

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
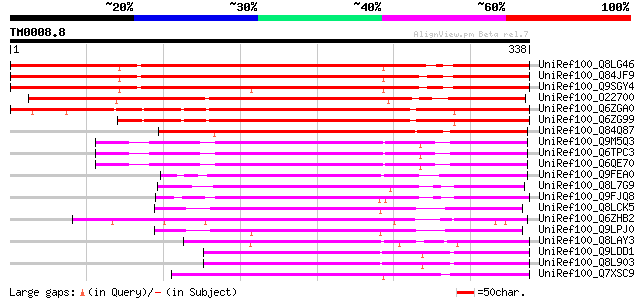
Query= TM0008.8
(338 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LG46 S-ribonuclease binding protein SBP1, putative [... 383 e-105
UniRef100_Q84JF9 Putative S-ribonuclease binding protein SBP1 [A... 382 e-104
UniRef100_Q9SGY4 F20B24.9 [Arabidopsis thaliana] 366 e-100
UniRef100_O22700 F8A5.13 protein [Arabidopsis thaliana] 352 6e-96
UniRef100_Q6ZGA0 Putative S-ribonuclease binding protein SBP1 [O... 236 5e-61
UniRef100_Q6ZG99 Putative S-ribonuclease binding protein SBP1 [O... 219 8e-56
UniRef100_Q84Q87 Hypothetical protein OJ1041F02.18 [Oryza sativa] 188 2e-46
UniRef100_Q9M5Q3 S-ribonuclease binding protein SBP1 [Petunia hy... 181 3e-44
UniRef100_Q6TPC3 S-RNase-binding protein [Solanum chacoense] 177 3e-43
UniRef100_Q6QE70 S-RNase binding protein 1 [Solanum chacoense] 177 5e-43
UniRef100_Q9FEA0 S-ribonuclease binding protein SBP1, putative [... 164 4e-39
UniRef100_Q8L7G9 Hypothetical protein At4g17680 [Arabidopsis tha... 145 1e-33
UniRef100_Q9FJQ8 Similarity to S-ribonuclease binding protein [A... 135 2e-30
UniRef100_Q8LCK5 Hypothetical protein [Arabidopsis thaliana] 133 8e-30
UniRef100_Q6ZHB2 S-ribonuclease binding protein SBP1-like [Oryza... 131 2e-29
UniRef100_Q9LPJ0 F6N18.12 [Arabidopsis thaliana] 125 2e-27
UniRef100_Q8LAY3 Inhibitor of apoptosis-like protein [Arabidopsi... 122 1e-26
UniRef100_Q9LDD1 Arabidopsis thaliana genomic DNA, chromosome 3,... 119 1e-25
UniRef100_Q8L903 Hypothetical protein [Arabidopsis thaliana] 119 1e-25
UniRef100_Q7XSC9 OSJNBb0118P14.4 protein [Oryza sativa] 119 2e-25
>UniRef100_Q8LG46 S-ribonuclease binding protein SBP1, putative [Arabidopsis
thaliana]
Length = 337
Score = 383 bits (983), Expect = e-105
Identities = 197/351 (56%), Positives = 256/351 (72%), Gaps = 27/351 (7%)
Query: 1 MFGGNNGNRLLPVFLDENQFQCQTNA-TNQLQLFGNLQAGRNVDPVSYIGNEHISSMIQP 59
M GNNGN PVF++ENQ Q QTN +NQL L GN+ G VDPV+Y N+++ MI+P
Sbjct: 1 MLSGNNGNTAPPVFMNENQLQYQTNLRSNQLHLLGNMGGGCTVDPVNYFANDNLVPMIRP 60
Query: 60 NKRSREMEDIS-----KKQRLQISLNYNVCQDDADWSASIPNPNALSTGLRLSYDDDERN 114
NKR RE E IS ++Q+LQ+SLNYN ++ +P N +STGLRLSYDDDE N
Sbjct: 61 NKRGREAESISNNIQRQQQKLQMSLNYNY--NNTSVREEVPKENLVSTGLRLSYDDDEHN 118
Query: 115 SSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMAS 174
SSVTSASGS+ A I SL D++R +L RQ++E D +IK+Q Q+++GVRDMKQ+H+AS
Sbjct: 119 SSVTSASGSILAASPIFQSLDDSLRIDLHRQKDEFDQFIKIQAAQMAKGVRDMKQRHIAS 178
Query: 175 LLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRN 234
LT++EKG+ KKL+EK+ EI +MN+KN+EL ERIKQVA+EAQNWHYRAKYNESVVN L+
Sbjct: 179 FLTTLEKGVSKKLQEKDHEINDMNKKNKELVERIKQVAMEAQNWHYRAKYNESVVNVLKA 238
Query: 235 NLQQAISH------GVEQGKEGFGDSEVDDAA-SYIDPNNFLSIPGAPMKSIHPRYQGME 287
NLQQA+SH +QGKEGFGDSE+DDAA SYIDPNN ++ IH R
Sbjct: 239 NLQQAMSHNNSVIAAADQGKEGFGDSEIDDAASSYIDPNNNNNM------GIHQR----- 287
Query: 288 NLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYLS 338
+ C+ C K+VS+L++PCRHLSLCK+CD F +CPVC+ +K++ V+V+ S
Sbjct: 288 -MRCKMCNVKEVSVLIVPCRHLSLCKECDVFTKICPVCKSLKSSCVQVFFS 337
>UniRef100_Q84JF9 Putative S-ribonuclease binding protein SBP1 [Arabidopsis thaliana]
Length = 339
Score = 382 bits (980), Expect = e-104
