

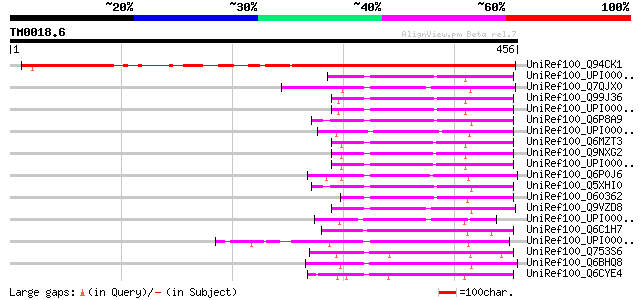
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0018.6
(456 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q94CK1 Hypothetical protein At5g12410 [Arabidopsis tha... 363 4e-99
UniRef100_UPI00003AF355 UPI00003AF355 UniRef100 entry 110 8e-23
UniRef100_Q7QJX0 ENSANGP00000021551 [Anopheles gambiae str. PEST] 106 2e-21
UniRef100_Q99J36 THUMP domain containing protein 1 [Mus musculus] 105 3e-21
UniRef100_UPI00001CEA23 UPI00001CEA23 UniRef100 entry 105 3e-21
UniRef100_Q6P8A9 Hypothetical protein MGC75804 [Xenopus tropicalis] 103 8e-21
UniRef100_UPI0000364297 UPI0000364297 UniRef100 entry 103 1e-20
UniRef100_Q6MZT3 Hypothetical protein DKFZp686C1054 [Homo sapiens] 102 2e-20
UniRef100_Q9NXG2 THUMP domain containing protein 1 [Homo sapiens] 102 2e-20
UniRef100_UPI000036A6A7 UPI000036A6A7 UniRef100 entry 102 2e-20
UniRef100_Q6P0J6 Hypothetical protein zgc:77221 [Brachydanio rerio] 102 2e-20
UniRef100_Q5XHI0 Hypothetical protein [Xenopus laevis] 102 3e-20
UniRef100_O60362 Hypothetical protein 44M2.1 [Homo sapiens] 101 5e-20
UniRef100_Q9VZD8 CG15014-PA [Drosophila melanogaster] 100 1e-19
UniRef100_UPI00004327AD UPI00004327AD UniRef100 entry 94 1e-17
UniRef100_Q6C1H7 Similar to sp|P53072 Saccharomyces cerevisiae Y... 92 2e-17
UniRef100_UPI000042C71A UPI000042C71A UniRef100 entry 85 4e-15
UniRef100_Q753S6 AFR250Cp [Ashbya gossypii] 84 1e-14
UniRef100_Q6BHQ8 Similar to CA5311|IPF2166 Candida albicans IPF2... 82 3e-14
UniRef100_Q6CYE4 Similar to sp|P53072 Saccharomyces cerevisiae Y... 80 1e-13
>UniRef100_Q94CK1 Hypothetical protein At5g12410 [Arabidopsis thaliana]
Length = 376
Score = 363 bits (933), Expect = 4e-99
Identities = 215/450 (47%), Positives = 277/450 (60%), Gaps = 82/450 (18%)
Query: 11 AAGGRKRKR---YLPHNKPVKK-GSYPLHPGVQGVFITCEGGREHQASREALNILESFYE 66
A+G + +KR Y P N+PVKK GSYPL PGVQG FI+C+GGRE QA++EA+N+++SF+E
Sbjct: 2 ASGDQSKKRKQHYRPQNRPVKKKGSYPLKPGVQGFFISCDGGREFQAAQEAINVIDSFFE 61
Query: 67 DLVDGEHLSVKE-LLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLK 125
+L+ G V L P NKK+TF+ S+ ++DEEE DE+N
Sbjct: 62 ELIQGTDSKVNPGLFGNPINKKVTFSYSE-------DEDEEE--------DESN------ 100
Query: 126 LDSSVDEHAGIGEKAVGANGAEKPKLDSSVDENAGIGGKTEGDEAKGDEKPKLDSSVDED 185
NG E +EN G G K E D
Sbjct: 101 ------------------NGEE--------EENKGDGDKAVVSEGGNDL----------- 123
Query: 186 AGIGEKAEGDEANGDKMPKMDSSVNENAGHDNEIGGKENPHKIDDVHAAAGNKTDDNEGD 245
+ EK E D++ ++ EI + + +V A N+T E D
Sbjct: 124 --VNEKEIASEGVNDQV------------NEKEIASEGSC----EVKQLAENETVKEEED 165
Query: 246 NDNIKTKTANELPVVKQCCKTNVPVQDSSNKVEKSIDTLIDEELRELGDKKKRRFVKLES 305
N K +E P K C + P+ + EKSID LI+ EL+ELGDK KRRF+KL+
Sbjct: 166 KGNQKNG-GDEPPRKKTCTEEANPLAKVNENAEKSIDKLIEAELKELGDKSKRRFMKLDP 224
Query: 306 GCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMSRFILRMLPIEVSCYASKEEISR 365
GCNG+VFIQM+K+DGD SPK+IV +TSAA+T+KHMSRFILR+LPIEVSCY S+EEISR
Sbjct: 225 GCNGLVFIQMKKRDGDPSPKEIVQHAMTSAAATKKHMSRFILRLLPIEVSCYPSEEEISR 284
Query: 366 AIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEIIDAVAKSVPGPHKVDLKNPDKTI 425
AIKPLVEQ FP+ T+ P KFAV+Y ARANTG+DRM+II+ +AKS+P PHKVDL NP+ TI
Sbjct: 285 AIKPLVEQYFPIETENPRKFAVLYGARANTGLDRMKIINTIAKSIPAPHKVDLSNPEMTI 344
Query: 426 VVEIARTVCLIGIIEKYKELSKYNLRELTS 455
VVEI +TVCLIG++EKYKEL+KYNLR+LTS
