

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0010.2
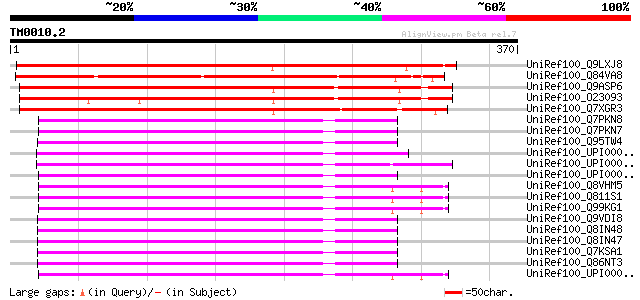
(370 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LXJ8 Putative RNA-binding protein [Arabidopsis thali... 418 e-115
UniRef100_Q84VA8 Putative RNA-binding protein [Oryza sativa] 382 e-105
UniRef100_Q9ASP6 AT4g00830/A_TM018A10_14 [Arabidopsis thaliana] 375 e-102
UniRef100_O23093 A_TM018A10.14 protein [Arabidopsis thaliana] 356 5e-97
UniRef100_Q7XGR3 Putative RNA-binding protein [Oryza sativa] 345 9e-94
UniRef100_Q7PKN8 ENSANGP00000023817 [Anopheles gambiae str. PEST] 176 1e-42
UniRef100_Q7PKN7 ENSANGP00000024050 [Anopheles gambiae str. PEST] 168 2e-40
UniRef100_Q95TW4 GH28335p [Drosophila melanogaster] 167 4e-40
UniRef100_UPI0000292EBE UPI0000292EBE UniRef100 entry 165 2e-39
UniRef100_UPI00003625C8 UPI00003625C8 UniRef100 entry 162 2e-38
UniRef100_UPI00004316F1 UPI00004316F1 UniRef100 entry 161 2e-38
UniRef100_Q8VHM5 Heterogeneous nuclear ribonucleoprotein R [Mus ... 161 2e-38
UniRef100_Q811S1 Heterogeneous nuclear ribonucleoprotein R [Ratt... 161 2e-38
UniRef100_Q99KG1 Hnrpr protein [Mus musculus] 161 2e-38
UniRef100_Q9VDI8 CG17838-PD, isoform D [Drosophila melanogaster] 161 2e-38
UniRef100_Q8IN48 CG17838-PE, isoform E [Drosophila melanogaster] 161 2e-38
UniRef100_Q8IN47 CG17838-PA, isoform A [Drosophila melanogaster] 161 2e-38
UniRef100_Q7KSA1 CG17838-PF, isoform F [Drosophila melanogaster] 161 2e-38
UniRef100_Q86NT3 AT01548p [Drosophila melanogaster] 161 2e-38
UniRef100_UPI00003AC553 UPI00003AC553 UniRef100 entry 161 3e-38
>UniRef100_Q9LXJ8 Putative RNA-binding protein [Arabidopsis thaliana]
Length = 471
Score = 418 bits (1075), Expect = e-115
Identities = 198/325 (60%), Positives = 262/325 (79%), Gaps = 5/325 (1%)
Query: 6 TNDEVEKKKHAELLALPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGE 65
T +E EKK+H ELLALPPH SEVY+GGI + +E DL+ FC S+GEV+EVRIM+ K+SG+
Sbjct: 72 TEEEEEKKRHVELLALPPHGSEVYLGGIPTDATEGDLKGFCGSIGEVTEVRIMREKDSGD 131
Query: 66 RKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKV 125
KGYAFV F++K+LA++AI+ LNN++F+GK+IKCS++QAKHRLF+GN+P+ W D+KK
Sbjct: 132 GKGYAFVTFRSKDLAAEAIDTLNNTDFRGKRIKCSTTQAKHRLFLGNVPRNWMESDIKKA 191
Query: 126 VADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSW 185
+GPGV +EL K+ Q+ GRNRGFAFIEYYNHACAEYS+QKMSN +FKL++NAPTVSW
Sbjct: 192 ANRIGPGVQIVELPKEPQNMGRNRGFAFIEYYNHACAEYSKQKMSNPSFKLDDNAPTVSW 251
Query: 186 AEPRN--SESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRF 243
AE R+ S+ SQVKA+Y+KNLP +ITQ+ LK LFEHHGKI KVV+PPAK G+E SR+
Sbjct: 252 AESRSGGGGDSSASQVKALYIKNLPRDITQERLKALFEHHGKILKVVIPPAKPGKEDSRY 311
Query: 244 GFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTS--NSQKPVVLPTYPH 301
GFVH+++R+S M+ALKNTE+YEIDG L+C+LAKP+ADQ+++ + N QK + P YP
Sbjct: 312 GFVHYAERTSVMRALKNTERYEIDGHMLDCTLAKPQADQKTNTNTVQNVQKSQLQPNYPP 371
Query: 302 RLGYGMVGGAYGGIGAGYGAAGFAQ 326
L YGM +G +G G+GA+ ++Q
Sbjct: 372 LLSYGMAPSPFGALG-GFGASAYSQ 395
>UniRef100_Q84VA8 Putative RNA-binding protein [Oryza sativa]
Length = 476
Score = 382 bits (982), Expect = e-105
Identities = 191/319 (59%), Positives = 255/319 (79%), Gaps = 12/319 (3%)
Query: 5 HTNDEVEKKKHAELLALPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESG 64
H D EK KHAELLALPPH SEVY+GGI+++VS EDL+ C+ VGEV EVR+M+ K+
Sbjct: 102 HQKDVTEKGKHAELLALPPHGSEVYVGGISSDVSSEDLKRLCEPVGEVVEVRMMRGKD-- 159
