

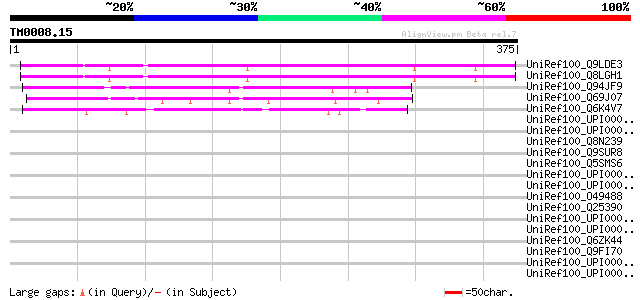
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.15
(375 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LDE3 F26F24.26 [Arabidopsis thaliana] 280 6e-74
UniRef100_Q8LGH1 Hypothetical protein [Arabidopsis thaliana] 272 1e-71
UniRef100_Q94JF9 P0684B02.9 protein [Oryza sativa] 135 1e-30
UniRef100_Q69J07 Kelch repeat-containing F-box-like protein [Ory... 125 2e-27
UniRef100_Q6K4V7 Kelch repeat-containing F-box-like protein [Ory... 106 1e-21
UniRef100_UPI000043623C UPI000043623C UniRef100 entry 45 0.003
UniRef100_UPI0000248B7A UPI0000248B7A UniRef100 entry 45 0.003
UniRef100_Q8N239 Hypothetical protein FLJ34960 [Homo sapiens] 45 0.003
UniRef100_Q9SUR8 Hypothetical protein F9D16.50 [Arabidopsis thal... 45 0.004
UniRef100_Q5SMS6 Fimbriata-like [Oryza sativa] 44 0.007
UniRef100_UPI00001D1754 UPI00001D1754 UniRef100 entry 41 0.047
UniRef100_UPI000027CD05 UPI000027CD05 UniRef100 entry 41 0.047
UniRef100_O49488 Hypothetical protein AT4g34170 [Arabidopsis tha... 40 0.081
UniRef100_Q25390 Alpha-scruin [Limulus polyphemus] 40 0.11
UniRef100_UPI000036328B UPI000036328B UniRef100 entry 40 0.14
UniRef100_UPI000035ED7D UPI000035ED7D UniRef100 entry 40 0.14
UniRef100_Q6ZK44 Kelch repeat-containing protein-like [Oryza sat... 40 0.14
UniRef100_Q9FI70 Arabidopsis thaliana genomic DNA, chromosome 5,... 40 0.14
UniRef100_UPI00001C66F3 UPI00001C66F3 UniRef100 entry 39 0.18
UniRef100_UPI0000363253 UPI0000363253 UniRef100 entry 39 0.24
>UniRef100_Q9LDE3 F26F24.26 [Arabidopsis thaliana]
Length = 394
Score = 280 bits (715), Expect = 6e-74
Identities = 160/380 (42%), Positives = 221/380 (58%), Gaps = 18/380 (4%)
Query: 9 EEAPIHGDVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLIILTQSTRAS 68
EE+ I GD+LE+ILS++PL+ L AC VS +W AVF SL +T+ PWL + Q
Sbjct: 16 EESSIDGDILESILSYLPLLDLDSACQVSKSWNRAVFYSLRRLKTM-PWLFVYNQRNSPP 74
Query: 69 HVIT--AHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSIDALH 126
+ + A AYDP+S W+E+ H S RSSHSTLLY LSPA F+FS DA H
Sbjct: 75 YTMATMAMAYDPKSEAWIELNTASSPVEHVSVA---RSSHSTLLYALSPARFSFSTDAFH 131
Query: 127 LDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMES--NTWVMCQSMP 184
L W H PRVWR DPIVA VG+ +++AGG CDFE+D AVE++D+ES W C+SMP
Sbjct: 132 LTWQHVAPPRVWRIDPIVAVVGRSLIIAGGVCDFEEDRFAVELFDIESGDGAWERCESMP 191
Query: 185 AMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPDQSVGYCVT-G 243
L S+ STWLSVAV+ E++ VTE+ SG+T F+ V+ W DLCP + Y + G
Sbjct: 192 DFLYESASSTWLSVAVSSEKMYVTEKRSGVTCSFNPVTRSWTKLLDLCPGECSLYSRSIG 251
Query: 244 MLGKRLMVAGVVGDGEDVKAVKLWAVKGGLGSGMT-EEVGEMPEELLEKFKGDSDE--LG 300
RL++AG++GD + ++LW V S + E +G MPE LEK +G + + L
