

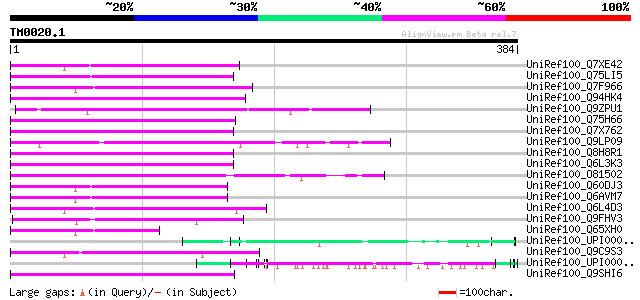
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0020.1
(384 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XE42 Putative mutator-like transposase [Oryza sativa] 132 1e-29
UniRef100_Q75LI5 Putative transposon protein [Oryza sativa] 129 1e-28
UniRef100_Q7F966 OSJNBa0091C07.2 protein [Oryza sativa] 118 3e-25
UniRef100_Q94HK4 Mutator-like transposase [Oryza sativa] 103 6e-21
UniRef100_Q9ZPU1 Mutator-like transposase [Arabidopsis thaliana] 95 3e-18
UniRef100_Q75H66 Hypothetical protein OSJNBb0007E22.22 [Oryza sa... 94 5e-18
UniRef100_Q7X762 OSJNBb0118P14.3 protein [Oryza sativa] 91 7e-17
UniRef100_Q9LP09 F9C16.9 [Arabidopsis thaliana] 89 2e-16
UniRef100_Q8H8R1 Putative mutator-like transposase [Oryza sativa] 89 3e-16
UniRef100_Q6L3K3 Putative transposon MuDR mudrA-like protein [So... 89 3e-16
UniRef100_O81502 F9D12.2 protein [Arabidopsis thaliana] 86 1e-15
UniRef100_Q60DJ3 MuDR family transposase protein [Oryza sativa] 86 2e-15
UniRef100_Q6AVM7 Putative polyprotein [Oryza sativa] 86 2e-15
UniRef100_Q6L4D3 Hypothetical protein OSJNBa0088M05.7 [Oryza sat... 84 7e-15
UniRef100_Q9FHV3 Mutator-like transposase-like protein [Arabidop... 83 1e-14
UniRef100_Q65XH0 Hypothetical protein OJ1126_B11.9 [Oryza sativa] 80 7e-14
UniRef100_UPI0000234ADD UPI0000234ADD UniRef100 entry 80 1e-13
UniRef100_Q9C9S3 Putative Mutator-like transposase; 12516-14947 ... 80 1e-13
UniRef100_UPI00003AF275 UPI00003AF275 UniRef100 entry 79 2e-13
UniRef100_Q9SHI6 Similar to mudrA protein [Arabidopsis thaliana] 79 2e-13
>UniRef100_Q7XE42 Putative mutator-like transposase [Oryza sativa]
Length = 1005
Score = 132 bits (333), Expect = 1e-29
Identities = 68/176 (38%), Positives = 102/176 (57%), Gaps = 3/176 (1%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMGYQGFLPP--VQKSRLEKEAAQKWTATW 58
I++ R KPIITMLE+IR V R I N+ + + P ++K + Q W
Sbjct: 655 IIESRFKPIITMLEDIRIQVTRRIQENRSNSERWTMTVCPNIIRKFNKIRHRTQFCHVLW 714
Query: 59 SGQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLT 118
+G ++ FEV R VDL + TC+CR+WQ++G+PC HACAA+ + P N +H +
Sbjct: 715 NG-DAGFEVRDKKWRFTVDLTSKTCSCRYWQVSGIPCQHACAALFKMAQEPNNCIHECFS 773
Query: 119 MGAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKKKAGRPKKNRRRDGTEEQVAG 174
+ Y+ TY+ +QP + W +P KP+PP +KK GRPKKNRR+D ++ +G
Sbjct: 774 LERYKKTYQHVLQPVEHESAWPVSPNPKPLPPRVKKMPGRPKKNRRKDPSKPVKSG 829
>UniRef100_Q75LI5 Putative transposon protein [Oryza sativa]
Length = 839
Score = 129 bits (325), Expect = 1e-28
Identities = 71/171 (41%), Positives = 99/171 (57%), Gaps = 3/171 (1%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMGY-QGFLPPV-QKSRLEKEAAQKWTATW 58
I++ R KPIITMLE+IR V R I NK + G P + +K + A Q W
Sbjct: 540 IIEARFKPIITMLEDIRMKVTRRIQENKTNSERWTMGICPNILKKINKIRHATQFCHVLW 599
Query: 59 SGQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLT 118
+G S FEV R VDL+A TC+CR+WQ++G+PC HACAA E P N+V+ +
Sbjct: 600 NGS-SGFEVREKKWRFTVDLSANTCSCRYWQISGIPCQHACAAYFKMAEEPNNHVNMCFS 658
Query: 119 MGAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKKKAGRPKKNRRRDGTE 169
+ YR+TY+ +QP + W + +P+PP +KK G PK+ RR+D TE
Sbjct: 659 IDQYRNTYQDVLQPVEHESVWPLSTNPRPLPPRVKKMPGSPKRARRKDPTE 709
>UniRef100_Q7F966 OSJNBa0091C07.2 protein [Oryza sativa]
Length = 879
Score = 118 bits (295), Expect = 3e-25
Identities = 65/186 (34%), Positives = 96/186 (50%), Gaps = 3/186 (1%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMGYQGFLPPVQKSRLEK--EAAQKWTATW 58
I+ R PII+M E IR V I N+ K G+ G + P +L K + + A W
Sbjct: 616 IIDIRAHPIISMFEGIRTKVYVRIQQNRSKAKGWLGRICPNILKKLNKYIDLSGNCEAIW 675
Query: 59 SGQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLT 118
