

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0017.9
(387 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
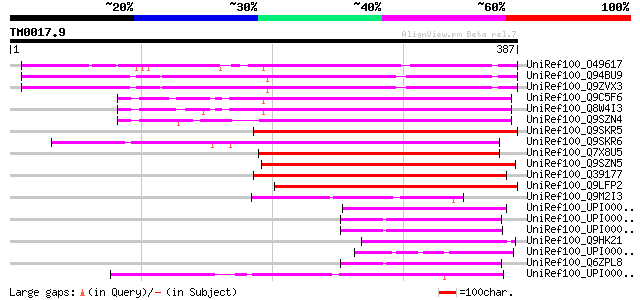
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O49617 Hypothetical protein M4E13.160 [Arabidopsis tha... 172 2e-41
UniRef100_Q94BU9 At2g16900/F12A24.8 [Arabidopsis thaliana] 164 3e-39
UniRef100_Q9ZVX3 Expressed protein [Arabidopsis thaliana] 164 4e-39
UniRef100_Q9C5F6 Putative phospholipase [Arabidopsis thaliana] 139 1e-31
UniRef100_Q8W4I3 Phospholipase like protein [Arabidopsis thaliana] 139 1e-31
UniRef100_Q9SZN4 Phospholipase like protein [Arabidopsis thaliana] 138 3e-31
UniRef100_Q9SKR5 PEARLI 4 protein [Arabidopsis thaliana] 135 2e-30
UniRef100_Q9SKR6 Hypothetical protein At2g20950 [Arabidopsis tha... 135 3e-30
UniRef100_Q7X8U5 Hypothetical protein At2g20950 [Arabidopsis tha... 134 4e-30
UniRef100_Q9SZN5 Phospholipase like protein [Arabidopsis thaliana] 132 2e-29
UniRef100_Q39177 PEARLI 4 protein [Arabidopsis thaliana] 131 3e-29
UniRef100_Q9LFP2 Hypothetical protein F2I11_30 [Arabidopsis thal... 128 3e-28
UniRef100_Q9M2I3 Hypothetical protein F9D24.240 [Arabidopsis tha... 59 3e-07
UniRef100_UPI00003C12B5 UPI00003C12B5 UniRef100 entry 54 7e-06
UniRef100_UPI00001D0F0B UPI00001D0F0B UniRef100 entry 53 2e-05
UniRef100_UPI00003AA891 UPI00003AA891 UniRef100 entry 52 3e-05
UniRef100_Q9HK21 Chromosome segregation protein related ptotein ... 52 3e-05
UniRef100_UPI0000363BD1 UPI0000363BD1 UniRef100 entry 51 5e-05
UniRef100_Q6ZPL8 MKIAA1601 protein [Mus musculus] 51 6e-05
UniRef100_UPI0000438431 UPI0000438431 UniRef100 entry 50 1e-04
>UniRef100_O49617 Hypothetical protein M4E13.160 [Arabidopsis thaliana]
Length = 386
Score = 172 bits (435), Expect = 2e-41
Identities = 126/404 (31%), Positives = 205/404 (50%), Gaps = 50/404 (12%)
Query: 10 SNPNCPHASNPYHQCTQACSKKTSHRTKPHAKNTSSSGAAPPPSNFDRRKKPGSRPEPPV 69
++P+C +A NP+H+C AC +K + SS +F R+KK S +PP
Sbjct: 7 AHPDCVYADNPFHECASACLEKIAQGHVKKNTKKQSSRLLSFSGSFGRKKKE-SNSQPPT 65
Query: 70 VAGAVPAAKVEAIQKSNASSPISNHS-----EIKKV---ESRN-----------DDHNHS 110
A P +N +SP +HS +KK E++N D NH
Sbjct: 66 PLSARPYQNGRG-GFANGNSPKVHHSVAPPASVKKKIVSETKNSFTSSSSGDPDDFFNHK 124
Query: 111 VPVSGQIHVPLPVPDVKHVHEEDQQKNGVEHLANPIQTKQEDKTASDVH----PGEGLAT 166
+PL ++ + K G++ I E + S + PG
Sbjct: 125 PEKKPSQTIPLSSNNLVDQSKVVSPKPGIQEHNGKIGEGGETRLFSFLSLPRSPG----- 179
Query: 167 TFKGGSIDFSFSGIIPQGNEDNDDKG---DTESVVSESRVPVGKYHVKESFASILESILD 223
K + DFS +E+N++ G D ESV+S++ V VGKY V+ ++IL ++++
Sbjct: 180 --KESNDDFS-----DDDDENNNEIGVELDLESVMSDTFVSVGKYRVRSGSSTILSAVIE 232
Query: 224 KYGDIGASCHLESLVLRSYYIECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLR 283
K+GDI +C LES +RS Y+EC+C ++QEL+S + L+K K+ E++A+LKD+ES +
Sbjct: 233 KHGDIAQNCKLESDSMRSRYLECLCSLMQELRSTPVGQLSKVKVKEMLAVLKDLESVNIE 292
Query: 284 VAWLRSALDEIAENVEVINQHQEVDAAKANTDREMESLREQLESEMESLAEKEQEVADIK 343
VAWLRS L+E A++ E +V+ K D +++ RE+LE++ L E+EV ++K
Sbjct: 293 VAWLRSVLEEFAQSQE------DVENEKERHDGLVKAKREELEAQETDLVRMEKEVVEVK 346
