

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
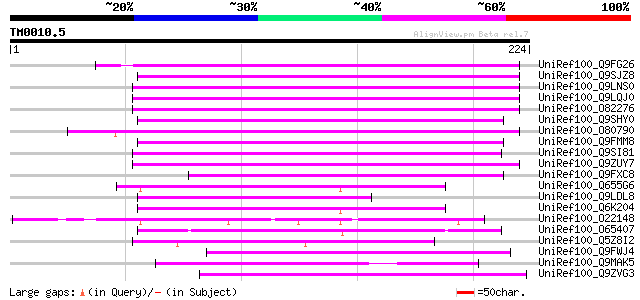
Query= TM0010.5
(224 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like... 125 6e-28
UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcrip... 122 7e-27
UniRef100_Q9LNS0 F1L3.4 [Arabidopsis thaliana] 116 5e-25
UniRef100_Q9LQJ0 F28G4.15 protein [Arabidopsis thaliana] 116 5e-25
UniRef100_O82276 Putative non-LTR retroelement reverse transcrip... 112 5e-24
UniRef100_Q9SHY0 F1E22.12 [Arabidopsis thaliana] 110 2e-23
UniRef100_O80790 Putative non-LTR retroelement reverse transcrip... 109 5e-23
UniRef100_Q9FMM8 Non-LTR retroelement reverse transcriptase-like... 107 2e-22
UniRef100_Q9SI81 F23N19.5 [Arabidopsis thaliana] 105 1e-21
UniRef100_Q9ZUY7 Putative reverse transcriptase [Arabidopsis tha... 104 2e-21
UniRef100_Q9FXC8 F12A21.24 [Arabidopsis thaliana] 90 4e-17
UniRef100_Q655G6 Hypothetical protein P0009H10.11 [Oryza sativa] 87 2e-16
UniRef100_Q9LDL8 Hypothetical protein AT4g08830 [Arabidopsis tha... 86 7e-16
UniRef100_Q6K204 Hypothetical protein B1469H02.35 [Oryza sativa] 85 1e-15
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 85 2e-15
UniRef100_O65407 Hypothetical protein F18E5.40 [Arabidopsis thal... 80 3e-14
UniRef100_Q5Z8I2 Hypothetical protein P0622F03.7 [Oryza sativa] 80 5e-14
UniRef100_Q9FWJ4 Hypothetical protein F21N10.9 [Arabidopsis thal... 80 5e-14
UniRef100_Q9MAK5 F27F5.12 [Arabidopsis thaliana] 76 7e-13
UniRef100_Q9ZVG3 T2P11.14 protein [Arabidopsis thaliana] 75 1e-12
>UniRef100_Q9FG26 Non-LTR retroelement reverse transcriptase-like [Arabidopsis
thaliana]
Length = 676
Score = 125 bits (315), Expect = 6e-28
Identities = 67/183 (36%), Positives = 100/183 (54%), Gaps = 5/183 (2%)
Query: 38 DATDDALIQRAHGTGDLRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFS 97
DA +D L++R + W P + W+ +NTDG+ A GGV+RD +G++LVGF+
Sbjct: 495 DAREDVLVERM-----IAWRKPAEGWVTMNTDGASHGNPGQATAGGVIRDEHGSWLVGFA 549
Query: 98 ALLGRCSINFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKN 157
+G CS AELWG+Y+G +A +RG+++V +E DSA V + G S P A LV+
Sbjct: 550 LNIGVCSAPLAELWGVYYGLVVAWERGWRRVRLEVDSALVVGFLQSGIGDSHPLAFLVRL 609
Query: 158 IKRTLSNFDQVHLKHVFREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCNTL 217
+S V + HV+RE N +AD A L F + D+ P +S L+ D T
Sbjct: 610 CHGFISKDWIVRITHVYREANRLADGLANYAFTLPFGFLLLDSCPEHVSSILLEDVMGTS 669
Query: 218 YHR 220
+ R
Sbjct: 670 FPR 672
>UniRef100_Q9SJZ8 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 321
Score = 122 bits (306), Expect = 7e-27
Identities = 62/165 (37%), Positives = 88/165 (52%)
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYH 115
WV P D W+K+NTDG+ A GGVLRDHNG ++ GF+ +G CS AELWG+Y+
Sbjct: 153 WVSPEDGWVKLNTDGASRGNPGFATAGGVLRDHNGAWIGGFAVNIGVCSAPLAELWGVYY 212