Sbjct: 252 FSVNRLIMAGIIGDEYNPTGIELWEVIDSDESHLKFESIGSMPETYLEKLRGINSDWPLT 311
Query: 301 SLEVTWVGNFVYLRNTLEI-EELVMCEVVNGSRCEWRSVRNAAV-----DGGTRMVVCGS 354
S+ + VG+ VY+ E E+V E+ G C+WR++ NA R++V S
Sbjct: 312 SIVLNAVGDMVYVHGAAENGGEIVAAEIEGGKLCKWRTLPNADATWKKSHAAERVIVACS 371
Query: 355 EVRLEDLQSAMVSGIQTCRT 374
V DL+ A + + T
Sbjct: 372 NVGFSDLKLAFRNNLSFLST 391
>UniRef100_Q8LGH1 Hypothetical protein [Arabidopsis thaliana]
Length = 394
Score = 272 bits (696), Expect = 1e-71
Identities = 158/380 (41%), Positives = 219/380 (57%), Gaps = 18/380 (4%)
Query: 9 EEAPIHGDVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLIILTQSTRAS 68
E+ I GD+LE+ILS++PL+ L A VS +W AVF SL +T+ PWL + Q
Sbjct: 16 EQRSIDGDILESILSYLPLLDLDSASQVSKSWNRAVFYSLRRLKTM-PWLFVYNQRNSPP 74
Query: 69 HVIT--AHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSIDALH 126
+ + A AYDP+S W+E+K H S RSSHSTLLY LSPA F+FS DA H
Sbjct: 75 YTMATMAMAYDPKSEAWIELKTASSPVEHVSVA---RSSHSTLLYALSPARFSFSTDAFH 131
Query: 127 LDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMES--NTWVMCQSMP 184
L W H PRVWR DPIVA VG+ +++AGG CDFE+D AVE++D+ES W C+SMP
Sbjct: 132 LTWQHVSPPRVWRIDPIVAVVGRSLIIAGGVCDFEEDRFAVELFDIESGDGAWERCESMP 191
Query: 185 AMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPDQSVGYCVT-G 243
L S+ STWLSVAV+ E++ VTE+ SG+T F+ V+ W DLCP + Y + G
Sbjct: 192 DFLYESASSTWLSVAVSSEKMYVTEKRSGVTCSFNPVTRXWTKLLDLCPGECSLYSRSIG 251
Query: 244 MLGKRLMVAGVVGDGEDVKAVKLWAVKGGLGSGMT-EEVGEMPEELLEKFKGDSDE--LG 300
RL++A ++GD + ++LW V S + E +G MPE LEK +G + + L
Sbjct: 252 FSVNRLIMAWIIGDEYNPTGIELWEVIDSDESHLKFESIGSMPETYLEKLRGINSDWPLT 311
Query: 301 SLEVTWVGNFVYLRNTLEI-EELVMCEVVNGSRCEWRSVRNAAV-----DGGTRMVVCGS 354
S+ + VG+ VY+ E E+V E+ G C+WR++ NA R++V S
Sbjct: 312 SIVLNAVGDMVYVHGAAENGGEIVAAEIEGGKLCKWRTLPNADATWKKSHAAERVIVACS 371
Query: 355 EVRLEDLQSAMVSGIQTCRT 374
V DL+ A + + T
Sbjct: 372 NVGFSDLKLAFRNNLSFLST 391
>UniRef100_Q94JF9 P0684B02.9 protein [Oryza sativa]
Length = 426
Score = 135 bits (341), Expect = 1e-30
Identities = 104/303 (34%), Positives = 140/303 (45%), Gaps = 23/303 (7%)
Query: 10 EAPIHGDVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLIILTQSTRASH 69
E ++GDVLE+++ VP L + VS W AV ++L PWL++ R
Sbjct: 41 EMDLYGDVLESVVERVPAADLAASARVSREWLRAVRAALRRRPRRLPWLVVHLHGRRRRT 100
Query: 70 VITAHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSIDALHLDW 129
AYDP S WV + P PS VR + + LS + A S D L D
Sbjct: 101 A----AYDPHSGAWVTV--PAARHDTPSHVRLVRGAGGDRVCALSLSGLAVSGDPLGKDV 154
Query: 130 HHA-PAPRVWRTDPIVARVGQRVVVAGGACDFE----DDPLAVEMYDMESNTWVMCQSMP 184
A AP VWR DP+ A VG RVV GGAC +D VE++ ES +W C MP
Sbjct: 155 CVALKAPGVWRVDPVFAAVGDRVVALGGACQLALGEGEDASVVEVH--ESGSWTACGPMP 212
