

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
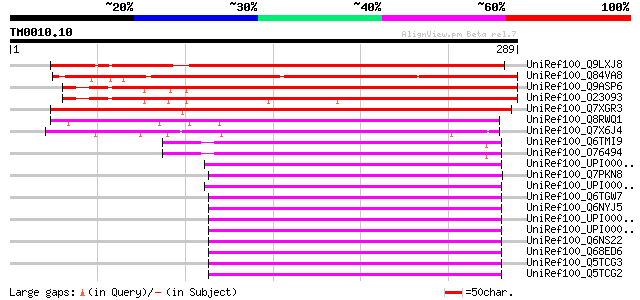
Query= TM0010.10
(289 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LXJ8 Putative RNA-binding protein [Arabidopsis thali... 322 9e-87
UniRef100_Q84VA8 Putative RNA-binding protein [Oryza sativa] 313 4e-84
UniRef100_Q9ASP6 AT4g00830/A_TM018A10_14 [Arabidopsis thaliana] 290 4e-77
UniRef100_O23093 A_TM018A10.14 protein [Arabidopsis thaliana] 271 1e-71
UniRef100_Q7XGR3 Putative RNA-binding protein [Oryza sativa] 242 7e-63
UniRef100_Q8RWQ1 At2g44720/F16B22.21 [Arabidopsis thaliana] 132 1e-29
UniRef100_Q7X6J4 OSJNBb0103I08.18 protein [Oryza sativa] 125 2e-27
UniRef100_Q6TMI9 RNA-binding protein [Dictyostelium discoideum] 122 1e-26
UniRef100_O76494 Putative RNA binding protein RNP [Dictyostelium... 121 2e-26
UniRef100_UPI0000292EBE UPI0000292EBE UniRef100 entry 120 4e-26
UniRef100_Q7PKN8 ENSANGP00000023817 [Anopheles gambiae str. PEST] 117 5e-25
UniRef100_UPI00003625C8 UPI00003625C8 UniRef100 entry 115 1e-24
UniRef100_Q6TGW7 NS1-associated protein 1 [Brachydanio rerio] 115 1e-24
UniRef100_Q6NYJ5 NS1-associated protein 1 [Brachydanio rerio] 115 1e-24
UniRef100_UPI00003AD068 UPI00003AD068 UniRef100 entry 114 2e-24
UniRef100_UPI00003AD067 UPI00003AD067 UniRef100 entry 114 2e-24
UniRef100_Q6NS22 MGC78820 protein [Xenopus laevis] 114 2e-24
UniRef100_Q68ED6 OTTHUMP00000016817 [Homo sapiens] 114 2e-24
UniRef100_Q5TCG3 OTTHUMP00000040572 [Homo sapiens] 114 2e-24
UniRef100_Q5TCG2 OTTHUMP00000016815 [Homo sapiens] 114 2e-24
>UniRef100_Q9LXJ8 Putative RNA-binding protein [Arabidopsis thaliana]
Length = 471
Score = 322 bits (824), Expect = 9e-87
Identities = 160/259 (61%), Positives = 203/259 (77%), Gaps = 11/259 (4%)
Query: 24 SEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEE 83
SE +S+E+VD DGDND EE +E EEVE EE VE+EE+EE+EEE EEE E EE EEE
Sbjct: 9 SEAHDSMESEERVDLDGDNDPEEILE-EEVEYEE-VEEEEIEEIEEEIEEEVEVEEEEEE 66
Query: 84 SKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGSEVYIGGIANNVSEEDLRVFCQSVGE 143
EEE EKK+H ELLALPPHGSEVY+GGI + +E DL+ FC S+GE
Sbjct: 67 EDAVATEEEE---------EKKRHVELLALPPHGSEVYLGGIPTDATEGDLKGFCGSIGE 117
Query: 144 VSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLFIG 203
V+EVRIM+ K+SG+GKGYAFV F++K+LA++AI+ LNN++F+GK+IKCS++QAKHRLF+G
Sbjct: 118 VTEVRIMREKDSGDGKGYAFVTFRSKDLAAEAIDTLNNTDFRGKRIKCSTTQAKHRLFLG 177
Query: 204 NIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSN 263
N+P+ W D+KK +GPGV +EL K+ Q+ GRNRGFAFIEYYNHACAEYS+QKMSN
Sbjct: 178 NVPRNWMESDIKKAANRIGPGVQIVELPKEPQNMGRNRGFAFIEYYNHACAEYSKQKMSN 237
Query: 264 SNFKLENNAPTVSWADPRN 282
+FKL++NAPTVSWA+ R+
Sbjct: 238 PSFKLDDNAPTVSWAESRS 256
>UniRef100_Q84VA8 Putative RNA-binding protein [Oryza sativa]
Length = 476
Score = 313 bits (801), Expect = 4e-84
Identities = 160/273 (58%), Positives = 216/273 (78%), Gaps = 16/273 (5%)
Query: 25 EPEKPTDSDEQVDFDGDNDQE---EEIEYEEVEEE---EEVEDEE--VEEVEEEEEEEEE 76
EPEK S+E ++FD D ++E EEIEYEE+EEE EEVE++E VEEVEE +EEE+E
Sbjct: 27 EPEK---SEECLEFDDDEEEEVEEEEIEYEEIEEEIEEEEVEEDEDVVEEVEEVDEEEDE 83
Query: 77 EEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGSEVYIGGIANNVSEEDLRV 136
EE EEES ++G + + H D EK KHAELLALPPHGSEVY+GGI+++VS EDL+
Sbjct: 84 EE--EEESDETEGVSKTKGVHQKDVTEKGKHAELLALPPHGSEVYVGGISSDVSSEDLKR 141
