

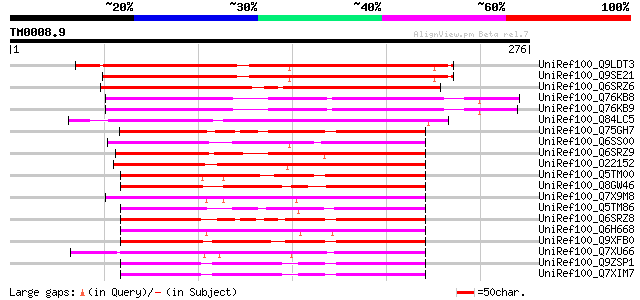
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.9
(276 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LDT3 F26F24.29 [Arabidopsis thaliana] 224 2e-57
UniRef100_Q9SE21 INNER NO OUTER [Arabidopsis thaliana] 221 2e-56
UniRef100_Q6SRZ6 YABBY-like transcription factor INNER NO OUTER-... 180 3e-44
UniRef100_Q76KB8 Putative transcription factor [Nymphaea colorata] 176 8e-43
UniRef100_Q76KB9 Putative transcription factor [Nymphaea alba] 172 7e-42
UniRef100_Q84LC5 YABBY transcription factor CDM51 [Chrysanthemum... 159 6e-38
UniRef100_Q75GH7 Putative yabby protein [Oryza sativa] 150 5e-35
UniRef100_Q6SS00 YABBY-like transcription factor GRAMINIFOLIA [A... 150 5e-35
UniRef100_Q6SRZ9 YABBY-like transcription factor PROLONGATA [Ant... 148 1e-34
UniRef100_O22152 Expressed protein [Arabidopsis thaliana] 146 7e-34
UniRef100_Q5TM00 Filamentous flower like protein [Amborella tric... 145 1e-33
UniRef100_Q8GW46 Hypothetical protein At2g26580/T9J22.25 [Arabid... 145 1e-33
UniRef100_Q7X9M8 Yabby15 protein [Zea mays] 145 1e-33
UniRef100_Q5TM86 YABBY2 like protein [Amborella trichopoda] 142 9e-33
UniRef100_Q6SRZ8 YABBY2-like transcription factor YAB2 [Antirrhi... 141 2e-32
UniRef100_Q6H668 Putative YABBY transcription factor CDM51 [Oryz... 141 2e-32
UniRef100_Q9XFB0 YABBY2 [Arabidopsis thaliana] 140 3e-32
UniRef100_Q7XU66 OSJNBb0020O11.2 protein [Oryza sativa] 135 1e-30
UniRef100_Q9ZSP1 Hypothetical protein [Oryza sativa] 128 1e-28
UniRef100_Q7XIM7 Putative MADS-box transcription factor CDM51 [O... 128 1e-28
>UniRef100_Q9LDT3 F26F24.29 [Arabidopsis thaliana]
Length = 262
Score = 224 bits (570), Expect = 2e-57
Identities = 123/214 (57%), Positives = 147/214 (68%), Gaps = 21/214 (9%)
Query: 36 THTYSHPHIQKDTMSTLNHLFDLPEQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCT 95
THT S + T +TLNHLFDLP QIC+VQCGFCTTIL+VSVP +SLSMVVTVRCGHCT
Sbjct: 27 THTLSMTKLPNMT-TTLNHLFDLPGQICHVQCGFCTTILLVSVPFTSLSMVVTVRCGHCT 85
Query: 96 SLLSVNMMKASFVPFHLLASLTHLQVIDYLTLTNSMFIYFSCSKEDANMTFN---SHSGA 152
SLLSVN+MKASF+P HLLASL+HL T + + E+ N +S
Sbjct: 86 SLLSVNLMKASFIPLHLLASLSHLDE------TGKEEVAATDGVEEEAWKVNQEKENSPT 139
Query: 153 SMMTYSDCEEEDLIPMSNFVNKPPEKRQRTPSAYNRFIKEEIKRLKAENPDMAHKEAFST 212
++++ SD E+ED+ + VNKPPEKRQR PSAYN FIKEEI+RLKA+NP MAHKEAFS
Sbjct: 140 TLVSSSDNEDEDVSRVYQVVNKPPEKRQRAPSAYNCFIKEEIRRLKAQNPSMAHKEAFSL 199