Query: 344 KRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRKSLIDELL 387
+RI E ++ E+E + +E+ ++E KS IDELL
Sbjct: 347 RRIEETRAQMVEIEAERLRMEKMGF----KMEKFKGKSFIDELL 386
>UniRef100_Q94BU9 At2g16900/F12A24.8 [Arabidopsis thaliana]
Length = 382
Score = 164 bits (416), Expect = 3e-39
Identities = 118/392 (30%), Positives = 193/392 (49%), Gaps = 30/392 (7%)
Query: 10 SNPNCPHASNPYHQCTQACSKKTSHRTKPHAKNTSSSGAAPPPSNFDRRKKPGSRPEPPV 69
++P+C ++SNP+H+C C +K S S P +F ++K P P
Sbjct: 7 AHPDCRYSSNPFHECASDCLEKISQGRGNKNSKKQGSKILSLPGSFGKKKTESQPPSPLS 66
Query: 70 VAGAVPAAKVEAIQKSNASSPISNHSEIKKVESRNDDHN-HSVPVSGQIHVPLPVPDVKH 128
A + + SP++ +KK + + HS+ G I + L +
Sbjct: 67 TRNYQNGAANNPKVRQSRPSPVA----MKKTPVPEANKSLHSLSSDG-ISIDLNGQNDSF 121
Query: 129 VHEEDQQKNGVEHLANPIQTKQEDKTASDV-HPGEGLATTFKGGSIDFSFSGIIPQGNED 187
H++++ V N + + + + H G T S+ S N+D
Sbjct: 122 NHKQEKPSRTVPLSPNSMADRGKPLSPRPQGHEHSGKNDTASEISLFNVVSPPRSCANDD 181
Query: 188 NDDKGDTE-----------SVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLES 236
+DD + SV+S+S V VGKY V S ++IL+SI+DK+GDI A+C LES
Sbjct: 182 DDDDDENNGYEEGVELDLISVMSDSCVSVGKYRVNSSVSTILQSIIDKHGDIAANCKLES 241
Query: 237 LVLRSYYIECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAE 296
+RS Y+EC+C ++QEL S + LT+ K+ E+VA+LKD+ES + V W+RS L+E A
Sbjct: 242 ASMRSRYLECLCSLMQELGSTPVGQLTELKVKEMVAVLKDLESVNIDVGWMRSVLEEFA- 300
Query: 297 NVEVINQHQE-VDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKE 355
Q+QE D+ K + + S ++++E + LA E+EVA+ + R+ E+ L E
Sbjct: 301 ------QYQENTDSEKERQEGLVRSKKQEMEIQEADLARIEKEVAEARLRVEEMKAELAE 354
Query: 356 LELKSAELEQSMLSSKSEVENLHRKSLIDELL 387
LE + +E+ +VE K+ +DELL
Sbjct: 355 LETERLRMEEMGF----KVEKYKGKTFLDELL 382
>UniRef100_Q9ZVX3 Expressed protein [Arabidopsis thaliana]
Length = 382
Score = 164 bits (415), Expect = 4e-39
Identities = 118/392 (30%), Positives = 193/392 (49%), Gaps = 30/392 (7%)
Query: 10 SNPNCPHASNPYHQCTQACSKKTSHRTKPHAKNTSSSGAAPPPSNFDRRKKPGSRPEPPV 69
++P+C ++SNP+H+C C +K S S P +F ++K P P
Sbjct: 7 AHPDCRYSSNPFHECASDCLEKISQGRGNKNSKKQGSKILSLPGSFGKKKTESQPPSPLS 66
Query: 70 VAGAVPAAKVEAIQKSNASSPISNHSEIKKVESRNDDHN-HSVPVSGQIHVPLPVPDVKH 128
A + + SP++ +KK + + HS+ G I + L +
Sbjct: 67 TRNYQNGAANTPKVRQSRPSPVA----MKKTPVPEANKSLHSLSSDG-ISIDLNGQNDSF 121
Query: 129 VHEEDQQKNGVEHLANPIQTKQEDKTASDV-HPGEGLATTFKGGSIDFSFSGIIPQGNED 187
H++++ V N + + + + H G T S+ S N+D
Sbjct: 122 NHKQEKPSRTVPLSPNSMADRGKPLSPRPQGHEHSGKNDTASEISLFNVVSPPRSCANDD 181
Query: 188 NDDKGDTE-----------SVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLES 236
+DD + SV+S+S V VGKY V S ++IL+SI+DK+GDI A+C LES
Sbjct: 182 DDDDDENNGYEEGVELDLISVMSDSCVSVGKYRVNSSVSTILQSIIDKHGDIAANCKLES 241
Query: 237 LVLRSYYIECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAE 296
+RS Y+EC+C ++QEL S + LT+ K+ E+VA+LKD+ES + V W+RS L+E A
Sbjct: 242 ASMRSRYLECLCSLMQELGSTPVGQLTELKVKEMVAVLKDLESVNIDVGWMRSVLEEFA- 300
Query: 297 NVEVINQHQE-VDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKE 355