Query: 65 ERKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKK 124
+ +GYAFV F+TK LA +A++ELNN++ KGK+I+ SSSQAK++LFIGN+P WT +D +K
Sbjct: 160 DSRGYAFVNFRTKGLALKAVKELNNAKLKGKRIRVSSSQAKNKLFIGNVPHSWTDDDFRK 219
Query: 125 VVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVS 184
VV +VGPGV+ +L+K + S+ RNRG+ F+EYYNHACAEY+RQ+MS+ FKL++NAPTVS
Sbjct: 220 VVEEVGPGVLKADLMK-VSSANRNRGYGFVEYYNHACAEYARQEMSSPTFKLDSNAPTVS 278
Query: 185 WAEPRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFG 244
WA+P+N++S++ SQVK+VYVKNLP+N+TQ LK+LFEHHG+I KVVLPP++ G + +R+G
Sbjct: 279 WADPKNNDSASTSQVKSVYVKNLPKNVTQAQLKRLFEHHGEIEKVVLPPSRGGHD-NRYG 337
Query: 245 FVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEA----DQRSSGTSNSQKPVVLPTYP 300
FVHF DRS AM+AL+NTE+YE+DG+ L+CSLAKP A D R S++ P +LP+YP
Sbjct: 338 FVHFKDRSMAMRALQNTERYELDGQVLDCSLAKPPAADKKDDRVPLPSSNGAP-LLPSYP 396
Query: 301 HRLGYGM--VGGAYGGIGA 317
LGYG+ V GAYG A
Sbjct: 397 -PLGYGIMSVPGAYGAAPA 414
>UniRef100_Q9ASP6 AT4g00830/A_TM018A10_14 [Arabidopsis thaliana]
Length = 495
Score = 375 bits (962), Expect = e-102
Identities = 181/324 (55%), Positives = 251/324 (76%), Gaps = 15/324 (4%)
Query: 8 DEVEKKKHAELLALPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERK 67
D+ +++K++ LL+LPPH SEV+IGG+ +V EEDLR C+ +GE+ EVR+MK ++SG+ K
Sbjct: 98 DDEDREKYSHLLSLPPHGSEVFIGGLPRDVGEEDLRDLCEEIGEIFEVRLMKDRDSGDSK 157
Query: 68 GYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVA 127
GYAFVAFKTK++A +AIEEL++ EFKGK I+CS S+ K+RLFIGNIPK WT ++ +KV+
Sbjct: 158 GYAFVAFKTKDVAQKAIEELHSKEFKGKTIRCSLSETKNRLFIGNIPKNWTEDEFRKVIE 217
Query: 128 DVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAE 187
DVGPGV +IEL+KD ++ RNRGFAF+ YYN+ACA+YSRQKM +SNFKLE NAPTV+WA+
Sbjct: 218 DVGPGVENIELIKDPTNTTRNRGFAFVLYYNNACADYSRQKMIDSNFKLEGNAPTVTWAD 277
Query: 188 PRNS--ESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGF 245
P++S S+A +QVKA+YVKN+PEN + + LK+LF+ HG++TK+V PP K G K FGF
Sbjct: 278 PKSSPEHSAAAAQVKALYVKNIPENTSTEQLKELFQRHGEVTKIVTPPGKGG--KRDFGF 335
Query: 246 VHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQR------SSGTSNSQKPVVLPTY 299
VH+++RSSA+KA+K+TE+YE++G+ LE LAKP+A+++ S G + + P V PT+
Sbjct: 336 VHYAERSSALKAVKDTERYEVNGQPLEVVLAKPQAERKHDPSSYSYGAAPTPAPFVHPTF 395
Query: 300 PHRLGYGMVGGAYGGIGAGYGAAG 323
G YG +GAG G AG
Sbjct: 396 G-----GFAAAPYGAMGAGLGIAG 414
>UniRef100_O23093 A_TM018A10.14 protein [Arabidopsis thaliana]
Length = 521
Score = 356 bits (914), Expect = 5e-97
Identities = 181/350 (51%), Positives = 251/350 (71%), Gaps = 41/350 (11%)
Query: 8 DEVEKKKHAELLALPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVR----------- 56
D+ +++K++ LL+LPPH SEV+IGG+ +V EEDLR C+ +GE+ EVR
Sbjct: 98 DDEDREKYSHLLSLPPHGSEVFIGGLPRDVGEEDLRDLCEEIGEIFEVRTAIIFVFHDIL 157
Query: 57 ---IMKAKESGERKGYAFVAFKTKELASQAIEELNNSEFK------------GKKIKCSS 101
+MK ++SG+ KGYAFVAFKTK++A +AIEEL++ EFK GK I+CS
Sbjct: 158 FVRLMKDRDSGDSKGYAFVAFKTKDVAQKAIEELHSKEFKASSTANCSLSLSGKTIRCSL 217
Query: 102 SQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHAC 161
S+ K+RLFIGNIPK WT ++ +KV+ DVGPGV +IEL+KD ++ RNRGFAF+ YYN+AC
Sbjct: 218 SETKNRLFIGNIPKNWTEDEFRKVIEDVGPGVENIELIKDPTNTTRNRGFAFVLYYNNAC 277
Query: 162 AEYSRQKMSNSNFKLENNAPTVSWAEPRNS--ESSAVSQVKAVYVKNLPENITQDSLKKL 219
A+YSRQKM +SNFKLE NAPTV+WA+P++S S+A +QVKA+YVKN+PEN + + LK+L
Sbjct: 278 ADYSRQKMIDSNFKLEGNAPTVTWADPKSSPEHSAAAAQVKALYVKNIPENTSTEQLKEL 337