Identities = 197/351 (56%), Positives = 254/351 (72%), Gaps = 25/351 (7%)
Query: 1 MFGGNNGNRLLPVFLDENQFQCQTNA-TNQLQLFGNLQAGRNVDPVSYIGNEHISSMIQP 59
M GNNGN PVF++ENQ Q QTN +NQL L GN+ G VDPV+Y N+++ MI+P
Sbjct: 1 MLSGNNGNTAPPVFMNENQLQYQTNLRSNQLHLLGNMGGGCTVDPVNYFANDNLVPMIRP 60
Query: 60 NKRSREMEDIS-----KKQRLQISLNYNVCQDDADWSASIPNPNALSTGLRLSYDDDERN 114
NKR RE E IS ++Q+LQ+SLNYN ++ +P N +STGLRLSYDDDE N
Sbjct: 61 NKRGREAESISNNIQRQQQKLQMSLNYNY--NNTSVREEVPKENLVSTGLRLSYDDDEHN 118
Query: 115 SSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMAS 174
SSVTSASGS+ A I SL D++R +L RQ++E D +IK+Q Q+++GVRDMKQ+H+AS
Sbjct: 119 SSVTSASGSILAASPIFQSLDDSLRIDLHRQKDEFDQFIKIQAAQMAKGVRDMKQRHIAS 178
Query: 175 LLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRN 234
LT++EKG+ KKL+EK+ EI +MN+KN+EL ERIKQVA+EAQNWHYRAKYNESVVN L+
Sbjct: 179 FLTTLEKGVSKKLQEKDHEINDMNKKNKELVERIKQVAMEAQNWHYRAKYNESVVNVLKA 238
Query: 235 NLQQAISH------GVEQGKEGFGDSEVDDAA-SYIDPNNFLSIPGAPMKSIHPRYQGME 287
NLQQA+SH +QGKEGFGDSE+DDAA SYIDPNN IH R
Sbjct: 239 NLQQAMSHNNSVIAAADQGKEGFGDSEIDDAASSYIDPNN----NNNNNMGIHQR----- 289
Query: 288 NLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYLS 338
+ C+ C K+VS+L++PCRHLSLCK+CD F +CPVC+ +K++ V+V+ S
Sbjct: 290 -MRCKMCNVKEVSVLIVPCRHLSLCKECDVFTKICPVCKSLKSSCVQVFFS 339
>UniRef100_Q9SGY4 F20B24.9 [Arabidopsis thaliana]
Length = 368
Score = 366 bits (940), Expect = e-100
Identities = 197/380 (51%), Positives = 254/380 (66%), Gaps = 54/380 (14%)
Query: 1 MFGGNNGNRLLPVFLDENQFQCQTNA-TNQLQLFGNLQAGRNVDPVSYIGNEHISSMIQP 59
M GNNGN PVF++ENQ Q QTN +NQL L GN+ G VDPV+Y N+++ MI+P
Sbjct: 1 MLSGNNGNTAPPVFMNENQLQYQTNLRSNQLHLLGNMGGGCTVDPVNYFANDNLVPMIRP 60
Query: 60 NKRSREMEDIS-----KKQRLQISLNYNVCQDDADWSASIPNPNALSTGLRLSYDDDERN 114
NKR RE E IS ++Q+LQ+SLNYN ++ +P N +STGLRLSYDDDE N
Sbjct: 61 NKRGREAESISNNIQRQQQKLQMSLNYNY--NNTSVREEVPKENLVSTGLRLSYDDDEHN 118
Query: 115 SSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQ------------------ 156
SSVTSASGS+ A I SL D++R +L RQ++E D +IK+Q
Sbjct: 119 SSVTSASGSILAASPIFQSLDDSLRIDLHRQKDEFDQFIKIQVLIVSACRLCFYVKRFFD 178
Query: 157 -----------KEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELA 205
Q+++GVRDMKQ+H+AS LT++EKG+ KKL+EK+ EI +MN+KN+EL
Sbjct: 179 SNVFVCFYVVQAAQMAKGVRDMKQRHIASFLTTLEKGVSKKLQEKDHEINDMNKKNKELV 238
Query: 206 ERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISH------GVEQGKEGFGDSEVDDA 259
ERIKQVA+EAQNWHYRAKYNESVVN L+ NLQQA+SH +QGKEGFGDSE+DDA
Sbjct: 239 ERIKQVAMEAQNWHYRAKYNESVVNVLKANLQQAMSHNNSVIAAADQGKEGFGDSEIDDA 298
Query: 260 A-SYIDPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGF 318
A SYIDPNN IH R + C+ C K+VS+L++PCRHLSLCK+CD F
Sbjct: 299 ASSYIDPNN----NNNNNMGIHQR------MRCKMCNVKEVSVLIVPCRHLSLCKECDVF 348
Query: 319 INVCPVCQMIKTASVEVYLS 338
+CPVC+ +K++ V+V+ S
Sbjct: 349 TKICPVCKSLKSSCVQVFFS 368
>UniRef100_O22700 F8A5.13 protein [Arabidopsis thaliana]
Length = 372
Score = 352 bits (904), Expect = 6e-96
Identities = 183/342 (53%), Positives = 250/342 (72%), Gaps = 34/342 (9%)
Query: 13 VFLDENQFQCQTNAT-NQLQLFGNLQAGRNVDPVSYIGNEHISSMIQPNK-RSREMED-- 68
V ++++QF+ Q+N + NQL L G ++AG +DPV+Y N++++ M++ N R RE E+
Sbjct: 45 VLMNDSQFRYQSNTSLNQLHLLGTMRAGCTIDPVNYFANDNLAPMMRLNSTRGRETENNN 104
Query: 69 -ISKKQRLQISLNYNVCQDDADWSASIPNPNALSTGLRLSYDDDERNSSVTSASGSMTAP 127
