

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
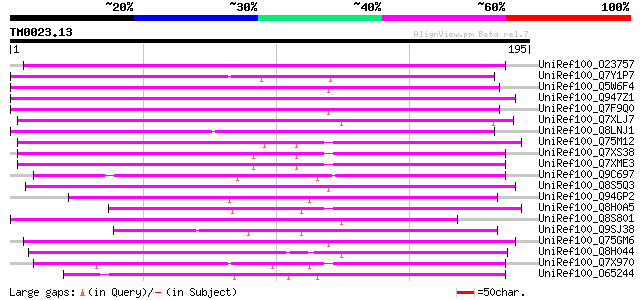
Query= TM0023.13
(195 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O23757 Reverse transcriptase [Beta vulgaris subsp. vul... 123 3e-27
UniRef100_Q7Y1P7 Putative reverse transcriptase [Oryza sativa] 105 5e-22
UniRef100_Q5W6F4 Hypothetical protein OSJNBb0006B22.8 [Oryza sat... 102 5e-21
UniRef100_Q947Z1 Putative retroelement [Oryza sativa] 101 1e-20
UniRef100_Q7F9Q0 OSJNBa0045O17.3 protein [Oryza sativa] 100 2e-20
UniRef100_Q7XLJ7 OSJNBa0009K15.13 protein [Oryza sativa] 90 4e-17
UniRef100_Q8LNJ1 Putative reverse transcriptase [Oryza sativa] 88 1e-16
UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa] 87 2e-16
UniRef100_Q7XS38 OSJNBa0081G05.2 protein [Oryza sativa] 87 2e-16
UniRef100_Q7XME3 OSJNBa0061G20.16 protein [Oryza sativa] 87 2e-16
UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [A... 87 3e-16
UniRef100_Q8S5Q3 Putative non-LTR retroelement reverse transcrip... 86 5e-16
UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa] 82 6e-15
UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa] 81 2e-14
UniRef100_Q8S801 Putative retroelement [Oryza sativa] 81 2e-14
UniRef100_Q9SJ38 Putative non-LTR retroelement reverse transcrip... 80 2e-14
UniRef100_Q75GM6 Putative non-LTR retroelement reverse transcrip... 80 2e-14
UniRef100_Q8H044 Putative reverse transcriptase [Oryza sativa] 79 6e-14
UniRef100_Q7X970 BZIP-like protein [Oryza sativa] 79 6e-14
UniRef100_O65244 F21E10.5 protein [Arabidopsis thaliana] 79 6e-14
>UniRef100_O23757 Reverse transcriptase [Beta vulgaris subsp. vulgaris]
Length = 507
Score = 123 bits (308), Expect = 3e-27
Identities = 62/181 (34%), Positives = 94/181 (51%)
Query: 6 QKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYF 65
+K++ LD L E ER WT L+ E SR++W+KEGD NT+YF
Sbjct: 168 EKIHFLDNIANSRLLCTNELQERDDAQLNLWTWLKRSESYWALNSRARWIKEGDSNTRYF 227
Query: 66 HSLINWRKRTNSIMGLMIDGNWVEDPQAIKTEVQNYFEQCFTNSLGTLVRLDEISFKSIS 125
H+L R+ N + + +G + P IK E YF+ F T + ++F +S
Sbjct: 228 HTLATIRRHKNHLQYIKTEGKNISKPGEIKEEAVAYFQHIFAEEFSTRPVFNGLNFNKLS 287
Query: 126 DADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNR 185
+++ L F EE+ AV C K+P PD FNFKFI+ W+++K D ++++EDFW
Sbjct: 288 ESECSSLIAPFTHEEIDEAVDSCDAQKAPGPDGFNFKFIKSSWEVIKKDIYKIVEDFWAS 347
Query: 186 G 186
G
Sbjct: 348 G 348
>UniRef100_Q7Y1P7 Putative reverse transcriptase [Oryza sativa]
Length = 1556
Score = 105 bits (263), Expect = 5e-22
Identities = 69/186 (37%), Positives = 99/186 (53%), Gaps = 5/186 (2%)
Query: 1 RNEVVQKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQ 60
+ E++ KL+ L +K E L+E E N + L D +LR E YQ+++ K L EGD