Query: 185 AMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPDQSV---GYCV 241
A L+ S+ +TWLSVA + V +T+ S+G FD +W L PD +V G
Sbjct: 213 AELRESAAATWLSVAATDQRVYLTDRSTGWASWFDPAKQQWGPTCRLRPDATVSTWGLAP 272
Query: 242 TGMLGKRLMVAGV--VGDGEDVKA---VKLWAVKG-GLGSGMTEEVGEMPEELLEK-FKG 294
+RL++ G G E K+ ++ W V G GL MP E+ E+ F
Sbjct: 273 GRGGAERLVLFGAKRCGRAEQAKSRVVIQAWEVDGDGLALSRGAAHDTMPGEMSERLFPR 332
Query: 295 DSD 297
D D
Sbjct: 333 DED 335
>UniRef100_Q69J07 Kelch repeat-containing F-box-like protein [Oryza sativa]
Length = 394
Score = 125 bits (314), Expect = 2e-27
Identities = 95/309 (30%), Positives = 136/309 (43%), Gaps = 27/309 (8%)
Query: 13 IHGDVLETILSHVPLIHLVPACHVSNTWKHAVFSSLAHTRTIKPWLIILTQSTRASHVIT 72
+HGDVLE+ + VP L A VS W AV ++L PWL++ R +
Sbjct: 4 LHGDVLESAVERVPAPDLAAAALVSREWLRAVRAALRRRMLRLPWLVVHVIHLRGQRRLA 63
Query: 73 AHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLY--------TLSPAEFAFSIDA 124
A AYDPRS W+ + P + H + + SH L+ LS + A + DA
Sbjct: 64 A-AYDPRSGAWLAVPTAPPAR-HGATSPPQPHSHVRLMRGASGDRVCALSLSGLAVARDA 121
Query: 125 LHLDWHHA----PAPRVWRTDPIVARVGQRVVVAGGACDFE----DDPLAVEMYDMESNT 176
L +D AP VWR DP++A VG RVV GGAC +D AVE++ E
Sbjct: 122 LGMDDDALVVALKAPGVWRVDPVLAAVGDRVVAMGGACRLALGDGEDTSAVEVH--ERGG 179
Query: 177 WVMCQSMPAMLKGS--SGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPD 234
W C ++P L+ S + +TWLS A + V V + ++G FD +W L PD
Sbjct: 180 WTHCGAVPTALRESAAAAATWLSTAATDQRVYVADRATGTASWFDPAKQQWGPTSRLRPD 239
Query: 235 QSVGY--CVTGMLGKRLMVAGVVGDGEDVKAVKLWAVKG---GLGSGMTEEVGEMPEELL 289
+V G G ++ V + ++ W V G L G MP E+
Sbjct: 240 ATVSTWGLAAGRAGAEKIILFGVKHADSRVVIRSWEVDGDSLSLSHGAAAAHDTMPSEMS 299
Query: 290 EKFKGDSDE 298
E+ D+
Sbjct: 300 ERLFPHGDD 308
>UniRef100_Q6K4V7 Kelch repeat-containing F-box-like protein [Oryza sativa]
Length = 401
Score = 106 bits (264), Expect = 1e-21
Identities = 97/305 (31%), Positives = 127/305 (40%), Gaps = 34/305 (11%)
Query: 10 EAPIHGDVLETILSHVPLIHLVPA-CHVSNTWKHAVFSSLAHTRTIK---PWLIILTQST 65
++ +HGD+LE +L VP L + VS W+ A + R + P L+
Sbjct: 8 DSVLHGDLLECVLLRVPHGELTASPALVSREWRRAAREAHQRHRRRRRHLPCLVAHVHGA 67
Query: 66 RASHVITAHAYDPRSHVWVE--MKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSID 123
A + H YDPR+ W ++ L A A +Y LS A A S D
Sbjct: 68 AAGVGRSTHVYDPRAGAWASDGWRVAGALPVRRCACAG-----GDRVYALSLASMAVSED 122
Query: 124 ALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSM 183
A+ W P PRVWR DP+VA VG VVV GG C VE+ D E W C M
Sbjct: 123 AVGAAWRELPPPRVWRVDPVVAAVGPHVVVLGGGCGATAAAGVVEVLD-EGAGWATCPPM 181
Query: 184 PAMLKGSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDL-CPD-------Q 235
