

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.7
(564 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
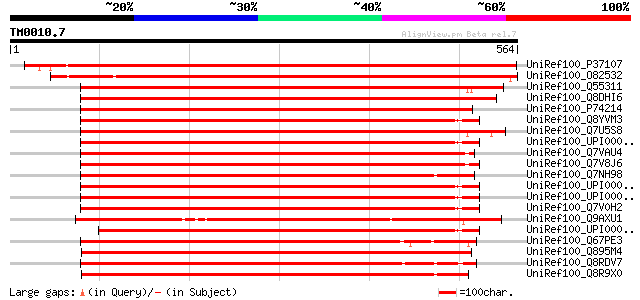
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P37107 Signal recognition particle 54 kDa protein, chl... 846 0.0
UniRef100_O82532 Signal recognition particle 54 kDa subunit [Pis... 835 0.0
UniRef100_Q55311 Signal sequence binding protein [Synechococcus ... 491 e-137
UniRef100_Q8DHI6 Signal recognition particle protein [Synechococ... 489 e-136
UniRef100_P74214 Signal recognition particle protein [Synechocys... 484 e-135
UniRef100_Q8YVM3 Signal recognition particle protein [Anabaena sp.] 483 e-135
UniRef100_Q7U5S8 Signal recognition particle protein [Synechococ... 465 e-129
UniRef100_UPI0000286CB5 UPI0000286CB5 UniRef100 entry 460 e-128
UniRef100_Q7VAU4 Signal recognition particle GTPase [Prochloroco... 456 e-127
UniRef100_Q7V8J6 Signal recognition particle protein [Prochloroc... 454 e-126
UniRef100_Q7NH98 Signal recognition particle protein SRP54 [Gloe... 442 e-122
UniRef100_UPI0000305F06 UPI0000305F06 UniRef100 entry 440 e-122
UniRef100_UPI000031BAD7 UPI000031BAD7 UniRef100 entry 439 e-121
UniRef100_Q7V0H2 Signal recognition particle protein [Prochloroc... 435 e-120
UniRef100_Q9AXU1 Chloroplast SRP54 [Chlamydomonas reinhardtii] 424 e-117
UniRef100_UPI0000330C1A UPI0000330C1A UniRef100 entry 417 e-115
UniRef100_Q67PE3 Signal recognition particle [Symbiobacterium th... 416 e-114
UniRef100_Q895M4 Signal recognition particle, subunit ffh/srp54 ... 415 e-114
UniRef100_Q8RDV7 Signal recognition particle, subunit FFH/SRP54 ... 412 e-113
UniRef100_Q8R9X0 Signal recognition particle GTPase [Thermoanaer... 412 e-113
>UniRef100_P37107 Signal recognition particle 54 kDa protein, chloroplast precursor
[Arabidopsis thaliana]
Length = 564
Score = 846 bits (2186), Expect = 0.0
Identities = 445/557 (79%), Positives = 496/557 (88%), Gaps = 11/557 (1%)
Query: 17 SSHNRTVCSSSPSPN---SLKLSSPWTNASI------SSRNSFTKEIWGWVQAKSVAVRR 67
SS NR C+ S + N SS +T +I SSRN T+EIW WV++K+V V
Sbjct: 7 SSVNRVPCTLSCTGNRRIKAAFSSAFTGGTINSASLSSSRNLSTREIWSWVKSKTV-VGH 65
Query: 68 RRDMPGVVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVR 127
R VRAEMFGQLT GLEAAW+KLKGEEVLTK+NI EPMRDIRRALLEADVSLPVVR
Sbjct: 66 GRYRRSQVRAEMFGQLTGGLEAAWSKLKGEEVLTKDNIAEPMRDIRRALLEADVSLPVVR 125
Query: 128 RFVQSVSDQAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQ 187
RFVQSVSDQAVG+GVIRGVKPDQQLVKIVH+ELVKLMGGEVSEL FAKSG TVILLAGLQ
Sbjct: 126 RFVQSVSDQAVGMGVIRGVKPDQQLVKIVHDELVKLMGGEVSELQFAKSGPTVILLAGLQ 185
Query: 188 GVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSE 247
GVGKTTVCAKL+ YLKKQGKSCML+AGD+YRPAAIDQL ILG+QVGVPVYTAGTDVKP++
Sbjct: 186 GVGKTTVCAKLACYLKKQGKSCMLIAGDVYRPAAIDQLVILGEQVGVPVYTAGTDVKPAD 245
Query: 248 IARQGLEEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAA 307
IA+QGL+EAKK +DVVI+DTAGRLQIDK MMDELK+VKK LNPTEVLLVVDAMTGQEAA
Sbjct: 246 IAKQGLKEAKKNNVDVVIMDTAGRLQIDKGMMDELKDVKKFLNPTEVLLVVDAMTGQEAA 305
Query: 308 ALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRM 367
ALVTTFN+EIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERM+DLEPFYPDRM
Sbjct: 306 ALVTTFNVEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRM 365
Query: 368 AGRILGMGDVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVM 427
AGRILGMGDVLSFVEKA EVM+QEDAE LQ+KIMSAKFDFNDFLKQTRAVA+MGS++RV+
Sbjct: 366 AGRILGMGDVLSFVEKATEVMRQEDAEDLQKKIMSAKFDFNDFLKQTRAVAKMGSMTRVL 425
Query: 428 GMIPGMSKVTPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTE 487
GMIPGM KV+PAQIREAEKNL MEAMIE MTPEERE+PELLAESP RRKR+A++SGKTE
Sbjct: 426 GMIPGMGKVSPAQIREAEKNLLVMEAMIEVMTPEERERPELLAESPERRKRIAKDSGKTE 485
Query: 488 QQVSQVVAQLFQMRVRMKNLMGVMQGGSMPTLSNLEEALKPEEKAPPGTARRRKRSEQRS 547
QQVS +VAQ+FQMRV+MKNLMGVM+GGS+P LS LE+ALK E+KAPPGTARR+++++ R
Sbjct: 486 QQVSALVAQIFQMRVKMKNLMGVMEGGSIPALSGLEDALKAEQKAPPGTARRKRKADSRK 545
Query: 548 LFADSA-ARPTPRGFGS 563
F +SA ++P PRGFGS
Sbjct: 546 KFVESASSKPGPRGFGS 562
>UniRef100_O82532 Signal recognition particle 54 kDa subunit [Pisum sativum]
Length = 525
Score = 835 bits (2158), Expect = 0.0
Identities = 443/524 (84%), Positives = 475/524 (90%), Gaps = 8/524 (1%)
Query: 46 SRNSFTKEIWGWVQAKSVAVRRRRDM--PGVVRAEMFGQLTTGLEAAWNKLKGEEVLTKE 103
+RNSF +EIWG VQ+KS+ RR DM P VVRAEMFGQLT+GLE+AWNKLK EEVLTKE
Sbjct: 4 TRNSFAREIWGLVQSKSLT-SRRTDMIRPVVVRAEMFGQLTSGLESAWNKLKREEVLTKE 62
Query: 104 NIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGVGVIRGVKPDQQLVKIVHEELVKL 163
NIVEPMRDIRRA ADVSLPVVRRFVQSV+D+AVGVGV RGVKPDQQLVKIVH+ELV+L
Sbjct: 63 NIVEPMRDIRRA--RADVSLPVVRRFVQSVTDKAVGVGVTRGVKPDQQLVKIVHDELVQL 120
Query: 164 MGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRPAAID 223
MGGEVSEL FA SG TVILLAGLQGVGKTTVCAKL+ Y KKQGKSCMLVAGD+YRPAAID