Query: 213 AAKNWANFPPTQ----------CKGDEEGVLHPC 236
AAKNWA+FPP C+ D +L PC
Sbjct: 200 AAKNWAHFPPAHNKRAASDQCFCEEDNNAIL-PC 232
>UniRef100_Q9SE21 INNER NO OUTER [Arabidopsis thaliana]
Length = 231
Score = 221 bits (562), Expect = 2e-56
Identities = 118/200 (59%), Positives = 141/200 (70%), Gaps = 20/200 (10%)
Query: 50 STLNHLFDLPEQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFVP 109
+TLNHLFDLP QIC+VQCGFCTTIL+VSVP +SLSMVVTVRCGHCTSLLSVN+MKASF+P
Sbjct: 9 TTLNHLFDLPGQICHVQCGFCTTILLVSVPFTSLSMVVTVRCGHCTSLLSVNLMKASFIP 68
Query: 110 FHLLASLTHLQVIDYLTLTNSMFIYFSCSKEDANMTFN---SHSGASMMTYSDCEEEDLI 166
HLLASL+HL T + + E+ N +S ++++ SD E+ED+
Sbjct: 69 LHLLASLSHLDE------TGKEEVAATDGVEEEAWKVNQEKENSPTTLVSSSDNEDEDVS 122
Query: 167 PMSNFVNKPPEKRQRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFPPTQ-- 224
+ VNKPPEKRQR PSAYN FIKEEI+RLKA+NP MAHKEAFS AAKNWA+FPP
Sbjct: 123 RVYQVVNKPPEKRQRAPSAYNCFIKEEIRRLKAQNPSMAHKEAFSLAAKNWAHFPPAHNK 182
Query: 225 --------CKGDEEGVLHPC 236
C+ D +L PC
Sbjct: 183 RAASDQCFCEEDNNAIL-PC 201
>UniRef100_Q6SRZ6 YABBY-like transcription factor INNER NO OUTER-like protein
[Antirrhinum majus]
Length = 235
Score = 180 bits (457), Expect = 3e-44
Identities = 97/182 (53%), Positives = 123/182 (67%), Gaps = 9/182 (4%)
Query: 49 MSTLNHLFDLPEQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFV 108
M TL+ LFD EQICYVQCGFCTTIL+VSVP + LSM VTVRCGHC+++LSVN+ A FV
Sbjct: 1 MLTLDSLFDFQEQICYVQCGFCTTILLVSVPKNCLSMAVTVRCGHCSTILSVNIPDAYFV 60
Query: 109 PFHLLASLTHLQVIDYLTLTNSMFIYFSCSKEDANMTFNSHSGASMMTYSDCEE-EDLIP 167
P H +S+ + + +CS E A + +G ++ SD EE ED +
Sbjct: 61 PLHFFSSINQQEQMSIQPKQE------ACSVEMAG--DHKKAGMTLCFSSDEEEYEDSLH 112
Query: 168 MSNFVNKPPEKRQRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFPPTQCKG 227
++ V+KPPEK+QR PSAYN FIK+EIKRLK E P+M HK+AFS AAKNWA+ P +Q K
Sbjct: 113 LNQLVHKPPEKKQRAPSAYNHFIKKEIKRLKIEYPNMTHKQAFSAAAKNWAHNPQSQYKR 172
Query: 228 DE 229
E
Sbjct: 173 GE 174
>UniRef100_Q76KB8 Putative transcription factor [Nymphaea colorata]
Length = 201
Score = 176 bits (445), Expect = 8e-43
Identities = 101/222 (45%), Positives = 131/222 (58%), Gaps = 32/222 (14%)
Query: 52 LNHLFDLPEQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFVPFH 111
++ L DL EQ+CYVQC FC TIL+VSVPCSSL VV VRCGHC++L SVNM+KASF+P
Sbjct: 1 MSQLLDLTEQLCYVQCSFCDTILLVSVPCSSLLKVVPVRCGHCSNLFSVNMLKASFLPLQ 60
Query: 112 LLASLTHLQVIDYLTLTNSMFIYFSCSKEDANMTFNSHSGASMMTYSDCEEEDLIPMSNF 171