Q+QE D+ K + + S ++++E + LA E+EVA+ + R+ E+ L E
Sbjct: 301 ------QYQENTDSEKERQEGLVRSKKQEMEIQEADLARIEKEVAEARLRVEEMKAELAE 354
Query: 356 LELKSAELEQSMLSSKSEVENLHRKSLIDELL 387
LE + +E+ +VE K+ +DELL
Sbjct: 355 LETERLRMEEMGF----KVEKYKGKTFLDELL 382
>UniRef100_Q9C5F6 Putative phospholipase [Arabidopsis thaliana]
Length = 612
Score = 139 bits (351), Expect = 1e-31
Identities = 85/303 (28%), Positives = 163/303 (53%), Gaps = 19/303 (6%)
Query: 83 QKSNASSPISNHSEIKKVESRNDDHNHSVPVSGQIHVPLPVPDVKHVHEEDQQKNGVEHL 142
QKS SP+ S +DD NHS + +I PL + E+ +
Sbjct: 324 QKSAPPSPVHG-----PYYSSSDDDNHSTYLYPEIRSPLR----SRIVSENSTPVHHNYQ 374
Query: 143 ANPIQTKQEDKTASDVHPGEGLATTFKGGSIDFSFSGIIPQGNEDNDDKG--DTESVVSE 200
+T ++DK P E S F+ I N + G +++S++SE
Sbjct: 375 IVAAETYEQDK---QFEPPE-----LPDESQSFTMQEITKMRGLKNYESGKEESQSMISE 426
Query: 201 SRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFVVQELQSDSIM 260
+ V V Y V++S + L++I+DK+GDI AS L+++ RSYY+E + VV EL+ +
Sbjct: 427 AYVSVANYRVRQSVSETLQAIIDKHGDIAASSKLQAMATRSYYLESLAAVVMELKKTVLR 486
Query: 261 HLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDREMES 320
LTK+++ E+ A++KD+ES ++ V+WL++A+ E+AE VE Q+ K +R++ +
Sbjct: 487 DLTKTRVAEIAAVVKDMESVKINVSWLKTAVTELAEAVEYFGQYDTAKVEKEVCERDLTA 546
Query: 321 LREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRK 380
+ ++E L ++E+E+ + ++++ ++ RL +LE+K ++L +++ +S+V +
Sbjct: 547 KKGEMEEMTAELVKREKEIKECREKVTVVAGRLGQLEMKGSKLNKNLDLFQSKVHKFQGE 606
Query: 381 SLI 383
+++
Sbjct: 607 AVL 609
>UniRef100_Q8W4I3 Phospholipase like protein [Arabidopsis thaliana]
Length = 612
Score = 139 bits (350), Expect = 1e-31
Identities = 87/305 (28%), Positives = 164/305 (53%), Gaps = 23/305 (7%)
Query: 83 QKSNASSPISNHSEIKKVESRNDDHNHSVPVSGQIHVPLPVPDVKHVHEEDQQKNGVEHL 142
QKS SP+ S +DD NHS + +I PL V + V H
Sbjct: 324 QKSAPPSPVHG-----PYYSSSDDDNHSTYLYPEIRSPLRSRIVS------ESSTPVHHN 372
Query: 143 ANPI--QTKQEDKTASDVHPGEGLATTFKGGSIDFSFSGIIPQGNEDNDDKG--DTESVV 198
+ +T ++DK P E S F+ I N + G +++S++
Sbjct: 373 YQIVAAETYEQDK---QFEPPE-----LPDESQSFTMQEITKMRGLKNYESGKEESQSMI 424
Query: 199 SESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFVVQELQSDS 258
SE+ V V Y V++S + L++I+DK+GDI AS L+++ RSYY+E + VV EL+
Sbjct: 425 SEAYVSVANYRVRQSVSETLQAIIDKHGDIAASSKLQAMATRSYYLESLAAVVMELKKTV 484
Query: 259 IMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDREM 318
+ LTK+++ E+ A++KD+ES ++ V+WL++A+ E+AE VE Q+ K +R++
Sbjct: 485 LRDLTKTRVAEIAAVVKDMESVKINVSWLKTAVTELAEAVEYFGQYDTAKVEKEVCERDL 544
Query: 319 ESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLH 378
+ + ++E L ++E+E+ + ++++ ++ RL +LE+K ++L +++ +S+V
Sbjct: 545 TAKKGEMEEMTAELVKREKEIKECREKVTVVAGRLGQLEMKGSKLNKNLDLFQSKVHKFQ 604
Query: 379 RKSLI 383
++++
Sbjct: 605 GEAVL 609
>UniRef100_Q9SZN4 Phospholipase like protein [Arabidopsis thaliana]
Length = 642
Score = 138 bits (347), Expect = 3e-31
Identities = 84/309 (27%), Positives = 162/309 (52%), Gaps = 38/309 (12%)
Query: 83 QKSNASSPISNHSEIKKVESRNDDHNHSVPVSGQIHVPLPVPDVK--------HVHEEDQ 134
QKS SP+ S +DD NHS + +I PL V +E+D+