Sbjct: 121 MGGEVSELTFANSGPTVILLAGLQGVGKTTVCAKLATYFKKQGKSCMLVAGDVYRPAAID 180
Query: 224 QLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKKKKIDVVIVDTAGRLQIDKAMMDELK 283
LTILGKQV VPVY AGTDVKPS IA+QG EEAKKKKIDVVIVDTAG LQIDKAMMDELK
Sbjct: 181 XLTILGKQVDVPVYAAGTDVKPSVIAKQGFEEAKKKKIDVVIVDTAGSLQIDKAMMDELK 240
Query: 284 EVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVS 343
EVK+ LNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVS
Sbjct: 241 EVKQVLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVS 300
Query: 344 GKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMQQEDAEKLQEKIMSA 403
GKPIKLVGRGERM+DLEPFYPDRMAGRILGMGDVLSFVEKAQEVM +EDAE +Q+KIMSA
Sbjct: 301 GKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMLEEDAEDMQKKIMSA 360
Query: 404 KFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTPAQIREAEKNLKNMEAMIEAMTPEER 463
KFDFND LKQTR VA+MGSVSRV+GMIPGM+KVTPAQIREAEKNL+ ME +IEAMTPEER
Sbjct: 361 KFDFNDLLKQTRTVAKMGSVSRVLGMIPGMAKVTPAQIREAEKNLQFMEVIIEAMTPEER 420
Query: 464 EKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLFQMRVRMKNLMGVMQGGSMPTLSNLE 523
KPELLAE PVRRK VAQESGKTEQQVSQ+VAQLFQMRVRMK LMGVM+GGSMPTLSNLE
Sbjct: 421 GKPELLAEFPVRRKSVAQESGKTEQQVSQLVAQLFQMRVRMKKLMGVMEGGSMPTLSNLE 480
Query: 524 EALKPEEKAPPGTARRRKRSEQRSLFADSAAR---PTPRGFGSK 564
EALK +KAPPGTARR+K+ + LF +++ P PRGFGSK
Sbjct: 481 EALKANKKAPPGTARRKKKGGLKRLFKGRSSKISLPAPRGFGSK 524
>UniRef100_Q55311 Signal sequence binding protein [Synechococcus sp.]
Length = 485
Score = 491 bits (1265), Expect = e-137
Identities = 256/483 (53%), Positives = 351/483 (72%), Gaps = 12/483 (2%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L LE AW KL+G++ +++ N+ E ++++RRALLEADV+L VV+ F+ V ++AV
Sbjct: 1 MFDALAERLEQAWTKLRGQDKISESNVQEALKEVRRALLEADVNLGVVKEFIAQVQEKAV 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G VI G++PDQQ +K+V++ELV++MG LA A TVIL+AGLQG GKTT AKL
Sbjct: 61 GTQVISGIRPDQQFIKVVYDELVQVMGETHVPLAQAAKAPTVILMAGLQGAGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+ +L+K+G+S +LVA D+YRPAAIDQL LGKQ+ VPV+ G+D P EIARQG+E+A++
Sbjct: 121 ALHLRKEGRSTLLVATDVYRPAAIDQLITLGKQIDVPVFELGSDANPVEIARQGVEKARQ 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+ ID VIVDTAGRLQID MM+EL +VK+++ P EVLLVVD+M GQEAA+L +F+ IG
Sbjct: 181 EGIDTVIVDTAGRLQIDTEMMEELAQVKEAIAPHEVLLVVDSMIGQEAASLTRSFHERIG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGAILTKLDGDSRGGAALS++ VSG+PIK VG GE+++ L+PF+P+RMA RILGMGDVL
Sbjct: 241 ITGAILTKLDGDSRGGAALSIRRVSGQPIKFVGLGEKVEALQPFHPERMASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGM-SKVT 437
+ VEKAQE + D EK+QEKI++AKFDF DFLKQ R + MGS++ + +IPG+ K+
Sbjct: 301 TLVEKAQEEIDLADVEKMQEKILAAKFDFTDFLKQMRLLKNMGSLAGFIKLIPGLGGKIN 360
Query: 438 PAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQL 497
Q+R+ E+ LK +EAMI +MTP+ER P+LL+ SP RR R+A+ G+TE ++ Q++ Q
Sbjct: 361 DEQLRQGERQLKRVEAMINSMTPQERRDPDLLSNSPSRRHRIAKGCGQTEAEIRQIIQQF 420
Query: 498 FQMRVRMKNL-------MGVMQ----GGSMPTLSNLEEALKPEEKAPPGTARRRKRSEQR 546
QMR M+ + MG M GG MP A +P + +++K S+++
Sbjct: 421 QQMRTMMQQMSQGGFPGMGGMGMPGFGGGMPGFGGGAPAPQPGFRGYGPPKKQKKGSKKK 480
Query: 547 SLF 549
F
Sbjct: 481 KGF 483
>UniRef100_Q8DHI6 Signal recognition particle protein [Synechococcus elongatus]
Length = 469
Score = 489 bits (1258), Expect = e-136
Identities = 250/464 (53%), Positives = 341/464 (72%), Gaps = 1/464 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L LE+AW L+G++ +T+ NI E +R++RRALLEADV+L V + F+++V +A+
Sbjct: 1 MFDALAERLESAWKTLRGQDKITENNIQEALREVRRALLEADVNLQVAKDFIENVRQRAI 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G VI GV+PDQQ +KIV++ LV++MG S LA + T+IL+AGLQG GKTT AKL
Sbjct: 61 GAEVIAGVRPDQQFIKIVYDALVEVMGESHSPLAHVEPPPTIILMAGLQGTGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+ +L+K+G+S +LVA D+YRPAAIDQL LGKQ+GVPV+ GT+V P EIARQG+ +AK+
Sbjct: 121 ALHLRKEGRSTLLVATDVYRPAAIDQLMTLGKQIGVPVFEMGTEVSPVEIARQGVAKAKE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+D VI+DTAGRLQID MM EL +K+ + P E LLVVDAMTGQEAA L F+ ++G
Sbjct: 181 LGVDTVIIDTAGRLQIDAEMMAELAAIKEGVQPHETLLVVDAMTGQEAANLTRAFHEQVG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGAILTKLDGD+RGGAALSV++VSG+PIK +G GE+++ L+PFYPDR+A RILGMGDVL
Sbjct: 241 ITGAILTKLDGDTRGGAALSVRQVSGQPIKFIGVGEKVEALQPFYPDRLASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQE + DAEK+ KI+ A+FDF+DFLKQ R + MGS++ ++ +IPGM+K++
Sbjct: 301 TLVEKAQEEVDLADAEKMSRKILEAQFDFDDFLKQLRLLKNMGSLAGIIKLIPGMNKIST 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
Q+++ E+ LK EAMI +MT EER PELLA SP RR+RVA+ SG V+++V+
Sbjct: 361 EQLQQGEQQLKRAEAMINSMTREERRNPELLASSPSRRERVAKGSGYQVADVTKLVSDFQ 420
Query: 499 QMRVRMKNL-MGVMQGGSMPTLSNLEEALKPEEKAPPGTARRRK 541
+MR M+ + G G MP ++ + P+ P ++RK
Sbjct: 421 RMRSLMQQMGQGGFPGMGMPGMNAMGRRGHPQTAKKPKKQKKRK 464
>UniRef100_P74214 Signal recognition particle protein [Synechocystis sp.]