PA L S W+S A + V V E +G FD + +W L P+ +
Sbjct: 182 PAPL----ASRWVSSAASERRVYVVERRTGWASWFDPAARQWGPARQLQLPEGNNTASVE 237
Query: 236 SVGYC-VT----GMLGKRLMVAGVVGDGEDVKAVKLWAVKGG-LGSGMTEEVGEMPEELL 289
S C VT G +RL+V G G+ V LW V G L MP E+
Sbjct: 238 SWAACGVTTSGGGGASERLLVLAGGGGGK----VSLWGVDGDTLLLDAEANNTSMPPEMS 293
Query: 290 EKFKG 294
E+ G
Sbjct: 294 ERLGG 298
>UniRef100_UPI000043623C UPI000043623C UniRef100 entry
Length = 617
Score = 45.1 bits (105), Expect = 0.003
Identities = 30/117 (25%), Positives = 45/117 (37%), Gaps = 14/117 (11%)
Query: 72 TAHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYT----LSPAEFAFSIDALHL 127
+ YD R + W + P P+ HST+ Y + E+ F D
Sbjct: 426 SVECYDARDNCWTAVS-PMPV---------AMEFHSTIEYKDRIYVLQGEYFFCFDPRKD 475
Query: 128 DWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSMP 184
W H P+ + RT + A + G C VE+YD+E NTW + +P
Sbjct: 476 YWGHLPSMNIPRTQGLAALHKNCIYYIAGICRNHQRTFTVEVYDIEQNTWCRKRDLP 532
>UniRef100_UPI0000248B7A UPI0000248B7A UniRef100 entry
Length = 576
Score = 45.1 bits (105), Expect = 0.003
Identities = 30/117 (25%), Positives = 45/117 (37%), Gaps = 14/117 (11%)
Query: 72 TAHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYT----LSPAEFAFSIDALHL 127
+ YD R + W + P P+ HST+ Y + E+ F D
Sbjct: 385 SVECYDARDNCWTAVS-PMPV---------AMEFHSTIEYKDRIYVLQGEYFFCFDPRKD 434
Query: 128 DWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSMP 184
W H P+ + RT + A + G C VE+YD+E NTW + +P
Sbjct: 435 YWGHLPSMNIPRTQGLAALHKNCIYYIAGICRNHQRTFTVEVYDIEQNTWCRKRDLP 491
>UniRef100_Q8N239 Hypothetical protein FLJ34960 [Homo sapiens]
Length = 644
Score = 45.1 bits (105), Expect = 0.003
Identities = 39/138 (28%), Positives = 56/138 (40%), Gaps = 25/138 (18%)
Query: 75 AYDPRSHVWVEM-KLPHPLKGHPSATAAVRSSHSTLLYTL---SPAEFAFS--------- 121
A+D +H W + +LP PL GH TA L+ L SP+ A S
Sbjct: 338 AFDVYNHRWRSLTQLPTPLLGHSVCTAG------NFLFVLGGESPSGSASSPLADDSRVV 391
Query: 122 ------IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESN 175
D W PA R R VG+R++ GG + +VEMYD+ +
Sbjct: 392 TAQVHRYDPRFHAWTEVPAMREARAHFWCGAVGERLLAVGGLGAGGEVLASVEMYDLRRD 451
Query: 176 TWVMCQSMPAMLKGSSGS 193
W ++P L G +G+
Sbjct: 452 RWTAAGALPRALHGHAGA 469
>UniRef100_Q9SUR8 Hypothetical protein F9D16.50 [Arabidopsis thaliana]
Length = 383
Score = 44.7 bits (104), Expect = 0.004
Identities = 46/156 (29%), Positives = 67/156 (42%), Gaps = 24/156 (15%)
Query: 129 WHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSMPAMLK 188
WH AP+ RV R P + R+ V GG CD D +E++D ++ TW Q +
Sbjct: 159 WHEAPSMRVGRVFPSACTLDGRIYVTGG-CDNLDTMNWMEIFDTKTQTWEFLQIPSEEI- 216
Query: 189 GSSGSTWLSVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPD------QSVGYCVT 242