+G++ FEVT +R VDL TC+CR+WQL G+PC HA A+ ++PE+Y+ +
Sbjct: 676 NGKDG-FEVTDKDKRYTVDLEKRTCSCRYWQLAGIPCAHAITALFVSSKQPEDYIADCYS 734
Query: 119 MGAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKKKAGRPKKNRRRDGTEEQVAGRARR 178
+ Y Y+ + P W T + KP PP K GRP+K RRRD E + + +
Sbjct: 735 VEVYNKIYDHCMMPMEGMMQWPITGHPKPGPPGYVKMPGRPRKERRRDPLEAKKGKKLSK 794
Query: 179 DATESQ 184
T+ +
Sbjct: 795 TGTKGR 800
>UniRef100_Q94HK4 Mutator-like transposase [Oryza sativa]
Length = 725
Score = 103 bits (258), Expect = 6e-21
Identities = 64/180 (35%), Positives = 88/180 (48%), Gaps = 2/180 (1%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMGYQGFLPPVQKSRLEKEAAQ-KWTATWS 59
IL R KPI+TM+E IR VM ++ + Q + P+ +LE E ++ +
Sbjct: 514 ILDARDKPIVTMVEHIRRKVMVRLSLKRQGGDAAQWEITPIVAGKLEMEKNHARYCWCYQ 573
Query: 60 GQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLTM 119
+ +EV C + AVD++A TC C WQL G+PC H A+ PE+YV +
Sbjct: 574 SNLTTWEVHCLDRSFAVDISARTCACHKWQLTGIPCKHDVCALYKAGHTPEDYVADYFRK 633
Query: 120 GAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKKKAGRPKKNRRRDGTE-EQVAGRARR 178
AY TY I P + W T PP K GRPKK+RRR E +V G AR+
Sbjct: 634 DAYMRTYTAVIYPVPDEHRWTKTDSPYIDPPKFDKHVGRPKKSRRRGPDEGPRVQGPARK 693
>UniRef100_Q9ZPU1 Mutator-like transposase [Arabidopsis thaliana]
Length = 597
Score = 95.1 bits (235), Expect = 3e-18
Identities = 75/285 (26%), Positives = 120/285 (41%), Gaps = 20/285 (7%)
Query: 5 RGKPIITMLEEIRCYVMRTIANNKMKLMGYQGFLPPVQKSRLEKEAAQKWTAT---WSGQ 61
R KP++ MLE+IR M + N K ++ Y+ +++ +E E A W +
Sbjct: 266 RRKPLLDMLEDIRRQCM--VRNEKRFVIAYRLKSRFTKRAHMEIEKMIDGAAVCDRWMAR 323
Query: 62 ESRFEVTCW-GQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLTMG 120
+R EV +VD+ TC CR WQ+ G+PC HA + I ++ E++V W T
Sbjct: 324 HNRIEVRVGLDDSFSVDMNDRTCGCRKWQMTGIPCIHAASVIIGNRQKVEDFVSDWYTTY 383
Query: 121 AYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKK-KAGRPKKNRRRDGTEEQVAGRARRD 179
++ TY I P + + W +PP ++ GRPK + RR G E R+
Sbjct: 384 MWKQTYYDGIMPVQGKMLWPIVNRVGVLPPPWRRGNPGRPKNHDRRKGIYESATASTSRE 443
Query: 180 ATESQLAGRGPTGSQTPLAPAAAAASYPTITT-----------PQGPPPLQPRVAYRAPI 228
++ + T + P P T QG P + + A +
Sbjct: 444 G-HNKAGCKNETVAPPPKRPRGRPRKDQDSRTILFGHGQSWQGSQGEPSVASQPAQPSQ- 501
Query: 229 AAAAAAPRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQ 273
+ + P QP QP ++ ++ A A P P P A +AP Q
Sbjct: 502 PSQPSQPSQPALSQPSLSQQSQRAPSAPSEPPAPPAPSAPSAPSQ 546
>UniRef100_Q75H66 Hypothetical protein OSJNBb0007E22.22 [Oryza sativa]
Length = 981
Score = 94.4 bits (233), Expect = 5e-18
Identities = 57/172 (33%), Positives = 87/172 (50%), Gaps = 1/172 (0%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMGYQGFLPPVQKSRLEKEAAQKWTA-TWS 59
IL+ R PI+TMLE+I+ +M N + + +QG + P + +L K A Q
Sbjct: 650 ILEAREMPILTMLEKIKGQLMTRFFNKQKEAQKWQGPICPKIRKKLLKIAEQANICYVLP 709
Query: 60 GQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLTM 119
+ F+V G + VD+ C CR W L G+PC HA A I H E+++ ++
Sbjct: 710 AGKGVFQVEERGTKYIVDVVTKHCDCRRWDLTGIPCCHAIACIREDHLSEEDFLPHCYSI 769
Query: 120 GAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKKKAGRPKKNRRRDGTEEQ 171
A+++ Y I P + WE + +PP +KK GRPKK+RR+ E Q
Sbjct: 770 NAFKAVYAENIIPCNDKANWEKMNGPQILPPVYEKKVGRPKKSRRKQPQEVQ 821
>UniRef100_Q7X762 OSJNBb0118P14.3 protein [Oryza sativa]
Length = 939
Score = 90.5 bits (223), Expect = 7e-17
Identities = 55/171 (32%), Positives = 82/171 (47%), Gaps = 2/171 (1%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKL-MGYQGFLPPVQKSRLEKEAAQKWTA-TW 58
IL R PI++MLE IR +M + + +L + + P K ++EK T +
Sbjct: 620 ILDAREMPILSMLERIRNQIMNRLYTKQKELERNWPCGICPKIKRKVEKNTEMANTCYVF 679
Query: 59 SGQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLT 118