Sbjct: 345 VVEIIKTVCLIGVVEKYKELAKYNLRQLTS 374
>UniRef100_UPI00003AF355 UPI00003AF355 UniRef100 entry
Length = 243
Score = 110 bits (275), Expect = 8e-23
Identities = 63/170 (37%), Positives = 101/170 (59%), Gaps = 9/170 (5%)
Query: 287 EELRELGDKKKRRFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMSRFI 346
E++R +++ RRF +ESG N VVFI+ R + P+++VH I+ +T+K +R I
Sbjct: 39 EQIRASTERQLRRFQSVESGANNVVFIRTRGIE----PENLVHHILKDIHTTKKKKTRVI 94
Query: 347 LRMLPIEVSCYASKEEISRAIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEIIDAV 406
LRMLPI +C A E++ + + E F P + F ++Y+AR N+ ++R E+I +
Sbjct: 95 LRMLPISGTCKAFLEDMKKYTETFFEPWFKAPNKST--FQIVYKARNNSHMNREEVIKEL 152
Query: 407 AK---SVPGPHKVDLKNPDKTIVVEIARTVCLIGIIEKYKELSKYNLREL 453
A S+ +KVDL NP T+VVEI + VC + ++ Y KYNL+E+
Sbjct: 153 AGIVGSLNPENKVDLNNPQYTVVVEIIKNVCCLSVVRDYVLFRKYNLQEV 202
>UniRef100_Q7QJX0 ENSANGP00000021551 [Anopheles gambiae str. PEST]
Length = 279
Score = 106 bits (264), Expect = 2e-21
Identities = 67/217 (30%), Positives = 114/217 (51%), Gaps = 13/217 (5%)
Query: 245 DNDNIKTKTANELPVVKQCCKTNVPVQDSSNKVEKSIDTLIDEELRELGDKKKR---RFV 301
D+ I + A+EL + + Q E+ I + +E G K+ RF
Sbjct: 22 DSYRILNEYADELYGPVETTRCEEENQPDGGSDEEDISVKLQKEAEAAGKKRNAASFRFQ 81
Query: 302 KLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMSRFILRMLPIEVSCYASKE 361
+ESG +FIQ D P +IV +++ ++T+KH SRFILRMLPI+ C A+ +
Sbjct: 82 SVESGAMNCLFIQTVLPD----PNEIVVKLMRDLSATKKHKSRFILRMLPIQAVCRANLK 137
Query: 362 EISRAIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEIIDAVAKSVPGP---HKVDL 418
+I + L +Q F ++P +A+++ R N + R ++I +A + +K +L
Sbjct: 138 DIMDVVGRLGDQYF---LKEPKTYAIVFNRRLNNDLSRDDVIRELADLITSKNAGNKANL 194
Query: 419 KNPDKTIVVEIARTVCLIGIIEKYKELSKYNLRELTS 455
KNP+ ++VE+ + +C IGI+ +Y L KYN+ EL +
Sbjct: 195 KNPELAVIVEVIKGLCCIGILPEYYPLRKYNVVELVA 231
>UniRef100_Q99J36 THUMP domain containing protein 1 [Mus musculus]
Length = 350
Score = 105 bits (262), Expect = 3e-21
Identities = 63/173 (36%), Positives = 101/173 (57%), Gaps = 15/173 (8%)
Query: 290 RELGD------KKKRRFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMS 343
+E+GD K+ RRF +ESG N VVFI+ + P+ +VH I+ T+K +
Sbjct: 101 KEVGDIKASTEKRLRRFQSVESGANNVVFIRTLGIE----PEKLVHHILQDMYKTKKKKT 156
Query: 344 RFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEII 403
R ILRMLPI +C A E++ + + +E F P + F ++Y++R N+ ++R E+I
Sbjct: 157 RVILRMLPISGTCKAFLEDMKKYAETFLEPWFKAPNKGT--FQIVYKSRNNSHMNREEVI 214
Query: 404 DAVAK---SVPGPHKVDLKNPDKTIVVEIARTVCLIGIIEKYKELSKYNLREL 453
+A S+ +KVDL NP+ T+VVEI + VC + +++ Y KYNL+E+
Sbjct: 215 KELAGIVGSLNSENKVDLTNPEYTVVVEIIKAVCCLSVVKDYVLFRKYNLQEV 267
Score = 45.1 bits (105), Expect = 0.004
Identities = 37/127 (29%), Positives = 62/127 (48%), Gaps = 19/127 (14%)
Query: 12 AGGRKRK-RYLPHNKPVKK---GSYPLHPGVQGVFITCEGGREHQASREALNILESFYED 67
AG RK K ++LP + + G L PG+QG+ ITC E + EA ++L + +D
Sbjct: 13 AGKRKGKSQFLPAKRARRGDAGGPRQLEPGLQGILITC-NMNERKCVEEAYSLLNEYGDD 71
Query: 68 LVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLD 127
+ E K+ S S G+D+D E ++ +VG+ +A+ +K+L+
Sbjct: 72 MYGPEKFIDKD-------------QQPSGSEGEDDDAEAALKKEVGD-IKASTEKRLRRF 117
Query: 128 SSVDEHA 134
SV+ A
Sbjct: 118 QSVESGA 124
>UniRef100_UPI00001CEA23 UPI00001CEA23 UniRef100 entry
Length = 353
Score = 105 bits (261), Expect = 3e-21