Sbjct: 361 QKSAPPSPVHG-----PYYSSSDDDNHSTYLYPEIRSPLRSRIVSENSTIVAAETYEQDK 415
Query: 135 QKNGVEHLANPIQTKQEDKTASDVHPGEGLATTFKGGSIDFSFSGIIPQGNEDNDDKGDT 194
Q E + + + T ++ GL G K ++
Sbjct: 416 QFEPPE-----LPDESQSFTMQEITKMRGLKNYESG--------------------KEES 450
Query: 195 ESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFVVQEL 254
+S++SE+ V V Y V++S + L++I+DK+GDI AS L+++ RSYY+E + VV EL
Sbjct: 451 QSMISEAYVSVANYRVRQSVSETLQAIIDKHGDIAASSKLQAMATRSYYLESLAAVVMEL 510
Query: 255 QSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANT 314
+ + LTK+++ E+ A++KD+ES ++ V+WL++A+ E+AE VE Q+ K
Sbjct: 511 KKTVLRDLTKTRVAEIAAVVKDMESVKINVSWLKTAVTELAEAVEYFGQYDTAKVEKEVC 570
Query: 315 DREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEV 374
+R++ + + ++E L ++E+E+ + ++++ ++ RL +LE+K ++L +++ +S+V
Sbjct: 571 ERDLTAKKGEMEEMTAELVKREKEIKECREKVTVVAGRLGQLEMKGSKLNKNLDLFQSKV 630
Query: 375 ENLHRKSLI 383
++++
Sbjct: 631 HKFQGEAVL 639
>UniRef100_Q9SKR5 PEARLI 4 protein [Arabidopsis thaliana]
Length = 748
Score = 135 bits (340), Expect = 2e-30
Identities = 67/201 (33%), Positives = 129/201 (63%)
Query: 187 DNDDKGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIEC 246
+N+ + +S++SES V VG+Y V+ S ++ L+ IL K+GDI + L+SL RSYY++
Sbjct: 548 NNEIRDVMQSILSESYVSVGQYKVRASVSTTLQHILQKHGDIASGSKLQSLATRSYYLDM 607
Query: 247 VCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQE 306
+ VV ELQ+ + +L +S++ E+VAI+KD+ES +++ WL+ L+EI E V+ ++H+
Sbjct: 608 LASVVFELQTTPLKYLKESRVVEMVAIVKDIESVKIKAGWLKPVLEEIVEAVKHYDEHKM 667
Query: 307 VDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQS 366
K +R++ +++ E +++ L EKE+++ + + ++ E++ +L +L++K A L +S
Sbjct: 668 SVVEKEVWERDVLLAKQETEKQVKELGEKEKKIKEWRAKMTEMAAKLGDLDMKRARLHKS 727
Query: 367 MLSSKSEVENLHRKSLIDELL 387
S+V+ K L+ +L
Sbjct: 728 FTFLSSKVDKFQGKPLLQGIL 748
Score = 35.0 bits (79), Expect = 3.6
Identities = 24/86 (27%), Positives = 37/86 (42%), Gaps = 16/86 (18%)
Query: 12 PNCPHASNPYHQCTQACSKKTS----HRTKP----HAKNTSSSGAAPPPSNFDRRKKPGS 63
P C +A NP+H+CT C ++ + H+ + K T S PP S + P
Sbjct: 10 PGCSNAGNPFHECTAICFERVNSPDVHKKEKKLFGFGKRTPSRDQTPPASPARGSRSP-- 67
Query: 64 RPEPPVVAGAVPAAKVEAIQKSNASS 89
+A KVE+ +S+ SS
Sbjct: 68 ------LASYFAKKKVESSAESSPSS 87
>UniRef100_Q9SKR6 Hypothetical protein At2g20950 [Arabidopsis thaliana]
Length = 657
Score = 135 bits (339), Expect = 3e-30
Identities = 94/361 (26%), Positives = 175/361 (48%), Gaps = 23/361 (6%)
Query: 33 SHRTKPHAKNTSSSGAA-PPPSNFDRRKKPGSRPEPPVVAGAVPAAKVEAIQKSNASSPI 91
S RT+ H + S N R +P S P P A+++ + +N
Sbjct: 245 SARTRTHERGRDYSRERYEAEGNVTSRNRPSSPFHPSQSRSPPPHARLQTQRYNNGK--- 301
Query: 92 SNHSEIKKVESRNDDHNHSVPVSG-QIHVPLPVPDVKHVHEEDQQKNGVEHLANPIQTKQ 150
+H E + +S P+S H P + +D+ N +L I T +
Sbjct: 302 -DHFEGMYEADADITPRNSPPMSPVHQHTSYSPPPPFYSSSDDEDDNNSTYLFPEIATGR 360
Query: 151 EDK----TASDVHPGEGLATT-------------FKGGSIDFSFSGIIPQGNEDNDDKGD 193
+ +++ VH + + S F+ I + K +
Sbjct: 361 RSRGVSGSSTPVHYKYQITSVETYEQERQFEPPELPDESQSFTMQEIARMRGLQSYGKEE 420
Query: 194 TESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFVVQE 253