Query: 116 GAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFR 175
G +A RG ++V +E DS V + G S P + L++ LS V + HV+R
Sbjct: 213 GLFIAWGRGARRVELEVDSKMVVGFLTTGIADSHPLSFLLRLCYDFLSKGWIVRISHVYR 272
Query: 176 EINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCNTLYHR 220
E N +AD A L + + ++ P +S L+ D Y R
Sbjct: 273 EANRLADGLANYAFSLSLGLHLLESRPDVVSSILLDDVAGVSYPR 317
>UniRef100_Q9LNS0 F1L3.4 [Arabidopsis thaliana]
Length = 253
Score = 116 bits (290), Expect = 5e-25
Identities = 60/167 (35%), Positives = 87/167 (51%)
Query: 54 LRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGI 113
+ W PP W K+NTDG+ LA GGV+RD +GN+ GFS +G CS AELWG
Sbjct: 83 IAWSPPRVGWFKLNTDGASRGNPRLATAGGVVRDGDGNWCYGFSLNIGICSAPLAELWGA 142
Query: 114 YHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
Y+G +A +RG ++ +E DS V + G S P + LV+ LS V + HV
Sbjct: 143 YYGLNIAWERGVTQLEMEIDSEMVVGFLRTGIDDSHPLSFLVRLCHGLLSKDWSVRISHV 202
Query: 174 FREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCNTLYHR 220
+RE N +AD A L + F ++++ P + + D + Y R
Sbjct: 203 YREANRLADGLANYAFFLPLGFHLFNSTPDIVMSIVHDDVAGSAYPR 249
>UniRef100_Q9LQJ0 F28G4.15 protein [Arabidopsis thaliana]
Length = 272
Score = 116 bits (290), Expect = 5e-25
Identities = 60/167 (35%), Positives = 87/167 (51%)
Query: 54 LRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGI 113
+ W PP W K+NTDG+ LA GGV+RD +GN+ GFS +G CS AELWG
Sbjct: 102 IAWSPPRVGWFKLNTDGASRGNPRLATAGGVVRDGDGNWCYGFSLNIGICSAPLAELWGA 161
Query: 114 YHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
Y+G +A +RG ++ +E DS V + G S P + LV+ LS V + HV
Sbjct: 162 YYGLNIAWERGVTQLEMEIDSEMVVGFLRTGIDDSHPLSFLVRLCHGLLSKDWSVRISHV 221
Query: 174 FREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCNTLYHR 220
+RE N +AD A L + F ++++ P + + D + Y R
Sbjct: 222 YREANRLADGLANYAFFLPLGFHLFNSTPDIVMSIVHDDVAGSAYPR 268
>UniRef100_O82276 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1231
Score = 112 bits (281), Expect = 5e-24
Identities = 57/167 (34%), Positives = 90/167 (53%)
Query: 54 LRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGI 113
+RW P+D W+KI TDG+ LAA GG +R+ G +L GF+ +G C+ AELWG
Sbjct: 1061 IRWQVPSDGWVKITTDGASRGNHGLAAAGGAIRNGQGEWLGGFALNIGSCAAPLAELWGA 1120
Query: 114 YHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
Y+G +A D+GF++V ++ D V ++ G + P + LV+ + + V + HV
Sbjct: 1121 YYGLLIAWDKGFRRVELDLDCKLVVGFLSTGVSNAHPLSFLVRLCQGFFTRDWLVRVSHV 1180
Query: 174 FREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCNTLYHR 220
+RE N +AD A L + +DA P + + L+ D T + R
Sbjct: 1181 YREANRLADGLANYAFTLPLGLHCFDACPEGVRLLLLADVNGTEFPR 1227
>UniRef100_Q9SHY0 F1E22.12 [Arabidopsis thaliana]
Length = 1055
Score = 110 bits (276), Expect = 2e-23
Identities = 60/158 (37%), Positives = 83/158 (51%)
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYH 115
WV P W+K+NTDG+ LA+ GGVLRD G + GFS +GRCS AELWG+Y+