Query: 220 FEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPE 279
F+ HG++TK+V PP K G K FGFVH+++RSSA+KA+K+TE+YE++G+ LE LAKP+
Sbjct: 338 FQRHGEVTKIVTPPGKGG--KRDFGFVHYAERSSALKAVKDTERYEVNGQPLEVVLAKPQ 395
Query: 280 ADQR------SSGTSNSQKPVVLPTYPHRLGYGMVGGAYGGIGAGYGAAG 323
A+++ S G + + P V PT+ G YG +GAG G AG
Sbjct: 396 AERKHDPSSYSYGAAPTPAPFVHPTFG-----GFAAAPYGAMGAGLGIAG 440
>UniRef100_Q7XGR3 Putative RNA-binding protein [Oryza sativa]
Length = 472
Score = 345 bits (886), Expect = 9e-94
Identities = 170/319 (53%), Positives = 237/319 (74%), Gaps = 11/319 (3%)
Query: 8 DEVEKKKHAELLALPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERK 67
DE EK+K ELLALPP SEV+IGG+ + +E+DL C++ GE+SEVR+MK KE+ E K
Sbjct: 91 DEEEKRKWDELLALPPQGSEVFIGGLPRDTTEDDLHELCEAFGEISEVRLMKDKETKENK 150
Query: 68 GYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVA 127
G+AFV F K+ A +AIE+L++ E KG+ ++CS SQAKHRLF+GN+PK + ++++ ++
Sbjct: 151 GFAFVTFTGKDGAQRAIEDLHDKEHKGRTLRCSLSQAKHRLFVGNVPKGLSEDELRNIIQ 210
Query: 128 DVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAE 187
GPGV++IE+ KDL RNRGF F+EYYNHACA+Y++QK+S NFK++ + TVSWAE
Sbjct: 211 GKGPGVVNIEMFKDLHDPSRNRGFLFVEYYNHACADYAKQKLSAPNFKVDGSQLTVSWAE 270
Query: 188 PRNS----ESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRF 243
P+ S SSA +QVK +YVKNLPEN +++ +K++FE HG++TKVVLPPAK+G ++ F
Sbjct: 271 PKGSSSSDSSSAAAQVKTIYVKNLPENASKEKIKEIFEIHGEVTKVVLPPAKAGNKRD-F 329
Query: 244 GFVHFSDRSSAMKALKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRL 303
GFVHF++RSSA+KA+K +EKYEIDG+ LE S+AKP D++ +S KP P +P
Sbjct: 330 GFVHFAERSSALKAVKGSEKYEIDGQVLEVSMAKPLGDKK---PDHSFKPGGAPNFPLPP 386
Query: 304 GYGMVG---GAYGGIGAGY 319
G +G GAYGG G G+
Sbjct: 387 YGGYMGDPYGAYGGGGPGF 405
>UniRef100_Q7PKN8 ENSANGP00000023817 [Anopheles gambiae str. PEST]
Length = 443
Score = 176 bits (445), Expect = 1e-42
Identities = 89/264 (33%), Positives = 157/264 (58%), Gaps = 10/264 (3%)
Query: 22 PPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELAS 81
P + EV+ G I ++ E++L + G++ ++R+M +G +GYAFV F +++ AS
Sbjct: 164 PGNGCEVFCGKIPKDMYEDELIPLFEKCGKIWDLRLMMDPMTGTNRGYAFVTFTSRDAAS 223
Query: 82 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 140
A+ ELN+ E + GKKI + S HRLF+GNIPK +++ + A PG++ + +
Sbjct: 224 NAVRELNDYEIRNGKKIGVTISFNNHRLFVGNIPKNRDRDELLEEFAKHAPGLVEVIIYS 283
Query: 141 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNS-ESSAVSQV 199
+NRGF F+EY +H A +++++ K+ N V WA+P+ + +S+V
Sbjct: 284 SPDDKKKNRGFCFLEYESHKAASLAKRRLGTGRIKVWNCDIIVDWADPQEEPDEQTMSKV 343
Query: 200 KAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALK 259
K +YV+NL ++ +++ LK+ FE G++ +V ++ + FVHF DR +A+KA+K
Sbjct: 344 KVLYVRNLTQDTSEEKLKESFEQFGRVERV--------KKIKDYAFVHFEDRDNAVKAMK 395
Query: 260 NTEKYEIDGKNLECSLAKPEADQR 283
+ + E+ G N+E SLAKP +D++
Sbjct: 396 DLDGKEVGGSNIEVSLAKPPSDKK 419
>UniRef100_Q7PKN7 ENSANGP00000024050 [Anopheles gambiae str. PEST]
Length = 443
Score = 168 bits (426), Expect = 2e-40
Identities = 87/264 (32%), Positives = 153/264 (57%), Gaps = 10/264 (3%)
Query: 22 PPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELAS 81
P + EV+ G I ++ E++L + G++ ++R+M +G +GYAFV F +++ AS
Sbjct: 164 PGNGCEVFCGKIPKDMYEDELIPLFEKCGKIWDLRLMMDPMTGTNRGYAFVTFTSRDAAS 223
Query: 82 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 140
A+ ELNN E K G +K + S RLF+GNIPK E++ + G++ + +