Length = 482
Score = 484 bits (1246), Expect = e-135
Identities = 248/437 (56%), Positives = 327/437 (74%), Gaps = 1/437 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L LE AW KL+G++ +++ NI E ++++RRALL ADV+L VV+ F++ V +A+
Sbjct: 1 MFDALADRLEDAWKKLRGQDKISESNIKEALQEVRRALLAADVNLQVVKGFIKDVEQKAL 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G VI GV P QQ +KIV++ELV LMG LA A+ TVIL+AGLQG GKTT AKL
Sbjct: 61 GADVISGVNPGQQFIKIVYDELVNLMGESNVPLAQAEQAPTVILMAGLQGTGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+ YL+KQ +S ++VA D+YRPAAIDQL LG+Q+ VPV+ G+D P EIARQG+E+AK+
Sbjct: 121 ALYLRKQKRSALMVATDVYRPAAIDQLKTLGQQIDVPVFDLGSDANPVEIARQGVEKAKE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+D V++DTAGRLQID MM EL E+K+ + P + LLVVDAMTGQEAA L TF+ +IG
Sbjct: 181 LGVDTVLIDTAGRLQIDPQMMAELAEIKQVVKPDDTLLVVDAMTGQEAANLTHTFHEQIG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGAILTKLDGD+RGGAALSV+++SG+PIK VG GE+++ L+PFYPDR+A RIL MGDVL
Sbjct: 241 ITGAILTKLDGDTRGGAALSVRQISGQPIKFVGVGEKVEALQPFYPDRLASRILNMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQE + D EKLQ KI+ A FDF+DF+KQ R + MGS+ ++ MIPGM+K++
Sbjct: 301 TLVEKAQEAIDVGDVEKLQNKILEATFDFDDFIKQMRFMKNMGSLGGLLKMIPGMNKLSS 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
I + EK LK EAMI +MT EER+ P+LLA+SP RR+R+AQ SG +E VS+++
Sbjct: 361 GDIEKGEKELKRTEAMISSMTTEERKNPDLLAKSPSRRRRIAQGSGHSETDVSKLITNFT 420
Query: 499 QMRVRMKNL-MGVMQGG 514
+MR M+ + MG M GG
Sbjct: 421 KMRTMMQQMGMGGMPGG 437
>UniRef100_Q8YVM3 Signal recognition particle protein [Anabaena sp.]
Length = 490
Score = 483 bits (1244), Expect = e-135
Identities = 250/444 (56%), Positives = 332/444 (74%), Gaps = 4/444 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L LE+AW KL+G++ +++ NI + +R++RRALLEADV+L VV+ F+ V +A
Sbjct: 1 MFDALADRLESAWKKLRGQDKISESNIQDALREVRRALLEADVNLQVVKDFISEVEAKAQ 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G VI GV+PDQQ +KIV++ELV++MG E LA + T++L+AGLQG GKTT AKL
Sbjct: 61 GAEVISGVRPDQQFIKIVYDELVQVMGEENVPLAEVEQKPTIVLMAGLQGTGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+ +L+K +SC+LVA D+YRPAAIDQL LGKQ+ VPV+ G+D P EIARQG+E AK
Sbjct: 121 ALHLRKLERSCLLVATDVYRPAAIDQLLTLGKQIDVPVFELGSDADPVEIARQGVERAKA 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+ ++ VI+DTAGRLQID+ MM EL +K+++ P E LLVVD+MTGQEAA L TF+ +IG
Sbjct: 181 EGVNTVIIDTAGRLQIDEDMMAELARIKETVQPHETLLVVDSMTGQEAANLTRTFHEQIG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGAILTK+DGDSRGGAALSV+++SG PIK VG GE+++ L+PFYPDRMA RILGMGDVL
Sbjct: 241 ITGAILTKMDGDSRGGAALSVRQISGAPIKFVGVGEKVEALQPFYPDRMASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQE DAEK+QEKI+SAKFDF DF+KQ R + MGS+ ++ MIPGM K++
Sbjct: 301 TLVEKAQEEFDLADAEKMQEKILSAKFDFTDFVKQLRMLKNMGSLGGLLKMIPGMGKLSD 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
Q+++ E LK EAMI +MT +ER P+LLA SP RR+R+A SG E V+++VA F
Sbjct: 361 DQLKQGETQLKRCEAMINSMTMQERRDPDLLASSPNRRRRIASGSGYKEADVNKLVAD-F 419
Query: 499 QMRVRMKNLMGVMQGGSMPTLSNL 522
Q +M++LM M G+ P + +
Sbjct: 420 Q---KMRSLMQQMGQGNFPGMPGM 440
>UniRef100_Q7U5S8 Signal recognition particle protein [Synechococcus sp.]