Sbjct: 561 WVSPCVGWVKVNTDGASRGNPGLASAGGVLRDCTGAWCGGFSLNIGRCSAPQAELWGVYY 620
Query: 116 GAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFR 175
G A ++ +V +E DS V + G S P + LV+ L V + HV+R
Sbjct: 621 GLYFAWEKKVPRVELEVDSEVIVGFLKTGISDSHPLSFLVRLCHGFLQKDWLVRIVHVYR 680
Query: 176 EINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDS 213
E N +AD A L + F +D +P +S L D+
Sbjct: 681 EANRLADGLANYAFSLSLGFHSFDLVPDAMSSLLREDT 718
>UniRef100_O80790 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 970
Score = 109 bits (273), Expect = 5e-23
Identities = 61/205 (29%), Positives = 102/205 (49%), Gaps = 10/205 (4%)
Query: 26 KKLSQDFLYTLTDATDDAL----------IQRAHGTGDLRWVPPNDDWIKINTDGSYSPV 75
+KL +D L + D ++ ++RA +RW P+D W+K+ TDG+
Sbjct: 762 RKLCRDRLKFIKDIAEEVRKAHVGTLNNHVKRARVERMIRWKAPSDRWVKLTTDGASRGH 821
Query: 76 SNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYHGAKLALDRGFKKVMIESDSA 135
LAA G + + G +L GF+ +G C AELWG Y+G +A D+GF++V + DS
Sbjct: 822 QGLAAASGAILNLQGEWLGGFALNIGSCDAPLAELWGAYYGLLIAWDKGFRRVELNLDSE 881
Query: 136 FAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADAFAKNGLGLLMDF 195
V ++ G + P + LV+ + + V + HV+RE N +AD A L + F
Sbjct: 882 LVVGFLSTGISKAHPLSFLVRLCQGFFTRDWLVRVSHVYREANRLADGLANYAFFLPLGF 941
Query: 196 RIYDAIPSFISVALMHDSCNTLYHR 220
++ P + + L+ D+ T + R
Sbjct: 942 HCFEICPEDVLLFLVEDANGTSFPR 966
>UniRef100_Q9FMM8 Non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 308
Score = 107 bits (268), Expect = 2e-22
Identities = 59/158 (37%), Positives = 84/158 (52%)
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYH 115
WV P W+K+NTDG+ LA+ GGVLRD G + GFS +GRCS AELWG+Y+
Sbjct: 140 WVLPCVGWVKVNTDGASRGNPGLASAGGVLRDCEGAWCGGFSLNIGRCSAQHAELWGVYY 199
Query: 116 GAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFR 175
G A ++ +V +E DS V + G S P + LV+ L V + +V+R
Sbjct: 200 GLYFAWEKKVPRVELEVDSEAIVGFLKTGISDSHPLSFLVRLCHNFLQKDWLVRIVYVYR 259
Query: 176 EINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDS 213
E N +AD A + L + F +D +P +S L D+
Sbjct: 260 EANCLADGLANYTILLSLGFHSFDFVPDAMSSLLKEDT 297
>UniRef100_Q9SI81 F23N19.5 [Arabidopsis thaliana]
Length = 233
Score = 105 bits (261), Expect = 1e-21
Identities = 56/159 (35%), Positives = 82/159 (51%)
Query: 54 LRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGI 113
+RW P+ W K+NTDG+ LA GG LR+ G + GF+ +GRC AELWG+
Sbjct: 63 VRWSKPSLGWCKLNTDGASHGNPGLATAGGALRNEYGEWCFGFALNIGRCLAPLAELWGV 122
Query: 114 YHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
Y+G +A DRG ++ +E DS V + G S P + LV+ LS V + HV
Sbjct: 123 YYGLFMAWDRGITRLELEVDSEMVVGFLRTGIGSSHPLSFLVRMCHGFLSRDWIVRIGHV 182
Query: 174 FREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHD 212
+RE N +AD A L + + + + P+ + L D
Sbjct: 183 YREANRLADGLANYAFDLPLGYHAFASPPNSLDSILRDD 221