F+V+ G + V+L C CR WQL G+PC HA + + + +PE+ V +
Sbjct: 680 PAGMGAFQVSDIGSQYIVELNVKRCDCRRWQLTGIPCNHAISCLRHERIKPEDVVSFCYS 739
Query: 119 MGAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKKKAGRPKKNRRRDGTE 169
Y Y + I P R +WE + PP +KK GRPKK RR+ E
Sbjct: 740 TRCYEQAYSYNIMPLRDSIHWEKMQGIEVKPPVYEKKVGRPKKTRRKQPQE 790
>UniRef100_Q9LP09 F9C16.9 [Arabidopsis thaliana]
Length = 946
Score = 89.0 bits (219), Expect = 2e-16
Identities = 83/314 (26%), Positives = 127/314 (40%), Gaps = 39/314 (12%)
Query: 1 ILKYRGKPIITMLEEIRCYVM-----RTIANNKMKLMGYQGFLPPVQKSRLEKEAAQKWT 55
I + R KP++ MLE+IR M R + ++++ + ++K + Q+W
Sbjct: 610 IREARRKPLLDMLEDIRRQCMVRNEKRYVIADRLRTRFTKRAHAEIEKMIAGSQVCQRWM 669
Query: 56 ATWSGQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHA 115
A + E + VD+ TC C WQ+ G+PC HA + I K ++ E+YV
Sbjct: 670 ARHNKHEIKVGPV---DSFTVDMNNNTCGCMKWQMTGIPCIHAASVIIGKRQKVEDYVSD 726
Query: 116 WLTMGAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKK-KAGRPKKNRRRDGTEEQVA- 173
W T ++ TY I P + + W +PP ++ GRPK + RR G E
Sbjct: 727 WYTTSMWKQTYNDGIGPVQGKLLWPTVNKVGVLPPPWRRGNPGRPKNHARRKGVFESSTA 786
Query: 174 --------GRARRDATESQLAGRGPTGSQTPLAPAAAAASYPTITTPQGPP--PLQPRVA 223
R R T S G G A P P+G P PR+
Sbjct: 787 SSSSTTELSRLHRVMTCSNCQGEGHNKQGCKNETVA-----PPAKRPRGRPRKDKDPRMV 841
Query: 224 Y-------RAPIAAAAAAPRQPPPLQPRVAYRAPIAVVA--APRQPPPLQPRAAAAPRQP 274
+ + P A A + P +V + AP++ + AP Q P Q A + QP
Sbjct: 842 FFGQGQTWQEPQAQGAPSSHGAP---SQVFHGAPLSQPSQEAPSQGAPSQ--GAQSLSQP 896
Query: 275 PPLAPAAAASFSTQ 288
AP+ AA T+
Sbjct: 897 SQEAPSQAAPTETR 910
>UniRef100_Q8H8R1 Putative mutator-like transposase [Oryza sativa]
Length = 746
Score = 88.6 bits (218), Expect = 3e-16
Identities = 54/170 (31%), Positives = 84/170 (48%), Gaps = 1/170 (0%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMGYQGFLPPVQKSRLEKEAAQKWTA-TWS 59
IL+ R I++MLE+IR +M I + + + + P K ++EK T
Sbjct: 496 ILEARELTILSMLEKIRSKLMNRIYTKQEECKKWVFDICPKIKQKVEKNIEMSNTCYALP 555
Query: 60 GQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLTM 119
+ F+VT ++ VD+ C CR WQL G+PC HA + + + +PE+ V T+
Sbjct: 556 SRMGVFQVTDRDKQFVVDIKNKQCDCRRWQLIGIPCNHAISCLRHERIKPEDEVSFCYTI 615
Query: 120 GAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKKKAGRPKKNRRRDGTE 169
+++ Y F I P R + +WE PP +KK GRPK RR+ E
Sbjct: 616 QSFKQAYMFNIMPVRDKTHWEKMNGVPVNPPVYEKKVGRPKTTRRKQPQE 665
>UniRef100_Q6L3K3 Putative transposon MuDR mudrA-like protein [Solanum demissum]
Length = 873
Score = 88.6 bits (218), Expect = 3e-16
Identities = 54/170 (31%), Positives = 79/170 (45%), Gaps = 1/170 (0%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMGYQG-FLPPVQKSRLEKEAAQKWTATWS 59
IL+ R KPII MLE IR +M + + + ++G F P + + + S
Sbjct: 512 ILEARAKPIIKMLENIRIKIMNRLQKLEEEGKNWKGDFSPYAMELYNDFNIIAQCCQVQS 571
Query: 60 GQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLTM 119
+ +EV R V+L CTCR W L G+ C HA A + P + + W +
Sbjct: 572 NGDQGYEVVEGEDRHVVNLNRKKCTCRTWDLTGILCPHAIKAYLHDKQEPLDQLSWWYSR 631
Query: 120 GAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKKKAGRPKKNRRRDGTE 169
AY Y IQP R +++W+ P PP + K GRPK R+R+ E
Sbjct: 632 EAYMLVYMHKIQPVRGEKFWKVDPSHAMEPPEIHKLVGRPKLKRKREKDE 681
>UniRef100_O81502 F9D12.2 protein [Arabidopsis thaliana]
Length = 940
Score = 86.3 bits (212), Expect = 1e-15
Identities = 80/298 (26%), Positives = 124/298 (40%), Gaps = 38/298 (12%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMGYQGFLPPVQKSRLE-KEAAQKWTATWS 59
I + R P++ MLEE+R M+ I+ K + PP LE + K+
Sbjct: 610 IREARKLPVVNMLEEVRRISMKRISKLCDKTAKCETRFPPKIMQILEGNRKSSKYCQVLK 669
Query: 60 GQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLTM 119