Length = 487
Score = 465 bits (1196), Expect = e-129
Identities = 242/486 (49%), Positives = 344/486 (69%), Gaps = 13/486 (2%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF +L+ E A LKG++ ++ N+ ++D+RRALLEADVSLPVV+ FV V ++AV
Sbjct: 1 MFDELSARFEDAVKGLKGQDKISDTNVEVALKDVRRALLEADVSLPVVKDFVADVREKAV 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G V+RGV PDQ+ +++VHE+LV++MGG+ + LA A TV+L+AGLQG GKTT AKL
Sbjct: 61 GAEVVRGVSPDQKFIQVVHEQLVEVMGGDNAPLAKAAEAPTVVLMAGLQGAGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+LK QG+ ++V D+YRPAAI+QL LG Q+GV V++ GT+ KP EIA GL +AK+
Sbjct: 121 GLHLKDQGRRALMVGADVYRPAAIEQLKTLGGQIGVEVFSLGTEAKPEEIAAAGLAKAKE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+ D ++VDTAGRLQID MM+E+ ++ ++ P EVLLVVD+M GQEAA L F+ ++G
Sbjct: 181 EGFDTLLVDTAGRLQIDTEMMEEMVRIRTAVQPDEVLLVVDSMIGQEAAELTRAFHDQVG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGA+LTKLDGDSRGGAALS+++VSG+PIK +G GE+++ L+PF+P+RMA RILGMGDVL
Sbjct: 241 ITGAVLTKLDGDSRGGAALSIRKVSGQPIKFIGTGEKVEALQPFHPERMASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQ+ ++ D EK+Q+K+ A FDF+DF++Q R + +MGS+ +M MIPGM+K+
Sbjct: 301 TLVEKAQKEVELADVEKMQKKLQEATFDFSDFVQQMRLIKRMGSLGGLMKMIPGMNKLDD 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
+++ E+ LK +EAMI +MTP+ERE P+LLA P RR+R+A SG V +V+A
Sbjct: 361 GILKQGEQQLKRIEAMIGSMTPKERENPDLLAGQPSRRRRIAAGSGHQPADVDKVLADFQ 420
Query: 499 QMRVRMKNL-------MGVMQG-GSMPTLSNLE--EALKPEEKAPP---GTARRRKRSEQ 545
+MR M+ + MG M G G MP + + + PP G+ +R+K +++
Sbjct: 421 KMRGFMQQMSRGGMPGMGGMPGMGGMPGMGGMPGMGGMPGGGGRPPGRGGSPKRQKPAKK 480
Query: 546 RSLFAD 551
R F D
Sbjct: 481 RRGFGD 486
>UniRef100_UPI0000286CB5 UPI0000286CB5 UniRef100 entry
Length = 463
Score = 460 bits (1184), Expect = e-128
Identities = 229/444 (51%), Positives = 325/444 (72%), Gaps = 4/444 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF +L+ E A L+G++ +++ N+ ++D+RRALLEADVSLPVV+ FV V D+AV
Sbjct: 1 MFDELSARFEDAVKGLRGQDSISETNVDVALKDVRRALLEADVSLPVVKEFVAEVRDKAV 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G V+RGV PDQ+ +++VHE+LV++MGG+ + LA A TV+L+AGLQG GKTT AKL
Sbjct: 61 GAEVVRGVSPDQKFIQVVHEQLVEVMGGDNAPLAKAAEAPTVVLMAGLQGAGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+LK QG+ ++V D+YRPAAI+QL LG Q+ V V++ G + KP +IA GL +AK+
Sbjct: 121 GLHLKDQGRRALMVGADVYRPAAIEQLKTLGAQIDVEVFSLGAEAKPEDIAAAGLAKAKE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+ D ++VDTAGRLQID MM+E+ ++ ++ P EVLLVVD+M GQEAA L F+ ++G
Sbjct: 181 EGFDTLLVDTAGRLQIDTEMMEEMVRIRTAVQPDEVLLVVDSMIGQEAAELTRAFHDQVG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGA+LTKLDGDSRGGAALS+++VSG+PIK +G GE+++ L+PF+P+RMA RILGMGDVL
Sbjct: 241 ITGAVLTKLDGDSRGGAALSIRKVSGQPIKFIGTGEKVEALQPFHPERMASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQ+ ++ D EK+Q+K+ A FDF+DF+KQ R + +MGS+ +M MIPGM+K+
Sbjct: 301 TLVEKAQKEVELADVEKMQKKLQEATFDFSDFVKQMRLIKRMGSLGGLMKMIPGMNKIDD 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
+++ E+ LK +EAMI +MT +ERE P+LLA P RR+R+A SG V +V+A F
Sbjct: 361 GMLKQGEQQLKRIEAMIGSMTQQERENPDLLASQPSRRRRIASGSGHQAADVDKVLAD-F 419
Query: 499 QMRVRMKNLMGVMQGGSMPTLSNL 522
Q +M+ M M G+MP + +
Sbjct: 420 Q---KMRGFMQQMSQGNMPGMGGM 440
>UniRef100_Q7VAU4 Signal recognition particle GTPase [Prochlorococcus marinus]
Length = 485
Score = 456 bits (1174), Expect = e-127
Identities = 232/439 (52%), Positives = 323/439 (72%), Gaps = 3/439 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF +L+ E A L+GE ++++N+ ++ +RRALLEADVSL VV++FVQ V D+A+
Sbjct: 1 MFDELSARFEDAVKGLRGENKISEDNVDIALKQVRRALLEADVSLSVVKQFVQEVRDKAI 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G V+RGV P Q+ +++VH+ELV++MGG S LA + +VIL+AGLQG GKTT AKL
Sbjct: 61 GAEVVRGVNPGQKFIEVVHQELVEVMGGANSPLADSTKKPSVILMAGLQGAGKTTAAAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
YLK +G ++VA D+YRPAAIDQL LGKQ+ V V++ G + KP +IA GL++A++
Sbjct: 121 GLYLKDKGLKPLMVAADVYRPAAIDQLNTLGKQINVEVFSLGKESKPEDIAASGLKKAQE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+ D +IVDTAGRLQID+ MM+E+ ++ +++P EVLLVVD+M GQEAA L F+ ++G
Sbjct: 181 EDFDTLIVDTAGRLQIDEEMMNEMVRIRSAVDPDEVLLVVDSMIGQEAADLTRAFHEKVG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITGA+LTKLDGDSRGGAALS+++VSG+PIK +G GE+++ L+PF+P+RMAGRILGMGDVL
Sbjct: 241 ITGAVLTKLDGDSRGGAALSIRKVSGQPIKFIGTGEKVEALQPFHPERMAGRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQ+ ++ DAE++Q K A FDF+DF+KQ R + +MGS+ +M MIPGM+K+
Sbjct: 301 TLVEKAQKEVEIADAEQMQRKFQEATFDFSDFVKQMRLIKRMGSLGGLMKMIPGMNKIDD 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
++ E LK +EAMI +MT E+ +PELLA P RRKRVA SG T +V +V+A
Sbjct: 361 GMLKSGEDQLKRIEAMIGSMTNAEQTQPELLAAQPSRRKRVAFGSGHTPVEVDKVLADFQ 420
Query: 499 QMRVRMKNLMGVMQGGSMP 517
+MR MK + GG +P
Sbjct: 421 KMRGLMKQM---SSGGGLP 436
>UniRef100_Q7V8J6 Signal recognition particle protein [Prochlorococcus marinus]
Length = 497
Score = 454 bits (1168), Expect = e-126
Identities = 226/444 (50%), Positives = 327/444 (72%), Gaps = 3/444 (0%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF +L+ E A L+G+ ++ N+ + ++ +RRALL ADVSL VVR FV+ V +AV
Sbjct: 1 MFEELSARFEDAVKGLRGQAKISDTNVEDALKQVRRALLGADVSLEVVREFVEEVRQKAV 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G V+RGV PDQ+ V++VH++LV++MGG+ + +A A+ TV+L+AGLQG GKTT AKL
Sbjct: 61 GAEVVRGVTPDQKFVQVVHQQLVEVMGGDNAPMAEAEDSPTVVLMAGLQGAGKTTATAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+LK QG+ ++VA D+YRPAAIDQL LG+Q+GV V++ G DVKP EIA GL +A++
Sbjct: 121 GLHLKDQGQRPLMVAADVYRPAAIDQLRTLGEQIGVDVFSLGDDVKPEEIAAAGLAKARE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+ D ++VDTAGRLQID MM+E+ ++ ++ P EVLLVVD+M GQEAA L F+ ++G
Sbjct: 181 EGFDTLLVDTAGRLQIDTEMMEEMVRIRSAVEPDEVLLVVDSMIGQEAAELTRAFHDKVG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
ITG++LTKLDGDSRGGAALS+++VSG+PIK +G GE+++ L+PF+P+RMA RILGMGDVL