>UniRef100_Q9ZUY7 Putative reverse transcriptase [Arabidopsis thaliana]
Length = 314
Score = 104 bits (259), Expect = 2e-21
Identities = 56/167 (33%), Positives = 83/167 (49%)
Query: 54 LRWVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGI 113
+ W P + W K+NTDG+ LA+ GGVLRD G + GF+ +G CS AELWG+
Sbjct: 144 IAWSKPEEGWWKLNTDGASRGNPGLASAGGVLRDEEGAWRGGFALNIGVCSAPLAELWGV 203
Query: 114 YHGAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
Y+G +A +R ++ IE DS V + G P + LV+ +S +V + HV
Sbjct: 204 YYGLYIAWERRVTRLEIEVDSEIVVGFLKIGINEVHPLSFLVRLCHDFISRDWRVRISHV 263
Query: 174 FREINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHDSCNTLYHR 220
+RE N +AD A L + F +P + L+ D+ R
Sbjct: 264 YREANRLADGLANYAFSLPLGFHSLSLVPDSLRFILLDDTSGATVSR 310
>UniRef100_Q9FXC8 F12A21.24 [Arabidopsis thaliana]
Length = 803
Score = 90.1 bits (222), Expect = 4e-17
Identities = 51/136 (37%), Positives = 69/136 (50%)
Query: 78 LAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYHGAKLALDRGFKKVMIESDSAFA 137
LA GGV+RD GN+ GF+ +GRCS AELWG+Y+G LA + +V +E DS
Sbjct: 657 LATAGGVIRDGAGNWCGGFALNIGRCSAPLAELWGVYYGFYLAWTKALTRVELEVDSELV 716
Query: 138 VNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADAFAKNGLGLLMDFRI 197
V + G P + LV+ LS V + HV+RE N +AD A L + F
Sbjct: 717 VGFLKTGIGDQHPLSFLVRLCHGLLSKDWIVRITHVYREANRLADGLANYAFSLPLGFHS 776
Query: 198 YDAIPSFISVALMHDS 213
+P + V L DS
Sbjct: 777 LIDVPDDLEVILHEDS 792
>UniRef100_Q655G6 Hypothetical protein P0009H10.11 [Oryza sativa]
Length = 240
Score = 87.4 bits (215), Expect = 2e-16
Identities = 49/144 (34%), Positives = 81/144 (56%), Gaps = 2/144 (1%)
Query: 47 RAHGTGDLR-WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSI 105
RA+ + R W P + W+K+N DGS + +A+ GGV RDH+G +L+G++ +G +
Sbjct: 69 RAYSSASARIWRKPEEGWMKLNFDGSSKHSTGIASIGGVYRDHDGAFLLGYAERIGTATS 128
Query: 106 NFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNLV-NKGCLPSKPAAALVKNIKRTLSN 164
+ AEL + G +LA+ G+++V E DS V++V ++ + S+ L + I L
Sbjct: 129 SVAELAALRRGLELAVRNGWRRVWAEGDSKAVVDVVCDRADVQSEEDLRLCREIAALLPQ 188
Query: 165 FDQVHLKHVFREINSVADAFAKNG 188
D + + HV R N VA FA+ G
Sbjct: 189 LDDMAVSHVRRGGNKVAHGFAELG 212
>UniRef100_Q9LDL8 Hypothetical protein AT4g08830 [Arabidopsis thaliana]
Length = 947
Score = 85.9 bits (211), Expect = 7e-16
Identities = 41/101 (40%), Positives = 60/101 (58%)
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYH 115
W PP+ +W+K+NTDG+ LA GGVLRD G++ GF+ +G CS AELWG+Y+
Sbjct: 841 WKPPDGEWVKLNTDGASRGNLGLATTGGVLRDGIGHWCGGFALDIGVCSAPLAELWGVYY 900
Query: 116 GAKLALDRGFKKVMIESDSAFAVNLVNKGCLPSKPAAALVK 156
G +A +R F +V +E DS V + G + + LV+
Sbjct: 901 GLYMAWERRFTRVELEVDSELVVGFLTTGISDTHSLSFLVR 941
>UniRef100_Q6K204 Hypothetical protein B1469H02.35 [Oryza sativa]
Length = 212
Score = 85.1 bits (209), Expect = 1e-15
Identities = 45/134 (33%), Positives = 74/134 (54%), Gaps = 1/134 (0%)