Query: 137 FCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQA 196
C+ VGEV EVR+M+ K+ + +GYAFV F+TK LA +A++ELNN++ KGK+I+ SSSQA
Sbjct: 142 LCEPVGEVVEVRMMRGKD--DSRGYAFVNFRTKGLALKAVKELNNAKLKGKRIRVSSSQA 199
Query: 197 KHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEY 256
K++LFIGN+P WT +D +KVV +VGPGV+ +L+K + S+ RNRG+ F+EYYNHACAEY
Sbjct: 200 KNKLFIGNVPHSWTDDDFRKVVEEVGPGVLKADLMK-VSSANRNRGYGFVEYYNHACAEY 258
Query: 257 SRQKMSNSNFKLENNAPTVSWADPRNSESSAVS 289
+RQ+MS+ FKL++NAPTVSWADP+N++S++ S
Sbjct: 259 ARQEMSSPTFKLDSNAPTVSWADPKNNDSASTS 291
Score = 42.4 bits (98), Expect = 0.014
Identities = 20/73 (27%), Positives = 37/73 (50%), Gaps = 1/73 (1%)
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
VY+ + NV++ L+ + GE+ +V ++ G Y FV FK + +A +A++
Sbjct: 296 VYVKNLPKNVTQAQLKRLFEHHGEIEKV-VLPPSRGGHDNRYGFVHFKDRSMAMRALQNT 354
Query: 180 NNSEFKGKKIKCS 192
E G+ + CS
Sbjct: 355 ERYELDGQVLDCS 367
>UniRef100_Q9ASP6 AT4g00830/A_TM018A10_14 [Arabidopsis thaliana]
Length = 495
Score = 290 bits (741), Expect = 4e-77
Identities = 153/292 (52%), Positives = 206/292 (70%), Gaps = 42/292 (14%)
Query: 31 DSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEE----------EEEEV 80
D+D++VDF EE Y E+E+E VE+E+VEE EEEEEE++ EE EV
Sbjct: 6 DNDDRVDF-------EEGSYSEMEDE--VEEEQVEEYEEEEEEDDDDDDVGNQNAEEREV 56
Query: 81 EEESKPSDGE-----EEMRVDHTN-----------------DEVEKKKHAELLALPPHGS 118
E+ G+ EE+ D N D+ +++K++ LL+LPPHGS
Sbjct: 57 EDYGDTKGGDMEDVQEEIAEDDDNHIDIETADDDEKPPSPIDDEDREKYSHLLSLPPHGS 116
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
EV+IGG+ +V EEDLR C+ +GE+ EVR+MK ++SG+ KGYAFVAFKTK++A +AIEE
Sbjct: 117 EVFIGGLPRDVGEEDLRDLCEEIGEIFEVRLMKDRDSGDSKGYAFVAFKTKDVAQKAIEE 176
Query: 179 LNNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSG 238
L++ EFKGK I+CS S+ K+RLFIGNIPK WT ++ +KV+ DVGPGV +IEL+KD ++
Sbjct: 177 LHSKEFKGKTIRCSLSETKNRLFIGNIPKNWTEDEFRKVIEDVGPGVENIELIKDPTNTT 236
Query: 239 RNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADPRNS-ESSAVS 289
RNRGFAF+ YYN+ACA+YSRQKM +SNFKLE NAPTV+WADP++S E SA +
Sbjct: 237 RNRGFAFVLYYNNACADYSRQKMIDSNFKLEGNAPTVTWADPKSSPEHSAAA 288
>UniRef100_O23093 A_TM018A10.14 protein [Arabidopsis thaliana]
Length = 521
Score = 271 bits (693), Expect = 1e-71
Identities = 153/318 (48%), Positives = 206/318 (64%), Gaps = 68/318 (21%)
Query: 31 DSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEE----------EEEEV 80
D+D++VDF EE Y E+E+E VE+E+VEE EEEEEE++ EE EV
Sbjct: 6 DNDDRVDF-------EEGSYSEMEDE--VEEEQVEEYEEEEEEDDDDDDVGNQNAEEREV 56
Query: 81 EEESKPSDG-----EEEMRVDHTN-----------------DEVEKKKHAELLALPPHGS 118
E+ G +EE+ D N D+ +++K++ LL+LPPHGS
Sbjct: 57 EDYGDTKGGDMEDVQEEIAEDDDNHIDIETADDDEKPPSPIDDEDREKYSHLLSLPPHGS 116
Query: 119 EVYIGGIANNVSEEDLRVFCQSVGEVSE--------------VRIMKAKESGEGKGYAFV 164
EV+IGG+ +V EEDLR C+ +GE+ E VR+MK ++SG+ KGYAFV
Sbjct: 117 EVFIGGLPRDVGEEDLRDLCEEIGEIFEVRTAIIFVFHDILFVRLMKDRDSGDSKGYAFV 176
Query: 165 AFKTKELASQAIEELNNSEFK------------GKKIKCSSSQAKHRLFIGNIPKKWTVE 212
AFKTK++A +AIEEL++ EFK GK I+CS S+ K+RLFIGNIPK WT +
Sbjct: 177 AFKTKDVAQKAIEELHSKEFKASSTANCSLSLSGKTIRCSLSETKNRLFIGNIPKNWTED 236
Query: 213 DMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNA 272
+ +KV+ DVGPGV +IEL+KD ++ RNRGFAF+ YYN+ACA+YSRQKM +SNFKLE NA
Sbjct: 237 EFRKVIEDVGPGVENIELIKDPTNTTRNRGFAFVLYYNNACADYSRQKMIDSNFKLEGNA 296