Sbjct: 241 ITGSVLTKLDGDSRGGAALSIRKVSGQPIKFIGTGEKVEALQPFHPERMASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ VEKAQ+ ++ D EK+Q+K+ A FDF+DFL+Q R + +MGS+ ++ M+PGM+K+
Sbjct: 301 TLVEKAQKEVELADVEKMQKKLQEASFDFSDFLQQMRLIKRMGSLGGLIKMMPGMNKLDD 360
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
+++ E+ LK +EAMI +MT +ER +PELLA P RR+R+A SG + V +V+A
Sbjct: 361 GMLKQGEQQLKRIEAMIGSMTADERNQPELLASQPSRRRRIAGGSGHSPADVDKVLADFQ 420
Query: 499 QMRVRMKNLMGVMQGGSMPTLSNL 522
+MR M+ + +GG MP + +
Sbjct: 421 KMRGFMQQM---SKGGGMPGMPGM 441
>UniRef100_Q7NH98 Signal recognition particle protein SRP54 [Gloeobacter violaceus]
Length = 497
Score = 442 bits (1136), Expect = e-122
Identities = 234/442 (52%), Positives = 316/442 (70%), Gaps = 5/442 (1%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L+ LE AW KL+G++ +T+ NI E +R++RRALLEADV+ V + FV V D A+
Sbjct: 1 MFESLSEKLEGAWRKLRGQDRITEGNIDEALREVRRALLEADVNFQVAKEFVADVRDDAL 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAF--AKSGTTVILLAGLQGVGKTTVCA 196
G V+ GV PDQQ +KIVH++LV LMG + LA AK V+L+AGLQG GKTT A
Sbjct: 61 GAEVVSGVTPDQQFIKIVHDQLVALMGEQNVPLAEPRAKGRPAVVLMAGLQGTGKTTASA 120
Query: 197 KLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEA 256
KL+ YL+K G+ +L A D+YRPAAIDQL LG Q+ VPV+T G + P +IAR LE A
Sbjct: 121 KLALYLQKNGQKPLLAAADVYRPAAIDQLQTLGGQIQVPVFTLGKEADPVDIARASLERA 180
Query: 257 KKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLE 316
+ DV+IVDTAGRL ID AMM EL+ +K +++P E+LLVVDAMTGQEAA L F+
Sbjct: 181 IADRHDVLIVDTAGRLSIDDAMMAELERIKAAIDPEEILLVVDAMTGQEAANLTRAFHDR 240
Query: 317 IGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGD 376
+GITGAILTKLDGD+RGGAALSV+++SG+PIK VG GE+++ L+PFYP+RMA RILGMGD
Sbjct: 241 LGITGAILTKLDGDTRGGAALSVRKISGQPIKFVGVGEKVEALQPFYPERMASRILGMGD 300
Query: 377 VLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKV 436
VL+ VEKAQE + DA K+++++++ +FDF F+KQ R + MG + V+ MIPGM+K+
Sbjct: 301 VLTLVEKAQEEIDFADAAKMEQQLLTGQFDFESFIKQMRMIKNMGPLGGVLKMIPGMNKI 360
Query: 437 TPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQ 496
+ QIR+ E LK EAMI +MT +ER P+L+ S R++R+++ SG + + V +++
Sbjct: 361 SDDQIRQGEVQLKKAEAMIGSMTTQERRNPDLINLS--RKRRISKGSGVSLEDVGKLLND 418
Query: 497 LFQMRVRMKNLMGVMQ-GGSMP 517
+MR MK + G GG MP
Sbjct: 419 FGKMRQMMKMMGGGGPFGGGMP 440
>UniRef100_UPI0000305F06 UPI0000305F06 UniRef100 entry
Length = 498
Score = 440 bits (1131), Expect = e-122
Identities = 225/445 (50%), Positives = 322/445 (71%), Gaps = 5/445 (1%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF +L++ E A L+GE +++ NI + +++++RALL+ADVSL VV+ F+ V D+A+
Sbjct: 1 MFDELSSRFEDAVKGLRGEAKISENNINDALKEVKRALLDADVSLSVVKEFISDVKDKAI 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G V+RGV P Q+ +++V++EL+ +MG E S L K+ TVIL+AGLQG GKTT KL
Sbjct: 61 GEEVVRGVNPGQKFIEVVNKELINIMGNENSPLNENKNSPTVILMAGLQGAGKTTATGKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGT-DVKPSEIARQGLEEAK 257
YLK++ K +LVA DIYRPAA++QL LG Q + V++A + KP EIA+Q L A
Sbjct: 121 GLYLKEKDKKVLLVAADIYRPAAVEQLKTLGSQYDLEVFSAKEKNSKPEEIAKQALNFAS 180
Query: 258 KKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEI 317
+ + +I+DTAGRLQID +MM+E+ +K+ NP EVLLVVD+M GQEAA L +F+ ++
Sbjct: 181 ENDFNTIIIDTAGRLQIDDSMMNEMVRIKEVSNPDEVLLVVDSMIGQEAAELTKSFHEKV 240
Query: 318 GITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDV 377
GI+GAILTKLDGDSRGGAALS++++SGKPIK +G GE+++ L+PF+P+RMA RILGMGDV
Sbjct: 241 GISGAILTKLDGDSRGGAALSIRKISGKPIKFIGVGEKIEALQPFHPERMASRILGMGDV 300
Query: 378 LSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVT 437
L+ VEKAQ+ ++ DAE +Q+K+ A FDFNDF+KQ R + +MGS+ ++ +IPGM+K+
Sbjct: 301 LTLVEKAQKEVELADAEAMQKKLQEATFDFNDFVKQMRLIKRMGSLGGLIKLIPGMNKID 360
Query: 438 PAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQL 497
I++ E LK +E+MI +MT EE++KPE+LA P RR+R+AQ SG + V +V+A
Sbjct: 361 DGMIKDGEDQLKKIESMISSMTLEEKQKPEVLAAQPSRRQRIAQGSGYEAKDVDKVLAD- 419
Query: 498 FQMRVRMKNLMGVMQGGSMPTLSNL 522
FQ RM+ M M G MP + +
Sbjct: 420 FQ---RMRGFMKQMSNGGMPGMGGM 441
>UniRef100_UPI000031BAD7 UPI000031BAD7 UniRef100 entry
Length = 489
Score = 439 bits (1128), Expect = e-121
Identities = 224/445 (50%), Positives = 321/445 (71%), Gaps = 5/445 (1%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF +L++ E A L+GE +++ NI + +++++RALL+ADVSL VV+ F+ V D+A+
Sbjct: 1 MFDELSSRFEDAVKGLRGEAKISENNINDALKEVKRALLDADVSLSVVKEFISDVKDKAI 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G V+RGV P Q+ +++V++EL+ +MG E S L ++ TVIL+AGLQG GKTT KL
Sbjct: 61 GEEVVRGVNPGQKFIEVVNKELINIMGNENSPLNENENSPTVILMAGLQGAGKTTATGKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTA-GTDVKPSEIARQGLEEAK 257
YLK++ K +LVA DIYRPAA++QL LG Q + V++A G + KP EIA+ L A
Sbjct: 121 GLYLKEKDKKVLLVAADIYRPAAVEQLKTLGSQYDLEVFSAKGKNSKPEEIAKDALNYAS 180
Query: 258 KKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEI 317
+ + +I+DTAGRLQID +MM E+ +K+ NP EVLLVVD+M GQEAA L +F+ ++
Sbjct: 181 ENNFNSIIIDTAGRLQIDDSMMSEMVRIKEVSNPDEVLLVVDSMIGQEAADLTKSFHEKV 240
Query: 318 GITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDV 377
GI+GAILTKLDGDSRGGAALS++++SGKPIK +G GE+++ L+PF+P+RMA RILGMGDV
Sbjct: 241 GISGAILTKLDGDSRGGAALSIRKISGKPIKFIGVGEKIEALQPFHPERMASRILGMGDV 300
Query: 378 LSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVT 437
L+ VEKAQ+ ++ DAE +Q+K+ A FDFNDF+KQ R + +MGS+ ++ +IPGM+K+
Sbjct: 301 LTLVEKAQKEVELADAEAMQKKLQEATFDFNDFVKQMRLIKRMGSLGGLIKLIPGMNKID 360
Query: 438 PAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQL 497
I++ E LK +E+MI +MT EE++KPE+LA P RR+R+AQ SG + V +V+A
Sbjct: 361 DGMIKDGEDQLKKIESMISSMTLEEKQKPEVLAAQPSRRQRIAQGSGYEAKDVDKVLAD- 419
Query: 498 FQMRVRMKNLMGVMQGGSMPTLSNL 522
FQ RM+ M M G MP + +
Sbjct: 420 FQ---RMRGFMKQMSNGGMPGMGGM 441
>UniRef100_Q7V0H2 Signal recognition particle protein [Prochlorococcus marinus subsp.