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYH 115
W P WIK+N DGS + +A+ GGV RDH G +++G++ +GR + + AEL +
Sbjct: 51 WRKPEAGWIKLNFDGSSKHATKIASIGGVYRDHEGAFVLGYAERIGRATSSVAELAALRR 110
Query: 116 GAKLALDRGFKKVMIESDSAFAVNLV-NKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVF 174
G +L + G+++V E DS V++V ++ + S+ + I L D + + HV+
Sbjct: 111 GLELVVRNGWRRVWAEGDSKTVVDVVCDRANVRSEEDLRQCREIAALLPLIDDMAVSHVY 170
Query: 175 REINSVADAFAKNG 188
R N VA FA+ G
Sbjct: 171 RSGNKVAHGFARLG 184
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 84.7 bits (208), Expect = 2e-15
Identities = 69/213 (32%), Positives = 111/213 (51%), Gaps = 20/213 (9%)
Query: 2 IWTARNELVFEKLGHTPAVVCSKIKKLSQDFLYTLTDATDDALIQRAHGTGDLR-----W 56
+W RN+LVF+ T V I K ++D DA ++ + T R W
Sbjct: 1164 LWKNRNDLVFKGREFTAPQV---ILKATEDM-----DAWNNRKEPQPQVTSSTRDRCVKW 1215
Query: 57 VPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYL-VGFSALLGRCSINFAELWGIYH 115
PP+ W+K NTDG++S G VLR+H G L +G AL + S+ E+ +
Sbjct: 1216 QPPSHGWVKCNTDGAWSKDLGNCGVGWVLRNHTGRLLWLGLRALPSQQSVLETEVEAL-R 1274
Query: 116 GAKLALDR-GFKKVMIESDSAFAVNLV-NKGCLPSKPAAALVKNIKRTLSNFDQVHLKHV 173
A L+L R +++V+ ESDS + V+L+ N+ +PS A +++I+ L +F++V +
Sbjct: 1275 WAVLSLSRFNYRRVIFESDSQYLVSLIQNEMDIPS--LAPRIQDIRNLLRHFEEVKFQFT 1332
Query: 174 FREINSVADAFAKNGLGLL-MDFRIYDAIPSFI 205
RE N+VAD A+ L L+ D ++Y P +I
Sbjct: 1333 RREGNNVADRTARESLSLMNYDPKMYSITPDWI 1365
>UniRef100_O65407 Hypothetical protein F18E5.40 [Arabidopsis thaliana]
Length = 229
Score = 80.5 bits (197), Expect = 3e-14
Identities = 50/158 (31%), Positives = 80/158 (49%), Gaps = 3/158 (1%)
Query: 56 WVPPNDDWIKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYH 115
W P + IK+N DGS A+ GGV R+H +L+G+S +G + AE +
Sbjct: 63 WKKPENGRIKLNFDGSRGREGQ-ASIGGVFRNHKAEFLLGYSESIGEATSTMAEFAALKR 121
Query: 116 GAKLALDRGFKKVMIESDSAFAVNLVN-KGCLPSKPAAALVKNIKRTLSNFDQVHLKHVF 174
G +LAL+ G + +E D+ +++++ +G L + V IK + + L HV+
Sbjct: 122 GLELALENGLTDLWLEGDAKIIMDIISRRGRLRCEKTNKHVNYIKVVMPELNNCVLSHVY 181
Query: 175 REINSVADAFAKNGLGLLMDFRIYDAIPSFISVALMHD 212
RE N VAD AK G D +++ P I + +MHD
Sbjct: 182 REGNRVADKLAKLG-HQFQDPKVWRVQPPEIVLPIMHD 218
>UniRef100_Q5Z8I2 Hypothetical protein P0622F03.7 [Oryza sativa]
Length = 217
Score = 79.7 bits (195), Expect = 5e-14
Identities = 44/132 (33%), Positives = 72/132 (54%), Gaps = 2/132 (1%)
Query: 54 LRWVPPNDDWIKINTDGS-YSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWG 112
+RW PP W+K+N DGS ++ S A+ GGV+RD +G L+ F+ +I E
Sbjct: 42 VRWTPPRHGWVKLNFDGSVHNDGSGRASIGGVIRDDHGRVLLAFAERTPHATIGVVEARA 101
Query: 113 IYHGAKLALDRGFK-KVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLK 171
+ G +LALD G+ ++++E D V L+ ++ A++ +I L +F ++
Sbjct: 102 LIRGLQLALDHGWNDRLLVEGDDLTLVRLLRCESTHTRIPPAMLDDILWLLDSFRVCEVQ 161
Query: 172 HVFREINSVADA 183