GS +LSV+ G V V + +T+ KW D+C + S YCV
Sbjct: 217 -CKGSEYLSVSYQG-TVYVKSDEKDVTYKMH--KGKW-READICMNNGWSLSSSSSYCVV 271
Query: 243 GMLGKRLMVAGVVGDGE----DVKAVKLWAVKGGLG 274
+ R +GE D+K + W + GLG
Sbjct: 272 ENVFYRYC------EGEIRWYDLKN-RAWTILKGLG 300
>UniRef100_Q5SMS6 Fimbriata-like [Oryza sativa]
Length = 411
Score = 43.9 bits (102), Expect = 0.007
Identities = 21/77 (27%), Positives = 39/77 (50%), Gaps = 7/77 (9%)
Query: 16 DVLETILSHVPLIHLVPACHVSNTWKHAVFSS---LAHTRTIKPWLIILTQSTRASHVIT 72
D+LE I + +P++ ++ + V W ++SS H +PW + T + A+
Sbjct: 55 DILERIFTFLPIVSMIRSTAVCKRWHDIIYSSRFLWTHMLPQRPWYFMFTSNESAA---- 110
Query: 73 AHAYDPRSHVWVEMKLP 89
+AYDP W +++LP
Sbjct: 111 GYAYDPILRKWYDLELP 127
>UniRef100_UPI00001D1754 UPI00001D1754 UniRef100 entry
Length = 645
Score = 41.2 bits (95), Expect = 0.047
Identities = 34/132 (25%), Positives = 54/132 (40%), Gaps = 13/132 (9%)
Query: 75 AYDPRSHVWVEM-KLPHPLKGHPSATA----------AVRSSHSTLLY--TLSPAEFAFS 121
++D +H W + +LP PL GH T ++ S S+ L + S
Sbjct: 339 SFDVYNHSWHTLTQLPTPLLGHSVCTIGNFLFVLGGESLSGSPSSSLADGSRSVTALVHR 398
Query: 122 IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQ 181
D W PA R R VG+ ++ GG + +VEMYD+ + WV
Sbjct: 399 YDPRFHTWTEVPAMREARAYFWCGVVGESLLAVGGLGTCGEALASVEMYDLRRDRWVAAG 458
Query: 182 SMPAMLKGSSGS 193
+P + G +G+
Sbjct: 459 ELPRAVHGHAGA 470
>UniRef100_UPI000027CD05 UPI000027CD05 UniRef100 entry
Length = 579
Score = 41.2 bits (95), Expect = 0.047
Identities = 29/113 (25%), Positives = 43/113 (37%), Gaps = 6/113 (5%)
Query: 72 TAHAYDPRSHVWVEMKLPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSIDALHLDWHH 131
+ YD R + W + P P+ +T R +Y L E+ F D W H
Sbjct: 388 SVECYDARDNCWTAVS-PMPVAMEFHSTLEYRDR----IYVLQ-GEYFFCFDPRKDYWSH 441
Query: 132 APAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSMP 184
V R+ + A + G C VE+YD+E NTW + +P
Sbjct: 442 LAPMSVPRSQGLAALYKNCIYYIAGICRNHQRTFTVEVYDIEKNTWSRRRDLP 494
>UniRef100_O49488 Hypothetical protein AT4g34170 [Arabidopsis thaliana]
Length = 293
Score = 40.4 bits (93), Expect = 0.081
Identities = 30/95 (31%), Positives = 45/95 (46%), Gaps = 4/95 (4%)
Query: 122 IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQ 181
+D+ W AP+ RV R P V + ++ V GG CD D +E++D ++ TW Q
Sbjct: 113 MDSRSHTWREAPSMRVPRMFPSVCTLDGKIYVMGG-CDNLDSTNWMEVFDTKTQTWEFLQ 171
Query: 182 SMPAMLKGSSGSTWLSVAVAGEEVLVTEESSGMTF 216
+ G GS + SV G V V E +T+
Sbjct: 172 IPSEEIFG--GSAYESVRYEG-TVYVWSEKKDVTY 203
>UniRef100_Q25390 Alpha-scruin [Limulus polyphemus]
Length = 918
Score = 40.0 bits (92), Expect = 0.11
Identities = 29/112 (25%), Positives = 44/112 (38%), Gaps = 6/112 (5%)
Query: 120 FSIDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVM 179