LLAS+ + +K++ + G + S CEEE +
Sbjct: 61 LLASINNE------------------TKQENFQNAPAKIGDTSFMESFCEEERKPAFT-- 100
Query: 172 VNKPPEKRQRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFPPTQCKGDEEG 231
VNKPPEKR R PSAYNRFIKEEI+RLK P ++H+EA STAAKNWA+ P Q K D E
Sbjct: 101 VNKPPEKRHRAPSAYNRFIKEEIQRLKTSEPSISHREALSTAAKNWAHLPRIQHKPDAES 160
Query: 232 VLHPCITSMCGTFRSMK--KDKVSEEERSQGTLSWKGHRLSE 271
G+ R KDK + E +G +++ ++S+
Sbjct: 161 ----------GSQRQSNKGKDKHVDREDKEGNQNFQQRKVSQ 192
>UniRef100_Q76KB9 Putative transcription factor [Nymphaea alba]
Length = 202
Score = 172 bits (437), Expect = 7e-42
Identities = 102/221 (46%), Positives = 130/221 (58%), Gaps = 32/221 (14%)
Query: 52 LNHLFDLPEQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFVPFH 111
++ L DL EQ+CYVQC FC TIL+VSVPCSSL VV VRCGHC++L SVNM+KASF+P
Sbjct: 1 MSQLLDLTEQLCYVQCSFCDTILLVSVPCSSLLKVVPVRCGHCSNLFSVNMLKASFLPLQ 60
Query: 112 LLASLTHLQVIDYLTLTNSMFIYFSCSKEDANMTFNSHSGASMMTYSDCEEEDLIPMSNF 171
LLAS+ + +K+D+ G + S EEE +
Sbjct: 61 LLASINNE------------------AKQDSFENAPVKIGDTTFMESLYEEERRPAFT-- 100
Query: 172 VNKPPEKRQRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFPPTQCKGDEEG 231
VNKPPEKR R PSAYNRFIKEEI+RLK P+++H+EAFSTAAKNWA+ P Q K D E
Sbjct: 101 VNKPPEKRHRAPSAYNRFIKEEIQRLKTSEPNISHREAFSTAAKNWAHMPRIQHKPDAES 160
Query: 232 VLHPCITSMCGTFRSMK--KDKVSEEERSQGTLSWKGHRLS 270
G+ R KDK + E +G ++ ++S
Sbjct: 161 ----------GSQRQSNKGKDKHVDREDKEGNQIFQQRKVS 191
>UniRef100_Q84LC5 YABBY transcription factor CDM51 [Chrysanthemum x morifolium]
Length = 220
Score = 159 bits (403), Expect = 6e-38
Identities = 96/217 (44%), Positives = 120/217 (55%), Gaps = 29/217 (13%)
Query: 32 SSSFTHTYSHPHIQKDTMSTLNHLFDLPEQICYVQCGFCTTILMVSVPCSSLSMVVTVRC 91
SSS + + H H +HL EQ+CYVQC FC T+L VSVPC SL VTVRC
Sbjct: 2 SSSASFSPDHHH---------HHLSPTSEQLCYVQCNFCDTVLAVSVPCGSLFTTVTVRC 52
Query: 92 GHCTSLLSVNMMKASFVPFHLLASLTHLQVIDYLTLTNSMFIYFSCSKEDANMTFNSH-S 150
GHCT+LLSVNM F AS+T + L ++ F S +E N N +
Sbjct: 53 GHCTNLLSVNMRALLFP-----ASVTTTAAANQFHLGHNFFSAQSLMEEMRNTPANLFLN 107
Query: 151 GASMMTYSDCEEEDLIPMSNFVNKPPEKRQRTPSAYNRFIKEEIKRLKAENPDMAHKEAF 210
+ + D +P N+PPEKRQR PSAYNRFIKEEI+R+KA NPD++H+EAF
Sbjct: 108 QPNPNDHFGPVRVDELPKPPVANRPPEKRQRVPSAYNRFIKEEIQRIKAGNPDISHREAF 167
Query: 211 STAAKNWANFP--------------PTQCKGDEEGVL 233
S AAKNWA+FP P C+ D E +L
Sbjct: 168 SAAAKNWAHFPHIHFGLMPDQPVKKPNVCQQDGEDLL 204
