

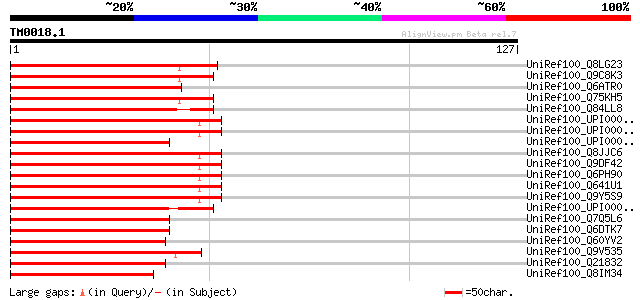
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0018.1
(127 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LG23 RNA-binding protein, putative [Arabidopsis thal... 76 2e-13
UniRef100_Q9C8K3 RNA-binding protein, putative; 35994-37391 [Ara... 75 3e-13
UniRef100_Q6ATR0 Putative RNA binding motif protein [Oryza sativa] 64 1e-09
UniRef100_Q75KH5 Hypothetical protein OJ1489_G03.1 [Oryza sativa] 63 1e-09
UniRef100_Q84LL8 Salt tolerance protein 4 [Beta vulgaris] 57 7e-08
UniRef100_UPI00001A08B4 UPI00001A08B4 UniRef100 entry 54 6e-07
UniRef100_UPI000035F04F UPI000035F04F UniRef100 entry 52 3e-06
UniRef100_UPI000031883B UPI000031883B UniRef100 entry 52 4e-06
UniRef100_Q8JJC6 RNA binding motif protein 8 [Oryzias latipes] 52 4e-06
UniRef100_Q9DF42 RNA-binding protein Y14 [Xenopus laevis] 51 7e-06
UniRef100_Q6PH90 MGC68591 protein [Xenopus laevis] 51 7e-06
UniRef100_Q641U1 Hypothetical protein [Xenopus laevis] 51 7e-06
UniRef100_Q9Y5S9 RNA-binding protein 8A [Homo sapiens] 51 7e-06
UniRef100_UPI000043458C UPI000043458C UniRef100 entry 50 9e-06
UniRef100_Q7Q5L6 ENSANGP00000013045 [Anopheles gambiae str. PEST] 50 1e-05
UniRef100_Q6DTK7 RNA binding motif protein 8B [Equus caballus] 50 1e-05
UniRef100_Q60YV2 Hypothetical protein CBG18074 [Caenorhabditis b... 50 1e-05
UniRef100_Q9V535 RNA-binding protein 8A [Drosophila melanogaster] 49 2e-05
UniRef100_Q21832 Hypothetical protein R07E5.14 [Caenorhabditis e... 49 3e-05
UniRef100_Q8IM34 RNA binding protein, putative [Plasmodium falci... 47 7e-05
>UniRef100_Q8LG23 RNA-binding protein, putative [Arabidopsis thaliana]
Length = 202
Score = 75.9 bits (185), Expect = 2e-13
Identities = 37/53 (69%), Positives = 45/53 (84%), Gaps = 1/53 (1%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPI-NESIRRKNARH 52
GYALIEYE+ EEA++AI +NG+ELLTQN+ VDWAFSSGP ES RRKN+R+
Sbjct: 137 GYALIEYEKKEEAQSAISAMNGAELLTQNVSVDWAFSSGPSGGESYRRKNSRY 189
>UniRef100_Q9C8K3 RNA-binding protein, putative; 35994-37391 [Arabidopsis thaliana]
Length = 191
Score = 75.1 bits (183), Expect = 3e-13
Identities = 37/52 (71%), Positives = 44/52 (84%), Gaps = 1/52 (1%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPI-NESIRRKNAR 51
GYALIEYE+ EEA++AI +NG+ELLTQN+ VDWAFSSGP ES RRKN+R
Sbjct: 137 GYALIEYEKKEEAQSAISAMNGAELLTQNVSVDWAFSSGPSGGESYRRKNSR 188
>UniRef100_Q6ATR0 Putative RNA binding motif protein [Oryza sativa]
Length = 197
Score = 63.5 bits (153), Expect = 1e-09
Identities = 27/43 (62%), Positives = 36/43 (82%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINE 43
GYAL+EYE EEA+ AI+ +NG++LLT+ +YVDWAFS GPI +
Sbjct: 132 GYALVEYESFEEAQTAIKAMNGTQLLTRTVYVDWAFSRGPIQK 174
>UniRef100_Q75KH5 Hypothetical protein OJ1489_G03.1 [Oryza sativa]
Length = 116
Score = 63.2 bits (152), Expect = 1e-09
Identities = 32/52 (61%), Positives = 42/52 (80%), Gaps = 1/52 (1%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPI-NESIRRKNAR 51
GYALIEYE EEA+ AI+ L+G+ELLTQ I VDWAFS+GP+ +IR+++ R
Sbjct: 55 GYALIEYETFEEAQAAIKALDGTELLTQIISVDWAFSNGPVKRRNIRKRSPR 106
>UniRef100_Q84LL8 Salt tolerance protein 4 [Beta vulgaris]
Length = 203