Sbjct: 1082 KKELLDKLDTLVKKAEHTILNEFELNVKHVLNDRLAELLREEEIKWYQRAKVKHLLEGDA 1141
Query: 61 NTKYFHSLINWRKRTNSIMGLMIDGNWVEDPQA-IKTEVQNYFEQCFTNSLGTLVRLDEI 119
NTKY+H L N R R I L DGN V A +K + YF+ F S + + LDE
Sbjct: 1142 NTKYYHLLANGRHRKTRIFQLQ-DGNEVITGDAQLKEHITKYFKTLFGPSQTSSIFLDES 1200
Query: 120 ---SFKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFH 176
+ +S +NE LT F EE+K A+++ + +P PD F +F Q FW ++KDD
Sbjct: 1201 QVDDIRQVSAEENESLTADFTEEEIKCAIFQMKHNTAPGPDGFPPEFYQAFWTIIKDDLL 1260
Query: 177 RVLEDF 182
+ DF
Sbjct: 1261 ALFSDF 1266
>UniRef100_Q5W6F4 Hypothetical protein OSJNBb0006B22.8 [Oryza sativa]
Length = 1237
Score = 102 bits (254), Expect = 5e-21
Identities = 59/187 (31%), Positives = 103/187 (54%), Gaps = 3/187 (1%)
Query: 1 RNEVVQKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQ 60
+ +++ KL+ LD+K E LS EE + R + + ++LR E YQ+++SK + EGD
Sbjct: 943 KKKILDKLDLLDKKAENSLLSPEELDIRWFCRNRLSSILREEEIKWYQRAKSKDILEGDS 1002
Query: 61 NTKYFHSLINWRKRTNSIMGLMIDGNWVEDPQAIKTEVQNYFEQCFTNSLGTLVRLDE-- 118
NTKYFH + N + R I L + ++ +K+ + Y++ F +L++ DE
Sbjct: 1003 NTKYFHLVANGKHRKTRIFQLQDSDHLIDGDANLKSHITTYYKGLFGPPDDSLLQFDESR 1062
Query: 119 -ISFKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHR 177
IS +NE LT F +E+K A+++ +K+ PD F +F Q FW+++K+D
Sbjct: 1063 VADIPQISLLENEALTQDFTEKEIKKAIFQMEHNKASGPDGFPAEFYQVFWNVIKNDLLD 1122
Query: 178 VLEDFWN 184
+ ++F N
Sbjct: 1123 LFKEFHN 1129
>UniRef100_Q947Z1 Putative retroelement [Oryza sativa]
Length = 427
Score = 101 bits (251), Expect = 1e-20
Identities = 60/190 (31%), Positives = 98/190 (51%)
Query: 1 RNEVVQKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQ 60
+ E++ K+ ++D+K E G++ E ER L +E V+R Q+ + K L EGD
Sbjct: 11 KQEIMGKIEDIDKKCEAYGMTILERKERGDLEEELKKVVREDRLKWMQRCKEKNLLEGDD 70
Query: 61 NTKYFHSLINWRKRTNSIMGLMIDGNWVEDPQAIKTEVQNYFEQCFTNSLGTLVRLDEIS 120
NTKY+H+ N RKR N I L + ++ + + N+++Q F L+
Sbjct: 71 NTKYYHAKANGRKRKNMIYSLDQEDGEIKGQSDLMKYITNFYKQLFGPPPENDFTLNLEG 130
Query: 121 FKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLE 180
+S+A+ E L MEE+K V+ +K+P PD F +F + FW L+KDD ++
Sbjct: 131 IAMLSEAEKERLIRPIEMEELKKVVFGMENNKAPGPDGFPVEFYKHFWYLIKDDLMELII 190
Query: 181 DFWNRGCGLK 190
DF R G++
Sbjct: 191 DFMKRKIGVE 200
>UniRef100_Q7F9Q0 OSJNBa0045O17.3 protein [Oryza sativa]
Length = 492
Score = 100 bits (250), Expect = 2e-20
Identities = 59/187 (31%), Positives = 98/187 (51%), Gaps = 3/187 (1%)
Query: 1 RNEVVQKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQ 60
+ E++ L+ LD+K E L EE + R + + ++LR E YQ+++SK L EGD
Sbjct: 74 KKEILDNLDLLDKKAETSLLRPEELDIRWFYRNRLSSILREEEIKWYQRAKSKDLLEGDS 133
Query: 61 NTKYFHSLINWRKRTNSIMGLMIDGNWVEDPQAIKTEVQNYFEQCFTNSLGTLVRLDE-- 118