Identities = 62/173 (35%), Positives = 101/173 (57%), Gaps = 15/173 (8%)
Query: 290 RELGD------KKKRRFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMS 343
+E+GD K+ RRF +ESG N VVFI+ + P+ +VH I+ T+K +
Sbjct: 101 KEVGDIKASTEKRLRRFQSVESGANNVVFIRTLGIE----PEKLVHHILQDMYKTKKKKT 156
Query: 344 RFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEII 403
R ILRMLP+ +C A E++ + + +E F P + F ++Y++R N+ ++R E+I
Sbjct: 157 RVILRMLPVSGTCKAFLEDMKKYAETFLEPWFKAPNKGT--FQIVYKSRNNSHMNREEVI 214
Query: 404 DAVAK---SVPGPHKVDLKNPDKTIVVEIARTVCLIGIIEKYKELSKYNLREL 453
+A S+ +KVDL NP+ T+VVEI + VC + +++ Y KYNL+E+
Sbjct: 215 KELAGIVGSLNSENKVDLTNPEYTVVVEIIKAVCCLSVVKDYVLFRKYNLQEV 267
Score = 46.2 bits (108), Expect = 0.002
Identities = 36/139 (25%), Positives = 67/139 (47%), Gaps = 20/139 (14%)
Query: 1 MASEGKGDAAAAGGRKR--KRYLPHNKPVK---KGSYPLHPGVQGVFITCEGGREHQASR 55
MA+ + + GG+++ ++LP + + G L PG+QG+ ITC E +
Sbjct: 1 MAATAQQSPPSVGGKRKGKSQFLPAKRARRCDAGGPRQLEPGLQGILITC-NMNERKCVE 59
Query: 56 EALNILESFYEDLVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEG 115
EA ++L + +D+ E K+ S S G+D+D E ++ +VG+
Sbjct: 60 EAYSLLNEYGDDMYGPEKFIDKD-------------QQPSGSEGEDDDAEAALKKEVGD- 105
Query: 116 DEANGDKKLKLDSSVDEHA 134
+A+ +K+L+ SV+ A
Sbjct: 106 IKASTEKRLRRFQSVESGA 124
>UniRef100_Q6P8A9 Hypothetical protein MGC75804 [Xenopus tropicalis]
Length = 290
Score = 103 bits (258), Expect = 8e-21
Identities = 61/185 (32%), Positives = 101/185 (53%), Gaps = 14/185 (7%)
Query: 272 DSSNKVEKSIDTLIDEELRELGDKKKRRFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRI 331
D+ ++K +D ++R +K RRF +ESG N V+FI+ + P+ +VH I
Sbjct: 84 DAEAALKKEVD-----QIRTSTEKNLRRFQSVESGANNVIFIRTLNVE----PEKLVHHI 134
Query: 332 VTSAASTRKHMSRFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPTQKPLKFAVMYEA 391
+ +T+K +R ILRMLP+ +C A E++ + + F P + F + Y+A
Sbjct: 135 LKDVHTTKKKKTRVILRMLPVSGTCKAFLEDLKKYAETFFAPWFKSPNKGT--FQIAYKA 192
Query: 392 RANTGIDRMEIIDAVAKSVPGPH---KVDLKNPDKTIVVEIARTVCLIGIIEKYKELSKY 448
R N ++R E+I +A V + KVDL NP+ T++VEI + VC +++ Y KY
Sbjct: 193 RNNNHMNRDEVIKELAGIVASQNPENKVDLSNPEYTVIVEIIKNVCCFSVVKDYTVFRKY 252
Query: 449 NLREL 453
NL+E+
Sbjct: 253 NLQEV 257
Score = 37.0 bits (84), Expect = 1.2
Identities = 33/125 (26%), Positives = 56/125 (44%), Gaps = 18/125 (14%)
Query: 13 GGRKRKRYLPH---NKPVKKGSYPLHPGVQGVFITCEGGREHQASREALNILESFYEDLV 69
G + +KRY N +K + L G+QG+ ITC E + EA ++L + + +
Sbjct: 5 GQKGKKRYKSQFCGNAKRQKRRHELEVGMQGILITC-NMNERKCVAEAYSLLNEYGDQMY 63
Query: 70 DGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSS 129
E LS K+ D S S +D+D E ++ +V + + +K L+ S
Sbjct: 64 GPEKLSEKD-------------DGLSESEEEDDDAEAALKKEVDQ-IRTSTEKNLRRFQS 109
Query: 130 VDEHA 134
V+ A
Sbjct: 110 VESGA 114
>UniRef100_UPI0000364297 UPI0000364297 UniRef100 entry
Length = 291
Score = 103 bits (257), Expect = 1e-20
Identities = 62/182 (34%), Positives = 105/182 (57%), Gaps = 12/182 (6%)
Query: 278 EKSIDTLIDEELREL---GDKKKRRFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTS 334
E+ + + +E+ +L G K++RRF L+SG N V+FI+ + DK +VH I++
Sbjct: 77 EEDVAAALKKEVAQLQGSGPKQERRFQALQSGANNVIFIKTHNLESDK----LVHHILSD 132
Query: 335 AASTRKHMSRFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPTQKPLKFAVMYEARAN 394
+T+K SR ILRMLP+ +C A E++ + + +E F P + A ++AR +
Sbjct: 133 LHTTKKKKSRVILRMLPVTGTCKAFPEDMLKYLTTFLEPWFKTPNSATYQIA--FKARNS 190
