

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.11
(265 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
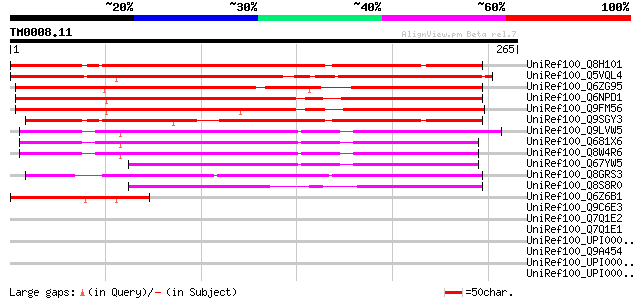
UniRef100_Q8H101 Hypothetical protein At1g10660 [Arabidopsis tha... 266 4e-70
UniRef100_Q5VQL4 Hypothetical protein P0583G08.26 [Oryza sativa] 225 8e-58
UniRef100_Q6ZG95 Hypothetical protein OJ1007_D04.9 [Oryza sativa] 224 1e-57
UniRef100_Q6NPD1 At5g62960 [Arabidopsis thaliana] 220 3e-56
UniRef100_Q9FM56 Gb|AAF17656.1 [Arabidopsis thaliana] 214 2e-54
UniRef100_Q9SGY3 F20B24.10 [Arabidopsis thaliana] 195 1e-48
UniRef100_Q9LVW5 Gb|AAF17656.1 [Arabidopsis thaliana] 177 2e-43
UniRef100_Q681X6 MRNA, complete cds, clone: RAFL21-92-C23 [Arabi... 177 3e-43
UniRef100_Q8W4R6 AT3g27760/MGF10_16 [Arabidopsis thaliana] 174 2e-42
UniRef100_Q67YW5 MRNA, complete cds, clone: RAFL24-05-D16 [Arabi... 135 8e-31
UniRef100_Q8GRS3 Hypothetical protein OJ1477_F01.109 [Oryza sativa] 134 2e-30
UniRef100_Q8S8R0 Predicted protein [Arabidopsis thaliana] 111 2e-23
UniRef100_Q6Z6B1 Hypothetical protein P0622F08.29 [Oryza sativa] 76 8e-13
UniRef100_Q9C6E3 Hypothetical protein T32G9.33 [Arabidopsis thal... 38 0.23
UniRef100_Q7Q1E2 ENSANGP00000015709 [Anopheles gambiae str. PEST] 35 2.0
UniRef100_Q7Q1E1 ENSANGP00000015887 [Anopheles gambiae str. PEST] 35 2.0
UniRef100_UPI00003AA3B7 UPI00003AA3B7 UniRef100 entry 35 2.6
UniRef100_Q9A454 Hypothetical protein CC2989 [Caulobacter cresce... 34 3.4
UniRef100_UPI000042D911 UPI000042D911 UniRef100 entry 34 4.4
UniRef100_UPI00002CCA07 UPI00002CCA07 UniRef100 entry 34 4.4
>UniRef100_Q8H101 Hypothetical protein At1g10660 [Arabidopsis thaliana]
Length = 320
Score = 266 bits (680), Expect = 4e-70
Identities = 134/249 (53%), Positives = 173/249 (68%), Gaps = 10/249 (4%)
Query: 1 METDTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQQQRAGLLYE 60
M DTTA SYWLNWR +CAL ++ + L + LIWKYE K R +R E Q++ G L++
Sbjct: 1 MAADTTASSYWLNWRVLLCALILLAPIVLAAVLIWKYE--GKRRRQR-ESQRELPGTLFQ 57
Query: 61 DELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGL 120
DE W TC KRIHP+WLLA+R+ SF+ +L LLI N+V DG GI YFYTQWTFTLVT+YFG
Sbjct: 58 DEAWTTCFKRIHPLWLLAFRVFSFVAMLTLLISNVVRDGAGIFYFYTQWTFTLVTLYFGY 117
Query: 121 ASFFSIYGCFFKHDKFEGNTVGSACL-NTERGTYVAP-TIDGDTDIPHMYKSTDAYQEPH 178
AS S+YGC + + GN + +TE+GTY P +DG+ + K+++ E
