

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0012b.1
(307 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
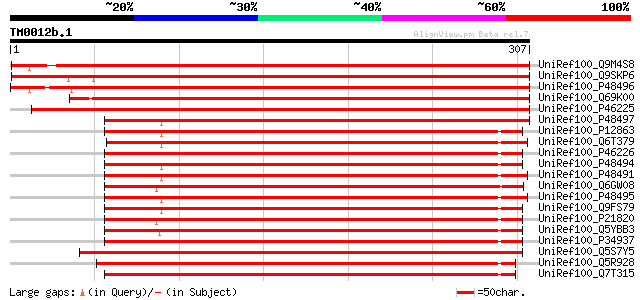
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9M4S8 Triosephosphate isomerase, chloroplast precurso... 500 e-140
UniRef100_Q9SKP6 Triosephosphate isomerase, chloroplast precurso... 496 e-139
UniRef100_P48496 Triosephosphate isomerase, chloroplast precurso... 484 e-135
UniRef100_Q69K00 Putative Triosephosphate isomerase, chloroplast... 443 e-123
UniRef100_P46225 Triosephosphate isomerase, chloroplast precurso... 440 e-122
UniRef100_P48497 Triosephosphate isomerase, cytosolic [Stellaria... 370 e-101
UniRef100_P12863 Triosephosphate isomerase, cytosolic [Zea mays] 321 2e-86
UniRef100_Q6T379 Triose phosphate isomerase cytosolic isoform [S... 319 7e-86
UniRef100_P46226 Triosephosphate isomerase, cytosolic [Secale ce... 317 2e-85
UniRef100_P48494 Triosephosphate isomerase, cytosolic [Oryza sat... 317 2e-85
UniRef100_P48491 Triosephosphate isomerase, cytosolic [Arabidops... 317 3e-85
UniRef100_Q6GW08 Triosephosphate isomerase [Glycine max] 316 4e-85
UniRef100_P48495 Triosephosphate isomerase, cytosolic [Petunia h... 316 6e-85
UniRef100_Q9FS79 Triosephosphat-isomerase [Triticum aestivum] 315 1e-84
UniRef100_P21820 Triosephosphate isomerase, cytosolic [Coptis ja... 313 4e-84
UniRef100_Q5YBB3 Triose phosphate isomerase [Helicosporidium sp.... 312 6e-84
UniRef100_P34937 Triosephosphate isomerase, cytosolic [Hordeum v... 311 1e-83
UniRef100_Q5S7Y5 Chloroplast triosephosphate isomerase [Chlamydo... 308 1e-82
UniRef100_Q5R928 Hypothetical protein DKFZp459E144 [Pongo pygmaeus] 307 3e-82
UniRef100_Q7T315 Triosephosphate isomerase 1b [Brachydanio rerio] 305 1e-81
>UniRef100_Q9M4S8 Triosephosphate isomerase, chloroplast precursor [Fragaria
ananassa]
Length = 314
Score = 500 bits (1287), Expect = e-140
Identities = 252/315 (80%), Positives = 280/315 (88%), Gaps = 14/315 (4%)
Query: 2 ATSLASQLS---------VGLRRPSPKLDSLNSQSTHSLFHSNLRLPISSSKPSRSVIAM 52
+TSLASQLS GLRR PKLD +HS +L L SS K SR+V+AM
Sbjct: 5 STSLASQLSGPKSLSQPYSGLRRSCPKLDQ-----SHSSLFQHLSLSSSSRKASRAVVAM 59
Query: 53 AGSGKFFVGGNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNSITDRIE 112
AG+GKFFVGGNWKCNGTKD ISKLV+DLNSAKLEPDVDVVVAPPF+Y+DQVK+S+TDRIE
Sbjct: 60 AGTGKFFVGGNWKCNGTKDLISKLVSDLNSAKLEPDVDVVVAPPFLYLDQVKSSLTDRIE 119
Query: 113 VSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEG 172
+S QNSWV+KGGAFTGEISVEQLKD G KWVILGHSERRH+IGE D+FIGKKAAYAL EG
Sbjct: 120 ISGQNSWVAKGGAFTGEISVEQLKDIGRKWVILGHSERRHVIGEDDQFIGKKAAYALNEG 179
Query: 173 LGVIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQ 232
LGVIACIGE LEEREAGKTFDVC++QLKA+ADAVPSW+NIV+AYEPVWAIGTGKVASP+Q
Sbjct: 180 LGVIACIGEKLEEREAGKTFDVCYEQLKAFADAVPSWENIVVAYEPVWAIGTGKVASPQQ 239
Query: 233 AQEVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPE 292
AQEVH A+R+WL+KNVSAEVASKTRIIYGGSVNGGNS+ELAK+EDIDGFLVGGASLKGPE
Sbjct: 240 AQEVHVAVREWLKKNVSAEVASKTRIIYGGSVNGGNSAELAKEEDIDGFLVGGASLKGPE 299
Query: 293 FATIINSVTSKKVAA 307
FATI+N+VTSKKVAA
Sbjct: 300 FATIVNAVTSKKVAA 314
>UniRef100_Q9SKP6 Triosephosphate isomerase, chloroplast precursor [Arabidopsis