NTKYFH + N + R I L + + +K+ + Y++ F +L++ DE
Sbjct: 134 NTKYFHLVANGKHRKTRIFQLHDGDHLIHGDANLKSHITTYYKGLFGPPEESLLQFDESK 193
Query: 119 -ISFKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHR 177
+S +NE LT F E+K A+++ +K+P PD F +F Q FW ++K+D
Sbjct: 194 VADIPQVSHLENEALTQEFIEMEIKEAIFQMEHNKAPGPDGFPAEFYQVFWSVIKNDLFD 253
Query: 178 VLEDFWN 184
+ +F N
Sbjct: 254 LFTEFHN 260
>UniRef100_Q7XLJ7 OSJNBa0009K15.13 protein [Oryza sativa]
Length = 779
Score = 89.7 bits (221), Expect = 4e-17
Identities = 54/196 (27%), Positives = 94/196 (47%), Gaps = 10/196 (5%)
Query: 4 VVQKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTK 63
+++++ + D + LS+ E +R L D+ + + E ++S KWL EGD NT
Sbjct: 19 LLKEMQKWDDAADSRNLSQGEWKQRYELEDQLNIIFQEEEIYWQERSGEKWLLEGDANTT 78
Query: 64 YFHSLINWRKRTNSIMGLMIDGNWVEDPQAIKTEVQNYFEQCFTNSLGTLVRLDEISFKS 123
+FH + N +KR +I L DG +E ++ + Y++ F + L V L + +
Sbjct: 79 FFHGVANGKKRIITIRSLEEDGRVIEGNSELREHITRYYKGLFGSELPPKVFLSQDMRRD 138
Query: 124 ---ISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLE 180
+ DN+ L F MEE++ A+ E + +P PD F ++FW+ LKD +L+
Sbjct: 139 RGRVDQEDNDFLVRPFSMEEIERALAEMKTNTAPGPDGLPVCFYKEFWEQLKDQIKEMLD 198
Query: 181 -------DFWNRGCGL 189
D W G+
Sbjct: 199 SLFEGRLDLWRLNYGV 214
>UniRef100_Q8LNJ1 Putative reverse transcriptase [Oryza sativa]
Length = 1151
Score = 87.8 bits (216), Expect = 1e-16
Identities = 49/182 (26%), Positives = 95/182 (51%), Gaps = 1/182 (0%)
Query: 1 RNEVVQKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQ 60
+ ++++K++ +D K E G++ + ER+ L + +R + +++S+ K EGD
Sbjct: 798 KEDILRKIDAIDIKVESAGMTSSDREERRSLEQKLCYTIREEKLKWFRRSKVKETLEGDC 857
Query: 61 NTKYFHSLINWRKRTNSIMGLMIDGNWVEDPQAIKTEVQNYFEQCFTNSLGTLVRLDEIS 120
NTKY+H+ N R+++ I L + +E + + +++ F +S + +D
Sbjct: 858 NTKYYHAKANGRRKSQ-IHSLSQEEGEIEGQSNLLQYITQFYKNLFGHSKENSITIDLNG 916
Query: 121 FKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLE 180
+S+ D L F +EEV+ V+ +K+P PD +F KFWDL+K+D ++
Sbjct: 917 TCKVSEEDKAKLVQPFCLEEVRTVVFYLKHNKAPGPDGLPGEFYVKFWDLIKNDLLDLIN 976
Query: 181 DF 182
DF
Sbjct: 977 DF 978
>UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa]
Length = 1936
Score = 87.4 bits (215), Expect = 2e-16
Identities = 59/194 (30%), Positives = 96/194 (49%), Gaps = 8/194 (4%)
Query: 4 VVQKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTK 63
V ++L + +K E+ S + + D +L E L Q+SR WLKEGD+NT+
Sbjct: 934 VGKELEKARKKLEDLIASNAARSSIRQATDHMNEMLYREEMLWLQRSRVNWLKEGDRNTR 993
Query: 64 YFHSLINWRKRTNSIMGLMIDGNWVEDPQAI-KTEVQNYFEQCF----TNSLGTLVRLDE 118
+FHS WR + N I L + + ++ +T YF+ + + + ++ RL +
Sbjct: 994 FFHSRAVWRAKKNKISKLRDENGAIHSTTSVLETMATEYFQGVYKADPSLNPESVTRLFQ 1053