E++FE+ +VDL TC CR W+L G+PC HA I + PE+YV L
Sbjct: 670 SGENKFEILEGSGYYSVDLLTRTCGCRQWELTGIPCPHAIYVITEHNRDPEDYVDRLLET 729
Query: 120 GAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKKKAGRPKKNRRRDGTEEQVAGRARRD 179
+++TY+ I+P ++ W+ + P+ + K GRPKK R + G + +
Sbjct: 730 QVWKATYKDNIEPMNGERLWKRRGKGRIEVPDKRGKRGRPKKFAR-----IKEPGESSTN 784
Query: 180 ATESQLAGRGPTGSQ-TPLAPAAAAASYPTITTPQGPPPLQ------------PRVAYRA 226
T+ G+ T S + + PT+ Q PP + P + A
Sbjct: 785 PTKLTREGKTVTCSNCKQIGHNKGSCKNPTV---QLAPPRKRGRPRKCVDNDDPWSIHNA 841
Query: 227 PIAAAAAAPRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAAS 284
P AA + PP VA + PP+ A + P Q P + A AA+
Sbjct: 842 PRRWRAAQSQSTPP-------------VAQSQSTPPV---AQSQPTQNPQHSSAPAAN 883
>UniRef100_Q60DJ3 MuDR family transposase protein [Oryza sativa]
Length = 1030
Score = 85.9 bits (211), Expect = 2e-15
Identities = 58/168 (34%), Positives = 87/168 (51%), Gaps = 4/168 (2%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMG-YQGFLPPVQKSRLEK--EAAQKWTAT 57
IL+ R PI+TMLE+I+ +M N + +L +QG + P + ++ K +AA A
Sbjct: 685 ILEARELPILTMLEKIKGQLMTRHFNKQKELADQFQGLICPKIRKKVLKNADAANTCYAL 744
Query: 58 WSGQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWL 117
+GQ F+V + VD+ A C CR W L G+PC HA + + + E+ +
Sbjct: 745 PAGQ-GIFQVHEREYQYIVDINAMYCDCRRWDLTGIPCNHAISCLRHERINAESILPNCY 803
Query: 118 TMGAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKKKAGRPKKNRRR 165
T A+ Y F I P + WE + PP +KKAGRPKK+RR+
Sbjct: 804 TTDAFSKAYGFNIWPCNDKSKWENVNGPEIKPPVYEKKAGRPKKSRRK 851
>UniRef100_Q6AVM7 Putative polyprotein [Oryza sativa]
Length = 1006
Score = 85.5 bits (210), Expect = 2e-15
Identities = 58/168 (34%), Positives = 87/168 (51%), Gaps = 4/168 (2%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMG-YQGFLPPVQKSRLEK--EAAQKWTAT 57
IL+ R PI+TMLE+I+ +M N + +L +QG + P + ++ K +AA A
Sbjct: 661 ILEARELPILTMLEKIKGQLMTRHFNKQKELADQFQGLICPKIRKKVLKNADAANTCYAL 720
Query: 58 WSGQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWL 117
+GQ F+V + VD+ A C CR W L G+PC HA + + + E+ +
Sbjct: 721 PAGQ-GIFQVHEREYQYIVDINAMHCDCRRWDLTGIPCNHAISCLRHERINAESILPNCY 779
Query: 118 TMGAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKKKAGRPKKNRRR 165
T A+ Y F I P + WE + PP +KKAGRPKK+RR+
Sbjct: 780 TTDAFSKAYGFNIWPCNDKSKWENINGPEIKPPVYEKKAGRPKKSRRK 827
>UniRef100_Q6L4D3 Hypothetical protein OSJNBa0088M05.7 [Oryza sativa]
Length = 1092
Score = 84.0 bits (206), Expect = 7e-15
Identities = 65/214 (30%), Positives = 94/214 (43%), Gaps = 21/214 (9%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMGYQGFLPP--VQKSRLEKEAAQKWTATW 58
IL+ R PIITMLE I VM I + K + + P ++K + + A
Sbjct: 750 ILESRFHPIITMLETIWRKVMVRINDQKAAGAKWTTVVCPGILKKLNVYITESAFCHAIC 809
Query: 59 SGQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLT 118
+G +S FEV R V L C+CR+WQL G+PC HA + I ++ + ++Y+
Sbjct: 810 NGGDS-FEVKHHDHRFTVHLDKKECSCRYWQLLGLPCPHAISCIFYRTNKLDDYIAPCYY 868
Query: 119 MGAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKKKAGRPKKNRRRDGTEE-------- 170
+ A+RSTY +QP W E P K GRPK RRR+ E
Sbjct: 869 VDAFRSTYVHCLQPLEGMSAWPQDDREPLNAPGYIKMPGRPKTERRREKHEPPKPTKMPK 928
Query: 171 ----------QVAGRARRDATESQLAGRGPTGSQ 194
+ G + ++ Q +G G GSQ
Sbjct: 929 YGTVIRCTRCKQVGHNKSSCSKHQSSGSGTVGSQ 962
>UniRef100_Q9FHV3 Mutator-like transposase-like protein [Arabidopsis thaliana]
Length = 599
Score = 83.2 bits (204), Expect = 1e-14
Identities = 60/188 (31%), Positives = 80/188 (41%), Gaps = 16/188 (8%)
Query: 3 KYRGKPIITMLEEIRCYVMRTIANNKMKLMGYQGFLPPVQKSRLEKE----AAQKWTATW 58
K R KP + MLE IR M IA ++ + G P L E AA K + +
Sbjct: 381 KAREKPFVAMLETIRRMAMVRIAKRSVESHTHTGVCTPYVTKFLAGEHKVAAAAKVSPST 440