thaliana]
Length = 315
Score = 496 bits (1278), Expect = e-139
Identities = 255/313 (81%), Positives = 277/313 (88%), Gaps = 7/313 (2%)
Query: 2 ATSLASQLSV-GLRRPSPKLDSLNSQSTHSLFH---SNLRLPISSSKPSRS---VIAMAG 54
ATSL + S GLRR SPKLD+ S S FH S+ RL SSS RS V+AMAG
Sbjct: 3 ATSLTAPPSFSGLRRISPKLDAAAVSSHQSFFHRVNSSTRLVSSSSSSHRSPRGVVAMAG 62
Query: 55 SGKFFVGGNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNSITDRIEVS 114
SGKFFVGGNWKCNGTKDSI+KL++DLNSA LE DVDVVV+PPFVYIDQVK+S+TDRI++S
Sbjct: 63 SGKFFVGGNWKCNGTKDSIAKLISDLNSATLEADVDVVVSPPFVYIDQVKSSLTDRIDIS 122
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
QNSWV KGGAFTGEISVEQLKD G KWVILGHSERRH+IGEKDEFIGKKAAYAL+EGLG
Sbjct: 123 GQNSWVGKGGAFTGEISVEQLKDLGCKWVILGHSERRHVIGEKDEFIGKKAAYALSEGLG 182
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VIACIGE LEEREAGKTFDVCF QLKA+ADAVPSWDNIV+AYEPVWAIGTGKVASP+QAQ
Sbjct: 183 VIACIGEKLEEREAGKTFDVCFAQLKAFADAVPSWDNIVVAYEPVWAIGTGKVASPQQAQ 242
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVH A+R WL+KNVS EVASKTRIIYGGSVNGGNS+ELAK+EDIDGFLVGGASLKGPEFA
Sbjct: 243 EVHVAVRGWLKKNVSEEVASKTRIIYGGSVNGGNSAELAKEEDIDGFLVGGASLKGPEFA 302
Query: 295 TIINSVTSKKVAA 307
TI+NSVTSKKVAA
Sbjct: 303 TIVNSVTSKKVAA 315
>UniRef100_P48496 Triosephosphate isomerase, chloroplast precursor [Spinacia
oleracea]
Length = 322
Score = 484 bits (1246), Expect = e-135
Identities = 246/321 (76%), Positives = 279/321 (86%), Gaps = 16/321 (4%)
Query: 1 MATSLASQLS----------VGLRRPSPKLDSLNSQSTHSLFHSN----LRLPISSSKPS 46
++TSLASQ++ GLRR KL+ NS ST S F N LRL SS +
Sbjct: 4 VSTSLASQITNPNSAVSTQFSGLRRSFLKLE--NSVSTQSSFFQNVDSHLRLSSSSRRCP 61
Query: 47 RSVIAMAGSGKFFVGGNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNS 106
R V+AMAGSGKFFVGGNWKCNGTK+SI+KLV+DLNSA LE DVDVVVAPPFVYIDQVK+S
Sbjct: 62 RGVVAMAGSGKFFVGGNWKCNGTKESITKLVSDLNSATLEADVDVVVAPPFVYIDQVKSS 121
Query: 107 ITDRIEVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAA 166
+T R+E+SAQN W+ KGGAFTGEISVEQLKD G +WVILGHSERRH+IGE++EFIGKKAA
Sbjct: 122 LTGRVEISAQNCWIGKGGAFTGEISVEQLKDLGCQWVILGHSERRHVIGEQNEFIGKKAA 181
Query: 167 YALTEGLGVIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGK 226
YAL +G+GVIACIGELLEEREAGKTFDVC+QQLKA+ADA+PSWDN+V+AYEPVWAIGTGK
Sbjct: 182 YALNQGVGVIACIGELLEEREAGKTFDVCYQQLKAFADALPSWDNVVVAYEPVWAIGTGK 241
Query: 227 VASPEQAQEVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGA 286
VASP+QAQEVH A+RDWL+KNVS EVASKTRIIYGGSVNGGN +ELAK+EDIDGFLVGGA
Sbjct: 242 VASPDQAQEVHVAVRDWLKKNVSEEVASKTRIIYGGSVNGGNCAELAKQEDIDGFLVGGA 301
Query: 287 SLKGPEFATIINSVTSKKVAA 307
SLKGPEFATI+NSVT+KKVAA
Sbjct: 302 SLKGPEFATIVNSVTAKKVAA 322
>UniRef100_Q69K00 Putative Triosephosphate isomerase, chloroplast [Oryza sativa]
Length = 304
Score = 443 bits (1139), Expect = e-123
Identities = 217/272 (79%), Positives = 245/272 (89%), Gaps = 1/272 (0%)
Query: 36 LRLPISSSKPSRSVIAMAGSGKFFVGGNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAP 95
LRL S + R V+AMAGSGKFFVGGNWKCNGTKDS+SKLV +LN+A LEPDVDVVVAP
Sbjct: 34 LRLGCSRRRAQR-VVAMAGSGKFFVGGNWKCNGTKDSVSKLVTELNAATLEPDVDVVVAP 92
Query: 96 PFVYIDQVKNSITDRIEVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIG 155
