 VLQLVHIGDDTLQSWGSKEFKVQVDLTEQDLSCGCKLFEHLGIICSHIIRVMVQYGFTEI 590
Query: 289 SECFIFKRWSKGAKDGVYSGKNIENDF 315
+ +I KRW+K A+D + K++E +
Sbjct: 591 PKKYILKRWTKDARDSI--PKHLEESY 615
Score = 42.4 bits (98), Expect = 0.028
Identities = 18/58 (31%), Positives = 31/58 (53%)
Query: 4 NDSVMTMQFMVELASKDDNYFCEHIADQINSLQHLFWRDGVGRLSYQVYGDVLAFDVT 61
ND T+ F E+ + N+F D + ++++FW + RLS+ +GD + FD T
Sbjct: 215 NDVQKTLNFFREMQCRSKNFFYTIQIDGASRIKNIFWCHSLSRLSFDHFGDAITFDTT 272
>UniRef100_Q75G83 Putative transposase [Oryza sativa]
Length = 813
Score = 92.4 bits (228), Expect = 2e-17
Identities = 88/345 (25%), Positives = 134/345 (38%), Gaps = 21/345 (6%)
Query: 58 FDVTEKYLCAWHLLRNANQNLGNPL------LTKGFKNCM--------FRKKWDELVLNL 103
F T LC WH+ N +NL + + FK C+ F K W EL+
Sbjct: 381 FRNTSHRLCLWHIYLNGGKNLSHVIHKHPNKFLADFKRCVYEDRSEYYFNKMWHELLSEY 440
Query: 104 GLQDNCWVKETYEKRSIWVAAHLRGKFFAGCRTTSRCEGFQSELGKHVHSRYNLTDFLQQ 163
L+DN W+ Y+ R W A R F A +T R EG + K + L++ L +
Sbjct: 441 NLEDNKWISNLYKLREKW-AIVFRNSFTADITSTQRSEGMNNVYKKRFRRKLGLSELLVE 499
Query: 164 LQNCVKHMRFREIEDDCHSIHGVPAMKT-NLESLEMSATKHFTNNVFLLFRPVLQHAPLM 222
+R E++ D S H P NL L+++A + + L Q L
Sbjct: 500 CDKVSATLRENELDADFKSRHSNPVTYIPNLPMLKIAAESYTRSMYSELEDEFKQQFTLS 559
Query: 223 KVLDCTDAVTCLNYTVAKLSFLAKEWHVSVSSDNKDFKYSCMKMESRGLACEHIVDVLAH 282
L T+ T L + V + + E V + SC K E GL C+H + V
Sbjct: 560 CKLLKTEGAT-LTFVVMPMEY-DHEATVVFNPTEMTITCSCRKYECIGLLCKHALRVFNM 617
Query: 283 LRIEELSECFIFKRWSKGAKDGVYSGKNIENDFWDFQKSSRCSALMDLYRALGYLNSNTA 342
++ L +I RW+K AK G Y K ++R S ++
Sbjct: 618 NKVFTLPSHYILNRWTKYAKSGFYIQKQGSEKETLKAHAARISRHATSVELKCSVSKELL 677
Query: 343 ADFNDATQKASELIEQSKAK---KSCQREGSLSLSQTNILNLKDP 384
D A K + S +K KSC+ + + + LN++DP
Sbjct: 678 DDLEQAINKLDLEADNSLSKMQEKSCEVPLNSNGCVKDTLNVEDP 722
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 775,465,072
Number of Sequences: 2790947
Number of extensions: 32274365
Number of successful extensions: 66257
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 89
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 65970
Number of HSP's gapped (non-prelim): 197
length of query: 463
length of database: 848,049,833
effective HSP length: 131
effective length of query: 332
effective length of database: 482,435,776
effective search space: 160168677632
effective search space used: 160168677632
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0023.16