+ ++Q+LQISLNYN ++ +P N +STGLRLSYDDDERNSSVTSA+GS+T P
Sbjct: 105 IMQRQQKLQISLNYNYNNNNTVVQDEVPKQNLVSTGLRLSYDDDERNSSVTSANGSITTP 164
Query: 128 PSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKL 187
+ SLGDNIR +L+RQ +ELD +IK + +Q+++GVRD+KQ+H+ S +T++EK + KKL
Sbjct: 165 --VYQSLGDNIRLDLNRQNDELDQFIKFRADQMAKGVRDIKQRHVTSFVTALEKDVSKKL 222
Query: 188 REKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVE-- 245
+EK+ EIE+MN+KNREL ++IKQVAVEAQNWHY+AKYNESVVNAL+ NLQQ +SHG +
Sbjct: 223 QEKDHEIESMNKKNRELVDKIKQVAVEAQNWHYKAKYNESVVNALKVNLQQVMSHGNDNN 282
Query: 246 ----------QGKEGFGDSEVDD-AASYIDPNNFLSIPGAPMKSIHPRYQGMENLSCRAC 294
Q KEGFGDSE+DD AASY N+L+IPG P + C+ C
Sbjct: 283 AVGGGVADHHQMKEGFGDSEIDDEAASY----NYLNIPGMP----------STGMRCKLC 328
Query: 295 KAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVY 336
K+VS+LL+PCRHLSLCKDCD F VCPVCQ +KT+SV+V+
Sbjct: 329 NVKNVSVLLVPCRHLSLCKDCDVFTGVCPVCQSLKTSSVQVF 370
>UniRef100_Q6ZGA0 Putative S-ribonuclease binding protein SBP1 [Oryza sativa]
Length = 343
Score = 236 bits (603), Expect = 5e-61
Identities = 141/346 (40%), Positives = 217/346 (61%), Gaps = 15/346 (4%)
Query: 1 MFGGNNGNRLLPV--FLDENQFQCQTNATN-QLQLFGN--LQAGRNVDPVSYIGNEHISS 55
MFG + P E+QF A+ QLQLFG+ + A + +YI N H+S+
Sbjct: 5 MFGSGHWGGSFPYASIPKESQFVFDAKASPLQLQLFGSAAVPAVGSTGYYNYIANNHLSA 64
Query: 56 MIQPNKRSREMEDISKKQRLQISLNYNVCQDDADWSASIPNPNALSTGLRLSYDDDERNS 115
M Q + ++ K+ LQ+SLNY ++ D A I NP+A+STGLRLSY+++E ++
Sbjct: 65 MNQERNTNNDVGH-EKQLNLQMSLNYFPVEN-LDRLARIGNPSAVSTGLRLSYENNE-HT 121
Query: 116 SVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASL 175
S+TS SG+M++ P I+ S D + ELD++ +E + Y LQ EQL + ++D+KQ+ M
Sbjct: 122 SITSGSGNMSSLP-IMASFVDEVMAELDKENKEFNCYFGLQVEQLVKCMKDVKQRQMVEF 180
Query: 176 LTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNN 235
L S+E+G+ KKL+EKE E+E MNRK++EL E+I+QVA+E Q+W A +N+SV N++++
Sbjct: 181 LASLERGVGKKLKEKELEVEAMNRKSKELNEQIRQVALEVQSWQSVALHNQSVANSMKSK 240
Query: 236 LQQAISHGVEQGKEGFGDSEVDDAASYIDPNNFLSIPGAPMKSIHPRYQGMEN---LSCR 292
L Q ++H +EG GDSEVD+ AS N ++PG +S M + +CR
Sbjct: 241 LMQMVAHSSNLTREGSGDSEVDNTAS---SQNVNAVPGVFFQSGLLGINSMADGGLGACR 297
Query: 293 ACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYLS 338
C+ K+ ++L++PCRHL LC DC+ +VCPVC+ K+ SVE+ +S
Sbjct: 298 LCRMKEAAVLVMPCRHLCLCADCEKNADVCPVCRFPKSCSVEINMS 343
>UniRef100_Q6ZG99 Putative S-ribonuclease binding protein SBP1 [Oryza sativa]
Length = 279
Score = 219 bits (558), Expect = 8e-56
Identities = 119/271 (43%), Positives = 183/271 (66%), Gaps = 9/271 (3%)
Query: 71 KKQRLQISLNYNVCQDDADWSASIPNPNALSTGLRLSYDDDERNSSVTSASGSMTAPPSI 130
K+ LQ+SLNY ++ D A I NP+A+STGLRLSY+++E ++S+TS SG+M++ P I
Sbjct: 15 KQLNLQMSLNYFPVEN-LDRLARIGNPSAVSTGLRLSYENNE-HTSITSGSGNMSSLP-I 71
Query: 131 ILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREK 190
+ S D + ELD++ +E + Y LQ EQL + ++D+KQ+ M L S+E+G+ KKL+EK
Sbjct: 72 MASFVDEVMAELDKENKEFNCYFGLQVEQLVKCMKDVKQRQMVEFLASLERGVGKKLKEK 131
Query: 191 EEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQGKEG 250
E E+E MNRK++EL E+I+QVA+E Q+W A +N+SV N++++ L Q ++H +EG
Sbjct: 132 ELEVEAMNRKSKELNEQIRQVALEVQSWQSVALHNQSVANSMKSKLMQMVAHSSNLTREG 191
Query: 251 FGDSEVDDAASYIDPNNFLSIPGAPMKSIHPRYQGMEN---LSCRACKAKDVSMLLIPCR 307