pastoris]
Length = 498
Score = 435 bits (1118), Expect = e-120
Identities = 225/445 (50%), Positives = 319/445 (71%), Gaps = 5/445 (1%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF +L++ E A L+GE +++ NI + ++ +++ALL+ADVSL VV+ FV V D+A+
Sbjct: 1 MFDELSSRFEDAVKGLRGEAKISENNIDDALKQVKKALLDADVSLSVVKEFVSDVKDKAI 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G V+RGV P Q+ +++V++EL+ +MG E S L ++ TVIL+AGLQG GKTT KL
Sbjct: 61 GEEVVRGVNPGQKFIEVVNKELINIMGNENSPLNKKETTPTVILMAGLQGAGKTTATGKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGT-DVKPSEIARQGLEEAK 257
YLK++ + +LVA DIYRPAA++QL +G Q + V++A + KP EIAR L A
Sbjct: 121 GLYLKEKEEKVLLVAADIYRPAAVEQLKTIGNQYDLEVFSAKEKNSKPEEIARDALSYAN 180
Query: 258 KKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEI 317
ID +IVDTAGRLQID++MM E+ +K+ P EVLLVVD+M GQEAA L +F+ ++
Sbjct: 181 ANNIDTLIVDTAGRLQIDESMMGEMVRIKQVTKPDEVLLVVDSMIGQEAADLTKSFHEKV 240
Query: 318 GITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDV 377
GITGAILTKLDGDSRGGAALS++++SGKPIK +G GE+++ L+PF+P+RMA RILGMGDV
Sbjct: 241 GITGAILTKLDGDSRGGAALSIRKISGKPIKFIGTGEKIEALQPFHPERMASRILGMGDV 300
Query: 378 LSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVT 437
L+ VEKAQ+ ++ DAE +Q+K+ A FDFNDF+KQ R + +MGS+ ++ +IPGM+K+
Sbjct: 301 LTLVEKAQKEVELADAEVMQKKLQEATFDFNDFVKQMRLMKRMGSLGGLIKLIPGMNKID 360
Query: 438 PAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQL 497
I+ E+ LK +E+MI +MT EE++KPE+LA P RR+R+AQ SG + V +V+A
Sbjct: 361 DGMIKNGEEQLKKIESMISSMTLEEKQKPEVLAAQPSRRQRIAQGSGYQAKDVDKVLAD- 419
Query: 498 FQMRVRMKNLMGVMQGGSMPTLSNL 522
FQ RM+ M M G MP + +
Sbjct: 420 FQ---RMRGFMKQMSNGGMPGMGGM 441
>UniRef100_Q9AXU1 Chloroplast SRP54 [Chlamydomonas reinhardtii]
Length = 549
Score = 424 bits (1089), Expect = e-117
Identities = 236/482 (48%), Positives = 337/482 (68%), Gaps = 17/482 (3%)
Query: 74 VVRAEMFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSV 133
V+R+ MF L+ +E A + LT EN+ EP++++RRALLEADVSLPVVRRF++ V
Sbjct: 61 VIRSAMFDSLSRSIEKAQRLIGKSGTLTAENMKEPLKEVRRALLEADVSLPVVRRFIKKV 120
Query: 134 SDQAVGVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTT 193
++A+G V+ GV PD Q +K V EL+ LMGG ++++ +IL+AGLQGVG+
Sbjct: 121 EERALGTKVLEGVTPDVQFIKTVSNELIDLMGGGNRGKTWSRASRKIILMAGLQGVGRH- 179
Query: 194 VCA-KLSNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQG 252
CA KL+ YL+ + +L+ D+Y AA QL+ G + VPV+ GTDV P IA++G
Sbjct: 180 -CAGKLALYLRARE---LLLLHDVY--AAHRQLSA-GAAIDVPVFEDGTDVSPVRIAKKG 232
Query: 253 LEEAKKKKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTT 312
+EEA++ +D VI+DTAGRLQ+D+ MM EL++VK ++ P++ LLVVDAMTGQEAA LV +
Sbjct: 233 VEEARRLGVDAVIIDTAGRLQVDEGMMAELRDVKSAVRPSDTLLVVDAMTGQEAANLVRS 292
Query: 313 FNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRIL 372
FN + I+GAILTK+DGDSRGGAALSV+EVSGKPIK VG GE+M+ LEPFYP+RMA RIL
Sbjct: 293 FNEAVDISGAILTKMDGDSRGGAALSVREVSGKPIKFVGVGEKMEALEPFYPERMASRIL 352
Query: 373 GMGDVLSFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPG 432
GMGDVL+ EKA+ +++EDA+K E++M KFDFNDFL Q +A+ MG + +++ M+PG
Sbjct: 353 GMGDVLTLYEKAEAAIKEEDAQKTMERLMEEKFDFNDFLNQWKAMNNMGGL-QMLKMMPG 411
Query: 433 MSKVTPAQIREAEKNLKNMEAMIEAMTPEEREKPELLAES-PVRRKRVAQESGKTEQQVS 491
+K++ + EAEK EA+I AM EER PE+L ++ +RR+RVAQ+SGK+E +V+
Sbjct: 412 FNKISEKHLYEAEKQFGVYEAIIGAMDEEERSNPEVLIKNLALRRRRVAQDSGKSEAEVT 471
Query: 492 QVVAQLFQMRVR---MKNLMGVMQGGSMPTLSN--LEEAL-KPEEKAPPGTARRRKRSEQ 545
+++A M+ + M L+ + + G+ P +N L+E + +K PG RR+K +
Sbjct: 472 KLMAAYTSMKAQVGGMSKLLKLQKAGADPQKANSLLQELVASAGKKVAPGKVRRKKEKDA 531
Query: 546 RS 547
S
Sbjct: 532 LS 533
>UniRef100_UPI0000330C1A UPI0000330C1A UniRef100 entry
Length = 478
Score = 417 bits (1071), Expect = e-115
Identities = 212/424 (50%), Positives = 308/424 (72%), Gaps = 5/424 (1%)
Query: 100 LTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGVGVIRGVKPDQQLVKIVHEE 159
+++ NI + ++++++ALL ADVSL VV+ F+ V +A+G V+RGV P Q+ +++V++E
Sbjct: 2 ISENNINDALKEVKKALLNADVSLSVVKDFISDVKTKAIGEEVVRGVNPGQKFIEVVNKE 61
Query: 160 LVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKLSNYLKKQGKSCMLVAGDIYRP 219
L+ +MG E S L + TVIL+AGLQG GKTT KL YLK++ K +LVA DIYRP
Sbjct: 62 LINIMGNENSPLIEKEEKPTVILMAGLQGAGKTTATGKLGLYLKEKEKKVLLVAADIYRP 121
Query: 220 AAIDQLTILGKQVGVPVYTAG-TDVKPSEIARQGLEEAKKKKIDVVIVDTAGRLQIDKAM 278
AA++QL +G Q + V++A + +P EIA+ L A + +D +IVDTAGRLQID++M