>UniRef100_Q75GH7 Putative yabby protein [Oryza sativa]
Length = 186
Score = 150 bits (378), Expect = 5e-35
Identities = 82/163 (50%), Positives = 104/163 (63%), Gaps = 16/163 (9%)
Query: 59 PEQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFVPFHLLASLTH 118
PE +CYV C FC TI VSVP +S+ +VTVRCGHCTSLLSVN+ L + H
Sbjct: 9 PEHVCYVHCNFCNTIFAVSVPSNSMLNIVTVRCGHCTSLLSVNLRGL----VQALPAEDH 64
Query: 119 LQVIDYLTLTNSMFIYFSCSKEDANMTFNSHSGASMMTYSDCEEEDLIPMSNFVNKPPEK 178
LQ D L + N F + + +S G M +S + E ++ + +PPEK
Sbjct: 65 LQ--DNLKMHNMSF-----RENYSEYGSSSRYGRVPMMFSKNDTEHMLHV-----RPPEK 112
Query: 179 RQRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFP 221
RQR PSAYNRFIKEEI+R+KA NPD++H+EAFSTAAKNWA+FP
Sbjct: 113 RQRVPSAYNRFIKEEIRRIKANNPDISHREAFSTAAKNWAHFP 155
>UniRef100_Q6SS00 YABBY-like transcription factor GRAMINIFOLIA [Antirrhinum majus]
Length = 211
Score = 150 bits (378), Expect = 5e-35
Identities = 84/174 (48%), Positives = 104/174 (59%), Gaps = 20/174 (11%)
Query: 53 NHLFDLPEQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFVPFHL 112
+H EQ+CYV C FC T+L VSVPC+SL VTVRCGHCT+LLSVNM
Sbjct: 11 HHHITPSEQLCYVHCNFCDTVLAVSVPCTSLIKTVTVRCGHCTNLLSVNMRGLL------ 64
Query: 113 LASLTHLQVIDYLTLTNSMFIYFSCSKEDANMTFN-----SHSGASMMTYSDCEEEDLIP 167
L + L L +S F + +E N N + SMM +E +P
Sbjct: 65 ------LPAANQLHLGHSFFSPQNLLEEIRNSPSNLLMNQPNPNDSMMPVRGLDE---LP 115
Query: 168 MSNFVNKPPEKRQRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFP 221
N+PPEKRQR PSAYNRFIK+EI+R+KA NPD++H+EAFS AAKNWA+FP
Sbjct: 116 KPPVANRPPEKRQRVPSAYNRFIKDEIQRIKAGNPDISHREAFSAAAKNWAHFP 169
>UniRef100_Q6SRZ9 YABBY-like transcription factor PROLONGATA [Antirrhinum majus]
Length = 186
Score = 148 bits (374), Expect = 1e-34
Identities = 81/167 (48%), Positives = 104/167 (61%), Gaps = 17/167 (10%)
Query: 57 DLPEQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFVPFHLLASL 116
D+ EQ+CY+ C FC+ +L VSVPCSSL VVTVRCGHCT+L SVNM A+ F L
Sbjct: 2 DVLEQLCYISCNFCSIVLAVSVPCSSLFDVVTVRCGHCTNLWSVNMAAAA--TFQSLQPH 59
Query: 117 THLQVIDYLTLTNSMFIYFSCSKEDANMTFNSHSGASMMTYSDCEEEDLI--PMSNFVNK 174
V+ + A+ +N G+S + + I P VN+
Sbjct: 60 WQDAVVHQ-------------APNHASTEYNVDLGSSSRWNNKMAVQPSITKPEQRIVNR 106
Query: 175 PPEKRQRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFP 221
PPEKRQR PSAYN+FIKEEI+R+KA NP+++H+EAFSTAAKNWA+FP
Sbjct: 107 PPEKRQRVPSAYNQFIKEEIQRIKANNPEISHREAFSTAAKNWAHFP 153