Query: 59 SGQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLT 118
+G +EV G VDL A TCTC WQ+ G+PC HA + I K PEN+V W
Sbjct: 441 NGM---YEVRHGGDLHRVDLQAYTCTCIKWQICGIPCEHAYSVIIHKKLEPENFVCCWFR 497
Query: 119 MGAYRSTYEFAIQPTRSQQYWE---------PTPYEKPIPPNMKKKAGRPKKNRRRDGTE 169
+R Y + P R ++W P P E M K + K T+
Sbjct: 498 TAMWRKNYTDGLFPQRGPKFWPESNGLRVYVPPPTEGEEDKKMTKAEKKRKNGVNESPTK 557
Query: 170 EQVAGRAR 177
+Q G+ R
Sbjct: 558 KQPKGKKR 565
>UniRef100_Q65XH0 Hypothetical protein OJ1126_B11.9 [Oryza sativa]
Length = 792
Score = 80.5 bits (197), Expect = 7e-14
Identities = 45/115 (39%), Positives = 65/115 (56%), Gaps = 3/115 (2%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMGYQGFLPPVQKSRLEK--EAAQKWTATW 58
I+ R PII+M E IR V I N+ K G+ G + P +L K + + A W
Sbjct: 616 IIDIRAHPIISMFEGIRTKVYVRIQQNRSKAKGWLGRICPNILKKLNKYIDLSGNCEAIW 675
Query: 59 SGQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYV 113
+G++ FEVT +R VDL TC+CR+WQL G+PC HA A+ ++PE+Y+
Sbjct: 676 NGKDG-FEVTDKDKRYTVDLEKRTCSCRYWQLAGIPCAHAITALFVSSKQPEDYI 729
>UniRef100_UPI0000234ADD UPI0000234ADD UniRef100 entry
Length = 1186
Score = 80.1 bits (196), Expect = 1e-13
Identities = 79/252 (31%), Positives = 99/252 (38%), Gaps = 45/252 (17%)
Query: 132 PTRSQQYWEPTPYEKPIPPNMKKKAGRPKKNRRRDGTEEQVAGRARRDATESQLAGRGPT 191
P +++ PTP P PP + +A P + AG+ + AG+ P
Sbjct: 305 PAKTEDDQAPTPPRDPSPPRPRFRAPPPIAD----------AGKY------AHTAGQAP- 347
Query: 192 GSQTPLAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPI 251
P P AA+ + TP PPP +P +AP+ A PPP Q AP
Sbjct: 348 ----PARPRAASGA-----TPGPPPPPRPP---KAPVDDAPPRFGVPPPFQGERKVSAPP 395
Query: 252 AVVA-APRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMN 310
A + +P PPP PR A+ P PP L P A FS+ PP PR S P
Sbjct: 396 APPSRSPAGPPPPPPRTAS-PATPPQLPPKVPA-FSSGPPPPPPRSPASQPPPPPPVPGA 453
Query: 311 QRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATHELPGGPIV 370
R P TA+ PP PP R P T PP PPP PA+ P P
Sbjct: 454 SRPVPPPTASVPPPP-------PPPARPTPTVTGPPP------PPPPPPASSGPPMPPPP 500
Query: 371 FMPTPSIQPRPP 382
P P PP
Sbjct: 501 PPPAPGSSAPPP 512
Score = 65.1 bits (157), Expect = 3e-09
Identities = 58/192 (30%), Positives = 66/192 (34%), Gaps = 33/192 (17%)
Query: 175 RARRDATESQLAGRGPT-GSQTPLAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAA 233
R AT QL + P S P P + AS P PPP P + P A+
Sbjct: 411 RTASPATPPQLPPKVPAFSSGPPPPPPRSPASQPP------PPPPVPGASRPVPPPTASV 464
Query: 234 APRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQ 293
P PPP +P V P PPP P ++ P PPP PA +S PP
Sbjct: 465 PPPPPPPARPT-------PTVTGPPPPPPPPPASSGPPMPPPPPPPAPGSSAPPPPPPPP 517
Query: 294 PRVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRR 353
P GAP PP G PP A PP G
Sbjct: 518 PGA----------------GAPPPPP---PPPGAGAPPPPPPPPGAGAPPPPPPPGAGAP 558
Query: 354 SPPPGPATHELP 365
PPPG A LP
Sbjct: 559 PPPPGGAAPPLP 570
Score = 62.8 bits (151), Expect = 2e-08
Identities = 67/225 (29%), Positives = 90/225 (39%), Gaps = 39/225 (17%)
Query: 168 TEEQVAGRARRDATESQLAGRGPTGSQTPLAPAA-AAASYPTITTPQGPPPLQPRVAYRA 226
TE+Q+A A D +S + + + ++ PA P P PPP V+ +
Sbjct: 226 TEDQIADNA--DFIKSYIEQKKASEGESATPPADDRKGKAPPPPPPSVPPPKTSAVSPQH 283
Query: 227 PIAAAAA---APRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAA- 282
+++ + AP PPP + A PR P P +PR R PPP+A A
Sbjct: 284 TGSSSGSKRGAPPPPPPSRKTPAKTEDDQAPTPPRDPSPPRPRF----RAPPPIADAGKY 339
Query: 283 ASFSTQAPPLQPRVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPAT 342
A + QAPP +PR TP G P PP RPP+ V+ A
Sbjct: 340 AHTAGQAPPARPRAASGATP----------GPP-------PPP------RPPKAPVDDAP 376
Query: 343 TTH--PPQSQGRR--SPPPGPATHELPGGPIVFMPTPSIQPRPPQ 383
PP QG R S PP P + P GP P + PPQ
Sbjct: 377 PRFGVPPPFQGERKVSAPPAPPSRS-PAGPPPPPPRTASPATPPQ 420