Sbjct: 118 ASVLSVYGCCIYNKEASGNMESYTSIGDTEQGTYRPPIALDGEGNTS---KASNRPSEAP 174
Query: 179 TQNTAGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFL 238
+ TAG W YIFQI+FQTCAGAVVLTD+VFW ++YPF K ++L VCMHS+NAVFL
Sbjct: 175 ARKTAGFWVYIFQILFQTCAGAVVLTDIVFWAIIYPF--TKGYKLSFLDVCMHSLNAVFL 232
Query: 239 LGETSLNGM 247
LG+TSLN +
Sbjct: 233 LGDTSLNSL 241
>UniRef100_Q5VQL4 Hypothetical protein P0583G08.26 [Oryza sativa]
Length = 333
Score = 225 bits (574), Expect = 8e-58
Identities = 117/263 (44%), Positives = 163/263 (61%), Gaps = 21/263 (7%)
Query: 1 METDTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQQQR------ 54
M DTTA YWLNWRF +CA+W+ M L FLIWKYE S+D G+ +
Sbjct: 1 MAVDTTASEYWLNWRFMLCAVWVYSCMVLACFLIWKYEG-PSSQDGNGDGGEDSEDARPP 59
Query: 55 ---AGLLYEDELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTF 111
+G++Y ++ W CL++IHP WLLA+R+VSF +L LL ++V DG + +YTQWTF
Sbjct: 60 RAASGVVYLEDCWKPCLEQIHPGWLLAFRVVSFFILASLLAVDVVVDGWSVFLYYTQWTF 119
Query: 112 TLVTIYFGLASFFSIYGCFFKHDKFEGNTVGSACLNTERGTY-VAPTIDGDTDIPHMYKS 170
LVT+YFGL S SIYGC+ K N G+ + GTY +AP G++ K+
Sbjct: 120 LLVTLYFGLGSVLSIYGCYQYSYKNGDNRSGA-----DHGTYIIAPA--GESVYDQSIKN 172
Query: 171 TDAYQEPH-TQNTAGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVC 229
Y + H + AG WGY+FQIMFQT AGAV++TDLVFW +LYPFLA + + +
Sbjct: 173 -PCYSKMHGGKEIAGFWGYLFQIMFQTNAGAVMITDLVFWFILYPFLAYNQYDMNFLLIG 231
Query: 230 MHSVNAVFLLGETSLNGMVKYLW 252
HS+N VF++G+T++N + ++ W
Sbjct: 232 THSINVVFMIGDTAMNSL-RFPW 253
>UniRef100_Q6ZG95 Hypothetical protein OJ1007_D04.9 [Oryza sativa]
Length = 362
Score = 224 bits (572), Expect = 1e-57
Identities = 114/257 (44%), Positives = 156/257 (60%), Gaps = 32/257 (12%)
Query: 4 DTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERG-------ERQQQRAG 56
DT+ L YW+ WR ++CAL ++ M + + L+W++E R G ++++R G
Sbjct: 37 DTSDLHYWMQWRAAVCALSVLACMAVAACLVWRHEGPGAERRPGGASGGGGGSKERRRPG 96
Query: 57 LLYEDELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTI 116
+LY+DE W CL+ IHP WLL YR++SF VLL LLI +++DG I Y+YTQWTF LVTI
Sbjct: 97 VLYDDEAWRPCLRDIHPAWLLGYRLISFFVLLSLLIVIVISDGGTIFYYYTQWTFILVTI 156
Query: 117 YFGLASFFSIYGCFFKHDKFEGNTVGSACLNTERGTYVA------PTIDGDTDIPHMYKS 170
YFGL + SIYGC K V + + E G+YVA P ++G+ D
Sbjct: 157 YFGLGTALSIYGC----SKLADENVVTERTDMELGSYVAHGAGTKPNLNGEDD------- 205
Query: 171 TDAYQEPHTQNTAGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCM 230
T AG WGY+ QI++QT AGAV+LTD VFW +++PFL KD+ L + M
Sbjct: 206 --------TGEIAGFWGYLLQIIYQTNAGAVMLTDCVFWFIIFPFLTVKDYNLNFLLIGM 257
Query: 231 HSVNAVFLLGETSLNGM 247
HSVNAVFLLGE +LN +
Sbjct: 258 HSVNAVFLLGEAALNSL 274
>UniRef100_Q6NPD1 At5g62960 [Arabidopsis thaliana]
Length = 347
Score = 220 bits (560), Expect = 3e-56
Identities = 109/248 (43%), Positives = 157/248 (62%), Gaps = 17/248 (6%)
Query: 4 DTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGE---RQQQRAGLLYE 60
+TT SYW NWR IC +W+ + +FLI+KYE F + R + GE +++ +G +YE
Sbjct: 28 NTTESSYWFNWRVMICCIWMAIATVITAFLIFKYEGFRRKRSDVGEVDGGEKEWSGNVYE 87
Query: 61 DELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGL 120
DE W CL+ IHP WLLA+R+V+F VLL +LI + DG I ++YTQWTF L+T+YFGL
Sbjct: 88 DETWRPCLRNIHPAWLLAFRVVAFFVLLVMLIVIGLVDGPTIFFYYTQWTFGLITLYFGL 147
Query: 121 ASFFSIYGCFFKHDKFEGNTVGS-ACLNTERGTYVAPTIDGDTDIPHMYKSTDAYQEPHT 179
S S++GC+ + + G+ V S +++ER A + D I Q ++
Sbjct: 148 GSLLSLHGCYQYNKRAAGDRVDSIEAIDSER----ARSKGADNTI---------QQSQYS 194
Query: 180 QNTAGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLL 239
N AG WGY+FQI+FQ AGAV+LTD VFW ++ PFL D+ L + + MHS+NA+FLL
Sbjct: 195 SNPAGFWGYVFQIIFQMNAGAVLLTDCVFWFIIVPFLEIHDYSLNVLVINMHSLNAIFLL 254
Query: 240 GETSLNGM 247
G+ +LN +
Sbjct: 255 GDAALNSL 262
>UniRef100_Q9FM56 Gb|AAF17656.1 [Arabidopsis thaliana]
Length = 317
Score = 214 bits (545), Expect = 2e-54
Identities = 110/257 (42%), Positives = 158/257 (60%), Gaps = 25/257 (9%)
Query: 4 DTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGE---RQQQRAGLLYE 60
+TT SYW NWR IC +W+ + +FLI+KYE F + R + GE +++ +G +YE
Sbjct: 28 NTTESSYWFNWRVMICCIWMAIATVITAFLIFKYEGFRRKRSDVGEVDGGEKEWSGNVYE 87
Query: 61 DELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFG- 119
DE W CL+ IHP WLLA+R+V+F VLL +LI + DG I ++YTQWTF L+T+YFG
Sbjct: 88 DETWRPCLRNIHPAWLLAFRVVAFFVLLVMLIVIGLVDGPTIFFYYTQWTFGLITLYFGV 147
Query: 120 -------LASFFSIYGCFFKHDKFEGNTVGSA-CLNTERGTYVAPTIDGDTDIPHMYKST 171
L S S++GC+ + + G+ V S +++ER A + D I
Sbjct: 148 MFCHKNLLGSLLSLHGCYQYNKRAAGDRVDSIEAIDSER----ARSKGADNTIQ------ 197
Query: 172 DAYQEPHTQNTAGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMH 231
Q ++ N AG WGY+FQI+FQ AGAV+LTD VFW ++ PFL D+ L + + MH
Sbjct: 198 ---QSQYSSNPAGFWGYVFQIIFQMNAGAVLLTDCVFWFIIVPFLEIHDYSLNVLVINMH 254