GDSEVD+ AS N ++PG +S M + +CR C+ K+ ++L++PCR
Sbjct: 192 SGDSEVDNTAS---SQNVNAVPGVFFQSGLLGINSMADGGLGACRLCRMKEAAVLVMPCR 248
Query: 308 HLSLCKDCDGFINVCPVCQMIKTASVEVYLS 338
HL LC DC+ +VCPVC+ K+ SVE+ +S
Sbjct: 249 HLCLCADCEKNADVCPVCRFPKSCSVEINMS 279
>UniRef100_Q84Q87 Hypothetical protein OJ1041F02.18 [Oryza sativa]
Length = 342
Score = 188 bits (478), Expect = 2e-46
Identities = 95/242 (39%), Positives = 150/242 (61%), Gaps = 7/242 (2%)
Query: 98 NALSTGLRLSYDDDERNSSVTSASGSMTAPPSIIL--SLGDNIRTELDRQQEELDHYIKL 155
+A+STGL LS +D + +G+ + ++L L D+I E+ R ++D +IK
Sbjct: 106 SAVSTGLALSLEDRRHGGGSGAGAGNSSGDSPLLLLPMLDDDISREVQRLDADMDRFIKA 165
Query: 156 QKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEA 215
Q E+L + + + Q L S+E I++K+R+KE E+EN+N++N EL ++IKQ+AVE
Sbjct: 166 QSERLRQSILEKVQAKQFEALASVEDKILRKIRDKEAEVENINKRNSELEDQIKQLAVEV 225
Query: 216 QNWHYRAKYNESVVNALRNNLQQAISHGVEQGKEGFGDSEVDDAASYIDPNNFLSIPGAP 275
W RAKYNES++NAL+ NL+Q +H + KEG GDSEVDD AS + ++ P
Sbjct: 226 GAWQQRAKYNESMINALKYNLEQVCAHQSKDFKEGCGDSEVDDTASCCN-GGAANLQLMP 284
Query: 276 MKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEV 335
++ H + + +CR CK+ + MLL+PCRHL LCK+C+ ++ CP+CQ K +E+
Sbjct: 285 KENRHSK----DLTACRVCKSSEACMLLLPCRHLCLCKECESKLSFCPLCQSSKILGMEI 340
Query: 336 YL 337
Y+
Sbjct: 341 YM 342
>UniRef100_Q9M5Q3 S-ribonuclease binding protein SBP1 [Petunia hybrida]
Length = 332
Score = 181 bits (459), Expect = 3e-44
Identities = 102/285 (35%), Positives = 160/285 (55%), Gaps = 35/285 (12%)
Query: 57 IQPNKRSREMEDISKKQRLQISLNYNVCQDDADWSASIPNPNALSTGLRLSYDDDERNSS 116
++P ++ + +D + QIS S P ++STGL LS D+ SS
Sbjct: 79 LEPKRKRPKEQDFLENNNSQIS------------SIDFLQPRSVSTGLGLSLDNGRLASS 126
Query: 117 VTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLL 176
SA + +GD+I EL RQ E+D YIK+Q ++L + + + Q + +
Sbjct: 127 GDSAFLGL---------VGDDIERELQRQDAEIDRYIKVQGDRLRQAILEKVQANQLQTV 177
Query: 177 TSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNL 236
T +E+ +++KLREKE E+E++N+KN EL R +Q+A+EA W RAKYNE+++N L+ NL
Sbjct: 178 TYVEEKVIQKLREKETEVEDINKKNMELELRTEQLALEANAWQQRAKYNENLINTLKVNL 237
Query: 237 QQAISHGVEQGKEGFGDSEVDDAASYIDPN----NFLSIPGAPMKSIHPRYQGMENLSCR 292
+ + KEG GDSEVDD AS + + L MK E ++C+
Sbjct: 238 EHVYAQS-RDSKEGCGDSEVDDTASCCNGRATDLHLLCRDSNEMK---------ELMTCK 287
Query: 293 ACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYL 337
C+ +VSMLL+PC+HL LCK+C+ +++CP+CQ K +E+Y+
Sbjct: 288 VCRVNEVSMLLLPCKHLCLCKECESKLSLCPLCQSTKYIGMEIYM 332
>UniRef100_Q6TPC3 S-RNase-binding protein [Solanum chacoense]
Length = 342
Score = 177 bits (450), Expect = 3e-43
Identities = 101/285 (35%), Positives = 159/285 (55%), Gaps = 35/285 (12%)
Query: 57 IQPNKRSREMEDISKKQRLQISLNYNVCQDDADWSASIPNPNALSTGLRLSYDDDERNSS 116
++P K+ + +D + QIS S + ++STGL LS D+ SS
Sbjct: 89 LEPKKKRPKEQDFMENNNSQIS------------SVDLLQRRSVSTGLGLSLDNGRLASS 136
Query: 117 VTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLL 176
SA + +GD+I EL RQ E+D YIK+Q ++L + V + Q + +
Sbjct: 137 CDSAFLGL---------VGDDIERELQRQDAEIDRYIKVQGDRLRQAVLEKVQANQIQAI 187
Query: 177 TSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNL 236
T +E+ +++KLRE++ E++++N+KN EL R++Q+ +EA W RAKYNE+++N L+ NL
Sbjct: 188 TYVEEKVLQKLRERDTEVDDINKKNMELELRMEQLDLEANAWQQRAKYNENLINTLKVNL 247