Score = 57.4 bits (137), Expect = 7e-08
Identities = 29/51 (56%), Positives = 37/51 (71%), Gaps = 3/51 (5%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIRRKNAR 51
GYALIEYE+ EEA+ AI+ +NG+++L Q I VDWAF +GP RR N R
Sbjct: 141 GYALIEYEKFEEAQAAIKEMNGAKMLEQPINVDWAFCNGPYR---RRGNRR 188
>UniRef100_UPI00001A08B4 UPI00001A08B4 UniRef100 entry
Length = 174
Score = 54.3 bits (129), Expect = 6e-07
Identities = 28/55 (50%), Positives = 35/55 (62%), Gaps = 2/55 (3%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIR--RKNARHP 53
GYAL+EYE +EA+ A+E LNG EL+ Q I VDW F GP R R+ +R P
Sbjct: 115 GYALVEYETYKEAQAAMEGLNGQELMGQPISVDWCFVRGPPKSKRRGGRRRSRSP 169
>UniRef100_UPI000035F04F UPI000035F04F UniRef100 entry
Length = 173
Score = 52.0 bits (123), Expect = 3e-06
Identities = 26/56 (46%), Positives = 35/56 (62%), Gaps = 3/56 (5%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIR---RKNARHP 53
GYAL+EYE +EA+ A+E LNG +++ Q I VDW F GP R R+ +R P
Sbjct: 113 GYALVEYETYKEAQAAMEGLNGQDMMGQPISVDWGFVRGPPKNKRRTGGRRRSRSP 168
>UniRef100_UPI000031883B UPI000031883B UniRef100 entry
Length = 175
Score = 51.6 bits (122), Expect = 4e-06
Identities = 22/40 (55%), Positives = 30/40 (75%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGP 40
GYAL+EYE +EA+ A+E LNG +++ Q+I VDW F GP
Sbjct: 115 GYALVEYETYKEAQAAMEGLNGQDMMGQSISVDWGFVRGP 154
>UniRef100_Q8JJC6 RNA binding motif protein 8 [Oryzias latipes]
Length = 173
Score = 51.6 bits (122), Expect = 4e-06
Identities = 26/56 (46%), Positives = 35/56 (62%), Gaps = 3/56 (5%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIR---RKNARHP 53
GYAL+EYE +EA+ A+E LN +L+ Q + VDWAF GP R R+ +R P
Sbjct: 113 GYALVEYETYKEAQAAMEGLNAQDLMGQPVSVDWAFVRGPPKNKRRTGGRRRSRSP 168
>UniRef100_Q9DF42 RNA-binding protein Y14 [Xenopus laevis]
Length = 174
Score = 50.8 bits (120), Expect = 7e-06
Identities = 26/55 (47%), Positives = 34/55 (61%), Gaps = 2/55 (3%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIR--RKNARHP 53
GYAL+EYE +EA A+E LNG +L+ Q + VDW F GP R R+ +R P
Sbjct: 115 GYALVEYETYKEALAAMEGLNGQDLMGQPVSVDWGFVRGPPKGKRRSGRRRSRSP 169
>UniRef100_Q6PH90 MGC68591 protein [Xenopus laevis]
Length = 174
Score = 50.8 bits (120), Expect = 7e-06
Identities = 26/55 (47%), Positives = 34/55 (61%), Gaps = 2/55 (3%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIR--RKNARHP 53
GYAL+EYE +EA A+E LNG +L+ Q + VDW F GP R R+ +R P
Sbjct: 115 GYALVEYETYKEALAAMEGLNGQDLMGQPVSVDWGFVRGPPKGKRRSGRRRSRSP 169
>UniRef100_Q641U1 Hypothetical protein [Xenopus laevis]
Length = 173
Score = 50.8 bits (120), Expect = 7e-06
Identities = 26/55 (47%), Positives = 34/55 (61%), Gaps = 2/55 (3%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIR--RKNARHP 53
GY L+EYE +EA+ A+E LNG +L+ Q I VDW F GP R R+ +R P
Sbjct: 114 GYTLVEYETYKEAQAAMEGLNGQDLMGQPISVDWCFVRGPPKGKRRGGRRRSRSP 168
>UniRef100_Q9Y5S9 RNA-binding protein 8A [Homo sapiens]
Length = 174
Score = 50.8 bits (120), Expect = 7e-06
Identities = 26/55 (47%), Positives = 34/55 (61%), Gaps = 2/55 (3%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIR--RKNARHP 53
GY L+EYE +EA+ A+E LNG +L+ Q I VDW F GP R R+ +R P
Sbjct: 115 GYTLVEYETYKEAQAAMEGLNGQDLMGQPISVDWCFVRGPPKGKRRGGRRRSRSP 169