>UniRef100_O22152 Expressed protein [Arabidopsis thaliana]
Length = 229
Score = 146 bits (368), Expect = 7e-34
Identities = 82/171 (47%), Positives = 108/171 (62%), Gaps = 11/171 (6%)
Query: 56 FDLPEQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFVPFHLLAS 115
F + +CYVQC FC TIL V+VP +SL VTVRCG CT+LLSVNM ++L +
Sbjct: 19 FSPSDHLCYVQCNFCQTILAVNVPYTSLFKTVTVRCGCCTNLLSVNMRS------YVLPA 72
Query: 116 LTHLQV-IDYLTLTNSMFIYFSCSKEDANMTF---NSHSGAS-MMTYSDCEEEDLIPMSN 170
LQ+ + + N I +NM N H + + ++ D ++ IP +
Sbjct: 73 SNQLQLQLGPHSYFNPQDILEELRDAPSNMNMMMMNQHPTMNDIPSFMDLHQQHEIPKAP 132
Query: 171 FVNKPPEKRQRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFP 221
VN+PPEKRQR PSAYNRFIKEEI+R+KA NPD++H+EAFS AAKNWA+FP
Sbjct: 133 PVNRPPEKRQRVPSAYNRFIKEEIQRIKAGNPDISHREAFSAAAKNWAHFP 183
>UniRef100_Q5TM00 Filamentous flower like protein [Amborella trichopoda]
Length = 201
Score = 145 bits (366), Expect = 1e-33
Identities = 80/169 (47%), Positives = 109/169 (64%), Gaps = 19/169 (11%)
Query: 60 EQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVN----MMKASFVPFHL--- 112
EQ+CYV C FC T+L VSVPCSSL +VTVRCGHCT++LSV+ + + HL
Sbjct: 5 EQLCYVHCNFCDTVLAVSVPCSSLFKMVTVRCGHCTNVLSVDTRGLLHPTAATQLHLGHA 64
Query: 113 LASLTHLQVIDYLTLTNSMFIYFSCSKEDANMTFNSHSGASMMTYSDCEEEDLIPMSNFV 172
S T ++D + +S+ + D + S++G++ + E + + V
Sbjct: 65 FFSPTPHNLLDECSPPSSLLL-------DHPLMTPSNTGSASTRLQENEA-----LHSPV 112
Query: 173 NKPPEKRQRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFP 221
++PPEKRQR PSAYNRFIKEEI+R+KA NPD+ H+EAFSTAAKNWA+FP
Sbjct: 113 SRPPEKRQRVPSAYNRFIKEEIQRIKAGNPDITHREAFSTAAKNWAHFP 161
>UniRef100_Q8GW46 Hypothetical protein At2g26580/T9J22.25 [Arabidopsis thaliana]
Length = 164
Score = 145 bits (365), Expect = 1e-33
Identities = 79/162 (48%), Positives = 101/162 (61%), Gaps = 23/162 (14%)
Query: 60 EQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFVPFHLLASLTHL 119
EQ+CY+ C FC IL V+VPCSSL +VTVRCGHCT+L SVNM A+L L
Sbjct: 9 EQLCYIPCNFCNIILAVNVPCSSLFDIVTVRCGHCTNLWSVNMA----------AALQSL 58
Query: 120 QVIDYLTLTNSMFIYFSCSKEDANMTFNSHSGASMMTYSDCEEEDLIPMSNFVNKPPEKR 179
++ ++ Y S S+ + S S T ++ VN+PPEKR
Sbjct: 59 SRPNFQATNYAVPEYGSSSRSHTKIP----SRISTRTITE---------QRIVNRPPEKR 105
Query: 180 QRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFP 221
QR PSAYN+FIKEEI+R+KA NPD++H+EAFSTAAKNWA+FP
Sbjct: 106 QRVPSAYNQFIKEEIQRIKANNPDISHREAFSTAAKNWAHFP 147
>UniRef100_Q7X9M8 Yabby15 protein [Zea mays]