Query: 273 PTVSWADPRNS-ESSAVS 289
PTV+WADP++S E SA +
Sbjct: 297 PTVTWADPKSSPEHSAAA 314
>UniRef100_Q7XGR3 Putative RNA-binding protein [Oryza sativa]
Length = 472
Score = 242 bits (618), Expect = 7e-63
Identities = 118/276 (42%), Positives = 180/276 (64%), Gaps = 13/276 (4%)
Query: 24 SEPEKPTDSDEQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEEVEEEEEEEEEEEEVEEE 83
++ ++ + +EQVD +GD+D ++ + E+ +D E E+ +E + E + E E
Sbjct: 2 ADRQQSEEPEEQVDLEGDDDIMDDDDGYRRHRREDSDDPEEEDPDERQGEGDGRREDAEG 61
Query: 84 SKPSDGEEEMRVDH-------------TNDEVEKKKHAELLALPPHGSEVYIGGIANNVS 130
+ G+ + DE EK+K ELLALPP GSEV+IGG+ + +
Sbjct: 62 PGGAGGDPAAGGEGGADVMDKVGGDAGPEDEEEKRKWDELLALPPQGSEVFIGGLPRDTT 121
Query: 131 EEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGKKIK 190
E+DL C++ GE+SEVR+MK KE+ E KG+AFV F K+ A +AIE+L++ E KG+ ++
Sbjct: 122 EDDLHELCEAFGEISEVRLMKDKETKENKGFAFVTFTGKDGAQRAIEDLHDKEHKGRTLR 181
Query: 191 CSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYN 250
CS SQAKHRLF+GN+PK + ++++ ++ GPGV++IE+ KDL RNRGF F+EYYN
Sbjct: 182 CSLSQAKHRLFVGNVPKGLSEDELRNIIQGKGPGVVNIEMFKDLHDPSRNRGFLFVEYYN 241
Query: 251 HACAEYSRQKMSNSNFKLENNAPTVSWADPRNSESS 286
HACA+Y++QK+S NFK++ + TVSWA+P+ S SS
Sbjct: 242 HACADYAKQKLSAPNFKVDGSQLTVSWAEPKGSSSS 277
Score = 35.4 bits (80), Expect = 1.8
Identities = 19/76 (25%), Positives = 41/76 (53%), Gaps = 1/76 (1%)
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
+Y+ + N S+E ++ + GEV++V + AK +G + + FV F + A +A++
Sbjct: 289 IYVKNLPENASKEKIKEIFEIHGEVTKVVLPPAK-AGNKRDFGFVHFAERSSALKAVKGS 347
Query: 180 NNSEFKGKKIKCSSSQ 195
E G+ ++ S ++
Sbjct: 348 EKYEIDGQVLEVSMAK 363
>UniRef100_Q8RWQ1 At2g44720/F16B22.21 [Arabidopsis thaliana]
Length = 809
Score = 132 bits (332), Expect = 1e-29
Identities = 93/282 (32%), Positives = 149/282 (51%), Gaps = 26/282 (9%)
Query: 24 SEPEKPTDS---DEQVDFDGDNDQEEEIEYEEVE-EEEEVEDEEVEEVEEEEEEEEEEEE 79
+E E+ D DE++D D + + E EY E EE E+ E+ E V EE EE EEE E
Sbjct: 96 TEVEESVDDFGKDERLDLDDNEPEYEAEEYGGEEFEERELGQEDHELVNEEGEELEEEIE 155
Query: 80 VEEES-----KPSDGEEEMRVDHTNDE--VEKKKHAELLALPPHGS-------------E 119
VEEE+ + DG E++ + +D+ +E+ KH E + + E
Sbjct: 156 VEEEAGEFADEIGDGAEDLDSEDDDDDHAIEEVKHGETVDVEEEEHHDVLHERRKRKEFE 215
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
+++G + SEEDL+ VGEV+EVRI+K ++ + KG AF+ F T E A +A++EL
Sbjct: 216 IFVGSLDKGASEEDLKKVFGHVGEVTEVRILKNPQTKKSKGSAFLRFATVEQAKRAVKEL 275
Query: 180 NNSEFKGKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVG-PGVISIELLKDLQSSG 238
+ GKK ++SQ LF+GNI K WT E +++ + G + I L++D +
Sbjct: 276 KSPMINGKKCGVTASQDNDTLFVGNICKIWTPEALREKLKHYGVENMDDITLVEDSNNVN 335
Query: 239 RNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPT-VSWAD 279
NRG+AF+E+ + + A + +++ + P VS+ D
Sbjct: 336 MNRGYAFLEFSSRSDAMDAHKRLVKKDVMFGVEKPAKVSFTD 377
Score = 44.7 bits (104), Expect = 0.003
Identities = 58/254 (22%), Positives = 109/254 (42%), Gaps = 35/254 (13%)
Query: 14 AQNPESPEGNSEPE---KPTDSD----EQVDFDGDNDQEEEIEYEEVEEEEEVEDEEVEE 66
AQNP + E + + SD ++V + D EEE + + + ++D E
Sbjct: 25 AQNPHVESSHDESVNIGELSGSDALEAKEVTPEVDKTVEEENPLDVPKSSDSIDDSEAAA 84
Query: 67 VEEEEEEEEEEEEVEEESKPSDGEEEMRVDHTNDEVEKKKHAELLALPPHGSEVYIGGIA 126
+ ++E EVEE +E + +D E E ++ +G E +
Sbjct: 85 NPHVDVPSKKETEVEESVDDFGKDERLDLDDNEPEYEAEE---------YGGEEF---EE 132