Query: 237 QQAISHGVEQGKEGFGDSEVDDAASYIDPN----NFLSIPGAPMKSIHPRYQGMENLSCR 292
Q + KEG GDSEVDD AS + + L MK E ++CR
Sbjct: 248 QHVYAQS-RDSKEGCGDSEVDDTASCCNGRATDLHLLCRDSKEMK---------ELMTCR 297
Query: 293 ACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYL 337
C+ +V ML +PC+HL LCK+C+ +++CP+CQ IK +EVY+
Sbjct: 298 VCRTNEVGMLWLPCKHLGLCKECESKLSLCPLCQSIKYIGMEVYM 342
>UniRef100_Q6QE70 S-RNase binding protein 1 [Solanum chacoense]
Length = 337
Score = 177 bits (448), Expect = 5e-43
Identities = 101/285 (35%), Positives = 159/285 (55%), Gaps = 35/285 (12%)
Query: 57 IQPNKRSREMEDISKKQRLQISLNYNVCQDDADWSASIPNPNALSTGLRLSYDDDERNSS 116
++P K+ + +D + QIS S + ++STGL LS D+ SS
Sbjct: 84 LEPKKKRPKEQDFMENNNSQIS------------SVDLFQRRSVSTGLGLSLDNGRLASS 131
Query: 117 VTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLL 176
SA + +GD+I EL RQ E+D YIK+Q ++L + V + Q + +
Sbjct: 132 CDSAFLGL---------VGDDIERELQRQDAEIDRYIKVQGDRLRQAVLEKVQANQIQAI 182
Query: 177 TSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNL 236
T +E+ +++KLRE++ E++++N+KN EL R++Q+ +EA W RAKYNE+++N L+ NL
Sbjct: 183 TYVEEKVLQKLRERDTEVDDINKKNMELELRMEQLDLEANAWQQRAKYNENLINTLKVNL 242
Query: 237 QQAISHGVEQGKEGFGDSEVDDAASYIDPN----NFLSIPGAPMKSIHPRYQGMENLSCR 292
Q + KEG GDSEVDD AS + + L MK E ++CR
Sbjct: 243 QHVYAQS-RDSKEGCGDSEVDDTASCCNGRATDLHLLCRDSKEMK---------ELMTCR 292
Query: 293 ACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYL 337
C+ +V MLL+PC+HL LCK+C+ +++CP+CQ K +EVY+
Sbjct: 293 VCRTNEVCMLLLPCKHLCLCKECESKLSLCPLCQSTKYIGMEVYM 337
>UniRef100_Q9FEA0 S-ribonuclease binding protein SBP1, putative [Arabidopsis
thaliana]
Length = 325
Score = 164 bits (414), Expect = 4e-39
Identities = 89/239 (37%), Positives = 142/239 (59%), Gaps = 22/239 (9%)
Query: 99 ALSTGLRLSYDDDERNSSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKE 158
++STGL LS D N+ V S+ GS +++ +GD+I EL RQ ++D ++K+Q +
Sbjct: 109 SVSTGLGLSLD----NARVASSDGS-----ALLSLVGDDIDRELQRQDADIDRFLKIQGD 159
Query: 159 QLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNW 218
QL + D ++ ++ +E+ +V+KLREK+EE+E +NRKN+EL R++Q+ +EA+ W
Sbjct: 160 QLRHAILDKIKRGQQKTVSLMEEKVVQKLREKDEELERINRKNKELEVRMEQLTMEAEAW 219
Query: 219 HYRAKYNESVVNALRNNLQQAISHGVEQGKEGFGDSEVDDAASYIDPNNFLSIPGAPMKS 278
RAKYNE+++ AL NL +A EG GDSEVDD AS + +
Sbjct: 220 QQRAKYNENMIAALNYNLDRAQGR-PRDSIEGCGDSEVDDTASCFNGRD----------- 267
Query: 279 IHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYL 337
+ + CR C +++ MLL+PC H+ LCK+C+ ++ CP+CQ K +EVY+
Sbjct: 268 -NSNNNTKTMMMCRFCGVREMCMLLLPCNHMCLCKECERKLSSCPLCQSSKFLGMEVYM 325
>UniRef100_Q8L7G9 Hypothetical protein At4g17680 [Arabidopsis thaliana]
Length = 314
Score = 145 bits (367), Expect = 1e-33
Identities = 85/248 (34%), Positives = 136/248 (54%), Gaps = 38/248 (15%)
Query: 97 PNALSTGLRLSYDDDERNSSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQ 156
PN +STGLRL +D + S SL ++ ++ RQ++ELD +I+ Q
Sbjct: 94 PNVVSTGLRLFHDQSQNQQQFFS-------------SLPGDVTGKIKRQRDELDRFIQTQ 140
Query: 157 KEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQ 216
E+L R + D +++ LL + E+ + +KLR+KE E+E R++ EL R+ + EA+
Sbjct: 141 GEELRRTLADNRERRYVELLCAAEEIVGRKLRKKEAELEKATRRHAELEARVAHIVEEAR 200
Query: 217 NWHYRAKYNESVVNALRNNLQQAISHGVEQ-------GKEGFGDSEVDDAAS-YIDPNNF 268
NW RA E+ V++L +LQQAI++ ++ G++G E +DA S Y+DP