Sbjct: 224 NAVRELNNYEIKPGNCLKINVSVPNLRLFVGNIPKSKGKEEILDEFGKLTAGLVEVIIYS 283
Query: 141 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNS-ESSAVSQV 199
+NRGF F+EY +H A +++++ K+ N V WA+P+ + +S+V
Sbjct: 284 SPDDKKKNRGFCFLEYESHKAASLAKRRLGTGRIKVWNCDIIVDWADPQEEPDEQTMSKV 343
Query: 200 KAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALK 259
K +YV+NL ++ +++ LK+ FE G++ +V ++ + FVHF DR +A+KA+K
Sbjct: 344 KVLYVRNLTQDTSEEKLKESFEQFGRVERV--------KKIKDYAFVHFEDRDNAVKAMK 395
Query: 260 NTEKYEIDGKNLECSLAKPEADQR 283
+ + E+ G N+E SLAKP +D++
Sbjct: 396 DLDGKEVGGSNIEVSLAKPPSDKK 419
>UniRef100_Q95TW4 GH28335p [Drosophila melanogaster]
Length = 529
Score = 167 bits (423), Expect = 4e-40
Identities = 86/265 (32%), Positives = 153/265 (57%), Gaps = 10/265 (3%)
Query: 21 LPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELA 80
+P + EV+ G I ++ E++L ++ G + ++R+M +G +GYAFV F +E A
Sbjct: 159 VPGNGCEVFCGKIPKDMYEDELIPLFENCGIIWDLRLMMDPMTGTNRGYAFVTFTNREAA 218
Query: 81 SQAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELL 139
A+ +LN+ E + GKKI + S HRLF+GNIPK +++ + + PG+ + +
Sbjct: 219 VNAVRQLNDFEIRTGKKIGVTISFNNHRLFVGNIPKNRDRDELIEEFSKHAPGLYEVIIY 278
Query: 140 KDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNS-ESSAVSQ 198
+NRGF F+EY +H A +++++ K+ V WA+P+ + +S+
Sbjct: 279 SSPDDKKKNRGFCFLEYESHKAASLAKRRLGTGRIKVWGCDIIVDWADPQEEPDEQTMSK 338
Query: 199 VKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKAL 258
VK +YV+NL +++++D LK+ FE +GK+ +V ++ + F+HF DR SA++A+
Sbjct: 339 VKVLYVRNLTQDVSEDKLKEQFEQYGKVERV--------KKIKDYAFIHFEDRDSAVEAM 390
Query: 259 KNTEKYEIDGKNLECSLAKPEADQR 283
+ EI N+E SLAKP +D++
Sbjct: 391 RGLNGKEIGASNIEVSLAKPPSDKK 415
>UniRef100_UPI0000292EBE UPI0000292EBE UniRef100 entry
Length = 636
Score = 165 bits (417), Expect = 2e-39
Identities = 88/274 (32%), Positives = 155/274 (56%), Gaps = 10/274 (3%)
Query: 20 ALPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKEL 79
A PP +E+++G I ++ E++L + G + ++R+M SG +GYAFV F +KE
Sbjct: 156 AQPPVGTEIFVGKIPRDLFEDELVPLFEKAGPIWDLRLMMDPLSGLNRGYAFVTFCSKEA 215
Query: 80 ASQAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIEL 138
A +A++ NN E + GK+I S A +RLF+G+IPK T E + + + V G+ + L
Sbjct: 216 AQEAVKLCNNHEIRPGKQIGVCISVANNRLFVGSIPKSKTKEQIVEEFSKVTEGLSDVIL 275
Query: 139 LKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEP-RNSESSAVS 197
Q +NRGF F+EY +H A +R+++ + K+ N TV WA+P + + ++
Sbjct: 276 YLQPQDKSKNRGFCFLEYEDHKTAAQARRRLMSGKVKVWGNMVTVEWADPMEDPDPEVMA 335
Query: 198 QVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKA 257
+VK ++V+NL ++T++ L+K F +G + +V ++ + F+HF +R A+KA
Sbjct: 336 KVKVLFVRNLANSVTEEILEKAFSEYGNLERV--------KKLKDYAFIHFEERDGAVKA 387
Query: 258 LKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQ 291
L+ E++G+ +E AKP +R + Q
Sbjct: 388 LEELNGKELEGEPIEIVFAKPPDQKRKERKAQRQ 421
>UniRef100_UPI00003625C8 UPI00003625C8 UniRef100 entry
Length = 540
Score = 162 bits (409), Expect = 2e-38
Identities = 96/306 (31%), Positives = 163/306 (52%), Gaps = 11/306 (3%)
Query: 20 ALPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKEL 79
A P +E+++G I ++ E++L + G + ++R+M SG +GYAFV F TKE
Sbjct: 161 AQPTIGTEIFVGKIPRDLFEDELVPLFEKAGPIWDLRLMMDPLSGLNRGYAFVTFCTKEA 220
Query: 80 ASQAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIEL 138
A QA++ NN+E + GK I S A +RLF+G+IPK T E + + + V G+ + L
Sbjct: 221 AQQAVKLCNNNEIRPGKHIGVCISVANNRLFVGSIPKSKTKEQIIEEFSKVTEGLNDVIL 280