Query: 127 NNVSEEDLRVFCQSVGEVSEVRIMKAKESGE-----GKGYAFVAFKTKELASQAIEELNN 181
+ +ED + + GE E I +E+GE G G + + + AIEE+ +
Sbjct: 133 RELGQEDHELVNEE-GEELEEEIEVEEEAGEFADEIGDGAEDLDSEDDD-DDHAIEEVKH 190
Query: 182 SEFKGKK-------IKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLKDL 234
E + + + + +F+G++ K + ED+KKV VG V + +LK+
Sbjct: 191 GETVDVEEEEHHDVLHERRKRKEFEIFVGSLDKGASEEDLKKVFGHVGE-VTEVRILKNP 249
Query: 235 QSSGRNRGFAFIEY 248
Q+ +++G AF+ +
Sbjct: 250 QTK-KSKGSAFLRF 262
>UniRef100_Q7X6J4 OSJNBb0103I08.18 protein [Oryza sativa]
Length = 774
Score = 125 bits (313), Expect = 2e-27
Identities = 88/274 (32%), Positives = 149/274 (54%), Gaps = 17/274 (6%)
Query: 21 EGNSEPEKPTDSDEQVDFDGDNDQEEE--IEYEEVE-EEEEVEDEEVEEVEEEEEE--EE 75
EG ++ D E+++F+ + + EEE ++Y+E + E+ E + E+ +EV E E+ EE
Sbjct: 82 EGTTKETYEEDKGERLEFEDEPEYEEEAAVDYDEKDLEQYEEQYEDGDEVVEYTEDVIEE 141
Query: 76 EEEEVEEESKPSD-----GEEEMRVDHTNDEVEKKKHAELLALPPHGSE--VYIGGIANN 128
E + V+EE D G E +H N +VE ++H E++ E V++GG+ +
Sbjct: 142 ETDMVDEELDGGDDGEGEGYENAEEEH-NVDVEDEEHHEMVKEHRKRKEFEVFVGGLDKD 200
Query: 129 VSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGKK 188
+E DLR VGE++EVR+M + + KG+AF+ + T E A +A+ EL N +GK+
Sbjct: 201 ATESDLRKVFGEVGEITEVRLMMNPVTKKNKGFAFLRYATVEQARRAVSELKNPSVRGKQ 260
Query: 189 IKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVG-PGVISIELLKDLQSSGRNRGFAFIE 247
+ S LF+GNI K WT E +K+ + G + L++D + G NRG+A +E
Sbjct: 261 CGVAPSHDNDTLFVGNICKTWTKEHLKEKLKSYGVENFDDLLLVEDSNNPGMNRGYALLE 320
Query: 248 YYN--HACAEYSRQKMSNSNFKLENNAPTVSWAD 279
+ A + R + + F ++ +A VS+AD
Sbjct: 321 FSTRPEAMDAFRRLQKRDVVFGVDRSA-KVSFAD 353
>UniRef100_Q6TMI9 RNA-binding protein [Dictyostelium discoideum]
Length = 551
Score = 122 bits (305), Expect = 1e-26
Identities = 67/197 (34%), Positives = 110/197 (55%), Gaps = 11/197 (5%)
Query: 88 DGEEEMRVDHTNDEVEKKKHAELLALPPHGSEVYIGGIANNVSEEDL-RVFCQ-SVGEVS 145
DG+ + +++ N+ + K + +E++ GG+ V+EE+L +F + S V
Sbjct: 69 DGDNNISINNNNNSISKSQRNA-------NNEIFFGGVNKTVNEEELLEIFNENSDNNVL 121
Query: 146 EVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLFIGNI 205
E+R+MK K +GE KG+ FV F + + I++LN KGK I+ S+ K +LFIGN+
Sbjct: 122 EIRLMKDKLTGESKGFGFVLFNDRSMCRNVIDKLNGKSIKGKIIEVKQSENKRKLFIGNL 181
Query: 206 PKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSN 265
PK + E + + G+ SI+ L +NRGFAFIEY +H A+ +R+ ++ S
Sbjct: 182 PKDLSKEQFISIFNEKTEGIESIDFLMSPDQPNKNRGFAFIEYQDHYLADTARRILTAST 241
Query: 266 FKLEN--NAPTVSWADP 280
KL + TV+W+DP
Sbjct: 242 VKLGDYITTLTVNWSDP 258
>UniRef100_O76494 Putative RNA binding protein RNP [Dictyostelium discoideum]
Length = 551
Score = 121 bits (303), Expect = 2e-26
Identities = 67/197 (34%), Positives = 110/197 (55%), Gaps = 11/197 (5%)
Query: 88 DGEEEMRVDHTNDEVEKKKHAELLALPPHGSEVYIGGIANNVSEEDL-RVFCQ-SVGEVS 145
DG+ + +++ N+ + K + +E++ GG+ V+EE+L +F + S V
Sbjct: 69 DGDNNISINNNNNSISKSQRNA-------NNEIFFGGVNKTVNEEELLEIFNENSDNNVL 121
Query: 146 EVRIMKAKESGEGKGYAFVAFKTKELASQAIEELNNSEFKGKKIKCSSSQAKHRLFIGNI 205
E+R+MK K +GE KG+ FV F + + I++LN KGK I+ S+ K +LFIGN+
Sbjct: 122 EIRLMKDKLTGESKGFGFVLFNDRSMCRNVIDKLNGKSIKGKIIEVKLSENKRKLFIGNL 181
Query: 206 PKKWTVEDMKKVVADVGPGVISIELLKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSN 265
PK + E + + G+ SI+ L +NRGFAFIEY +H A+ +R+ ++ S