Sbjct: 122 AAVEQLKTIGNQYDLEVFSAKENNNRPEEIAKDALNYATENDVDTLIVDTAGRLQIDESM 181
Query: 279 MDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALS 338
M+E+ +K+ NP EVLLVVD+M GQEAA L +F+ ++GI+GAILTKLDGDSRGGAALS
Sbjct: 182 MNEMVRIKEVTNPDEVLLVVDSMIGQEAADLTKSFHEKVGISGAILTKLDGDSRGGAALS 241
Query: 339 VKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMQQEDAEKLQE 398
++++SGKPIK +G GE+++ L+PF+P+RMA RILGMGDVL+ VEKAQ+ ++ DAE +Q+
Sbjct: 242 IRKISGKPIKFIGIGEKIEALQPFHPERMASRILGMGDVLTLVEKAQKEVELADAEVMQK 301
Query: 399 KIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTPAQIREAEKNLKNMEAMIEAM 458
K+ A FDFNDF+KQ R + +MGS+ ++ +IPGM+K+ I+ E+ LK +E+MI +M
Sbjct: 302 KLQEATFDFNDFVKQMRLMKRMGSLGGLIKLIPGMNKIDDGMIKNGEEQLKKIESMISSM 361
Query: 459 TPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLFQMRVRMKNLMGVMQGGSMPT 518
T EE++KPE+LA P RR+R+A+ SG + V +V+A FQ RM+ M M G MP
Sbjct: 362 TLEEKQKPEVLAAQPSRRQRIAKGSGYQAKDVDKVLAD-FQ---RMRGFMKQMSNGGMPG 417
Query: 519 LSNL 522
+ +
Sbjct: 418 MGGM 421
>UniRef100_Q67PE3 Signal recognition particle [Symbiobacterium thermophilum]
Length = 454
Score = 416 bits (1068), Expect = e-114
Identities = 218/448 (48%), Positives = 317/448 (70%), Gaps = 13/448 (2%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
MF L L+ A+ +L+G+ LT+ ++ E +R++R ALLEADV+ VV+ F+ V ++AV
Sbjct: 1 MFESLQERLQEAFKRLRGKGKLTESDVNEALREVRLALLEADVNFKVVKSFIARVKERAV 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G ++ + P Q ++KIV+EELV LMGGE L A T+I+L GLQG GKTT AKL
Sbjct: 61 GQEILESLNPAQMVIKIVYEELVALMGGESVGLNMADRPPTIIMLCGLQGAGKTTHAAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+ LKK+GK +LVA D+YRPAAI QL +LG+QVGVPVY+ G P +IA+ + A +
Sbjct: 121 ALKLKKEGKKPLLVAADVYRPAAIKQLEVLGEQVGVPVYSEGDQASPVDIAQHAVAYAAE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
++ DV+IVDTAGRL ID+ +M+EL+++K + P + LLV+DAM GQ+A F+ ++G
Sbjct: 181 ERRDVIIVDTAGRLTIDEELMEELQQIKARIQPHDTLLVLDAMIGQDAVTTAEHFHTKLG 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
I G ILTKLDGD+RGGAALSV+EV+G+PIK VG GE++D LEPFYPDR+A RILGMGDVL
Sbjct: 241 IDGVILTKLDGDTRGGAALSVREVTGRPIKFVGVGEKLDALEPFYPDRLASRILGMGDVL 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
+ +EKAQE + ++ AE++ K++ A+FDF DFL+Q RA+ +MG + ++ M+PG+
Sbjct: 301 TLIEKAQEQVDRKQAEEMARKMLKAQFDFEDFLEQLRALQRMGPLQDILKMLPGVG---- 356
Query: 439 AQIREA---EKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVA 495
AQ+++ EK LK++EA+I +MTP+ER P+++ + RR+R+A+ SG+ +V +++
Sbjct: 357 AQLKDINIDEKELKHIEAIILSMTPKERRNPDII--NVRRRERIARGSGRPIAEVHRLIK 414
Query: 496 QLFQMRVRMKNLMG----VMQGGSMPTL 519
Q Q R MK L G + +GG +P L
Sbjct: 415 QFEQTRKLMKQLGGMEKMLKRGGKLPRL 442
>UniRef100_Q895M4 Signal recognition particle, subunit ffh/srp54 [Clostridium tetani]
Length = 451
Score = 415 bits (1066), Expect = e-114
Identities = 205/436 (47%), Positives = 317/436 (72%), Gaps = 2/436 (0%)
Query: 80 FGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVG 139
F L + L+ + KL+G+ L++++I E MR++R ALLEADV+ +V+ FV+SVS++ G
Sbjct: 3 FDGLASKLQETFKKLRGKGKLSEKDIKEAMREVRLALLEADVNYKIVKDFVKSVSEKCNG 62
Query: 140 VGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKLS 199
V+ + P QQ+VKIV++EL++LMG + SE+ F + TVI+L GLQG GKTT+ KL+
Sbjct: 63 KEVLESLTPGQQVVKIVNDELIELMGSKESEINFEANKPTVIMLVGLQGAGKTTMAGKLA 122
Query: 200 NYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKKK 259
L+K+ K +LVA DIYRPAAI QL ++G Q+ VPV+T G P +I++ ++ AK+
Sbjct: 123 LQLRKKNKKPLLVACDIYRPAAIKQLQVVGSQIDVPVFTLGDKTNPVDISKAAIKHAKEN 182
Query: 260 KIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIGI 319
++ +I+DTAGRL ID+ +MDELK +K+++ P EVLLVVD+MTGQ+A + +FN ++ I
Sbjct: 183 NLNTIIIDTAGRLHIDENLMDELKSIKENIEPNEVLLVVDSMTGQDAINVAESFNEKLNI 242
Query: 320 TGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVLS 379
+G ILTKLDGD+RGGAALS+K ++GKPIK +G GE+M+DLE FYP+RMA RILGMGDVLS
Sbjct: 243 SGVILTKLDGDTRGGAALSIKAMTGKPIKFIGIGEKMNDLEVFYPERMASRILGMGDVLS 302
Query: 380 FVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMS--KVT 437
+EKA++ + +++AEK+ +IM+ +F+ DFL + + ++G +++++ M+PG + ++
Sbjct: 303 LIEKAEQEIDKDEAEKIGSRIMNQEFNLEDFLSAMQQMKKLGPLNKLVEMLPGANSKELK 362
Query: 438 PAQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQL 497
+ EK+LK +E++I +MT +ER+ P L++ SP R+KR+A SG T QQV++++
Sbjct: 363 GIDFDKGEKDLKRIESIISSMTIKERKNPSLVSGSPTRKKRIANGSGTTVQQVNKILKDF 422
Query: 498 FQMRVRMKNLMGVMQG 513
MR MK + G+ +G