Query: 395 TGIDRMEIIDAVAKSVPG---PHKVDLKNPDKTIVVEIARTVCLIGIIEKYKELSKYNLR 451
+ R EII ++A V +KVDL +P+ TI+VE+ + VC I +++ Y KYN++
Sbjct: 191 SHNKRDEIIKSIAGLVGKLNPKNKVDLTSPELTIIVEVIKAVCCISVVKDYTLYRKYNVQ 250
Query: 452 EL 453
E+
Sbjct: 251 EV 252
Score = 45.1 bits (105), Expect = 0.004
Identities = 32/95 (33%), Positives = 45/95 (46%), Gaps = 18/95 (18%)
Query: 15 RKRKRY-LPHNKPVKKGSYPLHPGVQGVFITCEGGREHQASREALNILESFYEDLVDGEH 73
R +KRY H+ KGS L G+QG+ ITC E + + EA N+L + E L E
Sbjct: 3 RSKKRYGAGHHSKRWKGSRELEVGMQGILITC-NMNERKCTAEAFNLLNEYAEKLYGPEK 61
Query: 74 LSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEV 108
+ D+D SSS ++E EE+V
Sbjct: 62 VK----------------DNDGSSSEEEEGGEEDV 80
>UniRef100_Q6MZT3 Hypothetical protein DKFZp686C1054 [Homo sapiens]
Length = 439
Score = 102 bits (255), Expect = 2e-20
Identities = 62/173 (35%), Positives = 99/173 (56%), Gaps = 15/173 (8%)
Query: 290 RELGDKKK------RRFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMS 343
+E+GD K RRF +ESG N VVFI+ + P+ +VH I+ T+K +
Sbjct: 187 KEVGDIKASTEMRLRRFQSVESGANNVVFIRTLGIE----PEKLVHHILQDMYKTKKKKT 242
Query: 344 RFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEII 403
R ILRMLPI +C A E++ + + +E F P + F ++Y++R N+ ++R E+I
Sbjct: 243 RVILRMLPISGTCKAFLEDMKKYAETFLEPWFKAPNKG--TFQIVYKSRNNSHVNREEVI 300
Query: 404 DAVAK---SVPGPHKVDLKNPDKTIVVEIARTVCLIGIIEKYKELSKYNLREL 453
+A ++ +KVDL NP T+VVEI + VC + +++ Y KYNL+E+
Sbjct: 301 RELAGIVCTLNSENKVDLTNPQYTVVVEIIKAVCCLSVVKDYMLFRKYNLQEV 353
Score = 41.6 bits (96), Expect = 0.047
Identities = 29/105 (27%), Positives = 51/105 (47%), Gaps = 15/105 (14%)
Query: 30 GSYPLHPGVQGVFITCEGGREHQASREALNILESFYEDLVDGEHLSVKELLNRPKNKKIT 89
G L PG+QG+ ITC E + EA ++L + +D+ E + K+
Sbjct: 121 GPRQLEPGLQGILITC-NMNERKCVEEAYSLLNEYGDDMYGPEKFTDKD----------- 168
Query: 90 FADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHA 134
S S G+D+D E ++ +VG+ +A+ + +L+ SV+ A
Sbjct: 169 --QQPSGSEGEDDDAEAALKKEVGD-IKASTEMRLRRFQSVESGA 210
>UniRef100_Q9NXG2 THUMP domain containing protein 1 [Homo sapiens]
Length = 353
Score = 102 bits (255), Expect = 2e-20
Identities = 62/173 (35%), Positives = 99/173 (56%), Gaps = 15/173 (8%)
Query: 290 RELGDKKK------RRFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMS 343
+E+GD K RRF +ESG N VVFI+ + P+ +VH I+ T+K +
Sbjct: 101 KEVGDIKASTEMRLRRFQSVESGANNVVFIRTLGIE----PEKLVHHILQDMYKTKKKKT 156
Query: 344 RFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEII 403
R ILRMLPI +C A E++ + + +E F P + F ++Y++R N+ ++R E+I
Sbjct: 157 RVILRMLPISGTCKAFLEDMKKYAETFLEPWFKAPNKG--TFQIVYKSRNNSHVNREEVI 214
Query: 404 DAVAK---SVPGPHKVDLKNPDKTIVVEIARTVCLIGIIEKYKELSKYNLREL 453
+A ++ +KVDL NP T+VVEI + VC + +++ Y KYNL+E+
Sbjct: 215 RELAGIVCTLNSENKVDLTNPQYTVVVEIIKAVCCLSVVKDYMLFRKYNLQEV 267
Score = 41.6 bits (96), Expect = 0.047
Identities = 29/105 (27%), Positives = 51/105 (47%), Gaps = 15/105 (14%)
Query: 30 GSYPLHPGVQGVFITCEGGREHQASREALNILESFYEDLVDGEHLSVKELLNRPKNKKIT 89
G L PG+QG+ ITC E + EA ++L + +D+ E + K+
Sbjct: 35 GPRQLEPGLQGILITC-NMNERKCVEEAYSLLNEYGDDMYGPEKFTDKD----------- 82
Query: 90 FADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHA 134
S S G+D+D E ++ +VG+ +A+ + +L+ SV+ A
Sbjct: 83 --QQPSGSEGEDDDAEAALKKEVGD-IKASTEMRLRRFQSVESGA 124