Sbjct: 201 NWQLRAATREAEVSSLHAHLQQAIANRLDTAAKQSTFGEDGGDAEEAEDAESVYVDPER- 259
Query: 269 LSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCD-GFINVCPVCQM 327
++ I P SCR C+ K +++ +PC+HL LC CD G + VCP+C
Sbjct: 260 -------IELIGP--------SCRICRRKSATVMALPCQHLILCNGCDVGAVRVCPICLA 304
Query: 328 IKTASVEV 335
+KT+ VEV
Sbjct: 305 VKTSGVEV 312
>UniRef100_Q9FJQ8 Similarity to S-ribonuclease binding protein [Arabidopsis thaliana]
Length = 300
Score = 135 bits (340), Expect = 2e-30
Identities = 85/253 (33%), Positives = 134/253 (52%), Gaps = 37/253 (14%)
Query: 96 NPNALSTGLRLSYDDDERNSSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKL 155
NPN +STGLRLS R S ++ P + ++ E+ Q +EL+ ++++
Sbjct: 75 NPNVVSTGLRLS-----REQSQNQEQRFLSFP------ITGDVAGEIKSQTDELNRFLQI 123
Query: 156 QKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEA 215
Q EQL R + + +++ LL + E+ + ++LREKE EIE R++ EL R Q+ EA
Sbjct: 124 QGEQLKRMLAENSERNYRELLRTTEESVRRRLREKEAEIEKATRRHVELEARATQIETEA 183
Query: 216 QNWHYRAKYNESVVNALRNNLQQA--ISHG------VE-QGKEGFGDSEVDDAAS-YIDP 265
+ W RA E+ +L+ L QA ++HG VE Q G E +DA S Y+DP
Sbjct: 184 RAWQMRAAAREAEATSLQAQLHQAVVVAHGGGVITTVEPQSGSVDGVDEAEDAESAYVDP 243
Query: 266 NNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVC 325
+ ++ I P CR C+ + ++L +PCRHL +C +CDG + +CP+C
Sbjct: 244 DR--------VEMIGP--------GCRICRRRSATVLALPCRHLVMCTECDGSVRICPLC 287
Query: 326 QMIKTASVEVYLS 338
K +SVEV+ S
Sbjct: 288 LSTKNSSVEVFYS 300
>UniRef100_Q8LCK5 Hypothetical protein [Arabidopsis thaliana]
Length = 312
Score = 133 bits (334), Expect = 8e-30
Identities = 81/254 (31%), Positives = 130/254 (50%), Gaps = 47/254 (18%)
Query: 95 PNPNALSTGLRLSYDDDERNSSVTSASGSMTAPPSIILSLGDNI-RTELDRQQEELDHYI 153
P N + TGLRL +D+ I L +++ ++RQ EELD ++
Sbjct: 88 PPSNMVHTGLRLFSGEDQAQK---------------ISHLSEDVFAAHINRQSEELDEFL 132
Query: 154 KLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAV 213
Q E+L R + + ++ H +LL ++E+ +V+KLREKE EIE R++ EL R Q+
Sbjct: 133 HAQAEELRRTLAEKRKMHYKALLGAVEESLVRKLREKEVEIERATRRHNELVARDSQLRA 192
Query: 214 EAQNWHYRAKYNESVVNALRNNLQQAI------------SHGVEQGKEGFGDSEVDDAAS 261
E Q W RAK +E +L++ LQQA+ S E+G S VDDA S
Sbjct: 193 EVQVWQERAKAHEDAAASLQSQLQQAVNQCAGGCVSAQDSRAAEEGLLCTTISGVDDAES 252
Query: 262 -YIDPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFIN 320
Y+DP + ++ +C+AC+ ++ +++++PCRHLS+C CD
Sbjct: 253 VYVDP------------------ERVKRPNCKACREREATVVVLPCRHLSICPGCDRTAL 294
Query: 321 VCPVCQMIKTASVE 334
CP+C ++ +SVE
Sbjct: 295 ACPLCLTLRNSSVE 308
>UniRef100_Q6ZHB2 S-ribonuclease binding protein SBP1-like [Oryza sativa]
Length = 342
Score = 131 bits (330), Expect = 2e-29
Identities = 99/323 (30%), Positives = 151/323 (46%), Gaps = 44/323 (13%)
Query: 42 VDPVSYIGNEHISSMIQPNKRSRE------------MEDISKKQRLQISLNYNVCQDDAD 89
V P Y + S + + NKRSRE + ++ Q Q + N+ Q
Sbjct: 36 VSPEVYFPSGGASGINRRNKRSREAIAMAPPPAKEELVNLFTLQPQQSTSFVNMAQLHNR 95
Query: 90 WSASIPNPNA--LSTGLRLSYDDDERNSSVTSASGSMTA-----PPSIILSLGDNIRTEL 142
SAS A +STGLRL+ D+ ++ S + A P +S D + ++
Sbjct: 96 VSASPSRAPAALVSTGLRLALDEQQQQQQQQQESKRLKALCYSSSPMPFVSFSDELAGQM 155
Query: 143 DRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNR 202
RQ EELD +IK Q EQL R + D ++H +LL + E+ ++LREK E E R+
Sbjct: 156 KRQDEELDRFIKEQGEQLRRAMADRVRRHNRALLVAAERSAARRLREKALEAEREARRGA 215