PF+YIDQVKNS+TDRIEVSAQN W+ KGGA+TGEIS EQL D G +WVILGHSERRH+IG
Sbjct: 93 PFIYIDQVKNSLTDRIEVSAQNVWIGKGGAYTGEISAEQLVDIGCQWVILGHSERRHVIG 152
Query: 156 EKDEFIGKKAAYALTEGLGVIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIA 215
E D+FIGKKAAYAL++ + VIACIGELLEEREAGKTFDVCF+Q+KA+AD++ +W ++VIA
Sbjct: 153 EDDQFIGKKAAYALSQNVKVIACIGELLEEREAGKTFDVCFKQMKAFADSITNWADVVIA 212
Query: 216 YEPVWAIGTGKVASPEQAQEVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKK 275
YEPVWAIGTGKVA+PEQAQEVHAA+RDWL+ NVS EVAS RIIYGGSVN N +ELAKK
Sbjct: 213 YEPVWAIGTGKVATPEQAQEVHAAVRDWLKTNVSPEVASGIRIIYGGSVNAANCAELAKK 272
Query: 276 EDIDGFLVGGASLKGPEFATIINSVTSKKVAA 307
EDIDGFLVGGASLKGP+FATIINSVTSKKVAA
Sbjct: 273 EDIDGFLVGGASLKGPDFATIINSVTSKKVAA 304
>UniRef100_P46225 Triosephosphate isomerase, chloroplast precursor [Secale cereale]
Length = 298
Score = 440 bits (1132), Expect = e-122
Identities = 220/295 (74%), Positives = 250/295 (84%), Gaps = 1/295 (0%)
Query: 14 RRPSPKLDSLNSQSTHSLFHSNLRLPISSSKPS-RSVIAMAGSGKFFVGGNWKCNGTKDS 72
RRPSP S S + S++ + + ++AMAGSGKFFVGGNWKCNGTK+S
Sbjct: 4 RRPSPPPASPPPPRPRSTTTTRTTSSASAAPAAAQRLVAMAGSGKFFVGGNWKCNGTKES 63
Query: 73 ISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNSITDRIEVSAQNSWVSKGGAFTGEISV 132
ISKLV+DLN+A LE DVDVVVAPPF+YIDQVK+S+TDRIEVSAQN+W+ KGGAFTGEIS
Sbjct: 64 ISKLVSDLNAATLESDVDVVVAPPFIYIDQVKSSLTDRIEVSAQNTWIGKGGAFTGEISA 123
Query: 133 EQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLGVIACIGELLEEREAGKTF 192
EQL D G +WVILGHSERRH+IGE DEFIGKKAAYAL++ L V+ACIGELLEEREAGKTF
Sbjct: 124 EQLVDIGCQWVILGHSERRHVIGEDDEFIGKKAAYALSQNLKVMACIGELLEEREAGKTF 183
Query: 193 DVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQEVHAAIRDWLQKNVSAEV 252
DVCF+Q+KA+AD + W N+VIAYEPVWAIGTGKVASPEQAQEVHAA+RDWL+ NVSA+V
Sbjct: 184 DVCFKQMKAFADNITDWTNVVIAYEPVWAIGTGKVASPEQAQEVHAAVRDWLKTNVSADV 243
Query: 253 ASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFATIINSVTSKKVAA 307
AS RIIYGGSVN N +ELAKKEDIDGFLVGGASLKGP+FATI NSVTSKKV A
Sbjct: 244 ASTVRIIYGGSVNAANCAELAKKEDIDGFLVGGASLKGPDFATICNSVTSKKVTA 298
>UniRef100_P48497 Triosephosphate isomerase, cytosolic [Stellaria longipes]
Length = 257
Score = 370 bits (951), Expect = e-101
Identities = 181/254 (71%), Positives = 213/254 (83%), Gaps = 3/254 (1%)
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKLEPD--VDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT++S+SK+V LN + + V VVV+PP+V++ + I+V+
Sbjct: 4 KFFVGGNWKCNGTQESVSKIVDTLNEPTIAANDVVTVVVSPPYVFLPDENAELKHEIQVA 63
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN WV KGGAFTGE+S + L + G+ WVILGHSERR ++GE +EF+GKKAAYA +EGLG
Sbjct: 64 AQNCWVKKGGAFTGEVSAQMLANLGITWVILGHSERRTLLGESNEFVGKKAAYAQSEGLG 123
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYAD-AVPSWDNIVIAYEPVWAIGTGKVASPEQA 233
VIACIGELLEEREAGKTFDVCF+QLK++A WDN+V+AYEPVWAIGTGKVASPEQA
Sbjct: 124 VIACIGELLEEREAGKTFDVCFKQLKSFARFPAKPWDNVVVAYEPVWAIGTGKVASPEQA 183
Query: 234 QEVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEF 293
QEVH A+RDWL+ NVS EVASKTRIIYGGSVNGGNS LA +ED+DGFLVGGASLKGPEF
Sbjct: 184 QEVHVAVRDWLKTNVSEEVASKTRIIYGGSVNGGNSLALAAQEDVDGFLVGGASLKGPEF 243
Query: 294 ATIINSVTSKKVAA 307
ATIINSVT+KKVAA
Sbjct: 244 ATIINSVTAKKVAA 257