H +RE N VADA
Sbjct: 162 HAYREGNQVADA 173
>UniRef100_Q9FWJ4 Hypothetical protein F21N10.9 [Arabidopsis thaliana]
Length = 256
Score = 79.7 bits (195), Expect = 5e-14
Identities = 42/131 (32%), Positives = 65/131 (49%)
Query: 86 RDHNGNYLVGFSALLGRCSINFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNLVNKGC 145
R+H GN+ GFS +G CS AELWG+Y+G +A ++ +V +E DS V + G
Sbjct: 37 REHQGNWCGGFSVSIGICSAPQAELWGVYYGLYIAWEKWVTRVELEVDSEIVVGFLKTGI 96
Query: 146 LPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADAFAKNGLGLLMDFRIYDAIPSFI 205
S P + L + +S V + HV+RE N +AD A L + F + + P +
Sbjct: 97 GDSHPLSFLARLCYGFISKDWIVRISHVYREANRLADGLANYAFTLPLGFHFFHSSPDVV 156
Query: 206 SVALMHDSCNT 216
+ D+ T
Sbjct: 157 DSVRLEDTSGT 167
>UniRef100_Q9MAK5 F27F5.12 [Arabidopsis thaliana]
Length = 205
Score = 75.9 bits (185), Expect = 7e-13
Identities = 42/139 (30%), Positives = 66/139 (47%), Gaps = 12/139 (8%)
Query: 64 IKINTDGSYSPVSNLAACGGVLRDHNGNYLVGFSALLGRCSINFAELWGIYHGAKLALDR 123
+K D S+ L GG++RD G++ GF+ +G CS AELWG+Y+G L +R
Sbjct: 58 VKARYDWSFHKNPVLTTAGGLIRDATGSWCGGFALNIGVCSAPLAELWGVYYGLYLTWER 117
Query: 124 GFKKVMIESDSAFAVNLVNKGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADA 183
++ +E DS V ++KG S P + ++ V HV+RE N +AD
Sbjct: 118 HITRLEVEVDSELVVGFLSKGISDSHPLSFVI------------VWTSHVYREANRLADE 165
Query: 184 FAKNGLGLLMDFRIYDAIP 202
L + F + +P
Sbjct: 166 LENYAFTLPLTFHMLKVVP 184
>UniRef100_Q9ZVG3 T2P11.14 protein [Arabidopsis thaliana]
Length = 158
Score = 75.5 bits (184), Expect = 1e-12
Identities = 44/141 (31%), Positives = 67/141 (47%)
Query: 83 GVLRDHNGNYLVGFSALLGRCSINFAELWGIYHGAKLALDRGFKKVMIESDSAFAVNLVN 142
G LRD G++ F+ LGRC+ AELWG+Y+G +A ++G ++ +E DS +
Sbjct: 17 GALRDEYGDWRGSFALNLGRCTAPLAELWGVYYGLVIAWEKGITRLELEVDSKLVAGFLT 76
Query: 143 KGCLPSKPAAALVKNIKRTLSNFDQVHLKHVFREINSVADAFAKNGLGLLMDFRIYDAIP 202
G S + LV+ S V + HV+RE N AD A LL+ F +D+
Sbjct: 77 TGIEDSHLLSFLVRLCYGFSSKDWIVRVSHVYREANRFADELANYAFSLLLGFLSFDSGL 136
Query: 203 SFISVALMHDSCNTLYHRGGP 223
F+ + D+ R P
Sbjct: 137 PFVDSIMREDAAGVAGARHVP 157
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.139 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 399,898,834
Number of Sequences: 2790947
Number of extensions: 16462257
Number of successful extensions: 33957
Number of sequences better than 10.0: 436
Number of HSP's better than 10.0 without gapping: 148
Number of HSP's successfully gapped in prelim test: 288
Number of HSP's that attempted gapping in prelim test: 33653
Number of HSP's gapped (non-prelim): 465
length of query: 224
length of database: 848,049,833
effective HSP length: 123
effective length of query: 101
effective length of database: 504,763,352
effective search space: 50981098552
effective search space used: 50981098552
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0010.5