>UniRef100_Q9C9S3 Putative Mutator-like transposase; 12516-14947 [Arabidopsis
thaliana]
Length = 761
Score = 80.1 bits (196), Expect = 1e-13
Identities = 58/197 (29%), Positives = 87/197 (43%), Gaps = 9/197 (4%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMGYQGFLPP--VQKSRLEKEAAQKWTATW 58
I K R K +I MLE IR M I K + ++G ++ LEK A + + +
Sbjct: 480 ITKARAKSLIPMLETIRRQGMTRIVKRNKKSLRHEGRFTKYALKMLALEKTDADR-SKVY 538
Query: 59 SGQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLT 118
FEV G VD+ C+C WQ++G+PC HA A+ ENY+ + +
Sbjct: 539 RCTHGVFEVYIDGNGHRVDIPKTQCSCGKWQISGIPCEHAYGAMIEAGLDAENYISEFFS 598
Query: 119 MGAYRSTYEFAIQPTRSQQYWEPTPY------EKPIPPNMKKKAGRPKKNRRRDGTEEQV 172
+R +YE A P R +YW + Y +PI P KK+ + K R G E
Sbjct: 599 TDLWRDSYETATMPLRGPKYWLNSSYGLVTAPPEPILPGRKKEKSKKGKFARIKGKNESP 658
Query: 173 AGRARRDATESQLAGRG 189
+ R+ +L +G
Sbjct: 659 KKKKRKKNEVEKLGKKG 675
>UniRef100_UPI00003AF275 UPI00003AF275 UniRef100 entry
Length = 721
Score = 79.0 bits (193), Expect = 2e-13
Identities = 71/223 (31%), Positives = 86/223 (37%), Gaps = 33/223 (14%)
Query: 180 ATESQLAGRGPTGSQTPLAPAA--AAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQ 237
A S A P G P APAA AA P TP P + A ++P+AA A P
Sbjct: 308 AAPSAPAAVVPAGPTNPAAPAATKAAPQSPVAATPAAPAAT--KAAPQSPVAATPAPPAS 365
Query: 238 PPPLQPRVAYRAPIAVVAAPRQPPPL------QPRAA---------AAPRQP------PP 276
P +P A +PIA AP PP P AA AAP+ P P
Sbjct: 366 PAATKP--APESPIAATPAPAAAPPATKAGPQSPIAATPAAPAATKAAPQSPVAATPAPA 423
Query: 277 LAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQ 336
APA AA +T+A P P+ + TP+ A + AP + A P
Sbjct: 424 TAPATAAPAATKAAPQTPQSPVAATPAPPAAPSAAKAAPQSPVAATPAPAAAP------A 477
Query: 337 RVEPATTTHPPQSQGRRSPPPGPATHELPGGPIVFMPTPSIQP 379
PA T PQS +P AT P PI P P P
Sbjct: 478 TAAPAATKAAPQSPVAATPAAPAATKAAPQSPIAATPAPPAAP 520
Score = 75.1 bits (183), Expect = 3e-12
Identities = 72/223 (32%), Positives = 88/223 (39%), Gaps = 32/223 (14%)
Query: 188 RGPTGSQTPL-APAAAAASYPTITTPQGPPPLQPRVAYRAPI------AAAAAAPRQPPP 240
R P GS P+ AP + S +P GPPP AP+ AA AAP P
Sbjct: 145 RPPPGSPIPVTAPVLPSPSPAAALSPSGPPPAAAAPLKAAPVIPVSLPAAVTAAPASSLP 204
Query: 241 LQPRVAYRA-PIAVVAAPRQP--PPLQPRA-AAAPRQPPPLAPAAAASFSTQAPPLQPRV 296
+ P +A A P A AP+ P PL P A AAA PP APAA + Q+P P
Sbjct: 205 VTPAMAAPATPPAAKGAPQSPVPAPLSPSAPAAAAPVVPPAAPAATKA-PPQSPVTTPSA 263
Query: 297 VYSLTPSVATAIMNQRGAPHA-TAAYRAPPV--RGKFFRP-----------------PRQ 336
++ P+ A A + AP + AA APP K P P
Sbjct: 264 PAAVVPAAAPAPAANKAAPKSPVAATPAPPAAPAAKKAAPQSPVAAPSAPAAVVPAGPTN 323
Query: 337 RVEPATTTHPPQSQGRRSPPPGPATHELPGGPIVFMPTPSIQP 379
PA T PQS +P AT P P+ P P P
Sbjct: 324 PAAPAATKAAPQSPVAATPAAPAATKAAPQSPVAATPAPPASP 366
Score = 74.3 bits (181), Expect = 5e-12
Identities = 77/221 (34%), Positives = 92/221 (40%), Gaps = 36/221 (16%)
Query: 168 TEEQVAGRARRDATESQLAGRGPTGSQTPLAPAA--AAASYPTITTPQGPP-PLQPRVAY 224
T A A ++ A + P + TP APAA AA P TP P P + A
Sbjct: 469 TPAPAAAPATAAPAATKAAPQSPVAA-TPAAPAATKAAPQSPIAATPAPPAAPPATKAAP 527
Query: 225 RAPIAAAAAAPR------QPPPLQPRVAYRAPIAVVAAPRQPP-PLQPRAA--------A 269
++PIAA AAP Q P AP A AAPR PP P P AA A
Sbjct: 528 QSPIAATPAAPAATKAAPQSPVAATPAPPAAPAATKAAPRTPPAPAAPPAAKKSPKSPTA 587
Query: 270 APRQPPPLAPAAAASFST-QAPPLQPRVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRG 328
AP PP APAA AS +T +APP P S P+ A AP ++AA +APP
Sbjct: 588 APPTPP--APAAPASSATAKAPPKSPTAAPSAPPAPA--------APASSAAAKAPPK-- 635
Query: 329 KFFRPPRQRVEPATTTHPPQSQGRRSPPPGP-ATHELPGGP 368
P P P S ++PP P A P P
Sbjct: 636 ---SPTAAPSAPPAPAAPASSAAAKAPPKSPTAALAAPAAP 673
Score = 70.5 bits (171), Expect = 8e-11
Identities = 60/200 (30%), Positives = 74/200 (37%), Gaps = 16/200 (8%)