>UniRef100_P12863 Triosephosphate isomerase, cytosolic [Zea mays]
Length = 252
Score = 321 bits (822), Expect = 2e-86
Identities = 155/249 (62%), Positives = 188/249 (75%), Gaps = 3/249 (1%)
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKLEPD--VDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT D + K+V LN ++ P V+VVV+PP+V++ VK+ + V+
Sbjct: 3 KFFVGGNWKCNGTTDQVEKIVKTLNEGQVPPSDVVEVVVSPPYVFLPVVKSQLRQEFHVA 62
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN WV KGGAFTGE+S E L + GV WVILGHSERR ++GE +EF+G K AYAL++GL
Sbjct: 63 AQNCWVKKGGAFTGEVSAEMLVNLGVPWVILGHSERRALLGESNEFVGDKVAYALSQGLK 122
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VIAC+GE LE+REAG T DV Q KA A+ + W N+V+AYEPVWAIGTGKVA+P QAQ
Sbjct: 123 VIACVGETLEQREAGSTMDVVAAQTKAIAEKIKDWSNVVVAYEPVWAIGTGKVATPAQAQ 182
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVHA++RDWL+ N S EVA TRIIYGGSV N ELA + D+DGFLVGGASLK PEF
Sbjct: 183 EVHASLRDWLKTNASPEVAESTRIIYGGSVTAANCKELAAQPDVDGFLVGGASLK-PEFI 241
Query: 295 TIINSVTSK 303
IIN+ T K
Sbjct: 242 DIINAATVK 250
>UniRef100_Q6T379 Triose phosphate isomerase cytosolic isoform [Solanum chacoense]
Length = 254
Score = 319 bits (817), Expect = 7e-86
Identities = 157/251 (62%), Positives = 191/251 (75%), Gaps = 3/251 (1%)
Query: 58 FFVGGNWKCNGTKDSISKLVADLNSAKLEPD--VDVVVAPPFVYIDQVKNSITDRIEVSA 115
FFVGGNWKCNGT + I K+VA LN+ ++ V+VVV+PP+V++ VKN + V+A
Sbjct: 5 FFVGGNWKCNGTSEEIKKIVATLNAGQVPSQDVVEVVVSPPYVFLPLVKNELRSDFHVAA 64
Query: 116 QNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLGV 175
QN WV KGGAFTGE+S + L + G+ WVILGHSERR I+GE +EF+G K AYAL++GL V
Sbjct: 65 QNCWVKKGGAFTGEVSADMLVNLGIPWVILGHSERRAILGESNEFVGDKVAYALSQGLRV 124
Query: 176 IACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQE 235
IAC+GE LE+RE+G T DV Q KA A+ V W N+V+AYEPVWAIGTGKVA+P QAQE
Sbjct: 125 IACVGETLEQRESGSTMDVVAAQTKAIAERVKDWSNVVVAYEPVWAIGTGKVATPAQAQE 184
Query: 236 VHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFAT 295
VHA +R WLQ NVSAEVA+ TRIIYGGSV+G N ELA + D+DGFLVGGASLK PEF
Sbjct: 185 VHAELRKWLQANVSAEVAASTRIIYGGSVSGANCKELAGQPDVDGFLVGGASLK-PEFID 243
Query: 296 IINSVTSKKVA 306
II + KK A
Sbjct: 244 IIKAAEVKKGA 254
>UniRef100_P46226 Triosephosphate isomerase, cytosolic [Secale cereale]
Length = 252
Score = 317 bits (813), Expect = 2e-85
Identities = 155/249 (62%), Positives = 190/249 (76%), Gaps = 3/249 (1%)
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKL-EPDV-DVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT + +V LN+ ++ PDV +VVV+PP+V++ VK+ + I+V+
Sbjct: 3 KFFVGGNWKCNGTVSQVETIVNTLNAGQIASPDVVEVVVSPPYVFLPTVKDKLRPEIQVA 62
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN WV KGGAFTGE+S E L + G+ WVILGHSERR ++ E EF+G+K AYAL +GL
Sbjct: 63 AQNCWVKKGGAFTGEVSAEMLVNLGIPWVILGHSERRSLLAESSEFVGEKVAYALAQGLK 122
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VIAC+GE LE+REAG T +V +Q KA AD + W N+V+AYEPVWAIGTGKVASP QAQ
Sbjct: 123 VIACVGETLEQREAGSTMEVVAEQTKAIADKIKDWTNVVVAYEPVWAIGTGKVASPAQAQ 182
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVHA +RDWL+ NVS EVA TRIIYGGSV G + ELA + D+DGFLVGGASLK PEF
Sbjct: 183 EVHANLRDWLKTNVSPEVAESTRIIYGGSVTGASCKELAAQPDVDGFLVGGASLK-PEFI 241