>UniRef100_UPI000036A6A7 UPI000036A6A7 UniRef100 entry
Length = 353
Score = 102 bits (254), Expect = 2e-20
Identities = 62/173 (35%), Positives = 99/173 (56%), Gaps = 15/173 (8%)
Query: 290 RELGDKKK------RRFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMS 343
+E+GD K RRF +ESG N VVFI+ + P+ +VH I+ T+K +
Sbjct: 101 KEVGDIKASTEMRLRRFQSVESGANNVVFIRTLGIE----PEKLVHHILHDMYKTKKKKT 156
Query: 344 RFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEII 403
R ILRMLPI +C A E++ + + +E F P + F ++Y++R N+ ++R E+I
Sbjct: 157 RVILRMLPISGTCKAFLEDMKKYAETFLEPWFKAPNKG--TFQIVYKSRNNSHVNREEVI 214
Query: 404 DAVAK---SVPGPHKVDLKNPDKTIVVEIARTVCLIGIIEKYKELSKYNLREL 453
+A ++ +KVDL NP T+VVEI + VC + +++ Y KYNL+E+
Sbjct: 215 RELAGIVCTLNSENKVDLTNPQYTVVVEIIKAVCCLSVVKDYMLFRKYNLQEV 267
Score = 41.6 bits (96), Expect = 0.047
Identities = 29/105 (27%), Positives = 51/105 (47%), Gaps = 15/105 (14%)
Query: 30 GSYPLHPGVQGVFITCEGGREHQASREALNILESFYEDLVDGEHLSVKELLNRPKNKKIT 89
G L PG+QG+ ITC E + EA ++L + +D+ E + K+
Sbjct: 35 GPRQLEPGLQGILITC-NMNERKCVEEAYSLLNEYGDDMYGPEKFTDKD----------- 82
Query: 90 FADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKKLKLDSSVDEHA 134
S S G+D+D E ++ +VG+ +A+ + +L+ SV+ A
Sbjct: 83 --QQPSGSEGEDDDAEAALKKEVGD-IKASTEMRLRRFQSVESGA 124
>UniRef100_Q6P0J6 Hypothetical protein zgc:77221 [Brachydanio rerio]
Length = 353
Score = 102 bits (254), Expect = 2e-20
Identities = 66/198 (33%), Positives = 109/198 (54%), Gaps = 16/198 (8%)
Query: 269 PVQDSSNKVEKSIDT----LIDEELRELGDKKK---RRFVKLESGCNGVVFIQMRKKDGD 321
P Q+S ++ E+ D + E+ +L K +RF ++SG N VVFI+ D
Sbjct: 62 PEQESLSEDEEQEDEDAGCALQREVSQLQSSSKGRQQRFSAVDSGANNVVFIRTHGVD-- 119
Query: 322 KSPKDIVHRIVTSAASTRKHMSRFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPTQK 381
P +VH I++ TRK SR ILRMLP+ +C A E++ + + +++ F P +
Sbjct: 120 --PAQLVHHILSDLHLTRKRKSRVILRMLPVSATCRAFPEDMQKLLSVFLQRWFLAP--R 175
Query: 382 PLKFAVMYEARANTGIDRMEIIDAVAKSVP--GP-HKVDLKNPDKTIVVEIARTVCLIGI 438
F + ++AR ++ R E+I AVA V P +KVDL NP+ +I++EI ++VC + +
Sbjct: 176 HATFQICFKARNSSHSKREEVITAVAGLVGQLNPLNKVDLTNPELSIIIEIIKSVCCVSV 235
Query: 439 IEKYKELSKYNLRELTSK 456
+ Y KYNL+E+ +
Sbjct: 236 VTDYMLFRKYNLQEVAKE 253
Score = 39.3 bits (90), Expect = 0.24
Identities = 22/64 (34%), Positives = 34/64 (52%), Gaps = 1/64 (1%)
Query: 15 RKRKRYLPHNKPVKKGSYPLHPGVQGVFITCEGGREHQASREALNILESFYEDLVDGEHL 74
R R+R+ H +KG+ L G QGV ITC E + + EA ++L + ++L E
Sbjct: 7 RTRRRFGGHAHKKQKGARELEVGAQGVLITC-NMNERKCTSEAFSLLSEYADELYGPEQE 65
Query: 75 SVKE 78
S+ E
Sbjct: 66 SLSE 69
>UniRef100_Q5XHI0 Hypothetical protein [Xenopus laevis]
Length = 283
Score = 102 bits (253), Expect = 3e-20
Identities = 61/185 (32%), Positives = 100/185 (53%), Gaps = 14/185 (7%)
Query: 272 DSSNKVEKSIDTLIDEELRELGDKKKRRFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRI 331
D+ ++K +D ++R +K RRF +ESG N VVFI+ + P+ VH I
Sbjct: 84 DAEAALKKEVD-----QIRTSTEKNLRRFQSVESGANNVVFIRTLNVE----PEKFVHHI 134
Query: 332 VTSAASTRKHMSRFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPTQKPLKFAVMYEA 391
+++K +R ILRMLP+ +C A E++ + + F P + F ++Y+A
Sbjct: 135 FKDVYTSKKKKTRVILRMLPVSGTCKAFLEDLKKYAETFFAPWFKSPNKGT--FQIVYKA 192
Query: 392 RANTGIDRMEIIDAVAKSVPGPH---KVDLKNPDKTIVVEIARTVCLIGIIEKYKELSKY 448
R N ++R E+I +A V + KVDL P+ TI+VEI + VC + +++ Y KY