>UniRef100_UPI000043458C UPI000043458C UniRef100 entry
Length = 56
Score = 50.4 bits (119), Expect = 9e-06
Identities = 25/51 (49%), Positives = 36/51 (70%), Gaps = 2/51 (3%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGPINESIRRKNAR 51
GYAL+EYE +EA+ A + LNG+E+L Q I VDW F GP + I++++ R
Sbjct: 6 GYALVEYETFKEAQAAKDALNGTEILGQPISVDWCFVKGP--KKIKKRSHR 54
>UniRef100_Q7Q5L6 ENSANGP00000013045 [Anopheles gambiae str. PEST]
Length = 163
Score = 50.1 bits (118), Expect = 1e-05
Identities = 24/40 (60%), Positives = 28/40 (70%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGP 40
GYAL+EYE ++A A E LNGSE+L Q I VDW F GP
Sbjct: 113 GYALVEYETYKQALAAKEALNGSEVLGQTIAVDWCFVKGP 152
>UniRef100_Q6DTK7 RNA binding motif protein 8B [Equus caballus]
Length = 133
Score = 49.7 bits (117), Expect = 1e-05
Identities = 22/40 (55%), Positives = 28/40 (70%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGP 40
GY L+EYE +EA+ A+E LNG +L+ Q I VDW F GP
Sbjct: 86 GYTLVEYETYKEAQAAMEGLNGQDLMGQPISVDWCFVRGP 125
>UniRef100_Q60YV2 Hypothetical protein CBG18074 [Caenorhabditis briggsae]
Length = 142
Score = 49.7 bits (117), Expect = 1e-05
Identities = 22/39 (56%), Positives = 27/39 (68%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSG 39
GYAL+EYE +EA AIE N ++LL QN+ VDW F G
Sbjct: 97 GYALVEYETQKEASEAIEKSNDTDLLGQNVKVDWCFIKG 135
>UniRef100_Q9V535 RNA-binding protein 8A [Drosophila melanogaster]
Length = 165
Score = 49.3 bits (116), Expect = 2e-05
Identities = 24/50 (48%), Positives = 34/50 (68%), Gaps = 2/50 (4%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSGP--INESIRRK 48
GYAL+EYE ++A A E LNG+E++ Q I VDW F GP + +S +R+
Sbjct: 115 GYALVEYETHKQALAAKEALNGAEIMGQTIQVDWCFVKGPKRVKKSEKRR 164
>UniRef100_Q21832 Hypothetical protein R07E5.14 [Caenorhabditis elegans]
Length = 142
Score = 48.5 bits (114), Expect = 3e-05
Identities = 21/39 (53%), Positives = 27/39 (68%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAFSSG 39
GYAL+EYE +EA AI+ N ++LL QN+ VDW F G
Sbjct: 97 GYALVEYETQKEANEAIDQSNDTDLLGQNVKVDWCFVKG 135
>UniRef100_Q8IM34 RNA binding protein, putative [Plasmodium falciparum]
Length = 107
Score = 47.4 bits (111), Expect = 7e-05
Identities = 20/36 (55%), Positives = 27/36 (74%)
Query: 1 GYALIEYERAEEAKNAIENLNGSELLTQNIYVDWAF 36
GYA +EYE +AK AI+ ++G+ LL Q I+VDWAF
Sbjct: 65 GYAFLEYENFVDAKRAIDEMDGTMLLNQEIHVDWAF 100
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.141 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 219,237,502
Number of Sequences: 2790947
Number of extensions: 8537389
Number of successful extensions: 21695
Number of sequences better than 10.0: 635
Number of HSP's better than 10.0 without gapping: 542
Number of HSP's successfully gapped in prelim test: 93
Number of HSP's that attempted gapping in prelim test: 20629
Number of HSP's gapped (non-prelim): 1158
length of query: 127
length of database: 848,049,833
effective HSP length: 103
effective length of query: 24
effective length of database: 560,582,292
effective search space: 13453975008
effective search space used: 13453975008
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0018.1