Sbjct: 182 PKDLSKEQFISIFNEKTEGIESIDFLMSPDQPNKNRGFAFIEYQDHYLADTARRILTAST 241
Query: 266 FKLEN--NAPTVSWADP 280
KL + TV+W+DP
Sbjct: 242 VKLGDYITTLTVNWSDP 258
>UniRef100_UPI0000292EBE UPI0000292EBE UniRef100 entry
Length = 636
Score = 120 bits (301), Expect = 4e-26
Identities = 63/170 (37%), Positives = 100/170 (58%), Gaps = 1/170 (0%)
Query: 112 ALPPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKEL 171
A PP G+E+++G I ++ E++L + G + ++R+M SG +GYAFV F +KE
Sbjct: 156 AQPPVGTEIFVGKIPRDLFEDELVPLFEKAGPIWDLRLMMDPLSGLNRGYAFVTFCSKEA 215
Query: 172 ASQAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIEL 230
A +A++ NN E + GK+I S A +RLF+G+IPK T E + + + V G+ + L
Sbjct: 216 AQEAVKLCNNHEIRPGKQIGVCISVANNRLFVGSIPKSKTKEQIVEEFSKVTEGLSDVIL 275
Query: 231 LKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADP 280
Q +NRGF F+EY +H A +R+++ + K+ N TV WADP
Sbjct: 276 YLQPQDKSKNRGFCFLEYEDHKTAAQARRRLMSGKVKVWGNMVTVEWADP 325
Score = 40.8 bits (94), Expect = 0.042
Identities = 22/71 (30%), Positives = 40/71 (55%), Gaps = 8/71 (11%)
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
+++ +AN+V+EE L G + V+ +K YAF+ F+ ++ A +A+EEL
Sbjct: 340 LFVRNLANSVTEEILEKAFSEYGNLERVKKLK--------DYAFIHFEERDGAVKALEEL 391
Query: 180 NNSEFKGKKIK 190
N E +G+ I+
Sbjct: 392 NGKELEGEPIE 402
>UniRef100_Q7PKN8 ENSANGP00000023817 [Anopheles gambiae str. PEST]
Length = 443
Score = 117 bits (292), Expect = 5e-25
Identities = 58/169 (34%), Positives = 98/169 (57%), Gaps = 1/169 (0%)
Query: 114 PPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELAS 173
P +G EV+ G I ++ E++L + G++ ++R+M +G +GYAFV F +++ AS
Sbjct: 164 PGNGCEVFCGKIPKDMYEDELIPLFEKCGKIWDLRLMMDPMTGTNRGYAFVTFTSRDAAS 223
Query: 174 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 232
A+ ELN+ E + GKKI + S HRLF+GNIPK +++ + A PG++ + +
Sbjct: 224 NAVRELNDYEIRNGKKIGVTISFNNHRLFVGNIPKNRDRDELLEEFAKHAPGLVEVIIYS 283
Query: 233 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADPR 281
+NRGF F+EY +H A +++++ K+ N V WADP+
Sbjct: 284 SPDDKKKNRGFCFLEYESHKAASLAKRRLGTGRIKVWNCDIIVDWADPQ 332
Score = 36.6 bits (83), Expect = 0.79
Identities = 21/73 (28%), Positives = 38/73 (51%), Gaps = 8/73 (10%)
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
+Y+ + + SEE L+ + G V V+ +K YAFV F+ ++ A +A+++L
Sbjct: 346 LYVRNLTQDTSEEKLKESFEQFGRVERVKKIK--------DYAFVHFEDRDNAVKAMKDL 397
Query: 180 NNSEFKGKKIKCS 192
+ E G I+ S
Sbjct: 398 DGKEVGGSNIEVS 410
>UniRef100_UPI00003625C8 UPI00003625C8 UniRef100 entry
Length = 540
Score = 115 bits (289), Expect = 1e-24
Identities = 63/170 (37%), Positives = 98/170 (57%), Gaps = 1/170 (0%)
Query: 112 ALPPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKEL 171
A P G+E+++G I ++ E++L + G + ++R+M SG +GYAFV F TKE
Sbjct: 161 AQPTIGTEIFVGKIPRDLFEDELVPLFEKAGPIWDLRLMMDPLSGLNRGYAFVTFCTKEA 220
Query: 172 ASQAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIEL 230
A QA++ NN+E + GK I S A +RLF+G+IPK T E + + + V G+ + L
Sbjct: 221 AQQAVKLCNNNEIRPGKHIGVCISVANNRLFVGSIPKSKTKEQIIEEFSKVTEGLNDVIL 280
Query: 231 LKDLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADP 280
+NRGF F+EY +H A +R+++ + K+ N TV WADP
Sbjct: 281 YLQPVDKKKNRGFCFLEYEDHKTAAQARRRLMSGKVKVWGNLVTVEWADP 330
Score = 34.3 bits (77), Expect = 3.9
Identities = 18/71 (25%), Positives = 40/71 (55%), Gaps = 8/71 (11%)
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