Query: 194 QTPLAPAAAAASYPTITTPQGPPPLQPRVAYRAPIAAAA--------AAPRQP---PPLQ 242
QTP +P AA + P + P P A AP AA A AAP+ P P
Sbjct: 439 QTPQSPVAATPAPPAAPSAAKAAPQSPVAATPAPAAAPATAAPAATKAAPQSPVAATPAA 498
Query: 243 PRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTP 302
P AP + +AA PP P AAP+ P PAA A +T+A P P + TP
Sbjct: 499 PAATKAAPQSPIAATPAPPAAPPATKAAPQSPIAATPAAPA--ATKAAPQSP---VAATP 553
Query: 303 SVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATH 362
+ A + AP A APP K + P P S PP T
Sbjct: 554 APPAAPAATKAAPRTPPAPAAPPAAKKSPKSPTAAPPTPPAPAAPASSATAKAPPKSPTA 613
Query: 363 ELPGGPIVFMPTPSIQPRPP 382
P P S + P
Sbjct: 614 APSAPPAPAAPASSAAAKAP 633
Score = 69.7 bits (169), Expect = 1e-10
Identities = 62/217 (28%), Positives = 81/217 (36%), Gaps = 20/217 (9%)
Query: 180 ATESQLAGRGPTGSQTPLAPAAAAASYPTITTPQGP-----PPLQPRVAYRAPIAAAAAA 234
AT G + PL+P+A AA+ P + P P PP P AP A AA
Sbjct: 213 ATPPAAKGAPQSPVPAPLSPSAPAAAAP-VVPPAAPAATKAPPQSPVTTPSAPAAVVPAA 271
Query: 235 PRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRA----AAAPRQPPPLAPAA----AASFS 286
P P + A ++P+A AP P + A AAP P + PA AA +
Sbjct: 272 --APAPAANKAAPKSPVAATPAPPAAPAAKKAAPQSPVAAPSAPAAVVPAGPTNPAAPAA 329
Query: 287 TQAPPLQPRVVYSLTPSVATAI----MNQRGAPHATAAYRAPPVRGKFFRPPRQRVEPAT 342
T+A P P P+ A + AP A+ A P P P
Sbjct: 330 TKAAPQSPVAATPAAPAATKAAPQSPVAATPAPPASPAATKPAPESPIAATPAPAAAPPA 389
Query: 343 TTHPPQSQGRRSPPPGPATHELPGGPIVFMPTPSIQP 379
T PQS +P AT P P+ P P+ P
Sbjct: 390 TKAGPQSPIAATPAAPAATKAAPQSPVAATPAPATAP 426
Score = 68.2 bits (165), Expect = 4e-10
Identities = 64/201 (31%), Positives = 72/201 (34%), Gaps = 17/201 (8%)
Query: 195 TPLAPAA--AAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIA 252
TP APAA AA P TP P P A A AA P+ P P AP A
Sbjct: 401 TPAAPAATKAAPQSPVAATPA--PATAPATAAPAATKAAPQTPQSPVAATPAPP-AAPSA 457
Query: 253 VVAAPRQPPPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQR 312
AAP+ P P AAAP P A AA A P P + S A
Sbjct: 458 AKAAPQSPVAATPAPAAAPATAAPAATKAAPQSPVAATPAAPAATKAAPQSPIAATPAPP 517
Query: 313 GAPHATAAYRAPPVRGKFFRPPRQRVEP----ATTTHPPQSQGR-----RSPP---PGPA 360
AP AT A P+ P + P A T PP + R+PP PA
Sbjct: 518 AAPPATKAAPQSPIAATPAAPAATKAAPQSPVAATPAPPAAPAATKAAPRTPPAPAAPPA 577
Query: 361 THELPGGPIVFMPTPSIQPRP 381
+ P P PTP P
Sbjct: 578 AKKSPKSPTAAPPTPPAPAAP 598
Score = 68.2 bits (165), Expect = 4e-10
Identities = 61/212 (28%), Positives = 77/212 (35%), Gaps = 28/212 (13%)
Query: 196 PLAPAAAAASYPT-ITTPQGPPPLQP---------RVAYRAPIAAAAAAPRQPP-----P 240
P APAA A + +TTP P + P + A ++P+AA A P P P
Sbjct: 244 PAAPAATKAPPQSPVTTPSAPAAVVPAAAPAPAANKAAPKSPVAATPAPPAAPAAKKAAP 303
Query: 241 LQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPPPLAPAAAAS---------FSTQAPP 291
P A AP AVV A P AAP+ P PAA A+ +T APP
Sbjct: 304 QSPVAAPSAPAAVVPAGPTNPAAPAATKAAPQSPVAATPAAPAATKAAPQSPVAATPAPP 363
Query: 292 LQPRVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRVEP----ATTTHPP 347
P S A AP AT A P+ P + P A T P
Sbjct: 364 ASPAATKPAPESPIAATPAPAAAPPATKAGPQSPIAATPAAPAATKAAPQSPVAATPAPA 423
Query: 348 QSQGRRSPPPGPATHELPGGPIVFMPTPSIQP 379
+ +P A + P P+ P P P
Sbjct: 424 TAPATAAPAATKAAPQTPQSPVAATPAPPAAP 455
Score = 66.2 bits (160), Expect = 1e-09
Identities = 73/248 (29%), Positives = 97/248 (38%), Gaps = 48/248 (19%)
Query: 142 TPYEKPIPPNMKKKAGRPKKNRRRDGTEEQVAGRARRDATESQLAGRGPTGS--QTPLAP 199
TP PP K P + A ++ AT + A T + +TP AP
Sbjct: 513 TPAPPAAPPATKAAPQSPIAATPAAPAATKAAPQSPVAATPAPPAAPAATKAAPRTPPAP 572
Query: 200 AAAAASYPTITTPQGPPPLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRAPIAVVA---- 255
AA A+ + +P PP P AP A A++A + PP P A AP A A
Sbjct: 573 AAPPAAKKSPKSPTAAPPTPP-----APAAPASSATAKAPPKSPTAAPSAPPAPAAPASS 627