Sbjct: 193 RNNNHMNREEVIKELAGIVASQNPENKVDLSKPENTIIVEIIKNVCCLSVVKDYTVFRKY 252
Query: 449 NLREL 453
NL+E+
Sbjct: 253 NLQEV 257
Score = 38.5 bits (88), Expect = 0.40
Identities = 35/134 (26%), Positives = 60/134 (44%), Gaps = 20/134 (14%)
Query: 1 MASEGKGDAAAAGGRKRKRYLPHNKPVKKGSYPLHPGVQGVFITCEGGREHQASREALNI 60
M+SEG+ G ++ K N +K + L G+QG+ ITC E + EA ++
Sbjct: 1 MSSEGQ-----KGKKRYKSQFCGNAKRQKRRHDLEVGMQGILITC-NMNERKCVAEAYSL 54
Query: 61 LESFYEDLVDGEHLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANG 120
L + + + E LS K+ D + S DD+D E ++ +V + +
Sbjct: 55 LNEYGDQMYGPEKLSEKD-------------DGLTESEEDDDDAEAALKKEVDQ-IRTST 100
Query: 121 DKKLKLDSSVDEHA 134
+K L+ SV+ A
Sbjct: 101 EKNLRRFQSVESGA 114
>UniRef100_O60362 Hypothetical protein 44M2.1 [Homo sapiens]
Length = 242
Score = 101 bits (251), Expect = 5e-20
Identities = 59/159 (37%), Positives = 92/159 (57%), Gaps = 9/159 (5%)
Query: 298 RRFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMSRFILRMLPIEVSCY 357
RRF +ESG N VVFI+ + P+ +VH I+ T+K +R ILRMLPI +C
Sbjct: 4 RRFQSVESGANNVVFIRTLGIE----PEKLVHHILQDMYKTKKKKTRVILRMLPISGTCK 59
Query: 358 ASKEEISRAIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEIIDAVAKSV---PGPH 414
A E++ + + +E F P + F ++Y++R N+ ++R E+I +A V +
Sbjct: 60 AFLEDMKKYAETFLEPWFKAPNKGT--FQIVYKSRNNSHVNREEVIRELAGIVCTLNSEN 117
Query: 415 KVDLKNPDKTIVVEIARTVCLIGIIEKYKELSKYNLREL 453
KVDL NP T+VVEI + VC + +++ Y KYNL+E+
Sbjct: 118 KVDLTNPQYTVVVEIIKAVCCLSVVKDYMLFRKYNLQEV 156
>UniRef100_Q9VZD8 CG15014-PA [Drosophila melanogaster]
Length = 324
Score = 99.8 bits (247), Expect = 1e-19
Identities = 59/169 (34%), Positives = 97/169 (56%), Gaps = 10/169 (5%)
Query: 290 RELGDKKKRRFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMSRFILRM 349
RE+ ++K RF +++ VFI+ + +D P + I+ A+T K MSRF+LR+
Sbjct: 114 REMLSQRKMRFQNVDTNTTNCVFIRTQLED----PVALGKHIINDIATTGKSMSRFVLRL 169
Query: 350 LPIEVSCYASKEEISRAIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEIIDAVAKS 409
+PIEV C A+ +I A L +++F ++P + +++ R N I R +II +A+
Sbjct: 170 VPIEVVCRANMPDIITAAGELFDKHF---LKEPTSYGIIFNHRYNQQIKRDQIITQLAEL 226
Query: 410 VPGPH---KVDLKNPDKTIVVEIARTVCLIGIIEKYKELSKYNLRELTS 455
V + KVDLK K+I+VE+ R CL+ +I+ Y E K+NL EL +
Sbjct: 227 VNSKNVGNKVDLKEAKKSIIVEVLRGWCLLSVIDNYLECKKFNLAELAN 275
>UniRef100_UPI00004327AD UPI00004327AD UniRef100 entry
Length = 242
Score = 93.6 bits (231), Expect = 1e-17
Identities = 56/173 (32%), Positives = 99/173 (56%), Gaps = 17/173 (9%)
Query: 275 NKVEKSIDTLIDEELRELGDK-----KKRRFVKLESGCNGVVFIQMRKKDGDKSPKDIVH 329
N E I ++++E+ EL + RRF +++G +VFI + +P ++V
Sbjct: 78 NDNEDDISVVLNKEINELKAEYSKPINARRFQVIDTGVKNIVFI----RSSLTNPLELVT 133
Query: 330 RIVTSAASTRKHMSRFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPTQKPLKFAVMY 389
+I+T +T++ +R++LR+LPIEV C A+ +I ++E+ F Q+P F++++
Sbjct: 134 KIITELYNTKQQRTRYLLRLLPIEVICKANMNDIKSKADAMLEKYF---AQEPKTFSIVF 190
Query: 390 EARANTGIDRMEIIDAVA----KSVPGPHKVDLKNPDKTIVVEIARTVCLIGI 438
+N I R EII+ +A K PG +K DLKNP+ ++VE+ R VCL+ +
Sbjct: 191 NRHSNNNIYRDEIIEDLAEIINKKNPG-NKADLKNPELAVIVEMVRGVCLMSV 242
Score = 38.5 bits (88), Expect = 0.40
Identities = 27/93 (29%), Positives = 43/93 (46%), Gaps = 13/93 (13%)
Query: 15 RKRKRYLPHN---KPVKKGSYPLHPGVQGVFITCEGGREHQASREALNILESFYEDLVDG 71
+KRK+ HN K+ + L PG++G TC E + R+A IL F +++