+ +SE+ V V Y V+ S A+ LE+I+DK+GDI AS L+S RS+Y+E + V E
Sbjct: 421 IQLAISETYVSVANYKVRMSIAATLEAIIDKHGDIAASSKLQSTSTRSFYLESLAAAVME 480
Query: 254 LQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKAN 313
L+S ++ LTK+++ E+ A++KD++S ++ V+WL++A+ E+AE VE ++ +
Sbjct: 481 LKSTALRDLTKTRVAEIAAVVKDMDSVKIDVSWLKTAVTELAEAVEYYGKYDTAKIVREE 540
Query: 314 TDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSE 373
DREM + ++++E E L +E+E + ++R+ E++ RL +LE+K +++++ +S+
Sbjct: 541 CDREMTAGKKEMEEMREELRRREKETKECRERVTELAGRLGQLEMKDVRVKKNLELYESK 600
Query: 374 V 374
V
Sbjct: 601 V 601
>UniRef100_Q7X8U5 Hypothetical protein At2g20950 [Arabidopsis thaliana]
Length = 520
Score = 134 bits (337), Expect = 4e-30
Identities = 64/184 (34%), Positives = 120/184 (64%)
Query: 191 KGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFV 250
K + + +SE+ V V Y V+ S A+ LE+I+DK+GDI AS L+S RS+Y+E +
Sbjct: 326 KEEIQLAISETYVSVANYKVRMSIAATLEAIIDKHGDIAASSKLQSTSTRSFYLESLAAA 385
Query: 251 VQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAA 310
V EL+S ++ LTK+++ E+ A++KD++S ++ V+WL++A+ E+AE VE ++
Sbjct: 386 VMELKSTALRDLTKTRVAEIAAVVKDMDSVKIDVSWLKTAVTELAEAVEYYGKYDTAKIV 445
Query: 311 KANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSS 370
+ DREM + ++++E E L +E+E + ++R+ E++ RL +LE+K +++++
Sbjct: 446 REECDREMTAGKKEMEEMREELRRREKETKECRERVTELAGRLGQLEMKDVRVKKNLELY 505
Query: 371 KSEV 374
+S+V
Sbjct: 506 ESKV 509
>UniRef100_Q9SZN5 Phospholipase like protein [Arabidopsis thaliana]
Length = 521
Score = 132 bits (332), Expect = 2e-29
Identities = 70/194 (36%), Positives = 118/194 (60%)
Query: 193 DTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFVVQ 252
+ +S++SES V VG Y V+ S +S L+ ILDK+GDI + L+SL +SY +E + VV
Sbjct: 327 EMKSMISESYVSVGSYKVRASVSSTLQKILDKHGDIASGSKLQSLRTKSYSLETLAAVVL 386
Query: 253 ELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKA 312
ELQS + L ++++ E+++++ D ES ++R WLR L+EI E + H+ K
Sbjct: 387 ELQSTPLKKLKQARVLEMLSVVIDAESVKIRAGWLREILNEILEAAHHYDGHETTVVEKE 446
Query: 313 NTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKS 372
+R+M RE+++ E + KE+E D +K + E++ RL EL++K A LE+ + S
Sbjct: 447 GRERDMLLEREEMKKIQEEVRLKEKEAKDFRKGVMEMAGRLGELKMKRARLEKRLAFLSS 506
Query: 373 EVENLHRKSLIDEL 386
+VE +SL++ +
Sbjct: 507 KVEKFEGESLLENV 520
>UniRef100_Q39177 PEARLI 4 protein [Arabidopsis thaliana]
Length = 766
Score = 131 bits (330), Expect = 3e-29
Identities = 64/193 (33%), Positives = 126/193 (65%)
Query: 187 DNDDKGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIEC 246
+N+ + +S++SES V VG+Y V+ S ++ L+ IL K+GDI + L+SL RSYY++
Sbjct: 548 NNEIRDVMQSILSESYVSVGQYKVRASVSTTLQHILQKHGDIASGSKLQSLATRSYYLDM 607
Query: 247 VCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQE 306
+ VV ELQ+ + +L +S++ E+VAI+KD+ES +++ WL+ L+EI E V+ ++H+
Sbjct: 608 LASVVFELQTTPLKYLKESRVVEMVAIVKDIESVKIKAGWLKPVLEEIVEAVKHYDEHKM 667
Query: 307 VDAAKANTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQS 366
K +R++ +++ E +++ L EKE+++ + + ++ E++ +L +L++K A L +S