Query: 232 SVNAVFLLGETSLNGMV 248
S+NA+FLLG+ +LN +V
Sbjct: 255 SLNAIFLLGDAALNSLV 271
>UniRef100_Q9SGY3 F20B24.10 [Arabidopsis thaliana]
Length = 409
Score = 195 bits (495), Expect = 1e-48
Identities = 110/244 (45%), Positives = 149/244 (60%), Gaps = 24/244 (9%)
Query: 9 SYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQQQRAGLLYEDELWNTCL 68
SYWLNWR +CAL ++ + L + LIWKYE K R +R E Q++ G L++DE W TC
Sbjct: 43 SYWLNWRVLLCALILLAPIVLAAVLIWKYE--GKRRRQR-ESQRELPGTLFQDEAWTTCF 99
Query: 69 KRIHPVWLLAYRIVSF---IVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLASFFS 125
KRIHP+WLLA+R+ + +L G +GN V + TLV AS S
Sbjct: 100 KRIHPLWLLAFRVDIYSCHTLLWGTSLGNSVVE-----------VTTLVLDTKSYASVLS 148
Query: 126 IYGCFFKHDKFEGNTVGSACL-NTERGTYVAP-TIDGDTDIPHMYKSTDAYQEPHTQNTA 183
+YGC + + GN + +TE+GTY P +DG+ + K+++ E + TA
Sbjct: 149 VYGCCIYNKEASGNMESYTSIGDTEQGTYRPPIALDGEGNTS---KASNRPSEAPARKTA 205
Query: 184 GAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLLGETS 243
G W YIFQI+FQTCAGAVVLTD+VFW ++YPF K ++L VCMHS+NAVFLLG+TS
Sbjct: 206 GFWVYIFQILFQTCAGAVVLTDIVFWAIIYPF--TKGYKLSFLDVCMHSLNAVFLLGDTS 263
Query: 244 LNGM 247
LN +
Sbjct: 264 LNSL 267
>UniRef100_Q9LVW5 Gb|AAF17656.1 [Arabidopsis thaliana]
Length = 343
Score = 177 bits (449), Expect = 2e-43
Identities = 96/256 (37%), Positives = 140/256 (54%), Gaps = 17/256 (6%)
Query: 6 TALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQQQRAG--LLYEDEL 63
T+ YW NWR +CA+W+I M + ++WKYE D + Q G +L D++
Sbjct: 10 TSFDYWFNWRVLLCAIWVIVPMIVSLLVLWKYE------DSSVQTQPSLNGNDVLCIDDV 63
Query: 64 WNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLASF 123
W C +RIHP WLL +R++ F LL I G I Y+YTQWTFTL+ IYFG+ S
Sbjct: 64 WRPCFERIHPGWLLGFRVLGFCFLLANNIARFANRGWRIYYYYTQWTFTLIAIYFGMGSL 123
Query: 124 FSIYGCF-FKHDKFEGNTVGSACLNTERGTYVAPTIDGDTDIPHMYKSTDAYQEPHTQNT 182
SIYGC +K G ++ E G + +P IDGD + + T
Sbjct: 124 LSIYGCLQYKKQGNTGLIADQVGIDAENG-FRSPLIDGDNMVSFEKRKTSG------SEA 176
Query: 183 AGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLLGET 242
++ ++FQI++Q AGA VLTD ++W V++PFL+ +D+ + V +H+ N V LL +T
Sbjct: 177 LKSYVHLFQIIYQMGAGAAVLTDSIYWTVIFPFLSLQDYEMSFMTVNLHTSNLVLLLIDT 236
Query: 243 SLNGMVKYLWL-SRLF 257
LN +V L L R+F
Sbjct: 237 FLNRLVLLLHLMDRMF 252
>UniRef100_Q681X6 MRNA, complete cds, clone: RAFL21-92-C23 [Arabidopsis thaliana]