Query: 295 TIINSVTSK 303
IIN+ T K
Sbjct: 242 DIINAATVK 250
>UniRef100_P48494 Triosephosphate isomerase, cytosolic [Oryza sativa]
Length = 253
Score = 317 bits (813), Expect = 2e-85
Identities = 154/249 (61%), Positives = 188/249 (74%), Gaps = 3/249 (1%)
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKLEPD--VDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT D + K+V LN ++ V+VVV+PP+V++ VK+ + I+V+
Sbjct: 4 KFFVGGNWKCNGTTDQVDKIVKILNEGQIASTDVVEVVVSPPYVFLPVVKSQLRPEIQVA 63
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN WV KGGAFTGE+S E L + + WVILGHSERR ++GE +EF+G K AYAL++GL
Sbjct: 64 AQNCWVKKGGAFTGEVSAEMLVNLSIPWVILGHSERRSLLGESNEFVGDKVAYALSQGLK 123
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VIAC+GE LE+RE+G T DV Q KA ++ + W N+V+AYEPVWAIGTGKVA+P+QAQ
Sbjct: 124 VIACVGETLEQRESGSTMDVVAAQTKAISERIKDWTNVVVAYEPVWAIGTGKVATPDQAQ 183
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVH +R WL NVSAEVA TRIIYGGSV G N ELA K D+DGFLVGGASLK PEF
Sbjct: 184 EVHDGLRKWLAANVSAEVAESTRIIYGGSVTGANCKELAAKPDVDGFLVGGASLK-PEFI 242
Query: 295 TIINSVTSK 303
IINS T K
Sbjct: 243 DIINSATVK 251
>UniRef100_P48491 Triosephosphate isomerase, cytosolic [Arabidopsis thaliana]
Length = 254
Score = 317 bits (811), Expect = 3e-85
Identities = 159/252 (63%), Positives = 188/252 (74%), Gaps = 3/252 (1%)
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKLEPD--VDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT + + K+V LN A++ V+VVV+PP+V++ VK+++ V+
Sbjct: 4 KFFVGGNWKCNGTAEEVKKIVNTLNEAQVPSQDVVEVVVSPPYVFLPLVKSTLRSDFFVA 63
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN WV KGGAFTGE+S E L + + WVILGHSERR I+ E EF+G K AYAL +GL
Sbjct: 64 AQNCWVKKGGAFTGEVSAEMLVNLDIPWVILGHSERRAILNESSEFVGDKVAYALAQGLK 123
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VIAC+GE LEEREAG T DV Q KA AD V +W N+VIAYEPVWAIGTGKVASP QAQ
Sbjct: 124 VIACVGETLEEREAGSTMDVVAAQTKAIADRVTNWSNVVIAYEPVWAIGTGKVASPAQAQ 183
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVH +R WL KNVSA+VA+ TRIIYGGSVNGGN EL + D+DGFLVGGASLK PEF
Sbjct: 184 EVHDELRKWLAKNVSADVAATTRIIYGGSVNGGNCKELGGQADVDGFLVGGASLK-PEFI 242
Query: 295 TIINSVTSKKVA 306
II + KK A
Sbjct: 243 DIIKAAEVKKSA 254
>UniRef100_Q6GW08 Triosephosphate isomerase [Glycine max]
Length = 253
Score = 316 bits (810), Expect = 4e-85
Identities = 157/250 (62%), Positives = 190/250 (75%), Gaps = 3/250 (1%)
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKL--EPDVDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT + + K+V LN AK+ E V+VVV+PPFV++ VK+ + VS
Sbjct: 4 KFFVGGNWKCNGTTEEVKKIVTTLNEAKVPGEDVVEVVVSPPFVFLPFVKSLLRPDFHVS 63
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN WV KGGA+TGE+S E L + G+ WVI+GHSERR ++ E +EF+G K AYAL +GL
Sbjct: 64 AQNCWVRKGGAYTGEVSAEMLVNLGIPWVIIGHSERRQLLNELNEFVGDKVAYALQQGLK 123
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VIACIGE LE+REAG T V +Q KA A + +WDN+V+AYEPVWAIGTGKVA+P QAQ
Sbjct: 124 VIACIGETLEQREAGTTTAVVAEQTKAIAAKISNWDNVVLAYEPVWAIGTGKVATPAQAQ 183
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVHA +R W+ NVSAEVA+ RIIYGGSVNGGN ELA + D+DGFLVGGASLK EF
Sbjct: 184 EVHADLRKWVHDNVSAEVAASVRIIYGGSVNGGNCKELAAQPDVDGFLVGGASLKA-EFV 242