Length = 250
Score = 145 bits (365), Expect = 1e-33
Identities = 87/195 (44%), Positives = 110/195 (55%), Gaps = 25/195 (12%)
Query: 52 LNHLFDLP-EQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMM------K 104
L+HL P EQ+CYV C C TIL V VPCSSL VTVRCGHC +LLSVN+
Sbjct: 7 LDHLAPSPTEQLCYVHCNCCDTILAVGVPCSSLFKTVTVRCGHCANLLSVNLRGLLLPPA 66
Query: 105 ASFVPFHL------LASLTHLQVIDYLTLTNSMFIYFSCSKEDANMTFNSHSG------- 151
A P HL L+ + ++D L L + + + + T S
Sbjct: 67 APAPPNHLNFAHSLLSPTSPHGLLDELALQQAPSFLMEQASANLSSTMTGRSSNSSCASN 126
Query: 152 ----ASMMTYSDCEEEDLIPMSN-FVNKPPEKRQRTPSAYNRFIKEEIKRLKAENPDMAH 206
A M ++E +P + VN+PPEKRQR PSAYNRFIK+EI+R+KA NPD+ H
Sbjct: 127 LPPPAPMPAAQPVQQEAELPKTAPSVNRPPEKRQRVPSAYNRFIKDEIQRIKAGNPDITH 186
Query: 207 KEAFSTAAKNWANFP 221
+EAFS AAKNWA+FP
Sbjct: 187 REAFSAAAKNWAHFP 201
>UniRef100_Q5TM86 YABBY2 like protein [Amborella trichopoda]
Length = 185
Score = 142 bits (358), Expect = 9e-33
Identities = 84/165 (50%), Positives = 99/165 (59%), Gaps = 25/165 (15%)
Query: 60 EQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFVPFHLLASLTHL 119
E +CYVQC C TIL VSVP S L +VTVRCGHCT+LLS+NM L
Sbjct: 8 EHVCYVQCNLCNTILAVSVPGSCLFGIVTVRCGHCTNLLSMNMGAL-------------L 54
Query: 120 QVIDYLTLTNSMFIYFSCSKEDANMTFNSHSGA---SMMTYSDCEEEDLIPMSNFVNKPP 176
Q I + L N +E M S S + S S+ EE IP N+PP
Sbjct: 55 QTIPFHDLQNQSV----APQERQRMEDGSSSKSIKDSETIPSENEEPRTIP-----NRPP 105
Query: 177 EKRQRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFP 221
EKRQR PSAYNRFIKEEI+R+KA NP++ H+EAFSTAAKNWA+FP
Sbjct: 106 EKRQRVPSAYNRFIKEEIQRIKARNPEITHREAFSTAAKNWAHFP 150
>UniRef100_Q6SRZ8 YABBY2-like transcription factor YAB2 [Antirrhinum majus]
Length = 186
Score = 141 bits (355), Expect = 2e-32
Identities = 78/162 (48%), Positives = 105/162 (64%), Gaps = 22/162 (13%)
Query: 60 EQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFVPFHLLASLTHL 119
E +CYV C FC TIL VSVPCS++ +VTVRCGHC +LLSVNM L HL
Sbjct: 8 ECVCYVHCNFCNTILAVSVPCSNMFTIVTVRCGHCANLLSVNM--------GALLQSVHL 59
Query: 120 QVIDYLTLTNSMFIYFSCSKEDANMTFNSHSGASMMTYSDCEEEDLIPMSNFVNKPPEKR 179
Q D+ ++ + +K++ + +S S ++ E+ + P+ +PPEKR
Sbjct: 60 Q--DFQKQQHAE----AAAKDNGS---SSKSNRYAPLQAEHEQPKMPPI-----RPPEKR 105
Query: 180 QRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFP 221
QR PSAYNRFIKEEI+R+KA NP+++H+EAFSTAAKNWA+FP
Sbjct: 106 QRVPSAYNRFIKEEIQRIKAGNPEISHREAFSTAAKNWAHFP 147