+++ +A++V+EE L G++ V+ +K YAF+ F+ ++ A +A+ +L
Sbjct: 345 LFVRNLASSVTEELLEKAFSQFGKLERVKKLK--------DYAFIHFEERDGAVKALADL 396
Query: 180 NNSEFKGKKIK 190
N + +G+ I+
Sbjct: 397 NGKDLEGEHIE 407
>UniRef100_Q6TGW7 NS1-associated protein 1 [Brachydanio rerio]
Length = 558
Score = 115 bits (288), Expect = 1e-24
Identities = 61/168 (36%), Positives = 97/168 (57%), Gaps = 1/168 (0%)
Query: 114 PPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELAS 173
P G+E+++G I ++ E++L + G + ++R+M SG +GYAFV F TKE A
Sbjct: 159 PTIGTEIFVGKIPRDLFEDELVPLFEKAGPIWDLRLMMDPLSGLNRGYAFVTFCTKEAAQ 218
Query: 174 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 232
+A++ NN+E + GK I S A +RLF+G+IPK T + + + A V G+ + L
Sbjct: 219 KAVKLCNNNEIRPGKHIGVCISVANNRLFVGSIPKSKTKDQIVEEFAKVTEGLNDVILYH 278
Query: 233 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADP 280
+NRGF F+EY +H A +R+++ + K+ N TV WADP
Sbjct: 279 QPDDKKKNRGFCFLEYEDHKTAAQARRRLMSGKVKVWGNVVTVEWADP 326
Score = 34.7 bits (78), Expect = 3.0
Identities = 21/72 (29%), Positives = 43/72 (59%), Gaps = 10/72 (13%)
Query: 120 VYIGGIANNVSEEDL-RVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
+++ +A+ V+EE L + FCQ G++ V+ +K YAF+ F+ ++ A +A+ E
Sbjct: 341 LFVRNLASTVTEELLEKTFCQ-FGKLERVKKLK--------DYAFIHFEERDGAVKALAE 391
Query: 179 LNNSEFKGKKIK 190
L+ + +G+ I+
Sbjct: 392 LHGKDLEGEPIE 403
>UniRef100_Q6NYJ5 NS1-associated protein 1 [Brachydanio rerio]
Length = 560
Score = 115 bits (288), Expect = 1e-24
Identities = 61/168 (36%), Positives = 97/168 (57%), Gaps = 1/168 (0%)
Query: 114 PPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELAS 173
P G+E+++G I ++ E++L + G + ++R+M SG +GYAFV F TKE A
Sbjct: 159 PTIGTEIFVGKIPRDLFEDELVPLFEKAGPIWDLRLMMDPLSGLNRGYAFVTFCTKEAAQ 218
Query: 174 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 232
+A++ NN+E + GK I S A +RLF+G+IPK T + + + A V G+ + L
Sbjct: 219 KAVKLCNNNEIRPGKHIGVCISVANNRLFVGSIPKSKTKDQIVEEFAKVTEGLNDVILYH 278
Query: 233 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADP 280
+NRGF F+EY +H A +R+++ + K+ N TV WADP
Sbjct: 279 QPDDKKKNRGFCFLEYEDHKTAAQARRRLMSGKVKVWGNVVTVEWADP 326
Score = 34.7 bits (78), Expect = 3.0
Identities = 21/72 (29%), Positives = 43/72 (59%), Gaps = 10/72 (13%)
Query: 120 VYIGGIANNVSEEDL-RVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEE 178
+++ +A+ V+EE L + FCQ G++ V+ +K YAF+ F+ ++ A +A+ E
Sbjct: 341 LFVRNLASTVTEELLEKTFCQ-FGKLERVKKLK--------DYAFIHFEERDGAVKALAE 391
Query: 179 LNNSEFKGKKIK 190
L+ + +G+ I+
Sbjct: 392 LHGKDLEGEPIE 403
>UniRef100_UPI00003AD068 UPI00003AD068 UniRef100 entry
Length = 532
Score = 114 bits (286), Expect = 2e-24
Identities = 60/168 (35%), Positives = 96/168 (56%), Gaps = 1/168 (0%)
Query: 114 PPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELAS 173
P G+E+++G I ++ E++L + G + ++R+M +G +GYAFV F TKE A
Sbjct: 67 PSVGTEIFVGKIPRDLFEDELVPLFEKAGPIWDLRLMMDPLTGLNRGYAFVTFCTKEAAQ 126
Query: 174 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 232
+A++ NN E + GK I S A +RLF+G+IPK T E + + + V G+ + L
Sbjct: 127 EAVKLYNNHEIRSGKHIGVCISVANNRLFVGSIPKSKTKEQIVEEFSKVTEGLTDVILYH 186
Query: 233 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADP 280
+NRGF F+EY +H A +R+++ + K+ N TV WADP
Sbjct: 187 QPDDKKKNRGFCFLEYEDHKTAAQARRRLMSGKVKVWGNVVTVEWADP 234