Query: 295 TIINSVTSKK 304
IIN+ T KK
Sbjct: 243 DIINAATVKK 252
>UniRef100_P48495 Triosephosphate isomerase, cytosolic [Petunia hybrida]
Length = 254
Score = 316 bits (809), Expect = 6e-85
Identities = 156/252 (61%), Positives = 188/252 (73%), Gaps = 3/252 (1%)
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKLEPD--VDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT + + K++A LN+A + V+VVV+PP+V++ VKN + V+
Sbjct: 4 KFFVGGNWKCNGTAEEVKKILATLNAADVPSQDVVEVVVSPPYVFLPLVKNELRPDFHVA 63
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN WV KGGAFTGE+S E L + + WVILGHSERR ++GE +EF+G K AYAL++GL
Sbjct: 64 AQNCWVKKGGAFTGEVSAEMLVNLSIPWVILGHSERRALLGESNEFVGDKVAYALSQGLK 123
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VIAC+GE LEERE+G T DV Q KA AD V W N+V+AYEPVWAIGTGKVASP QAQ
Sbjct: 124 VIACVGETLEERESGSTMDVVAAQTKAIADRVKDWTNVVVAYEPVWAIGTGKVASPAQAQ 183
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVHA +R WL NVS EVA+ TRIIYGGSVNG N EL + D+DGFLVGGASLK PEF
Sbjct: 184 EVHAELRKWLAANVSPEVAASTRIIYGGSVNGANCKELGGQPDVDGFLVGGASLK-PEFI 242
Query: 295 TIINSVTSKKVA 306
II + K+ A
Sbjct: 243 DIIKAAEVKRNA 254
>UniRef100_Q9FS79 Triosephosphat-isomerase [Triticum aestivum]
Length = 253
Score = 315 bits (807), Expect = 1e-84
Identities = 154/249 (61%), Positives = 187/249 (74%), Gaps = 3/249 (1%)
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKLEPD--VDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT + + +V LN+ ++ V+VVV+PP+V++ VK + I+V+
Sbjct: 4 KFFVGGNWKCNGTVEQVESIVNTLNAGQIASTDVVEVVVSPPYVFLPTVKGKLRPEIQVA 63
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN WV KGGAFTGE+S E L + GV WVILGHSERR ++GE EF+G+K AYAL +GL
Sbjct: 64 AQNCWVKKGGAFTGEVSAEMLVNLGVPWVILGHSERRSLMGESSEFVGEKVAYALAQGLK 123
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VIAC+GE LE+REAG T V +Q KA AD + W N+V+AYEPVWAIGTGKVASP QAQ
Sbjct: 124 VIACVGETLEQREAGSTMAVVAEQTKAIADKIKDWTNVVVAYEPVWAIGTGKVASPAQAQ 183
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVHA +RDWL+ NVS EVA TRIIYGGSV G + ELA + D+DGFLVGGASLK PEF
Sbjct: 184 EVHANLRDWLKTNVSPEVAESTRIIYGGSVTGASCKELAAQPDVDGFLVGGASLK-PEFI 242
Query: 295 TIINSVTSK 303
IIN+ K
Sbjct: 243 DIINAAAVK 251
>UniRef100_P21820 Triosephosphate isomerase, cytosolic [Coptis japonica]
Length = 253
Score = 313 bits (802), Expect = 4e-84
Identities = 156/249 (62%), Positives = 187/249 (74%), Gaps = 3/249 (1%)
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKL--EPDVDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT + + K+V LN A++ E V+VVV+PP+V++ VKN + V+
Sbjct: 4 KFFVGGNWKCNGTSEEVKKIVTLLNEAEVPSEDVVEVVVSPPYVFLPFVKNLLRADFHVA 63
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN WV KGGAFTGE+S E L + G+ WVILGHSERR ++ E +EF+G K AYAL++GL
Sbjct: 64 AQNCWVKKGGAFTGEVSAEMLVNLGIPWVILGHSERRALLNESNEFVGDKTAYALSQGLK 123
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VIAC+GE LE+REAG T V Q KA A+ V W NIV+AYEPVWAIGTGKVASP QAQ
Sbjct: 124 VIACVGETLEQREAGSTISVVAAQTKAIAEKVSDWTNIVVAYEPVWAIGTGKVASPAQAQ 183
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVH +R W+++NV A+VA RIIYGGSVNG NS ELA + DIDGFLVGGASLK PEF
Sbjct: 184 EVHFELRKWIKENVGADVAGSVRIIYGGSVNGANSKELAGQPDIDGFLVGGASLK-PEFV 242