Sbjct: 668 SVVEKEVWERDVLLAKQETEKQVKELGEKEKKIKEWRAKMTEMAAKLGDLDMKRARLHKS 727
Query: 367 MLSSKSEVENLHR 379
S+V+ + +
Sbjct: 728 FTFLSSKVDQVSK 740
Score = 35.0 bits (79), Expect = 3.6
Identities = 24/86 (27%), Positives = 37/86 (42%), Gaps = 16/86 (18%)
Query: 12 PNCPHASNPYHQCTQACSKKTS----HRTKP----HAKNTSSSGAAPPPSNFDRRKKPGS 63
P C +A NP+H+CT C ++ + H+ + K T S PP S + P
Sbjct: 10 PGCSNAGNPFHECTAICFERVNSPDVHKKEKKLFGFGKRTPSRDQTPPASPARGSRSP-- 67
Query: 64 RPEPPVVAGAVPAAKVEAIQKSNASS 89
+A KVE+ +S+ SS
Sbjct: 68 ------LASYFAKKKVESSAESSPSS 87
>UniRef100_Q9LFP2 Hypothetical protein F2I11_30 [Arabidopsis thaliana]
Length = 241
Score = 128 bits (321), Expect = 3e-28
Identities = 64/185 (34%), Positives = 119/185 (63%)
Query: 203 VPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFVVQELQSDSIMHL 262
V VG+Y VK S +S L+SI DKYGDI ++ L+SL R+Y++E + VV ELQS + L
Sbjct: 57 VSVGEYKVKASLSSTLQSIFDKYGDITSNSKLQSLSTRTYHLETLAEVVIELQSTPLRRL 116
Query: 263 TKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDREMESLR 322
++S+ E++AI+ D+E+A++RV WLRS L+E+ E ++ + K + + +
Sbjct: 117 SESRATEILAIVDDIETAKIRVGWLRSVLEEVLEATRYFDRCEMAVMEKKAGEHRLLLAK 176
Query: 323 EQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRKSL 382
+++E ++ LAEKE+E+ + ++++ + + +L LE+K L++ ++ +S+VE +S+
Sbjct: 177 QEMELSLKKLAEKEKEMKEFREKLMKTTGKLGSLEMKRTCLDKRLVFLRSKVEKFQGQSV 236
Query: 383 IDELL 387
++L
Sbjct: 237 FQDIL 241
>UniRef100_Q9M2I3 Hypothetical protein F9D24.240 [Arabidopsis thaliana]
Length = 173
Score = 58.5 bits (140), Expect = 3e-07
Identities = 43/166 (25%), Positives = 82/166 (48%), Gaps = 9/166 (5%)
Query: 185 NEDNDDKGDTESVVSESRVPVGKYHVKESFASILESILDKYGDIGASCHLESLVLRSYYI 244
++D D +S E V V +HV S +++ +L K+ D+ ++ L++ L++ Y+
Sbjct: 12 SDDGDFSPSQDSSEDEETVEVNGFHVLPSQENLVSQMLKKHPDLTSNFDLKNQQLKNAYM 71
Query: 245 ECVCFVVQELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQH 304
+ V + E S S L+ +++ + L D+ A L+V WLR LDE + +
Sbjct: 72 D-VLVDISETLSQSTKALSMEDLDKAESTLFDLTKAGLKVGWLRQKLDE----AYLKKEK 126
Query: 305 QEVDAAKANTDREMESLREQLESEMESLAEKEQ----EVADIKKRI 346
Q + + E + R+ S++ES +KE+ +A +KKR+
Sbjct: 127 QRISGVRIRELEEQVNKRKLTLSDLESDLKKEKANQLSMAQLKKRM 172
>UniRef100_UPI00003C12B5 UPI00003C12B5 UniRef100 entry
Length = 2328
Score = 53.9 bits (128), Expect = 7e-06
Identities = 34/126 (26%), Positives = 66/126 (51%), Gaps = 1/126 (0%)
Query: 255 QSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSAL-DEIAENVEVINQHQEVDAAKAN 313
Q + +S + + A+ + A R A + S+L D IA+N + +QH+E++ A
Sbjct: 372 QVKELKQQAESSRSSITALEGKLAEATDREAKVNSSLKDMIAKNSTLASQHEELEKKHAK 431
Query: 314 TDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSE 373
T+ +++ +++E +SLA+ E+ A +K R +L ++ +S L+ S+ K +
Sbjct: 432 TEADVQIWTKKVEQHTQSLAKSEEAAASVKDRANSAEKQLAAVQKESDLLDSSLSDVKQQ 491
Query: 374 VENLHR 379
VE L R
Sbjct: 492 VETLTR 497
Score = 34.3 bits (77), Expect = 6.1
Identities = 29/114 (25%), Positives = 56/114 (48%), Gaps = 10/114 (8%)
Query: 261 HLTKSKINELVAILKDVESAQLRVAWLRSALDEIA--------ENVEVINQHQEVDAAKA 312