Query: 256 APRQPPPLQPRAAAAPRQPP-PLAPAAAASFSTQAPPLQPRVVYSLTPSVATAIMNQRGA 314
A + PP P AAP PP P APA++A + +APP P TA + A
Sbjct: 628 AAAKAPPKSP--TAAPSAPPAPAAPASSA--AAKAPPKSP-----------TAALAAPAA 672
Query: 315 PHATAAYRAPPVRGKFFRPPRQRVEPATTTHPPQSQGRRSPPPGPATHELPGGPIVFMPT 374
P A +A +A PV + G P PG A PG P+ +P
Sbjct: 673 PSAPSAAKAAPVSS-------------------AAPGTLPPAPGAAV-SAPGAPVA-LPK 711
Query: 375 PSIQPRPP 382
P+ P
Sbjct: 712 PAADRAAP 719
Score = 60.5 bits (145), Expect = 8e-08
Identities = 68/239 (28%), Positives = 87/239 (35%), Gaps = 46/239 (19%)
Query: 189 GPTGSQTPLAPAAAAASYPTITTPQGPP--------PLQPRVAYRAPIAAAA-------A 233
G + PLAP AA PQ PP PL P A P AAA +
Sbjct: 21 GAAPAALPLAPPPAATPAGATALPQSPPAVPVHPAAPLPPPAAPLGPALAAAISPPIFLS 80
Query: 234 APRQPPPLQPRVAYRAPIAVVAAPRQPPPLQPRAAAAPRQPP-----PLAPAAAASFSTQ 288
AP PL P P + A+PR P P AP P P+ PA AAS +
Sbjct: 81 APMALAPLLP--GGTVPCSAAASPRGPMSATPVCPLAPAAPSGAVTCPVGPALAASGTPA 138
Query: 289 AP----------PLQPRVVYSLTPSVATAIMNQRGAPHATAAYRAPPVRGKFFRPPRQRV 338
P P+ +PS A A+ P A A +A PV P
Sbjct: 139 VPHSVARPPPGSPIPVTAPVLPSPSPAAALSPSGPPPAAAAPLKAAPVI-PVSLPAAVTA 197
Query: 339 EPATT----------THPPQSQGR-RSPPPGPATHELP--GGPIVFMPTPSIQPRPPQN 384
PA++ PP ++G +SP P P + P P+V P+ PPQ+
Sbjct: 198 APASSLPVTPAMAAPATPPAAKGAPQSPVPAPLSPSAPAAAAPVVPPAAPAATKAPPQS 256
Score = 50.8 bits (120), Expect = 6e-05
Identities = 60/206 (29%), Positives = 74/206 (35%), Gaps = 26/206 (12%)
Query: 196 PLAPAAAAASYPTITTPQGPP------PLQPRVAYRAPIAAAAAAPRQPPPLQPRVAYRA 249
P +P AAAS P P+ P PL P A A A A A P+ PP + A A
Sbjct: 1 PPSPTGAAASAPHPAPPKSPGAAPAALPLAPPPA--ATPAGATALPQSPPAVPVHPA--A 56
Query: 250 PIAVVAAPRQP-------PPLQPRAAAAPRQPPPLAPAAAASFSTQAPPLQPRVVYSLTP 302
P+ AAP P PP+ +AP PL P S A P P + P
Sbjct: 57 PLPPPAAPLGPALAAAISPPI---FLSAPMALAPLLPGGTVPCSAAASPRGPMSATPVCP 113
Query: 303 SVATAIMNQRGAP--HATAAYRAPPVRGKFFRPPRQRVEPATT----THPPQSQGRRSPP 356
A P A AA P V RPP P T + P + S P
Sbjct: 114 LAPAAPSGAVTCPVGPALAASGTPAVPHSVARPPPGSPIPVTAPVLPSPSPAAALSPSGP 173
Query: 357 PGPATHELPGGPIVFMPTPSIQPRPP 382
P A L P++ + P+ P
Sbjct: 174 PPAAAAPLKAAPVIPVSLPAAVTAAP 199
>UniRef100_Q9SHI6 Similar to mudrA protein [Arabidopsis thaliana]
Length = 622
Score = 79.0 bits (193), Expect = 2e-13
Identities = 53/172 (30%), Positives = 81/172 (46%), Gaps = 2/172 (1%)
Query: 1 ILKYRGKPIITMLEEIRCYVMRTIANNKMKLMGYQGFLPPVQKSRLEKEAAQ-KWTATWS 59
I + R KP++ +LE+IR M A + + P + +EK A + T +
Sbjct: 273 IRQARRKPLLDLLEDIRRQCMVRTAKRFIIADRCKTKYTPRAHAEIEKMIAGFQNTQRYM 332
Query: 60 GQESRFEVTCWGQRVAVDLAAGTCTCRWWQLNGMPCFHACAAIGWKHERPENYVHAWLTM 119
+++ E+ G VD+ TC CR WQ+ G+PC HA I K E+ E+YV+ + T
Sbjct: 333 SRDNLHEIYVNGVGYFVDMDLKTCGCRKWQMVGIPCVHATCVIIGKKEKVESYVNDYYTR 392
Query: 120 GAYRSTYEFAIQPTRSQQYWEPTPYEKPIPPNMKK-KAGRPKKNRRRDGTEE 170
+R TY I+P + W +PP ++ AGRP RR G E
Sbjct: 393 NRWRETYFRGIRPVQGMPLWGRLNRLPVLPPPWRRGNAGRPSNYARRKGRNE 444
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.132 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 742,424,101
Number of Sequences: 2790947
Number of extensions: 37307710
Number of successful extensions: 377953
Number of sequences better than 10.0: 11716
Number of HSP's better than 10.0 without gapping: 1384
Number of HSP's successfully gapped in prelim test: 11061
Number of HSP's that attempted gapping in prelim test: 235015
Number of HSP's gapped (non-prelim): 55669
length of query: 384
length of database: 848,049,833
effective HSP length: 129
effective length of query: 255
effective length of database: 488,017,670
effective search space: 124444505850
effective search space used: 124444505850
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0020.1