Query: 119 ISFKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRV 178
+ ++DA NE L F EE+ A+++ G KSP PD F +F Q+ W LK D
Sbjct: 1054 ---EKVTDAMNEKLCQEFKEEEIAQAIFQIGPLKSPRPDGFPARFYQRNWGTLKSDIILA 1110
Query: 179 LEDFWNRGCGLKDV 192
+ +F+ G K V
Sbjct: 1111 VRNFFQSGLMPKGV 1124
>UniRef100_Q7XS38 OSJNBa0081G05.2 protein [Oryza sativa]
Length = 1711
Score = 87.4 bits (215), Expect = 2e-16
Identities = 57/188 (30%), Positives = 94/188 (49%), Gaps = 8/188 (4%)
Query: 4 VVQKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTK 63
V ++L + +K E+ S + + D +L E L Q+SR WLKEGD+NT+
Sbjct: 846 VGRELEKARKKLEDLIASNAARSSIRQATDHMNEMLYREEMLWLQRSRVNWLKEGDRNTR 905
Query: 64 YFHSLINWRKRTNSIMGLMIDGNWVED-PQAIKTEVQNYFEQCF----TNSLGTLVRLDE 118
+FHS WR + N I L + + ++T YF++ + + + ++ RL +
Sbjct: 906 FFHSRAVWRAKKNKISKLRDENGAIHSMTSVLETMATEYFQEVYKADPSLNPESVTRLFQ 965
Query: 119 ISFKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRV 178
+ ++DA NE L F EE+ A+++ G KSP PD F +F Q+ W LK D
Sbjct: 966 ---EKVTDAMNEKLCQEFKEEEIAQAIFQIGPLKSPGPDGFPARFYQRNWGTLKSDIILA 1022
Query: 179 LEDFWNRG 186
+ +F+ G
Sbjct: 1023 VRNFFQSG 1030
>UniRef100_Q7XME3 OSJNBa0061G20.16 protein [Oryza sativa]
Length = 1285
Score = 87.4 bits (215), Expect = 2e-16
Identities = 57/188 (30%), Positives = 94/188 (49%), Gaps = 8/188 (4%)
Query: 4 VVQKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTK 63
V ++L + +K E+ S + + D +L E L Q+SR WLKEGD+NT+
Sbjct: 846 VGRELEKARKKLEDLIASNAARSSIRQATDHMNEMLYREEMLWLQRSRVNWLKEGDRNTR 905
Query: 64 YFHSLINWRKRTNSIMGLMIDGNWVED-PQAIKTEVQNYFEQCF----TNSLGTLVRLDE 118
+FHS WR + N I L + + ++T YF++ + + + ++ RL +
Sbjct: 906 FFHSRAVWRAKKNKISKLRDENGAIHSMTSVLETMATEYFQEVYKADPSLNPESVTRLFQ 965
Query: 119 ISFKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRV 178
+ ++DA NE L F EE+ A+++ G KSP PD F +F Q+ W LK D
Sbjct: 966 ---EKVTDAMNEKLCQEFKEEEIAQAIFQIGPLKSPGPDGFPARFYQRNWGTLKSDIILA 1022
Query: 179 LEDFWNRG 186
+ +F+ G
Sbjct: 1023 VRNFFQSG 1030
>UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [Arabidopsis thaliana]
Length = 1142
Score = 86.7 bits (213), Expect = 3e-16
Identities = 61/180 (33%), Positives = 89/180 (48%), Gaps = 7/180 (3%)
Query: 10 ELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLI 69
ELD + ++ S EE E L E + R E YQKSRS W+K GD N+K+FH+L
Sbjct: 87 ELDVAQRDDDRSREEITELTLRLKEAY---RDEEQYWYQKSRSLWMKLGDNNSKFFHALT 143
Query: 70 NWRKRTNSIMGLMID-GNWVEDPQAIKTEVQNYFEQCFTNSLGTLV--RLDEISFKSISD 126
R+ N I GL + G W + I+ +YF+ FT + + L E+ I+D
Sbjct: 144 KQRRARNRITGLHDENGIWSIEDDDIQNIAVSYFQNLFTTANPQVFDEALGEVQV-LITD 202
Query: 127 ADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNRG 186
N+LLT EV+ A++ +K+P PD F QK W ++K D ++ F G