Score = 38.1 bits (87), Expect = 0.27
Identities = 20/71 (28%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
+++ +AN V+EE L G++ V+ +K YAF+ F ++ A +A+EE+
Sbjct: 249 LFVRNLANTVTEEILEKAFSQFGKLERVKKLK--------DYAFIHFDERDGAVKAMEEM 300
Query: 180 NNSEFKGKKIK 190
N + +G+ I+
Sbjct: 301 NGKDLEGENIE 311
>UniRef100_UPI00003AD067 UPI00003AD067 UniRef100 entry
Length = 566
Score = 114 bits (286), Expect = 2e-24
Identities = 60/168 (35%), Positives = 96/168 (56%), Gaps = 1/168 (0%)
Query: 114 PPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELAS 173
P G+E+++G I ++ E++L + G + ++R+M +G +GYAFV F TKE A
Sbjct: 162 PSVGTEIFVGKIPRDLFEDELVPLFEKAGPIWDLRLMMDPLTGLNRGYAFVTFCTKEAAQ 221
Query: 174 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 232
+A++ NN E + GK I S A +RLF+G+IPK T E + + + V G+ + L
Sbjct: 222 EAVKLYNNHEIRSGKHIGVCISVANNRLFVGSIPKSKTKEQIVEEFSKVTEGLTDVILYH 281
Query: 233 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADP 280
+NRGF F+EY +H A +R+++ + K+ N TV WADP
Sbjct: 282 QPDDKKKNRGFCFLEYEDHKTAAQARRRLMSGKVKVWGNVVTVEWADP 329
Score = 38.1 bits (87), Expect = 0.27
Identities = 20/71 (28%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
+++ +AN V+EE L G++ V+ +K YAF+ F ++ A +A+EE+
Sbjct: 344 LFVRNLANTVTEEILEKAFSQFGKLERVKKLK--------DYAFIHFDERDGAVKAMEEM 395
Query: 180 NNSEFKGKKIK 190
N + +G+ I+
Sbjct: 396 NGKDLEGENIE 406
>UniRef100_Q6NS22 MGC78820 protein [Xenopus laevis]
Length = 624
Score = 114 bits (286), Expect = 2e-24
Identities = 60/168 (35%), Positives = 96/168 (56%), Gaps = 1/168 (0%)
Query: 114 PPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELAS 173
P G+E+++G I ++ E++L + G + ++R+M +G +GYAFV F TKE A
Sbjct: 162 PSVGTEIFVGKIPRDLFEDELVPLFEKAGSIWDLRLMMDPLTGLNRGYAFVTFCTKEAAQ 221
Query: 174 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 232
+A++ NN E + GK I S A +RLF+G+IPK T E + + + V G+ + L
Sbjct: 222 EAVKLYNNYEIRPGKHIGVCISVANNRLFVGSIPKSKTKEQIVEEFSKVTEGLTDVILYH 281
Query: 233 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADP 280
+NRGF F+EY +H A +R+++ + K+ N TV WADP
Sbjct: 282 QPDDKKKNRGFCFLEYEDHKTAAQARRRLMSGKVKVWGNVVTVEWADP 329
Score = 35.8 bits (81), Expect = 1.3
Identities = 20/71 (28%), Positives = 38/71 (53%), Gaps = 8/71 (11%)
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
+++ +AN V+EE L G++ V+ +K YAF+ F + A +A++E+
Sbjct: 344 LFVRNLANTVTEEILEKAFGQFGKLERVKKLK--------DYAFIHFDERVGAVKAMDEM 395
Query: 180 NNSEFKGKKIK 190
N E +G+ I+
Sbjct: 396 NGKELEGENIE 406
>UniRef100_Q68ED6 OTTHUMP00000016817 [Homo sapiens]
Length = 561
Score = 114 bits (286), Expect = 2e-24
Identities = 60/168 (35%), Positives = 96/168 (56%), Gaps = 1/168 (0%)
Query: 114 PPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELAS 173
P G+E+++G I ++ E++L + G + ++R+M +G +GYAFV F TKE A
Sbjct: 158 PSVGTEIFVGKIPRDLFEDELVPLFEKAGPIWDLRLMMDPLTGLNRGYAFVTFCTKEAAQ 217
Query: 174 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 232
+A++ NN E + GK I S A +RLF+G+IPK T E + + + V G+ + L
Sbjct: 218 EAVKLYNNHEIRSGKHIGVCISVANNRLFVGSIPKSKTKEQILEEFSKVTEGLTDVILYH 277
Query: 233 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADP 280
+NRGF F+EY +H A +R+++ + K+ N TV WADP
Sbjct: 278 QPDDKKKNRGFCFLEYEDHKTAAQARRRLMSGKVKVWGNVGTVEWADP 325
Score = 38.1 bits (87), Expect = 0.27