Query: 295 TIINSVTSK 303
II S T K
Sbjct: 243 DIIKSATVK 251
>UniRef100_Q5YBB3 Triose phosphate isomerase [Helicosporidium sp. ex Simulium
jonesii]
Length = 252
Score = 312 bits (800), Expect = 6e-84
Identities = 157/249 (63%), Positives = 185/249 (74%), Gaps = 3/249 (1%)
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKLEP--DVDVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGTK+S+S LV L++AKL VDVVVAP F+++ Q +S+ R+EVS
Sbjct: 4 KFFVGGNWKCNGTKESVSALVKALSAAKLPSADKVDVVVAPTFLHLSQALSSLPSRVEVS 63
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN W+ GGAFTGEIS EQ+ D G KWVILGHSERRHI+GE D + K AYAL++GLG
Sbjct: 64 AQNCWIKGGGAFTGEISAEQIVDLGAKWVILGHSERRHIMGESDATVADKVAYALSKGLG 123
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VI CIGE L EREA T DVC +Q+ A A + W IV+AYEPVWAIGTGKVA+P+QAQ
Sbjct: 124 VIFCIGEQLSEREANHTMDVCTRQMAALASKISDWSRIVVAYEPVWAIGTGKVATPDQAQ 183
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVH R W+ ++VS VA TRIIYGGSV GN +LA + DIDGFLVGGASLK PEF
Sbjct: 184 EVHGLCRRWVAQHVSPAVAETTRIIYGGSVTAGNCRDLAAQPDIDGFLVGGASLK-PEFV 242
Query: 295 TIINSVTSK 303
I+ S K
Sbjct: 243 DIVASAELK 251
>UniRef100_P34937 Triosephosphate isomerase, cytosolic [Hordeum vulgare]
Length = 252
Score = 311 bits (798), Expect = 1e-83
Identities = 154/249 (61%), Positives = 188/249 (74%), Gaps = 3/249 (1%)
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKL-EPDV-DVVVAPPFVYIDQVKNSITDRIEVS 114
KFFVGGNWKCNGT + + +V LN+ ++ PDV +VVV+PP+V++ VK + I+V+
Sbjct: 3 KFFVGGNWKCNGTVEQVEAIVQTLNAGQIVSPDVVEVVVSPPYVFLPIVKAKLRPEIQVA 62
Query: 115 AQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLG 174
AQN WV KGGAFTGE+S E L + GV WVILGHSERR ++GE EF+G+K AYAL +GL
Sbjct: 63 AQNCWVKKGGAFTGEVSAEMLANLGVPWVILGHSERRSLLGESSEFVGEKVAYALAQGLK 122
Query: 175 VIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQ 234
VIAC+GE LE+REAG T +V +Q KA A + W N V+AYEPVWAIGTGKVA+P QAQ
Sbjct: 123 VIACVGETLEQREAGSTMEVVAEQTKAIAGKIKDWSNGVVAYEPVWAIGTGKVATPAQAQ 182
Query: 235 EVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFA 294
EVHA +RDWL+ NVS EVA TRIIYGGSV G + ELA + D+DGFLVGGASLK PEF
Sbjct: 183 EVHANLRDWLKTNVSPEVAESTRIIYGGSVTGASCKELAAQADVDGFLVGGASLK-PEFI 241
Query: 295 TIINSVTSK 303
IIN+ K
Sbjct: 242 DIINAAAVK 250
>UniRef100_Q5S7Y5 Chloroplast triosephosphate isomerase [Chlamydomonas reinhardtii]
Length = 282
Score = 308 bits (789), Expect = 1e-82
Identities = 152/263 (57%), Positives = 188/263 (70%), Gaps = 1/263 (0%)
Query: 42 SSKPSRSVIAMAGSGKFFVGGNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPFVYID 101
+S+ R + A S KFFVGGNWKCNG+ +++KLV +LN+ + VDVVVAPPF+YID
Sbjct: 17 ASRSQRVEVVCASSAKFFVGGNWKCNGSVANVAKLVDELNAGTIPRGVDVVVAPPFIYID 76
Query: 102 QVKNSIT-DRIEVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEF 160
V + D+ ++SAQN+W+ GAFTGE+S EQL D GV WVILGHSERR + GE +E
Sbjct: 77 YVMQHLDRDKYQLSAQNAWIGGNGAFTGEVSAEQLTDFGVPWVILGHSERRSLFGESNEV 136
Query: 161 IGKKAAYALTEGLGVIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVW 220
+ KK ++AL GLGVIACIGE LE+R +G F V Q+ A D V W +V+AYEPVW
Sbjct: 137 VAKKTSHALAAGLGVIACIGETLEQRNSGSVFKVLDAQMDALVDEVKDWTKVVLAYEPVW 196
Query: 221 AIGTGKVASPEQAQEVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDG 280