H SK+++ A K++E+A+ ++ L S L + E E QE++
Sbjct: 1284 HAEHSKVSQ--AQTKELEAAKAKIDDLSSELSASSAAYANVKTEMEEKTTLAQELEHKLQ 1341
Query: 313 NTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQS 366
+ E+E L E+ + +SL K++E ++ + E + +LK LE + A ++S
Sbjct: 1342 TSITEIEKLTERATAGEQSLIAKQEEFDTLQGQADEQAKKLKALETELAAAQKS 1395
Score = 34.3 bits (77), Expect = 6.1
Identities = 33/123 (26%), Positives = 64/123 (51%), Gaps = 10/123 (8%)
Query: 264 KSKINE----LVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDREME 319
K+K+ E L +DV SAQ R+ L S L+ A++ E+ + E D KA ++ +E
Sbjct: 1127 KAKLEESDVKLSQSTEDVASAQARIQELHSQLE--AKSSELNAKTSESDQYKAKVEQLVE 1184
Query: 320 SLREQLESEMESLAEKEQEVADIKKRIPEISD-RLKELELKSAEL--EQSMLSSKSEVEN 376
L E + + +L +K +E A + ++ + + E E AE+ ++++++ K++
Sbjct: 1185 QL-ETAQQQQSNLQDKLKEAATAHVDLSKLHEQKTAEHEAAQAEIKEQRTLVTKKTKDHE 1243
Query: 377 LHR 379
L R
Sbjct: 1244 LAR 1246
>UniRef100_UPI00001D0F0B UPI00001D0F0B UniRef100 entry
Length = 794
Score = 52.8 bits (125), Expect = 2e-05
Identities = 37/123 (30%), Positives = 59/123 (47%), Gaps = 2/123 (1%)
Query: 253 ELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKA 312
+LQ+ S S ELVA+ +D S + S + E +EV+ + EV+ + +
Sbjct: 230 QLQACSKNQTEDSLRKELVALQEDKHSYETTAK--ESLRRVLQEKIEVVRKLSEVERSLS 287
Query: 313 NTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKS 372
NT+ E LRE E E L E + I ++SD+LK E K E++Q + K
Sbjct: 288 NTEDECTHLREMNERTQEELRELANKYNGAVNEIKDLSDKLKAAEGKQEEIQQKGQAEKK 347
Query: 373 EVE 375
E++
Sbjct: 348 ELQ 350
>UniRef100_UPI00003AA891 UPI00003AA891 UniRef100 entry
Length = 259
Score = 52.0 bits (123), Expect = 3e-05
Identities = 35/124 (28%), Positives = 61/124 (48%), Gaps = 2/124 (1%)
Query: 253 ELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKA 312
+LQ+ S S EL+A+ +D + + S + E +EV+ + EV+ + +
Sbjct: 87 QLQACSKNQTEDSIRKELIALQEDKHNYETTAK--ESLRRVLQEKIEVVRKLSEVERSLS 144
Query: 313 NTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKS 372
NT+ E L+E E E L E + I ++SD+LK E K E++Q L+ K
Sbjct: 145 NTEDECTHLKEMNERTQEELRELANKYNGAVNEIKDLSDKLKVAEGKQEEIQQKGLAEKK 204
Query: 373 EVEN 376
E+++
Sbjct: 205 ELQH 208
>UniRef100_Q9HK21 Chromosome segregation protein related ptotein [Thermoplasma
acidophilum]
Length = 1140
Score = 52.0 bits (123), Expect = 3e-05
Identities = 31/119 (26%), Positives = 64/119 (53%), Gaps = 3/119 (2%)
Query: 269 ELVAILKDVESAQLRVAWLRSALDEIAENVEVINQH-QEVDAAKANTDREMESLREQLES 327
E+ + D+ S ++ +A +R +DE N+E + + + ++ + +TDRE+E L Q+E
Sbjct: 283 EMNRVKTDLHSVEVDIAKIRGIIDEKNRNMEKLEETIAKYESERDSTDREIEDLDRQIEE 342
Query: 328 EMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRKSLIDEL 386
+ + E AD+KKR ++ R + + +AE + + +++ L R+ I+EL
Sbjct: 343 KAKRKRALEDRYADLKKRYDDLFSRAQAEAVDAAETRRKSKEYQEKIDGLGRE--IEEL 399
>UniRef100_UPI0000363BD1 UPI0000363BD1 UniRef100 entry
Length = 227
Score = 51.2 bits (121), Expect = 5e-05
Identities = 37/121 (30%), Positives = 63/121 (51%), Gaps = 8/121 (6%)
Query: 264 KSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDREMESLRE 323