Sbjct: 203 RINDLLTADATECEVRAALFMIHPEKAPGPDGMTALFFQKSWAIIKSDLLSLVNSFLQEG 262
>UniRef100_Q8S5Q3 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1228
Score = 85.9 bits (211), Expect = 5e-16
Identities = 53/187 (28%), Positives = 91/187 (48%), Gaps = 3/187 (1%)
Query: 7 KLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFH 66
K+ ELD+K E L+ +E N+R ++ + + E +++ +W+ EGD+NT++FH
Sbjct: 824 KIEELDKKALTEELTSDEWNQRYSWENDLEHIYEMEELYWHKRCGEQWILEGDRNTEFFH 883
Query: 67 SLINWRKRTNSIMGLMIDGNWVEDPQAIKTEVQNYFEQCFTNSLGTLVRLDE---ISFKS 123
+ N RKR I LM + I+ + Y++ F + + L + + +
Sbjct: 884 RVANGRKRKCHITSLMDGETELTSKSEIREHIVEYYKTLFRDVPTANIHLSQGVWTNHLN 943
Query: 124 ISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFW 183
+SDA E LT F M E+ + E + +P PD F+ F + FW +K+D +L
Sbjct: 944 LSDASKENLTRPFVMAELDKVINEAKLNTAPGPDGFSIPFYRAFWPQVKNDLFEMLLMLH 1003
Query: 184 NRGCGLK 190
N LK
Sbjct: 1004 NEDLDLK 1010
>UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa]
Length = 1833
Score = 82.4 bits (202), Expect = 6e-15
Identities = 53/163 (32%), Positives = 85/163 (51%), Gaps = 2/163 (1%)
Query: 23 EENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLINWRKRTNSIMGL- 81
+ +NE + E +L E Q+SR WLKEGD NT+YFH +WR + N I L
Sbjct: 298 DTDNEVHSVKKELEEMLHREEIWWKQRSRITWLKEGDLNTRYFHLKASWRAKKNKIKKLK 357
Query: 82 MIDGNWVEDPQAIKTEVQNYFEQCFTNSLG-TLVRLDEISFKSISDADNELLTGRFGMEE 140
DG+ + + +K +++F+Q +T V L + + IS+ N L F EE
Sbjct: 358 KNDGSTTMNKKEMKEINRSFFQQLYTKDDNLNPVNLLNMFHEKISEQMNADLIKPFTNEE 417
Query: 141 VKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFW 183
+ A+++ G K+P PD F +F+Q+ W LLK + + +F+
Sbjct: 418 ISDALFQIGPLKAPGPDGFPARFLQRNWGLLKGEVIAAVRNFF 460
>UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa]
Length = 1557
Score = 80.9 bits (198), Expect = 2e-14
Identities = 49/160 (30%), Positives = 81/160 (50%), Gaps = 8/160 (5%)
Query: 38 VLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLINWRKRTNSIMGLM-IDGNWVEDPQAIKT 96
+L E L Q+SR WLKEGD+NTK+FH+ + WR + N I L G + ++
Sbjct: 716 LLYREEMLWLQRSRVNWLKEGDRNTKFFHNRVAWRAKKNKITKLRDAAGTFHRSKMVMEQ 775
Query: 97 EVQNYFEQCFTN----SLGTLVRLDEISFKSISDADNELLTGRFGMEEVKGAVWECGGDK 152
YF+ FT + RL + +++ N L F +E+ A+++ G K
Sbjct: 776 MATQYFKDMFTADPNLDHSPVTRLIQ---PKVTEEMNNRLCSEFSEDEIANALFQIGPLK 832
Query: 153 SPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNRGCGLKDV 192
+P D F +F Q+ W +K+D R +++F++ G L+ V
Sbjct: 833 APGTDGFLARFYQRNWGCIKNDIVRAVKEFFHSGIMLEGV 872
>UniRef100_Q8S801 Putative retroelement [Oryza sativa]
Length = 288
Score = 80.9 bits (198), Expect = 2e-14
Identities = 48/171 (28%), Positives = 85/171 (49%), Gaps = 3/171 (1%)