Length = 315
Score = 177 bits (448), Expect = 3e-43
Identities = 92/243 (37%), Positives = 134/243 (54%), Gaps = 16/243 (6%)
Query: 6 TALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQQQRAG--LLYEDEL 63
T+ YW NWR +CA+W+I M + ++WKYE D + Q G +L D++
Sbjct: 10 TSFDYWFNWRVLLCAIWVIVPMIVSLLVLWKYE------DSSVQTQPSLNGNDVLCIDDV 63
Query: 64 WNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLASF 123
W C +RIHP WLL +R++ F LL I G I Y+YTQWTFTL+ IYFG+ S
Sbjct: 64 WRPCFERIHPGWLLGFRVLGFCFLLANNIARFANRGWRIYYYYTQWTFTLIAIYFGMGSL 123
Query: 124 FSIYGCF-FKHDKFEGNTVGSACLNTERGTYVAPTIDGDTDIPHMYKSTDAYQEPHTQNT 182
SIYGC +K G G ++ E G + +P IDGD + + T
Sbjct: 124 LSIYGCLQYKKQGNTGLIAGQVGIDAENG-FRSPLIDGDNMVSFEKRKTSG------SEA 176
Query: 183 AGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLLGET 242
++ ++FQI++Q AGA VLTD ++W V++PFL+ +D+ + V +H+ N V LL +T
Sbjct: 177 LKSYVHLFQIIYQMGAGAAVLTDSIYWTVIFPFLSLQDYEMSFMTVNLHTSNLVLLLIDT 236
Query: 243 SLN 245
LN
Sbjct: 237 FLN 239
>UniRef100_Q8W4R6 AT3g27760/MGF10_16 [Arabidopsis thaliana]
Length = 315
Score = 174 bits (441), Expect = 2e-42
Identities = 91/243 (37%), Positives = 133/243 (54%), Gaps = 16/243 (6%)
Query: 6 TALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQQQRAG--LLYEDEL 63
T+ YW NWR +CA+W+I M + ++WKYE D + Q G +L D++
Sbjct: 10 TSFDYWFNWRVLLCAIWVIVPMIVSLLVLWKYE------DSSVQTQPSLNGNDVLCIDDV 63
Query: 64 WNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLASF 123
W C +RIHP WLL +R++ F LL I G I Y+YTQWTFTL+ IYFG+ S
Sbjct: 64 WRPCFERIHPGWLLGFRVLGFCFLLANNIARFANRGWRIYYYYTQWTFTLIAIYFGMGSL 123
Query: 124 FSIYGCF-FKHDKFEGNTVGSACLNTERGTYVAPTIDGDTDIPHMYKSTDAYQEPHTQNT 182
SIYGC +K G ++ E G + +P IDGD + + T
Sbjct: 124 LSIYGCLQYKKQGNTGLIADQVGIDAENG-FRSPLIDGDNMVSFEKRKTSG------SEA 176
Query: 183 AGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLLGET 242
++ ++FQI++Q AGA VLTD ++W V++PFL+ +D+ + V +H+ N V LL +T
Sbjct: 177 LKSYVHLFQIIYQMGAGAAVLTDSIYWTVIFPFLSLQDYEMSFMTVNLHTSNLVLLLIDT 236
Query: 243 SLN 245
LN
Sbjct: 237 FLN 239
>UniRef100_Q67YW5 MRNA, complete cds, clone: RAFL24-05-D16 [Arabidopsis thaliana]
Length = 272
Score = 135 bits (341), Expect = 8e-31
Identities = 71/184 (38%), Positives = 103/184 (55%), Gaps = 8/184 (4%)
Query: 63 LWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLAS 122
L+ C +RIHP WLL +R++ F LL I G I Y+YTQWTFTL+ IYFG+ S