Sbjct: 1 QKRKKNYFHNHGNNKKKRKQFNLEPGMKGFLCTCNFS-EKECVRDAYKILNEFADEIYG- 58
Query: 72 EHLSVKELLNRPKNKKITFADSDSSSSGDDEDD 104
L+ KN K++ D + D+EDD
Sbjct: 59 --------LDSIKNIKVSDIKKDETKYNDNEDD 83
>UniRef100_Q6C1H7 Similar to sp|P53072 Saccharomyces cerevisiae YGL232w [Yarrowia
lipolytica]
Length = 372
Score = 92.4 bits (228), Expect = 2e-17
Identities = 59/179 (32%), Positives = 97/179 (53%), Gaps = 9/179 (5%)
Query: 281 IDTLIDEELRELGDKKKRRFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRK 340
ID + E+ + D KK L GC ++F++ RK P D V I T K
Sbjct: 82 IDKAMAAEVAAMKDTKKDLITPLPLGCECMLFVRTRKP---VVPVDFVKAICQGVKDTGK 138
Query: 341 HMSRFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPT-QKPLKFAVMYEARANTGIDR 399
+RF+ RM P+ ++C ASK E+ + ++ +F + QKPLK+A+ R G++R
Sbjct: 139 KSTRFVQRMTPVTLTCSASKPELEKLCDIVLGPHFHLKEGQKPLKYAIRPTMRNFDGMNR 198
Query: 400 MEIIDAVAKSV--PGPHKVDLKNPDKTIVVEIART---VCLIGIIEKYKELSKYNLREL 453
E+I +VA V H VDLKN DK I+V+ + + ++G +E+Y+ L+++N+ +L
Sbjct: 199 DEVIQSVASRVGQDHGHSVDLKNYDKLILVDCFKASIGMSVVGNVEEYEGLARFNIEQL 257
>UniRef100_UPI000042C71A UPI000042C71A UniRef100 entry
Length = 409
Score = 85.1 bits (209), Expect = 4e-15
Identities = 82/283 (28%), Positives = 126/283 (43%), Gaps = 37/283 (13%)
Query: 186 AGIGEKAEGDEANGDKMPKMDSSVNENAGHD-----NEIGGKENPHKIDDVHAAAGNKTD 240
AG G+K G NG + D NE H ++ K P K+ + + T
Sbjct: 27 AGGGDKGNG---NGGEKGNQDDQSNEGKEHPKVFPLDKYREKPLPEKLS-LPGILISTTK 82
Query: 241 DNEGDNDNIKTKTANELPVVKQCCK-TNVPVQDSSNKVEKSIDTLID---------EELR 290
D E A EL ++K K + +SS E S D +D E ++
Sbjct: 83 DKE---------KAAELEIIKHLEKIADELYHESSEGQEDSHDGNLDFEEMLKRDLESMK 133
Query: 291 ELGDKKKR-RFVKLESGCNGVVFIQMRKKDGDKSPKDIVHRIVTSAASTRKHMSRFILRM 349
+ K +R R E C V + + P +V I+ AAST K R R+
Sbjct: 134 DQSTKSQRFRLCSREGFCLIYVIVLPPLQ-----PHRLVEYILEHAASTGKCPLRHCKRL 188
Query: 350 LPIEVSCYASKEEISRAIKPLVEQNFPVPTQKPLKFAVMYEARANTGIDRMEIIDAVAKS 409
+PI + A+ ++S +V+ F P + KFAV +R++ ++RME+I AVA+
Sbjct: 189 IPIPATARATLRQLSEVTASVVKSGFESPDGQAFKFAVNANSRSSDKLERMEMIRAVAEQ 248
Query: 410 VP---GPHKVDLKNPDKTIVVEIARTVCLIGIIEKYKELSKYN 449
V G H VDLKN DKTI+VE+ + + ++ Y++ KYN
Sbjct: 249 VAMLGGGHTVDLKNADKTILVEVYKNNLGVTVLNDYEKYKKYN 291
>UniRef100_Q753S6 AFR250Cp [Ashbya gossypii]
Length = 277
Score = 83.6 bits (205), Expect = 1e-14
Identities = 61/199 (30%), Positives = 102/199 (50%), Gaps = 18/199 (9%)
Query: 270 VQDSSNKVEKSIDTLIDEELREL-------GDKKKRRFVKLESGCNGVVFIQMRKKDGDK 322
+ D ++ E S++ + +EL +L KKK +++ GC +VFI+ R+
Sbjct: 72 LSDKEDEEELSVEEQVQKELEQLKKGSGPVDTKKKPVLQEIQLGCECMVFIKTRRPI--- 128
Query: 323 SPKDIVHRIVTSAASTRK--HMSRFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPTQ 380
P+ V R+V AS+ +SR++ R+ PI SC AS E+ + + ++ +F +
Sbjct: 129 KPECFVKRLVQELASSENTTKVSRYVQRLTPITDSCNASLTELEKLCRRVLAPHFHTDKE 188
Query: 381 KPLKFAVMYEARANTGIDRMEIIDAVAKSVPGP----HKVDLKNPDKTIVVEIARTVCLI 436
KFAV R ID+M+II VAK V H VDLK+ DK ++V+ + +
Sbjct: 189 IKYKFAVEVVKRNFNTIDKMDIIKLVAKEVGKSGELGHSVDLKDYDKLVIVQCYKNNIGM 248
Query: 437 GIIEK--YKELSKYNLREL 453
+++K L KYNL+E+
Sbjct: 249 SVVDKDYSVALKKYNLQEI 267
Score = 35.0 bits (79), Expect = 4.4
Identities = 27/103 (26%), Positives = 46/103 (44%), Gaps = 9/103 (8%)
Query: 16 KRKRYLPHNKPVKKGSY---PLHPGVQGVFITCEGGREHQASREALNILESFYEDLVDGE 72
KR R + P KK + P G++ TC E QASRE ++IL+ E
Sbjct: 3 KRARSGDSSGPKKKYKVVHATIDPNTSGIYATCARRHEKQASRELMSILQE------KAE 56