Query: 203 ELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQGKE---GFGDSEVDDA 259
EL ER+ ++ EA W +A ++ +L LQQA + G E G +
Sbjct: 216 ELEERLARLRSEAAAWQAKALSEQAAAVSLHAQLQQAAAAARASGDELRGGEAGPAESSS 275
Query: 260 ASYIDPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDC--DG 317
++Y+D PR G + +C C+ + +++L+PCRHLSLC DC G
Sbjct: 276 SAYVD----------------PRRSGSDR-ACLTCRLRPATVVLLPCRHLSLCGDCFAAG 318
Query: 318 FINV---CPVCQMIKTASVEVYL 337
++V CPVC ++T VE L
Sbjct: 319 DVDVAMACPVCHCVRTGGVEAIL 341
>UniRef100_Q9LPJ0 F6N18.12 [Arabidopsis thaliana]
Length = 277
Score = 125 bits (314), Expect = 2e-27
Identities = 81/263 (30%), Positives = 130/263 (48%), Gaps = 56/263 (21%)
Query: 95 PNPNALSTGLRLSYDDDERNSSVTSASGSMTAPPSIILSLGDNI-RTELDRQQEELDHYI 153
P N + TGLRL +D+ I L +++ ++RQ EELD ++
Sbjct: 44 PPSNMVHTGLRLFSGEDQAQK---------------ISHLSEDVFAAHINRQSEELDEFL 88
Query: 154 KLQ---------KEQLSRGVRDMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNREL 204
Q E+L R + + ++ H +LL ++E+ +V+KLREKE EIE R++ EL
Sbjct: 89 HAQVLISYETIWAEELRRTLAEKRKMHYKALLGAVEESLVRKLREKEVEIERATRRHNEL 148
Query: 205 AERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAI------------SHGVEQGKEGFG 252
R Q+ E Q W RAK +E +L++ LQQA+ S E+G
Sbjct: 149 VARDSQLRAEVQVWQERAKAHEDAAASLQSQLQQAVNQCAGGCVSAQDSRAAEEGLLCTT 208
Query: 253 DSEVDDAAS-YIDPNNFLSIPGAPMKSIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSL 311
S VDDA S Y+DP + ++ +C+AC+ ++ +++++PCRHLS+
Sbjct: 209 ISGVDDAESVYVDP------------------ERVKRPNCKACREREATVVVLPCRHLSI 250
Query: 312 CKDCDGFINVCPVCQMIKTASVE 334
C CD CP+C ++ +SVE
Sbjct: 251 CPGCDRTALACPLCLTLRNSSVE 273
>UniRef100_Q8LAY3 Inhibitor of apoptosis-like protein [Arabidopsis thaliana]
Length = 358
Score = 122 bits (307), Expect = 1e-26
Identities = 77/243 (31%), Positives = 129/243 (52%), Gaps = 24/243 (9%)
Query: 114 NSSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKL---QKEQLSRGVRDMKQK 170
+S V +A+ + T P L +I + +++QQ E+D ++ L Q E++ + + +++
Sbjct: 121 SSCVNAATTTTTTTPFSFLGQDIDISSHMNQQQHEIDRFVSLHLYQMERVKYEIEEKRKR 180
Query: 171 HMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVN 230
+++ +IE+G+VK+LR KEEE E + + N L ER+K +++E Q W A+ NE+ N
Sbjct: 181 QARTIMEAIEQGLVKRLRVKEEERERIGKVNHALEERVKSLSIENQIWRDLAQTNEATAN 240
Query: 231 ALRNNLQQAISHGVEQGKEGFG----DSEVDDAASYIDPNNFLSIPGAPMKSIHPRYQGM 286
LR NL+ ++ V+ G G +E DDA S S G +++ R G+
Sbjct: 241 HLRTNLEHVLAQ-VKDVSRGAGLEKNMNEEDDAESCCGS----SCGGGGEETVRRRV-GL 294
Query: 287 ENLS-----------CRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEV 335
E + CR C ++ +LL+PCRHL LC C ++ CP+C K ASV V
Sbjct: 295 EREAQDKAERRRRRMCRNCGEEESCVLLLPCRHLCLCGVCGSSVHTCPICTSPKNASVHV 354
Query: 336 YLS 338
+S
Sbjct: 355 NMS 357
>UniRef100_Q9LDD1 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone: MGH6
[Arabidopsis thaliana]
Length = 335
Score = 119 bits (298), Expect = 1e-25
Identities = 68/219 (31%), Positives = 118/219 (53%), Gaps = 10/219 (4%)
Query: 127 PPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKK 186
P ++ LG ++ + + + ++D I E++ + + ++ ++ ++E+G++K
Sbjct: 119 PTDPLMFLGQDLSSNVQQHHFDIDRLISNHVERMRMEIEEKRKTQGRRIVEAVEQGLMKT 178
Query: 187 LREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQ 246