Query: 139 LKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEP-RNSESSAVS 197
+NRGF F+EY +H A +R+++ + K+ N TV WA+P + + ++
Sbjct: 281 YLQPVDKKKNRGFCFLEYEDHKTAAQARRRLMSGKVKVWGNLVTVEWADPLEDPDPEVMA 340
Query: 198 QVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKA 257
+VK ++V+NL ++T++ L+K F GK+ +V ++ + F+HF +R A+KA
Sbjct: 341 KVKVLFVRNLASSVTEELLEKAFSQFGKLERV--------KKLKDYAFIHFEERDGAVKA 392
Query: 258 LKNTEKYEIDGKNLECSLAKPEADQRSSGTSNSQKPVVLPTYPHRLGYGMVGGAYGGIGA 317
L + +++G+++E AKP DQ+ ++ Y YG G
Sbjct: 393 LADLNGKDLEGEHIEIVFAKP-PDQKRKERKAQRQAAKTHMYDEYYYYGPPHMPPPSRGR 451
Query: 318 GYGAAG 323
G GA G
Sbjct: 452 GRGARG 457
>UniRef100_UPI00004316F1 UPI00004316F1 UniRef100 entry
Length = 428
Score = 161 bits (408), Expect = 2e-38
Identities = 86/264 (32%), Positives = 149/264 (55%), Gaps = 10/264 (3%)
Query: 22 PPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELAS 81
P EV+ G I ++ E++L + G++ ++R+M +G +GYAF+ F +E A
Sbjct: 159 PGTGCEVFCGKIPKDMYEDELIPLFEKCGKIWDLRLMMDPMAGCNRGYAFITFTNREAAQ 218
Query: 82 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 140
QA+ EL+N E K GK +K + S RLF+GNIPK E++ + + G+ + +
Sbjct: 219 QAVRELDNHEIKPGKNLKVNISVPNLRLFVGNIPKSKGKEEILEEFGKLTAGLTEVIIYS 278
Query: 141 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNS-ESSAVSQV 199
+NRGF F+EY +H A +++++S K+ V WA+P+ + +S+V
Sbjct: 279 SPDDKKKNRGFCFLEYESHKAASLAKRRLSTGRIKVWGCDIIVDWADPQEEPDEQTMSKV 338
Query: 200 KAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALK 259
+ +YVKNL ++ +++ LK+ FE +G I +V ++ + FVHF +R +A+KA+
Sbjct: 339 RVLYVKNLTQDCSEEKLKESFEQYGNIERV--------KKIKDYAFVHFEERDNAVKAMN 390
Query: 260 NTEKYEIDGKNLECSLAKPEADQR 283
EI G ++E SLAKP +D++
Sbjct: 391 ELNGKEIGGSHIEVSLAKPPSDKK 414
Score = 42.0 bits (97), Expect = 0.027
Identities = 26/92 (28%), Positives = 47/92 (50%), Gaps = 1/92 (1%)
Query: 188 PRNSESSAVSQVKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVH 247
P N E V+ +P+++ +D L LFE GKI + L + F+
Sbjct: 151 PPNWEGPTPGTGCEVFCGKIPKDMYEDELIPLFEKCGKIWDLRLMMDPMAGCNRGYAFIT 210
Query: 248 FSDRSSAMKALKNTEKYEI-DGKNLECSLAKP 278
F++R +A +A++ + +EI GKNL+ +++ P
Sbjct: 211 FTNREAAQQAVRELDNHEIKPGKNLKVNISVP 242
>UniRef100_Q8VHM5 Heterogeneous nuclear ribonucleoprotein R [Mus musculus]
Length = 632
Score = 161 bits (408), Expect = 2e-38
Identities = 97/307 (31%), Positives = 166/307 (53%), Gaps = 17/307 (5%)
Query: 22 PPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELAS 81
P +EV++G I ++ E++L + G + ++R+M SG+ +GYAF+ F KE A
Sbjct: 161 PGIGTEVFVGKIPRDLYEDELVPLFEKAGPIWDLRLMMDPLSGQNRGYAFITFCGKEAAQ 220
Query: 82 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 140
+A++ ++ E + GK + S A +RLF+G+IPK T E++ + + V G++ + L
Sbjct: 221 EAVKLCDSYEIRPGKHLGVCISVANNRLFVGSIPKNKTKENILEEFSKVTEGLVDVILYH 280
Query: 141 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEP-RNSESSAVSQV 199
+NRGF F+EY +H A +R+++ + K+ N TV WA+P + +++V
Sbjct: 281 QPDDKKKNRGFCFLEYEDHKSAAQARRRLMSGKVKVWGNVVTVEWADPVEEPDPEVMAKV 340
Query: 200 KAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALK 259
K ++V+NL +T++ L+K F GK+ +V ++ + FVHF DR +A+KA+
Sbjct: 341 KVLFVRNLATTVTEEILEKSFSEFGKLERV--------KKLKDYAFVHFEDRGAAVKAMD 392
Query: 260 NTEKYEIDGKNLECSLAKP---EADQRSSGTSNSQKPVVLPTY---PHRLGYGMVGGAYG 313
EI+G+ +E LAKP + +R + S+ Y P R+ M G G
Sbjct: 393 EMNGKEIEGEEIEIVLAKPPDKKRKERQAARQASRSTAYEDYYYHPPPRMPPPMRGRGRG 452