>UniRef100_Q6H668 Putative YABBY transcription factor CDM51 [Oryza sativa]
Length = 256
Score = 141 bits (355), Expect = 2e-32
Identities = 81/184 (44%), Positives = 107/184 (58%), Gaps = 22/184 (11%)
Query: 60 EQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMM-------------KAS 106
EQ+CYV C C TIL V VPCSSL VTVRCGHC +LLSVN+ +
Sbjct: 23 EQLCYVHCNCCDTILAVGVPCSSLFKTVTVRCGHCANLLSVNLRGLLLPAPAPAPANQLH 82
Query: 107 FVPFHLLASLTHLQVIDYLTLTNSMFIYFSCSKEDANMTFNSHSGAS-------MMTYSD 159
F P L + H + + T S+ + + S +++T S S + M
Sbjct: 83 FGPSLLSPTSPHGLLDEVAFQTPSLLMEQAASASLSSITGRSSSSCASNAPAMQMPPAKP 142
Query: 160 CEEEDLIPMSN--FVNKPPEKRQRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNW 217
++E +P + N+PPEKRQR PSAYNRFIK+EI+R+KA NPD++H+EAFS AAKNW
Sbjct: 143 VQQEPELPKNAPASANRPPEKRQRVPSAYNRFIKDEIQRIKAGNPDISHREAFSAAAKNW 202
Query: 218 ANFP 221
A+FP
Sbjct: 203 AHFP 206
>UniRef100_Q9XFB0 YABBY2 [Arabidopsis thaliana]
Length = 184
Score = 140 bits (354), Expect = 3e-32
Identities = 80/163 (49%), Positives = 104/163 (63%), Gaps = 17/163 (10%)
Query: 60 EQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFVPFHLL-ASLTH 118
E++CYV C FCTTIL VSVP +SL +VTVRCGHCT+LLS+N+ V H A H
Sbjct: 8 ERVCYVHCSFCTTILAVSVPYASLFTLVTVRCGHCTNLLSLNIG----VSLHQTSAPPIH 63
Query: 119 LQVIDYLTLTNSMFIYFSCSKEDANMTFNSHSGASMMTYSDCEEEDLIPMSNFVNKPPEK 178
+ + T S+ C+ +S S ++ D E + P+ +PPEK
Sbjct: 64 QDLQPHRQHTTSLVTRKDCAS-------SSRSTNNLSENIDREAPRMPPI-----RPPEK 111
Query: 179 RQRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFP 221
RQR PSAYNRFIKEEI+R+KA NP+++H+EAFSTAAKNWA+FP
Sbjct: 112 RQRVPSAYNRFIKEEIQRIKACNPEISHREAFSTAAKNWAHFP 154
>UniRef100_Q7XU66 OSJNBb0020O11.2 protein [Oryza sativa]
Length = 265
Score = 135 bits (340), Expect = 1e-30
Identities = 86/214 (40%), Positives = 114/214 (53%), Gaps = 29/214 (13%)
Query: 33 SSFTHTYSHPHIQKDTMSTLNHLFDLPEQICYVQCGFCTTILMVSVPCSSLSMVVTVRCG 92
SS T+S H+ + L + EQ+CYV C FC TIL V VPCSSL VTVRCG
Sbjct: 2 SSAPETFSLDHLSQHQQQQPPPLAE-QEQLCYVHCNFCDTILAVGVPCSSLFKTVTVRCG 60
Query: 93 HCTSLLSVNM---------MKASFVPF-----------HLLASLTHLQVIDYLTLTNSMF 132
HC +LLSVN+ A+ +PF LL + Q L +
Sbjct: 61 HCANLLSVNLRGLLLPAAASTANQLPFGQALLSPTSPHGLLDEVPSFQAPASLMTEQASP 120
Query: 133 IYFSCSKEDANMTFNS-----HSGASMMTYSDCEEEDLIPMSNFVNKPPEKRQRTPSAYN 187
S + +++ N+ S A+ T + ++ P + N+ EKRQR PSAYN
Sbjct: 121 NVSSITSSNSSCANNAPATSMASAANKATQREPQQPKNAPSA---NRTSEKRQRVPSAYN 177
Query: 188 RFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFP 221