Identities = 20/71 (28%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
+++ +AN V+EE L G++ V+ +K YAF+ F ++ A +A+EE+
Sbjct: 340 LFVRNLANTVTEEILEKSFSQFGKLERVKKLK--------DYAFIHFDERDGAVKAMEEM 391
Query: 180 NNSEFKGKKIK 190
N + +G+ I+
Sbjct: 392 NGKDLEGENIE 402
>UniRef100_Q5TCG3 OTTHUMP00000040572 [Homo sapiens]
Length = 562
Score = 114 bits (286), Expect = 2e-24
Identities = 60/168 (35%), Positives = 96/168 (56%), Gaps = 1/168 (0%)
Query: 114 PPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELAS 173
P G+E+++G I ++ E++L + G + ++R+M +G +GYAFV F TKE A
Sbjct: 158 PSVGTEIFVGKIPRDLFEDELVPLFEKAGPIWDLRLMMDPLTGLNRGYAFVTFCTKEAAQ 217
Query: 174 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 232
+A++ NN E + GK I S A +RLF+G+IPK T E + + + V G+ + L
Sbjct: 218 EAVKLYNNHEIRSGKHIGVCISVANNRLFVGSIPKSKTKEQILEEFSKVTEGLTDVILYH 277
Query: 233 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADP 280
+NRGF F+EY +H A +R+++ + K+ N TV WADP
Sbjct: 278 QPDDKKKNRGFCFLEYEDHKTAAQARRRLMSGKVKVWGNVGTVEWADP 325
Score = 38.1 bits (87), Expect = 0.27
Identities = 20/71 (28%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
+++ +AN V+EE L G++ V+ +K YAF+ F ++ A +A+EE+
Sbjct: 340 LFVRNLANTVTEEILEKAFSQFGKLERVKKLK--------DYAFIHFDERDGAVKAMEEM 391
Query: 180 NNSEFKGKKIK 190
N + +G+ I+
Sbjct: 392 NGKDLEGENIE 402
>UniRef100_Q5TCG2 OTTHUMP00000016815 [Homo sapiens]
Length = 623
Score = 114 bits (286), Expect = 2e-24
Identities = 60/168 (35%), Positives = 96/168 (56%), Gaps = 1/168 (0%)
Query: 114 PPHGSEVYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELAS 173
P G+E+++G I ++ E++L + G + ++R+M +G +GYAFV F TKE A
Sbjct: 158 PSVGTEIFVGKIPRDLFEDELVPLFEKAGPIWDLRLMMDPLTGLNRGYAFVTFCTKEAAQ 217
Query: 174 QAIEELNNSEFK-GKKIKCSSSQAKHRLFIGNIPKKWTVEDMKKVVADVGPGVISIELLK 232
+A++ NN E + GK I S A +RLF+G+IPK T E + + + V G+ + L
Sbjct: 218 EAVKLYNNHEIRSGKHIGVCISVANNRLFVGSIPKSKTKEQILEEFSKVTEGLTDVILYH 277
Query: 233 DLQSSGRNRGFAFIEYYNHACAEYSRQKMSNSNFKLENNAPTVSWADP 280
+NRGF F+EY +H A +R+++ + K+ N TV WADP
Sbjct: 278 QPDDKKKNRGFCFLEYEDHKTAAQARRRLMSGKVKVWGNVGTVEWADP 325
Score = 38.1 bits (87), Expect = 0.27
Identities = 20/71 (28%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query: 120 VYIGGIANNVSEEDLRVFCQSVGEVSEVRIMKAKESGEGKGYAFVAFKTKELASQAIEEL 179
+++ +AN V+EE L G++ V+ +K YAF+ F ++ A +A+EE+
Sbjct: 340 LFVRNLANTVTEEILEKAFSQFGKLERVKKLK--------DYAFIHFDERDGAVKAMEEM 391
Query: 180 NNSEFKGKKIK 190
N + +G+ I+
Sbjct: 392 NGKDLEGENIE 402
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.303 0.125 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 537,574,709
Number of Sequences: 2790947
Number of extensions: 26837893
Number of successful extensions: 1211243
Number of sequences better than 10.0: 25260
Number of HSP's better than 10.0 without gapping: 16537
Number of HSP's successfully gapped in prelim test: 9496
Number of HSP's that attempted gapping in prelim test: 435746
Number of HSP's gapped (non-prelim): 231525
length of query: 289
length of database: 848,049,833
effective HSP length: 126
effective length of query: 163
effective length of database: 496,390,511
effective search space: 80911653293
effective search space used: 80911653293
T: 11
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.8 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0010.10