Query: 314 GIGAGYG 320
G G GYG
Sbjct: 453 GRG-GYG 458
>UniRef100_Q811S1 Heterogeneous nuclear ribonucleoprotein R [Rattus norvegicus]
Length = 632
Score = 161 bits (408), Expect = 2e-38
Identities = 97/307 (31%), Positives = 166/307 (53%), Gaps = 17/307 (5%)
Query: 22 PPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELAS 81
P +EV++G I ++ E++L + G + ++R+M SG+ +GYAF+ F KE A
Sbjct: 161 PGIGTEVFVGKIPRDLYEDELVPLFEEAGPIWDLRLMMDPLSGQNRGYAFITFCGKEAAQ 220
Query: 82 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 140
+A++ ++ E + GK + S A +RLF+G+IPK T E++ + + V G++ + L
Sbjct: 221 EAVKLCDSYEIRPGKHLGVCISVANNRLFVGSIPKNKTKENILEEFSKVTEGLVDVILYH 280
Query: 141 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEP-RNSESSAVSQV 199
+NRGF F+EY +H A +R+++ + K+ N TV WA+P + +++V
Sbjct: 281 QPDDKKKNRGFCFLEYEDHKSAAQARRRLMSGKVKVWGNVVTVEWADPVEEPDPEVMAKV 340
Query: 200 KAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALK 259
K ++V+NL +T++ L+K F GK+ +V ++ + FVHF DR +A+KA+
Sbjct: 341 KVLFVRNLATTVTEEILEKSFSEFGKLERV--------KKLKDYAFVHFEDRGAAVKAMD 392
Query: 260 NTEKYEIDGKNLECSLAKP---EADQRSSGTSNSQKPVVLPTY---PHRLGYGMVGGAYG 313
EI+G+ +E LAKP + +R + S+ Y P R+ M G G
Sbjct: 393 EMNGKEIEGEEIEIVLAKPPDKKRKERQAARQASRSTAYEDYYYHPPPRMPPPMRGRGRG 452
Query: 314 GIGAGYG 320
G G GYG
Sbjct: 453 GRG-GYG 458
>UniRef100_Q99KG1 Hnrpr protein [Mus musculus]
Length = 601
Score = 161 bits (408), Expect = 2e-38
Identities = 97/307 (31%), Positives = 166/307 (53%), Gaps = 17/307 (5%)
Query: 22 PPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELAS 81
P +EV++G I ++ E++L + G + ++R+M SG+ +GYAF+ F KE A
Sbjct: 130 PGIGTEVFVGKIPRDLYEDELVPLFEKAGPIWDLRLMMDPLSGQNRGYAFITFCGKEAAQ 189
Query: 82 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 140
+A++ ++ E + GK + S A +RLF+G+IPK T E++ + + V G++ + L
Sbjct: 190 EAVKLCDSYEIRPGKHLGVCISVANNRLFVGSIPKNKTKENILEEFSKVTEGLVDVILYH 249
Query: 141 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEP-RNSESSAVSQV 199
+NRGF F+EY +H A +R+++ + K+ N TV WA+P + +++V
Sbjct: 250 QPDDKKKNRGFCFLEYEDHKSAAQARRRLMSGKVKVWGNVVTVEWADPVEEPDPEVMAKV 309
Query: 200 KAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALK 259
K ++V+NL +T++ L+K F GK+ +V ++ + FVHF DR +A+KA+
Sbjct: 310 KVLFVRNLATTVTEEILEKSFSEFGKLERV--------KKLKDYAFVHFEDRGAAVKAMD 361
Query: 260 NTEKYEIDGKNLECSLAKP---EADQRSSGTSNSQKPVVLPTY---PHRLGYGMVGGAYG 313
EI+G+ +E LAKP + +R + S+ Y P R+ M G G
Sbjct: 362 EMNGKEIEGEEIEIVLAKPPDKKRKERQAARQASRSTAYEDYYYHPPPRMPPPMRGRGRG 421
Query: 314 GIGAGYG 320
G G GYG
Sbjct: 422 GRG-GYG 427
>UniRef100_Q9VDI8 CG17838-PD, isoform D [Drosophila melanogaster]
Length = 707
Score = 161 bits (408), Expect = 2e-38
Identities = 84/265 (31%), Positives = 151/265 (56%), Gaps = 10/265 (3%)
Query: 21 LPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELA 80
+P + EV+ G I ++ E++L ++ G + ++R+M +G +GYAFV F +E A
Sbjct: 201 VPGNGCEVFCGKIPKDMYEDELIPLFENCGIIWDLRLMMDPMTGTNRGYAFVTFTNREAA 260
Query: 81 SQAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELL 139
A+ +L+N E K GK +K + S RLF+GNIPK +++ + + G+ + +
Sbjct: 261 VNAVRQLDNHEIKPGKCLKINISVPNLRLFVGNIPKSKGKDEILEEFGKLTAGLYEVIIY 320
Query: 140 KDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNS-ESSAVSQ 198
+NRGF F+EY +H A +++++ K+ V WA+P+ + +S+
Sbjct: 321 SSPDDKKKNRGFCFLEYESHKAASLAKRRLGTGRIKVWGCDIIVDWADPQEEPDEQTMSK 380