F + W R R V + +R+ V GG + AVEMY+ E + W
Sbjct: 158 FQLTVQTKQWRKRADMRYARAHHNVTVMDERIFVFGGRDSNGEILAAVEMYEPEMDQWTT 217
Query: 180 CQSMPAMLKGSSGSTWLSVAVAGEEVLVTEES------SGMTFCFDTVSMKW 225
S+P + GS+ + + V +E S FCFD ++ KW
Sbjct: 218 LASIPEPMMGSAIANNEGIIYVIGGVTTNKEKKPEGNLSNKIFCFDPLNNKW 269
Score = 37.4 bits (85), Expect = 0.69
Identities = 27/78 (34%), Positives = 35/78 (44%), Gaps = 10/78 (12%)
Query: 155 GGACDFEDDPLAVEMYDMESNTWVMCQSMPAMLKGSS-----GSTWLSVAVAGEEVLVTE 209
GG D +VE YD ESN W M +SMP+ G + G WL + G + TE
Sbjct: 701 GGRDDIGRLVSSVESYDHESNEWTMEKSMPSPRMGMAAVAHGGYIWL---LGGLTSMTTE 757
Query: 210 ESSGM--TFCFDTVSMKW 225
E + C+D V W
Sbjct: 758 EPPVLDDVLCYDPVFKHW 775
>UniRef100_UPI000036328B UPI000036328B UniRef100 entry
Length = 579
Score = 39.7 bits (91), Expect = 0.14
Identities = 28/114 (24%), Positives = 44/114 (38%), Gaps = 8/114 (7%)
Query: 72 TAHAYDPRSHVWVEMK-LPHPLKGHPSATAAVRSSHSTLLYTLSPAEFAFSIDALHLDWH 130
+ YD R + W + +P ++ H + R +Y L E+ F D W
Sbjct: 388 SVECYDARDNCWTAVSPMPVAMEFHSALEYRDR------IYVLQ-GEYFFCFDPRKDYWS 440
Query: 131 HAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQSMP 184
H V R+ + A + G C VE+YD+E NTW + +P
Sbjct: 441 HLAPMSVPRSQGLAALYKNCIYYIAGICRNHQRTFTVEVYDIERNTWSRKRDLP 494
>UniRef100_UPI000035ED7D UPI000035ED7D UniRef100 entry
Length = 599
Score = 39.7 bits (91), Expect = 0.14
Identities = 34/125 (27%), Positives = 53/125 (42%), Gaps = 8/125 (6%)
Query: 143 IVARVGQRVVVAGGACDFEDDPLAVEMY----DMESNTWVMCQSMPAM--LKGSSGSTWL 196
++ + + V+ G D ED ++ Y D S+ W MP+ L L
Sbjct: 333 LITKENKLHVLGGLFVDEEDKETPLQCYHYQLDALSSDWTALPPMPSSRCLFAMGECESL 392
Query: 197 SVAVAGEEVLVTEESSGMTFCFDTVSMKWDGPYDLCPDQSVGYCVTGMLGKRLMVAGVVG 256
AVAG++ L + ES C+DT MKW+ L P + G+CV G + G
Sbjct: 393 IFAVAGKD-LQSNESHDTVLCYDTEKMKWNETKKL-PLRIHGHCVVSENGLVYSIGGKTD 450
Query: 257 DGEDV 261
D + +
Sbjct: 451 DNKTI 455
>UniRef100_Q6ZK44 Kelch repeat-containing protein-like [Oryza sativa]
Length = 410
Score = 39.7 bits (91), Expect = 0.14
Identities = 43/162 (26%), Positives = 62/162 (37%), Gaps = 18/162 (11%)
Query: 122 IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQ 181
I A DW PA V R D ++G + V G + + V++Y+ SNTW
Sbjct: 78 IPAPRWDWEEMPAAPVPRLDGYSVQIGDLLYVFAGYENLDHVHSHVDVYNFTSNTWTGRF 137
Query: 182 SMPAMLKGSSGSTWLSVAVAGEEVLVTEESSG--------MTFCFDTVSMKWDGPYDLCP 233
MP + S L +A G + G F DTV+ +W ++L P
Sbjct: 138 DMPKEMANSH----LGIATDGRYIYALTGQFGPQCRSPINRNFVVDTVTKEW---HELPP 190
Query: 234 DQSVGYCVTGMLGK-RLMVAGVVGDGEDVKAVKLW--AVKGG 272
Y L + RL V G + ++ W AVK G
Sbjct: 191 LPVPRYAPATQLWRGRLHVMGGGKEDRHEPGLEHWSLAVKDG 232
>UniRef100_Q9FI70 Arabidopsis thaliana genomic DNA, chromosome 5, TAC clone:K19E20