Query: 1 RNEVVQKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQ 60
+ E++ ++ ++D + E++ L+ EE R L + E +S KW+ EGD
Sbjct: 114 KEELISQIQKIDEEVEKKELTCEEWGHRYELEGALNQIFISEEIYWQGRSGEKWMLEGDA 173
Query: 61 NTKYFHSLINWRKRTNSIMGLMIDGNWVEDPQAIKTEVQNYFEQCFTNSLGTLVRLDEIS 120
N +FH + N RKR +I L + +E+ +K + ++++ F L L
Sbjct: 174 NAAFFHGVANGRKRKTTIRSLEGEEGPIENADELKEHIYGFYKKLFGMELPPKFCLSHNM 233
Query: 121 FKS---ISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFW 168
++ +S DN LT F MEE++GA+ E + +P PD F+ F ++FW
Sbjct: 234 WEGRGRLSSEDNIELTKPFSMEEIEGALREMKTNTAPGPDGFSVSFYKEFW 284
>UniRef100_Q9SJ38 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1229
Score = 80.5 bits (197), Expect = 2e-14
Identities = 51/147 (34%), Positives = 74/147 (49%), Gaps = 4/147 (2%)
Query: 40 RLHE*LLYQKSRSKWLKEGDQNTKYFHSLINWRKRTNSIMGLMIDGNWV--EDPQAIKTE 97
+L E Q+SR WL GD+NT YFH++ +RT + + +M D N V + I
Sbjct: 225 KLEEQFWKQRSRVLWLHSGDRNTGYFHAVTR-NRRTQNRLTVMEDINGVAQHEEHQISQI 283
Query: 98 VQNYFEQCFTN-SLGTLVRLDEISFKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEP 156
+ YF+Q FT+ S G +DE +S DN+ LT EEVK AV+ K+P P
Sbjct: 284 ISGYFQQIFTSESDGDFSVVDEAIEPMVSQGDNDFLTRIPNDEEVKDAVFSINASKAPGP 343
Query: 157 DEFNFKFIQKFWDLLKDDFHRVLEDFW 183
D F F +W ++ D R + F+
Sbjct: 344 DGFTAGFYHSYWHIISTDVGREIRLFF 370
>UniRef100_Q75GM6 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1614
Score = 80.5 bits (197), Expect = 2e-14
Identities = 49/188 (26%), Positives = 88/188 (46%), Gaps = 3/188 (1%)
Query: 6 QKLNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYF 65
+KL E+D K + L+ +E R L D + L E +++ +WL EGD+NT+Y+
Sbjct: 925 KKLEEIDSKAVDAELNADEWRHRYELEDALEHIYELEELYWHKRCGEQWLLEGDRNTEYY 984
Query: 66 HSLINWRKRTNSIMGLMIDGNWVEDPQAIKTEVQNYFEQCFTNSLGTLVRLDE---ISFK 122
H + N RK+ +I LM + + +K + Y++ F + + + L + +
Sbjct: 985 HRIANGRKKRCTITSLMDGDRELTTKEDLKHHIVEYYKSLFRSESSSSIHLSQGVWVDSL 1044
Query: 123 SISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDF 182
+ + D L F +EE+ + E + + PD F+ F + FW L+ D +L
Sbjct: 1045 CLKEEDKNTLMRPFTIEELYKVLREAKPNTAFGPDGFSIPFYRAFWPQLRPDLFEMLLML 1104
Query: 183 WNRGCGLK 190
+N LK
Sbjct: 1105 YNEELDLK 1112
>UniRef100_Q8H044 Putative reverse transcriptase [Oryza sativa]
Length = 1183
Score = 79.0 bits (193), Expect = 6e-14
Identities = 56/182 (30%), Positives = 85/182 (45%), Gaps = 5/182 (2%)
Query: 8 LNELDRKEEEEGLSEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFHS 67
+ LD +E LS +E R L + + + Q SR ++EGD NTK+FH
Sbjct: 37 IQRLDVAQEHRDLSPQEMWLRAALKRKILGLASIERARKKQASRVTHIREGDANTKFFHL 96
Query: 68 LINWRKRTNSIMGLMIDGNWVEDPQAIKTEVQNYFEQCFTNSLGTLVRLDEISFKS--IS 125
IN RKR N+I L D W ++ + ++F F G R +E+++++ I
Sbjct: 97 KINVRKRKNTIQRLRKDNGWAVTHGEKESTIHDHFAS-FMGRSGP--RPNELNWEALDIM 153