Query: 199 VKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKAL 258
VK +YV+NL +++++D LK+ FE +GK+ +V ++ + F+HF DR SA++A+
Sbjct: 381 VKVLYVRNLTQDVSEDKLKEQFEQYGKVERV--------KKIKDYAFIHFEDRDSAVEAM 432
Query: 259 KNTEKYEIDGKNLECSLAKPEADQR 283
+ EI N+E SLAKP +D++
Sbjct: 433 RGLNGKEIGASNIEVSLAKPPSDKK 457
>UniRef100_Q8IN48 CG17838-PE, isoform E [Drosophila melanogaster]
Length = 571
Score = 161 bits (408), Expect = 2e-38
Identities = 84/265 (31%), Positives = 151/265 (56%), Gaps = 10/265 (3%)
Query: 21 LPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELA 80
+P + EV+ G I ++ E++L ++ G + ++R+M +G +GYAFV F +E A
Sbjct: 201 VPGNGCEVFCGKIPKDMYEDELIPLFENCGIIWDLRLMMDPMTGTNRGYAFVTFTNREAA 260
Query: 81 SQAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELL 139
A+ +L+N E K GK +K + S RLF+GNIPK +++ + + G+ + +
Sbjct: 261 VNAVRQLDNHEIKPGKCLKINISVPNLRLFVGNIPKSKGKDEILEEFGKLTAGLYEVIIY 320
Query: 140 KDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNS-ESSAVSQ 198
+NRGF F+EY +H A +++++ K+ V WA+P+ + +S+
Sbjct: 321 SSPDDKKKNRGFCFLEYESHKAASLAKRRLGTGRIKVWGCDIIVDWADPQEEPDEQTMSK 380
Query: 199 VKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKAL 258
VK +YV+NL +++++D LK+ FE +GK+ +V ++ + F+HF DR SA++A+
Sbjct: 381 VKVLYVRNLTQDVSEDKLKEQFEQYGKVERV--------KKIKDYAFIHFEDRDSAVEAM 432
Query: 259 KNTEKYEIDGKNLECSLAKPEADQR 283
+ EI N+E SLAKP +D++
Sbjct: 433 RGLNGKEIGASNIEVSLAKPPSDKK 457
>UniRef100_Q8IN47 CG17838-PA, isoform A [Drosophila melanogaster]
Length = 563
Score = 161 bits (408), Expect = 2e-38
Identities = 84/265 (31%), Positives = 151/265 (56%), Gaps = 10/265 (3%)
Query: 21 LPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELA 80
+P + EV+ G I ++ E++L ++ G + ++R+M +G +GYAFV F +E A
Sbjct: 193 VPGNGCEVFCGKIPKDMYEDELIPLFENCGIIWDLRLMMDPMTGTNRGYAFVTFTNREAA 252
Query: 81 SQAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELL 139
A+ +L+N E K GK +K + S RLF+GNIPK +++ + + G+ + +
Sbjct: 253 VNAVRQLDNHEIKPGKCLKINISVPNLRLFVGNIPKSKGKDEILEEFGKLTAGLYEVIIY 312
Query: 140 KDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNS-ESSAVSQ 198
+NRGF F+EY +H A +++++ K+ V WA+P+ + +S+
Sbjct: 313 SSPDDKKKNRGFCFLEYESHKAASLAKRRLGTGRIKVWGCDIIVDWADPQEEPDEQTMSK 372
Query: 199 VKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKAL 258
VK +YV+NL +++++D LK+ FE +GK+ +V ++ + F+HF DR SA++A+
Sbjct: 373 VKVLYVRNLTQDVSEDKLKEQFEQYGKVERV--------KKIKDYAFIHFEDRDSAVEAM 424
Query: 259 KNTEKYEIDGKNLECSLAKPEADQR 283
+ EI N+E SLAKP +D++
Sbjct: 425 RGLNGKEIGASNIEVSLAKPPSDKK 449
>UniRef100_Q7KSA1 CG17838-PF, isoform F [Drosophila melanogaster]
Length = 529
Score = 161 bits (408), Expect = 2e-38
Identities = 84/265 (31%), Positives = 151/265 (56%), Gaps = 10/265 (3%)
Query: 21 LPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELA 80
+P + EV+ G I ++ E++L ++ G + ++R+M +G +GYAFV F +E A
Sbjct: 159 VPGNGCEVFCGKIPKDMYEDELIPLFENCGIIWDLRLMMDPMTGTNRGYAFVTFTNREAA 218
Query: 81 SQAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELL 139
A+ +L+N E K GK +K + S RLF+GNIPK +++ + + G+ + +
Sbjct: 219 VNAVRQLDNHEIKPGKCLKINISVPNLRLFVGNIPKSKGKDEILEEFGKLTAGLYEVIIY 278
Query: 140 KDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNS-ESSAVSQ 198
+NRGF F+EY +H A +++++ K+ V WA+P+ + +S+
Sbjct: 279 SSPDDKKKNRGFCFLEYESHKAASLAKRRLGTGRIKVWGCDIIVDWADPQEEPDEQTMSK 338
Query: 199 VKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKAL 258