[Arabidopsis thaliana]
Length = 372
Score = 39.7 bits (91), Expect = 0.14
Identities = 25/93 (26%), Positives = 47/93 (49%), Gaps = 8/93 (8%)
Query: 91 PLKGHPSATAAVRSSHSTLLYTL------SPAEFAFSIDALHLDWHHAPAPRVWRTDPIV 144
P+ H S ++++ + S + Y + +P+ +D WH AP+ R+ R P
Sbjct: 114 PIPNHHSHSSSIVAIGSNI-YAIGGSIENAPSSKVSILDCRSHTWHEAPSMRMKRNYPAA 172
Query: 145 ARVGQRVVVAGGACDFEDDPLAVEMYDMESNTW 177
V ++ VAGG +F D +E++D+++ TW
Sbjct: 173 NVVDGKIYVAGGLEEF-DSSKWMEVFDIKTQTW 204
>UniRef100_UPI00001C66F3 UPI00001C66F3 UniRef100 entry
Length = 644
Score = 39.3 bits (90), Expect = 0.18
Identities = 32/132 (24%), Positives = 50/132 (37%), Gaps = 13/132 (9%)
Query: 75 AYDPRSHVWVEM-KLPHPLKGHPSATAAV------------RSSHSTLLYTLSPAEFAFS 121
++D +H W + +LP PL GH T S S + S
Sbjct: 338 SFDVYNHSWHSLTQLPTPLLGHSVCTTGNFLFVLGGESLPGSPSSSLAAGSRSITALVHR 397
Query: 122 IDALHLDWHHAPAPRVWRTDPIVARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVMCQ 181
D W PA R R VG+ ++ GG + +VEMYD+ + W+
Sbjct: 398 YDPRFHTWTQVPAMREARAYFWCGVVGESLLAVGGLGICGEALASVEMYDLRRDRWMAAG 457
Query: 182 SMPAMLKGSSGS 193
+P + G +G+
Sbjct: 458 QLPRAVHGHAGA 469
>UniRef100_UPI0000363253 UPI0000363253 UniRef100 entry
Length = 423
Score = 38.9 bits (89), Expect = 0.24
Identities = 55/199 (27%), Positives = 77/199 (38%), Gaps = 17/199 (8%)
Query: 88 LPHPLKGHPSATAAVRSSHSTLLYT---LSPAEFAFSIDALHLDWHHAPAPRVWRTDPIV 144
LP S VR + TLL S E A S D W H + R RT +
Sbjct: 177 LPFQQNTLQSRRTQVRGAQQTLLTIGGRPSLTERALSYDPRFNTWLHLASMRQRRTHFSL 236
Query: 145 ARVGQRVVVAGGACDFEDDPLAVEMYDMESNTWVM--CQSMPAMLKGSSGSTWLSVAVAG 202
A G R+ GG + E E Y SNTW M MP SS ++ V G
Sbjct: 237 AASGGRLYAIGGR-NVEGLLATTESYLPSSNTWQMRAPMEMPRCCHSSSTLPSGNILVTG 295
Query: 203 EEVLVTEESSGMTFCFDTVSMKWDGPYDLCPDQSV-GYCVTGMLGKRLMVA-----GVVG 256
+ S C+ S W DL P ++ G+ + LG ++ V G G
Sbjct: 296 GYINCAYSRS--VACYQVESDTW---VDLAPMETPRGWHCSATLGGKVYVVGGSQLGPGG 350
Query: 257 DGEDVKAVKLWAVKGGLGS 275
+ DV +V++++ + G S
Sbjct: 351 ERVDVVSVEVFSPESGTWS 369
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.133 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 653,315,387
Number of Sequences: 2790947
Number of extensions: 27469901
Number of successful extensions: 59515
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 113
Number of HSP's that attempted gapping in prelim test: 59363
Number of HSP's gapped (non-prelim): 230
length of query: 375
length of database: 848,049,833
effective HSP length: 129
effective length of query: 246
effective length of database: 488,017,670
effective search space: 120052346820
effective search space used: 120052346820
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0008.15