Query: 126 DADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNR 185
D L F EEV A+ E DK+P PD F F + W+++K+D V +N
Sbjct: 154 PVDLATLGKPFSEEEVHRAIKEMPADKAPGPDGFTGAFFKACWEIIKEDILLVFNSIFNL 213
Query: 186 GC 187
C
Sbjct: 214 RC 215
>UniRef100_Q7X970 BZIP-like protein [Oryza sativa]
Length = 2367
Score = 79.0 bits (193), Expect = 6e-14
Identities = 58/186 (31%), Positives = 88/186 (47%), Gaps = 13/186 (6%)
Query: 10 ELDRKEEEEGLSEEENNERKWL---LDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFH 66
EL++ ++ E N +R + D +L E L Q+SR WLK+ D+NTK+FH
Sbjct: 1031 ELEKARKKLAELIESNADRTVIRNATDHMNELLYREEMLWLQRSRVNWLKDEDRNTKFFH 1090
Query: 67 SLINWRKRTNSIMGLMIDGNWVEDPQAIKTE--VQNYFEQCFTNSLG----TLVRLDEIS 120
S WR + N I L D N +K E YF+ +T T+ RL +
Sbjct: 1091 SRAVWRAKKNKISKLR-DANETVHSSTMKLESMATEYFQDVYTADPNLNPETVTRLIQ-- 1147
Query: 121 FKSISDADNELLTGRFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLE 180
+ ++D NE L F +E+ A+++ G KSP PD F +F Q+ W +K D +
Sbjct: 1148 -EKVTDIMNEKLCEDFTEDEISQAIFQIGPLKSPGPDGFPARFYQRNWGTIKADIIGAVR 1206
Query: 181 DFWNRG 186
F+ G
Sbjct: 1207 RFFQTG 1212
>UniRef100_O65244 F21E10.5 protein [Arabidopsis thaliana]
Length = 928
Score = 79.0 bits (193), Expect = 6e-14
Identities = 52/172 (30%), Positives = 84/172 (48%), Gaps = 9/172 (5%)
Query: 21 SEEENNERKWLLDEFWTVLRLHE*LLYQKSRSKWLKEGDQNTKYFHSLINWRKRTNSIMG 80
S +E NE D + L E L Q+S+ WL GD+N K FH + R+ NSI
Sbjct: 223 SMQEENEAYAKWDH---IAVLEEKFLKQRSKLHWLDIGDRNNKAFHRAVVAREAQNSIRE 279
Query: 81 LMI-DGNWVEDPQAIKTEVQNYFE---QCFTNSLGTLV--RLDEISFKSISDADNELLTG 134
++ DG+ + IKTE +++F Q N + L ++ SD+D E+LT
Sbjct: 280 IICHDGSVASQEEKIKTEAEHHFREFLQLIPNDFEGIAVEELQDLLPYRCSDSDKEMLTN 339
Query: 135 RFGMEEVKGAVWECGGDKSPEPDEFNFKFIQKFWDLLKDDFHRVLEDFWNRG 186
EE+ V+ DKSP PD + +F + W+++ +F ++ F+ +G
Sbjct: 340 HVSAEEIHKVVFSMPNDKSPGPDGYTAEFYKGAWNIIGAEFILAIQSFFAKG 391
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.139 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 368,471,309
Number of Sequences: 2790947
Number of extensions: 16002038
Number of successful extensions: 55175
Number of sequences better than 10.0: 347
Number of HSP's better than 10.0 without gapping: 201
Number of HSP's successfully gapped in prelim test: 146
Number of HSP's that attempted gapping in prelim test: 54780
Number of HSP's gapped (non-prelim): 387
length of query: 195
length of database: 848,049,833
effective HSP length: 121
effective length of query: 74
effective length of database: 510,345,246
effective search space: 37765548204
effective search space used: 37765548204
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0023.13