VK +YV+NL +++++D LK+ FE +GK+ +V ++ + F+HF DR SA++A+
Sbjct: 339 VKVLYVRNLTQDVSEDKLKEQFEQYGKVERV--------KKIKDYAFIHFEDRDSAVEAM 390
Query: 259 KNTEKYEIDGKNLECSLAKPEADQR 283
+ EI N+E SLAKP +D++
Sbjct: 391 RGLNGKEIGASNIEVSLAKPPSDKK 415
>UniRef100_Q86NT3 AT01548p [Drosophila melanogaster]
Length = 626
Score = 161 bits (408), Expect = 2e-38
Identities = 84/265 (31%), Positives = 151/265 (56%), Gaps = 10/265 (3%)
Query: 21 LPPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELA 80
+P + EV+ G I ++ E++L ++ G + ++R+M +G +GYAFV F +E A
Sbjct: 120 VPGNGCEVFCGKIPKDMYEDELIPLFENCGIIWDLRLMMDPMTGTNRGYAFVTFTNREAA 179
Query: 81 SQAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELL 139
A+ +L+N E K GK +K + S RLF+GNIPK +++ + + G+ + +
Sbjct: 180 VNAVRQLDNHEIKPGKCLKINISVPNLRLFVGNIPKSKGKDEILEEFGKLTAGLYEVIIY 239
Query: 140 KDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEPRNS-ESSAVSQ 198
+NRGF F+EY +H A +++++ K+ V WA+P+ + +S+
Sbjct: 240 SSPDDKKKNRGFCFLEYESHKAASLAKRRLGTGRIKVWGCDIIVDWADPQEEPDEQTMSK 299
Query: 199 VKAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKAL 258
VK +YV+NL +++++D LK+ FE +GK+ +V ++ + F+HF DR SA++A+
Sbjct: 300 VKVLYVRNLTQDVSEDKLKEQFEQYGKVERV--------KKIKDYAFIHFEDRDSAVEAM 351
Query: 259 KNTEKYEIDGKNLECSLAKPEADQR 283
+ EI N+E SLAKP +D++
Sbjct: 352 RGLNGKEIGASNIEVSLAKPPSDKK 376
>UniRef100_UPI00003AC553 UPI00003AC553 UniRef100 entry
Length = 510
Score = 161 bits (407), Expect = 3e-38
Identities = 96/307 (31%), Positives = 166/307 (53%), Gaps = 17/307 (5%)
Query: 22 PPHSSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGERKGYAFVAFKTKELAS 81
P +EV++G I ++ E++L + G + ++R+M SG+ +GYAF+ F +K+ A
Sbjct: 56 PGIGTEVFVGKIPRDLYEDELVPLFEKAGPIWDLRLMMDPLSGQNRGYAFITFCSKDAAQ 115
Query: 82 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 140
+A++ +N E + GK + S A +RLF+G+IPK T E++ + V G++ + L
Sbjct: 116 EAVKLCDNYEIRPGKHLGVCISVANNRLFVGSIPKNKTKENILEEFNKVTEGLVDVILYH 175
Query: 141 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWAEP-RNSESSAVSQV 199
+NRGF F+EY +H A +R+++ + K+ N TV WA+P + +++V
Sbjct: 176 QPDDKKKNRGFCFLEYEDHKSAAQARRRLMSGKVKVWGNVVTVEWADPVEEPDPEVMAKV 235
Query: 200 KAVYVKNLPENITQDSLKKLFEHHGKITKVVLPPAKSGQEKSRFGFVHFSDRSSAMKALK 259
K ++V+NL +T++ L+K F GK+ +V ++ + FVHF DR +A+KA+
Sbjct: 236 KVLFVRNLATTVTEEILEKSFSEFGKLERV--------KKLKDYAFVHFEDRGAAVKAMN 287
Query: 260 NTEKYEIDGKNLECSLAKP---EADQRSSGTSNSQKPVVLPTY---PHRLGYGMVGGAYG 313
EI+G+ +E LAKP + +R + S+ Y P R+ + G G
Sbjct: 288 EMNGKEIEGEEIEIVLAKPPDKKRKERQAARQASRSTAYEDYYYYPPPRMPPPIRGRGRG 347
Query: 314 GIGAGYG 320
G G GYG
Sbjct: 348 GRG-GYG 353
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.132 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 617,627,770
Number of Sequences: 2790947
Number of extensions: 25401416
Number of successful extensions: 120624
Number of sequences better than 10.0: 4627
Number of HSP's better than 10.0 without gapping: 2980
Number of HSP's successfully gapped in prelim test: 1656
Number of HSP's that attempted gapping in prelim test: 103398
Number of HSP's gapped (non-prelim): 13263
length of query: 370
length of database: 848,049,833
effective HSP length: 129
effective length of query: 241
effective length of database: 488,017,670
effective search space: 117612258470
effective search space used: 117612258470
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0010.2