AIGTG VASPEQAQEVHA +R + K + A VA K RIIYGGSV+ N +L+K+EDIDG
Sbjct: 197 AIGTGVVASPEQAQEVHAYLRQYCAKKLGAAVADKLRIIYGGSVSDTNCKDLSKQEDIDG 256
Query: 281 FLVGGASLKGPEFATIINSVTSK 303
FLVGGASLKG F TI N+ K
Sbjct: 257 FLVGGASLKGAAFVTICNAAGPK 279
>UniRef100_Q5R928 Hypothetical protein DKFZp459E144 [Pongo pygmaeus]
Length = 249
Score = 307 bits (786), Expect = 3e-82
Identities = 152/248 (61%), Positives = 180/248 (72%), Gaps = 1/248 (0%)
Query: 52 MAGSGKFFVGGNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNSITDRI 111
MA S KFFVGGNWK NG K S+ +L+ LN+AK+ D +VV APP YID + + +I
Sbjct: 1 MAPSRKFFVGGNWKMNGRKQSLGELIGTLNAAKVPADTEVVCAPPTAYIDFARQKLDPKI 60
Query: 112 EVSAQNSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTE 171
V+AQN + GAFTGEIS +KD+G WV+LGHSERRH+ GE DE IG+K A+AL E
Sbjct: 61 AVAAQNCYKVTNGAFTGEISPGMIKDYGATWVVLGHSERRHVFGESDELIGQKVAHALAE 120
Query: 172 GLGVIACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPE 231
GLGVIACIGE L+EREAG T V F+Q K AD V W +V+AYEPVWAIGTGK A+P+
Sbjct: 121 GLGVIACIGEKLDEREAGITEKVVFEQTKVIADNVKDWSKVVLAYEPVWAIGTGKTATPQ 180
Query: 232 QAQEVHAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGP 291
QAQEVH +R WL+ NVS VA TRIIYGGSV G ELA + D+DGFLVGGASLK P
Sbjct: 181 QAQEVHEKLRGWLKSNVSDAVAQSTRIIYGGSVTGATCKELASQPDVDGFLVGGASLK-P 239
Query: 292 EFATIINS 299
EF IIN+
Sbjct: 240 EFVDIINA 247
>UniRef100_Q7T315 Triosephosphate isomerase 1b [Brachydanio rerio]
Length = 248
Score = 305 bits (781), Expect = 1e-81
Identities = 153/243 (62%), Positives = 178/243 (72%), Gaps = 1/243 (0%)
Query: 57 KFFVGGNWKCNGTKDSISKLVADLNSAKLEPDVDVVVAPPFVYIDQVKNSITDRIEVSAQ 116
KFFVGGNWK NG K SI +L LNSAKL PD +VV P +Y+D ++ + I+V+AQ
Sbjct: 5 KFFVGGNWKMNGDKKSIEELANTLNSAKLNPDTEVVCGAPTIYLDYARSKLNPNIDVAAQ 64
Query: 117 NSWVSKGGAFTGEISVEQLKDHGVKWVILGHSERRHIIGEKDEFIGKKAAYALTEGLGVI 176
N + GAFTGEIS +KD GVKWVILGHSERRH+ GE DE IG+K A+AL GLGVI
Sbjct: 65 NCYKVAKGAFTGEISPAMIKDCGVKWVILGHSERRHVFGESDELIGQKVAHALENGLGVI 124
Query: 177 ACIGELLEEREAGKTFDVCFQQLKAYADAVPSWDNIVIAYEPVWAIGTGKVASPEQAQEV 236
ACIGE L+EREAG T V F Q K AD V W +V+AYEPVWAIGTGK ASP+QAQEV
Sbjct: 125 ACIGEKLDEREAGITEKVVFAQTKFIADNVKDWSKVVLAYEPVWAIGTGKTASPQQAQEV 184
Query: 237 HAAIRDWLQKNVSAEVASKTRIIYGGSVNGGNSSELAKKEDIDGFLVGGASLKGPEFATI 296
H +R WL+ NVS VA+ RIIYGGSV GG ELA ++D+DGFLVGGASLK PEF I
Sbjct: 185 HDKLRQWLKTNVSEAVANSVRIIYGGSVTGGTCKELASQKDLDGFLVGGASLK-PEFIDI 243
Query: 297 INS 299
IN+
Sbjct: 244 INA 246
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 503,375,399
Number of Sequences: 2790947
Number of extensions: 21070495
Number of successful extensions: 41736
Number of sequences better than 10.0: 1007
Number of HSP's better than 10.0 without gapping: 943
Number of HSP's successfully gapped in prelim test: 64
Number of HSP's that attempted gapping in prelim test: 39465
Number of HSP's gapped (non-prelim): 1012
length of query: 307
length of database: 848,049,833
effective HSP length: 127
effective length of query: 180
effective length of database: 493,599,564
effective search space: 88847921520
effective search space used: 88847921520
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0012b.1