RFIK+EI+R+KA NPD+ H+EAFS AAKNWA+FP
Sbjct: 178 RFIKDEIQRIKASNPDITHREAFSAAAKNWAHFP 211
>UniRef100_Q9ZSP1 Hypothetical protein [Oryza sativa]
Length = 169
Score = 128 bits (322), Expect = 1e-28
Identities = 69/162 (42%), Positives = 96/162 (58%), Gaps = 18/162 (11%)
Query: 60 EQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFVPFHLLASLTHL 119
E +CYV C +C TIL+V+VP + +VTVRCGHCT +LS+++ PFH ++
Sbjct: 8 EHVCYVNCNYCNTILVVNVPNNCSYNIVTVRCGHCTMVLSMDL-----APFHQARTVQDH 62
Query: 120 QVIDYLTLTNSMFIYFSCSKEDANMTFNSHSGASMMTYSDCEEEDLIPMSNFVNKPPEKR 179
QV + N+ Y S+ T M ++ + P+ +PPEKR
Sbjct: 63 QVQNRGFQGNNFGSYDIASRNQRTST--------AMYPMPTSQQQVSPI-----RPPEKR 109
Query: 180 QRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFP 221
QR PSAYNRFIKEEI+R+K NP+++H+EAFS AAKNWA+ P
Sbjct: 110 QRVPSAYNRFIKEEIQRIKTSNPEISHREAFSAAAKNWAHLP 151
>UniRef100_Q7XIM7 Putative MADS-box transcription factor CDM51 [Oryza sativa]
Length = 169
Score = 128 bits (322), Expect = 1e-28
Identities = 69/162 (42%), Positives = 96/162 (58%), Gaps = 18/162 (11%)
Query: 60 EQICYVQCGFCTTILMVSVPCSSLSMVVTVRCGHCTSLLSVNMMKASFVPFHLLASLTHL 119
E +CYV C +C TIL+V+VP + +VTVRCGHCT +LS+++ PFH ++
Sbjct: 8 EHVCYVNCNYCNTILVVNVPNNCSYNIVTVRCGHCTMVLSMDL-----APFHQARTVQDH 62
Query: 120 QVIDYLTLTNSMFIYFSCSKEDANMTFNSHSGASMMTYSDCEEEDLIPMSNFVNKPPEKR 179
QV + N+ Y S+ T M ++ + P+ +PPEKR
Sbjct: 63 QVQNRGFQGNNFGSYDIASRNQRTST--------AMYPMPTSQQQVSPI-----RPPEKR 109
Query: 180 QRTPSAYNRFIKEEIKRLKAENPDMAHKEAFSTAAKNWANFP 221
QR PSAYNRFIKEEI+R+K NP+++H+EAFS AAKNWA+ P
Sbjct: 110 QRVPSAYNRFIKEEIQRIKTSNPEISHREAFSAAAKNWAHLP 151
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.129 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 449,635,583
Number of Sequences: 2790947
Number of extensions: 17553427
Number of successful extensions: 51348
Number of sequences better than 10.0: 98
Number of HSP's better than 10.0 without gapping: 62
Number of HSP's successfully gapped in prelim test: 37
Number of HSP's that attempted gapping in prelim test: 51191
Number of HSP's gapped (non-prelim): 125
length of query: 276
length of database: 848,049,833
effective HSP length: 125
effective length of query: 151
effective length of database: 499,181,458
effective search space: 75376400158
effective search space used: 75376400158
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0008.9