+S+++ELV L+ ++S +R WL + AE EV + E D + R +E L
Sbjct: 13 RSQLDELVLQLQHLKSKAMRERWLLQGMS--AEEAEVRQKQLEQDEERG---RSLEDLIH 67
Query: 324 QLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKSEVENLHRKSLI 383
+LESE+ +L EQE + I + + +RLKE E +L++S++S + + L
Sbjct: 68 RLESEIGAL---EQEESQISAKEQVLRERLKETERSIEDLQKSLMSQDGDATSCVSAPLS 124
Query: 384 D 384
D
Sbjct: 125 D 125
>UniRef100_Q6ZPL8 MKIAA1601 protein [Mus musculus]
Length = 856
Score = 50.8 bits (120), Expect = 6e-05
Identities = 35/123 (28%), Positives = 59/123 (47%), Gaps = 2/123 (1%)
Query: 253 ELQSDSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKA 312
+LQ+ S S EL+A+ +D S + S + E +EV+ + EV+ + +
Sbjct: 258 QLQACSKNQTEDSLRKELIALQEDKHSYETTAK--ESLRRVLQEKIEVVRKLSEVERSLS 315
Query: 313 NTDREMESLREQLESEMESLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKS 372
NT+ E L+E E E L E + I ++SD+LK E K E++Q + K
Sbjct: 316 NTEDECTHLKEMNERTQEELRELANKYNGAVNEIKDLSDKLKVAEGKQEEIQQKGQAEKK 375
Query: 373 EVE 375
E++
Sbjct: 376 ELQ 378
>UniRef100_UPI0000438431 UPI0000438431 UniRef100 entry
Length = 836
Score = 50.1 bits (118), Expect = 1e-04
Identities = 53/305 (17%), Positives = 126/305 (40%), Gaps = 27/305 (8%)
Query: 78 KVEAIQKSNASSPISNHSEIKKVESRNDDHNHSVPVSGQIHVPLPVPDVKHVHEEDQQKN 137
K++ +Q+ ++ +S +K+ ++ ++ + + ++H L + +++ ++++
Sbjct: 382 KLQQLQRKLQAALVSRKEALKEKQALKEEQAAAEKIKLELHQKLELIEMELNKSREEREK 441
Query: 138 GVEHLANPIQTKQEDKTASDVHPGEGLATTFKGGSIDFSFSGIIPQGNEDNDDKGDTESV 197
+E + + Q + + S+ + G++ NE + K +
Sbjct: 442 LIEEVDRTLLENQSLSASCE--------------SLKLAMEGVL---NEKDACKRQADIA 484
Query: 198 VSESRVPVGKYHVK-ESFASILESILDKYGDIGASCHLESLVLRSYYIECVCFVVQELQS 256
ES + K ++ ES+L Y ++ VL + E QEL +
Sbjct: 485 KEESEQVCKQLEEKVQNMKEEYESLLKSYENVSDEAERVRKVLEAARQER-----QELAT 539
Query: 257 DSIMHLTKSKINELVAILKDVESAQLRVAWLRSALDEIAENVEVINQHQEVDAAKANTDR 316
+ H + E + D E ++ S E E+ + ++ + N +
Sbjct: 540 KARAHEAARQEAERNEQILDAELTSIKQQLQESLEREDIHKEEISKKENQLQELRMNLEA 599
Query: 317 EMESLREQLESEME----SLAEKEQEVADIKKRIPEISDRLKELELKSAELEQSMLSSKS 372
E + L E+L +++ S+A +QE +D + R+ ++ L++LE + AELE +LS +
Sbjct: 600 ERDDLEERLMNQLAQLNGSIAGYQQEASDSRDRLTDMQRELEKLERERAELEAEVLSERD 659
Query: 373 EVENL 377
+
Sbjct: 660 RAARM 664
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.308 0.126 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 631,674,213
Number of Sequences: 2790947
Number of extensions: 27256749
Number of successful extensions: 173676
Number of sequences better than 10.0: 4694
Number of HSP's better than 10.0 without gapping: 930
Number of HSP's successfully gapped in prelim test: 3908
Number of HSP's that attempted gapping in prelim test: 150888
Number of HSP's gapped (non-prelim): 21312
length of query: 387
length of database: 848,049,833
effective HSP length: 129
effective length of query: 258
effective length of database: 488,017,670
effective search space: 125908558860
effective search space used: 125908558860
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0017.9