Sbjct: 423 ENMRKMMKQMNGIQKG 438
>UniRef100_Q8RDV7 Signal recognition particle, subunit FFH/SRP54 [Fusobacterium
nucleatum]
Length = 444
Score = 412 bits (1060), Expect = e-113
Identities = 212/441 (48%), Positives = 309/441 (69%), Gaps = 8/441 (1%)
Query: 79 MFGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAV 138
M L + + K++G L++ NI + +R+++ +LLEADV+ VV+ F +S++A+
Sbjct: 1 MLENLGNRFQDIFKKIRGHGKLSETNIKDALREVKMSLLEADVNYKVVKDFTNRISEKAI 60
Query: 139 GVGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKL 198
G VIRGV P QQ +K+V++ELV+L+GG S+L T+I+LAGLQG GKTT AKL
Sbjct: 61 GTEVIRGVNPAQQFIKLVNDELVELLGGTSSKLTKGLRNPTIIMLAGLQGAGKTTFAAKL 120
Query: 199 SNYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKK 258
+ +LKKQ + +LV D+YRPAAI QL +LG+Q+GV VY+ + IA + +E+AK+
Sbjct: 121 AKFLKKQNEKLLLVGVDVYRPAAIKQLQVLGQQIGVDVYSEEDNKDVVGIATRAIEKAKE 180
Query: 259 KKIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 318
+IVDTAGRL +D+ +MDELKE+KK++ P E+LLVVDAM GQ+A L +FN +
Sbjct: 181 INATYMIVDTAGRLHVDETLMDELKELKKAIKPQEILLVVDAMIGQDAVNLAESFNNALS 240
Query: 319 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVL 378
+ G ILTKLDGD+RGGAALS+K V GKPIK +G GE+++D+E F+PDR+ RILGMGDV+
Sbjct: 241 VDGVILTKLDGDTRGGAALSIKAVVGKPIKFIGVGEKLNDIEIFHPDRLVSRILGMGDVV 300
Query: 379 SFVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTP 438
S VEKAQEV+ + +A+ L+EKI S KFD NDFLKQ + + ++GS+ ++ +IPGM K+
Sbjct: 301 SLVEKAQEVIDENEAKSLEEKIKSQKFDLNDFLKQLQTIKRLGSLGGILKLIPGMPKID- 359
Query: 439 AQIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLF 498
+ AEK +K +EA+I++MT EER+KP++L S R+ R+A+ SG V++++ Q
Sbjct: 360 -DLAPAEKEMKKVEAIIQSMTKEERKKPDILKAS--RKIRIAKGSGTDVSDVNKLLKQFD 416
Query: 499 QMRVRMKNLMGVMQGGSMPTL 519
Q MK++M + G MP +
Sbjct: 417 Q----MKSMMKMFSSGKMPNI 433
>UniRef100_Q8R9X0 Signal recognition particle GTPase [Thermoanaerobacter
tengcongensis]
Length = 447
Score = 412 bits (1059), Expect = e-113
Identities = 209/431 (48%), Positives = 305/431 (70%), Gaps = 2/431 (0%)
Query: 80 FGQLTTGLEAAWNKLKGEEVLTKENIVEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVG 139
F L+ L+ + KL+G+ LT+++I E MR+++ ALLEADV+ VV+ F+ SV+++A+G
Sbjct: 4 FESLSERLQGVFKKLRGKGKLTEKDIKEAMREVKVALLEADVNFKVVKDFINSVTEKALG 63
Query: 140 VGVIRGVKPDQQLVKIVHEELVKLMGGEVSELAFAKSGTTVILLAGLQGVGKTTVCAKLS 199
V+ + P QQ++KIVHEEL+KLMG S L VI++ GLQG GKTT C KL+
Sbjct: 64 QEVMESLTPAQQVIKIVHEELIKLMGSVESRLNLGSKVPAVIMMVGLQGSGKTTACGKLA 123
Query: 200 NYLKKQGKSCMLVAGDIYRPAAIDQLTILGKQVGVPVYTAGTDVKPSEIARQGLEEAKKK 259
N LKKQGK+ +LVA D RPAAI QL +LG + VPV+T G V ++IA+ ++ AK
Sbjct: 124 NLLKKQGKNPLLVACDTVRPAAIKQLQVLGANINVPVFTMGDKVDTADIAKASIDYAKSH 183
Query: 260 KIDVVIVDTAGRLQIDKAMMDELKEVKKSLNPTEVLLVVDAMTGQEAAALVTTFNLEIGI 319
+DVVI+DTAGRL ID +M+EL +K++++P E+LLV DAMTGQ+A + ++FN + I
Sbjct: 184 NVDVVIIDTAGRLHIDDELMEELVRIKEAVHPDEILLVADAMTGQDAVNVASSFNERLDI 243
Query: 320 TGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMDDLEPFYPDRMAGRILGMGDVLS 379
TG ILTKLDGD+RGGAALS+K V+ KPIK VG GE++ D+EPFYPDRMA RILGMGDVL+
Sbjct: 244 TGVILTKLDGDTRGGAALSIKAVTQKPIKYVGIGEKLGDIEPFYPDRMASRILGMGDVLT 303
Query: 380 FVEKAQEVMQQEDAEKLQEKIMSAKFDFNDFLKQTRAVAQMGSVSRVMGMIPGMSKVTPA 439
+EKAQ + ++ A ++ +KI+S +F DFL+Q R++ MG + +++ MIPG++K
Sbjct: 304 LIEKAQAAIDEKKALEMGQKILSKQFTLEDFLEQLRSLKNMGPLDQLLAMIPGVNKSVLK 363
Query: 440 QIREAEKNLKNMEAMIEAMTPEEREKPELLAESPVRRKRVAQESGKTEQQVSQVVAQLFQ 499
+ +EK+LK +EA+I +MT EER+ P ++ S R++R+A+ SG T Q+V+ ++ Q +
Sbjct: 364 NVEVSEKDLKRIEAIILSMTKEERQNPSIINGS--RKRRIARGSGTTIQEVNNLLKQFEE 421
Query: 500 MRVRMKNLMGV 510
+ MK +
Sbjct: 422 TKKMMKRFADI 432
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.130 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 853,086,999
Number of Sequences: 2790947
Number of extensions: 34294015
Number of successful extensions: 129514
Number of sequences better than 10.0: 2109
Number of HSP's better than 10.0 without gapping: 1606
Number of HSP's successfully gapped in prelim test: 505
Number of HSP's that attempted gapping in prelim test: 124980
Number of HSP's gapped (non-prelim): 2495
length of query: 564
length of database: 848,049,833
effective HSP length: 133
effective length of query: 431
effective length of database: 476,853,882
effective search space: 205524023142
effective search space used: 205524023142
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0010.7