Query: 73 HLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEG 115
V EL + + ++ + + S +++ +E Q++ G G
Sbjct: 57 EYYVDELKAIAETELLSDKEDEEELSVEEQVQKELEQLKKGSG 99
>UniRef100_Q6BHQ8 Similar to CA5311|IPF2166 Candida albicans IPF2166 unknown function
[Debaryomyces hansenii]
Length = 297
Score = 82.0 bits (201), Expect = 3e-14
Identities = 56/196 (28%), Positives = 102/196 (51%), Gaps = 9/196 (4%)
Query: 267 NVPVQDSSNKVEKSIDTLIDEELRELGDKK--KRRFVK-LESGCNGVVFIQMRKKDGDKS 323
N + N+ E SI+ I +EL ++ + K K+ +K ++ GC +VFI+ R+
Sbjct: 65 NEDSETEQNEKELSIEEKIQKELNDMQETKGTKKEILKPIDLGCECLVFIKTRRPI---E 121
Query: 324 PKDIVHRIVTSAASTRKHMSRFILRMLPIEVSCYASKEEISRAIKPLVEQNFPVPT-QKP 382
P V ++ + ++ +R+ ++ PI S + EEI + + ++ +F T QKP
Sbjct: 122 PATFVEKLCEEFSQSKVKTTRYTQKLSPITFSVTPTMEEIKKLAQRVLGPHFHNETNQKP 181
Query: 383 LKFAVMYEARANTGIDRMEIIDAVAKSVPGPH--KVDLKNPDKTIVVEIARTVCLIGIIE 440
KFA+ R + + +II A+A+ V H VDLKN DK I+VE ++ + ++
Sbjct: 182 YKFAIQVSKRNFNTLQKPDIIKAIAECVGRDHGHSVDLKNYDKLIMVECYKSNIGMSVVN 241
Query: 441 KYKELSKYNLRELTSK 456
Y L ++NL+++ K
Sbjct: 242 NYDRLERFNLQQIFEK 257
Score = 40.8 bits (94), Expect = 0.081
Identities = 26/111 (23%), Positives = 48/111 (42%), Gaps = 23/111 (20%)
Query: 13 GGRKRKRYLPHNKPVKKGSYPLHPGVQGVFITCEGGREHQASREALNILESFYEDLVDGE 72
GG K K++ + S + P G++ TC G+E Q +E LN+ E+ + +
Sbjct: 11 GGNKNKKF--------RTSGFIDPSTSGIYATCNRGKEQQCRKELLNLFSDKAEEYFNLD 62
Query: 73 HLSVKELLNRPKNKKITFADSDSSSSGDDEDDEEEVQVQVGEGDEANGDKK 123
L+ DS++ + + EE++Q ++ + E G KK
Sbjct: 63 ELN---------------EDSETEQNEKELSIEEKIQKELNDMQETKGTKK 98
>UniRef100_Q6CYE4 Similar to sp|P53072 Saccharomyces cerevisiae YGL232w singleton
[Kluyveromyces lactis]
Length = 309
Score = 80.1 bits (196), Expect = 1e-13
Identities = 64/202 (31%), Positives = 108/202 (52%), Gaps = 21/202 (10%)
Query: 269 PVQDSSNKVEKSIDTLIDEELRELG-------DKKKRRFV--KLESGCNGVVFIQMRKKD 319
PV S K E SI+ + +EL L D KK + + +L+ +VF++ RK
Sbjct: 84 PVTTESKK-EISIEEELQQELEGLKNHKTTIVDTKKNKPILEQLDLQTECLVFVKTRKPI 142
Query: 320 GDKSPKDIVHRIVTSAASTR--KHMSRFILRMLPIEVSCYASKEEISRAIKPLVEQNFPV 377
P+ VHRI+ A + +RF+ ++ PI SC+AS EE+++ + ++ Q+F
Sbjct: 143 ---VPEKFVHRIIRDLADPHDLRKRTRFVQKLTPITDSCHASMEELAKLCERVLPQHFHK 199
Query: 378 PTQKPLKFAVMYEARANTGIDRMEIIDAVA----KSVPGPHKVDLKNPDKTIVVEIARTV 433
P+KFAV R + +D+ME+I +A K HKVDLKN DK ++++ +
Sbjct: 200 EEMVPVKFAVEINKRNFSTMDKMEMIKYIAGFVGKQGSLEHKVDLKNYDKLVLLQCFKNN 259
Query: 434 CLIGIIEK-YK-ELSKYNLREL 453
+ +++K Y+ + KYN++EL
Sbjct: 260 IGMCVVDKDYRTDYKKYNVQEL 281
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.308 0.131 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 809,602,286
Number of Sequences: 2790947
Number of extensions: 38186321
Number of successful extensions: 171812
Number of sequences better than 10.0: 3175
Number of HSP's better than 10.0 without gapping: 228
Number of HSP's successfully gapped in prelim test: 3182
Number of HSP's that attempted gapping in prelim test: 146836
Number of HSP's gapped (non-prelim): 12148
length of query: 456
length of database: 848,049,833
effective HSP length: 131
effective length of query: 325
effective length of database: 482,435,776
effective search space: 156791627200
effective search space used: 156791627200
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0018.6