LR K++EI ++ + N L E++K + VE Q W A+ NE+ VNALR+NLQQ ++ VE+
Sbjct: 179 LRAKDDEINHIGKLNLFLEEKVKSLCVENQIWRDVAQSNEATVNALRSNLQQVLA-AVER 237
Query: 247 GKEGFGDSEVDDAASYIDPNN-------FLSIPGAPMKSIHPRYQGMENLSCRACKAKDV 299
+ + DDA S N+ + G + GM CR+C +
Sbjct: 238 NRWEEPPTVADDAQSCCGSNDEGDSEEERWKLAGEAQDTKKMCRVGMS--MCRSCGKGEA 295
Query: 300 SMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYLS 338
S+LL+PCRH+ LC C +N CP+C+ KTAS+ V LS
Sbjct: 296 SVLLLPCRHMCLCSVCGSSLNTCPICKSPKTASLHVNLS 334
>UniRef100_Q8L903 Hypothetical protein [Arabidopsis thaliana]
Length = 335
Score = 119 bits (298), Expect = 1e-25
Identities = 68/219 (31%), Positives = 118/219 (53%), Gaps = 10/219 (4%)
Query: 127 PPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVRDMKQKHMASLLTSIEKGIVKK 186
P ++ LG ++ + + + ++D I E++ + + ++ ++ ++E+G++K
Sbjct: 119 PTDPLMFLGQDLSSNVQQHHFDIDRLISNHVERMRMEIEEKRKTQGRRIVEAVEQGLMKT 178
Query: 187 LREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYNESVVNALRNNLQQAISHGVEQ 246
LR K++EI ++ + N L E++K + VE Q W A+ NE+ VNALR+NLQQ ++ VE+
Sbjct: 179 LRAKDDEINHIGKLNLFLEEKVKSLCVENQIWRDVAQSNEATVNALRSNLQQVLA-AVER 237
Query: 247 GKEGFGDSEVDDAASYIDPNN-------FLSIPGAPMKSIHPRYQGMENLSCRACKAKDV 299
+ + DDA S N+ + G + GM CR+C +
Sbjct: 238 NRWEEPPTVADDAQSCYGSNDEGDSEEERWKLAGEAQDTKKMCRVGMS--MCRSCGKGEA 295
Query: 300 SMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYLS 338
S+LL+PCRH+ LC C +N CP+C+ KTAS+ V LS
Sbjct: 296 SVLLLPCRHMCLCSVCGSSLNTCPICKSPKTASLHVNLS 334
>UniRef100_Q7XSC9 OSJNBb0118P14.4 protein [Oryza sativa]
Length = 347
Score = 119 bits (297), Expect = 2e-25
Identities = 70/241 (29%), Positives = 118/241 (48%), Gaps = 13/241 (5%)
Query: 106 LSYDDDERNSSVTSASGSMTAPPSIILSLGDNIRTELDRQQEELDHYIKLQKEQLSRGVR 165
L + D ++ +V + S + L + ++L Q E+D ++L+ E++ G+
Sbjct: 112 LVFPGDVQSRAVGCGAASTSGRAGNAAGLSQGLLSQLYHQGVEIDALVRLESERMRAGLE 171
Query: 166 DMKQKHMASLLTSIEKGIVKKLREKEEEIENMNRKNRELAERIKQVAVEAQNWHYRAKYN 225
+ +++H+ ++++++E+ +LR E E+E +N EL ER++Q+ E Q W AK +
Sbjct: 172 EARRRHVRAVVSTVERAAAGRLRAAEAELERARCRNMELEERLRQMTAEGQAWLSVAKSH 231
Query: 226 ESVVNALRNNLQQAISH--------GVEQGKEGFGDSEVDDAASYIDPNNFLSIPGAPMK 277
E+V LR L Q + G GD+E + Y P +
Sbjct: 232 EAVAAGLRATLDQLLQSPCAALAVAGAAGAGGAEGDAEDAQSCCYETPCGGDNAGADDAA 291
Query: 278 SIHPRYQGMENLSCRACKAKDVSMLLIPCRHLSLCKDCDGFINVCPVCQMIKTASVEVYL 337
S P C+AC A + SMLL+PCRHL LC+ C+ ++ CPVC K AS+ V L
Sbjct: 292 SKTP-----AAALCKACGAGEASMLLLPCRHLCLCRGCEAAVDACPVCAATKNASLHVLL 346
Query: 338 S 338
S
Sbjct: 347 S 347
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.131 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 552,647,129
Number of Sequences: 2790947
Number of extensions: 22853909
Number of successful extensions: 106178
Number of sequences better than 10.0: 1568
Number of HSP's better than 10.0 without gapping: 411
Number of HSP's successfully gapped in prelim test: 1175
Number of HSP's that attempted gapping in prelim test: 104080
Number of HSP's gapped (non-prelim): 2937
length of query: 338
length of database: 848,049,833
effective HSP length: 128
effective length of query: 210
effective length of database: 490,808,617
effective search space: 103069809570
effective search space used: 103069809570
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0008.8