Sbjct: 20 LYGPCFERIHPGWLLGFRVLGFCFLLANNIARFANRGWRIYYYYTQWTFTLIAIYFGMGS 79
Query: 123 FFSIYGCF-FKHDKFEGNTVGSACLNTERGTYVAPTIDGDTDIPHMYKSTDAYQEPHTQN 181
SIYGC +K G ++ E G + +P IDGD + + T
Sbjct: 80 LLSIYGCLQYKKQGNTGLIADQVGIDAENG-FRSPLIDGDNMVSFEKRKTSG------SE 132
Query: 182 TAGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLLGE 241
++ ++FQI++Q AGA VLTD ++W V++PFL+ +D+ + V +H+ N V LL +
Sbjct: 133 ALKSYVHLFQIIYQMGAGAAVLTDSIYWTVIFPFLSLQDYEMSFMTVNLHTSNLVLLLID 192
Query: 242 TSLN 245
T LN
Sbjct: 193 TFLN 196
>UniRef100_Q8GRS3 Hypothetical protein OJ1477_F01.109 [Oryza sativa]
Length = 390
Score = 134 bits (337), Expect = 2e-30
Identities = 80/240 (33%), Positives = 117/240 (48%), Gaps = 17/240 (7%)
Query: 9 SYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQQQRAGLLYEDELWNTCL 68
+YWL W+ +C I+ + L+ R ++ A L +LW C
Sbjct: 84 AYWLRWQVFVCGALIVLPTAAAAALL--------------PRLRRAAAPLRGTDLWVPCW 129
Query: 69 KRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLASFFSIYG 128
R+HP WLL YR + + LL+ +V G+ + +FYT WTF LVTIYF A+ S +G
Sbjct: 130 ARLHPGWLLGYRAFALAAAVALLVRLLVGHGIDVFFFYT-WTFLLVTIYFAFATAISAHG 188
Query: 129 CF-FKHDKFEGNTVGSACLNTERGTYVAPTIDGDTDIPHMYKSTDAYQEPHTQNTAGAWG 187
C+ + + L+ + T G+ K T+ +++ + AG WG
Sbjct: 189 CWVYSKKNLKKADESHEFLSDDVENREFSTSSGEMKRDEE-KITNYHEQIANEKRAGLWG 247
Query: 188 YIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLLGETSLNGM 247
QI++QT AGA +LTD+ FW +L PF F L L MHS+NAV LL +T LN M
Sbjct: 248 RCMQIIYQTSAGATMLTDITFWGLLVPFFYRDKFGLSLITDGMHSLNAVLLLIDTFLNNM 307
>UniRef100_Q8S8R0 Predicted protein [Arabidopsis thaliana]
Length = 289
Score = 111 bits (277), Expect = 2e-23
Identities = 65/185 (35%), Positives = 88/185 (47%), Gaps = 37/185 (20%)
Query: 63 LWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLAS 122
LW +C R+HP WLL R SF+ + LL +++ I +YT+WTF LV IYF +
Sbjct: 59 LWASCWTRLHPGWLLFTRSTSFLSMAALLAWDVIKWDASIFVYYTEWTFMLVIIYFAMGI 118
Query: 123 FFSIYGCFFKHDKFEGNTVGSACLNTERGTYVAPTIDGDTDIPHMYKSTDAYQEPHTQNT 182
S+YGC + T++ D D+
Sbjct: 119 VASVYGCLIHLKEL--------------------TLETDEDV-----------------V 141
Query: 183 AGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLLGET 242
G F+ +T AGAVVLTD+VFWLV+ PFL+ F L +CMH+ NA FLL ET
Sbjct: 142 VEKVGDEFRRRLETSAGAVVLTDIVFWLVIVPFLSTTRFGLNTLTICMHTANAGFLLLET 201
Query: 243 SLNGM 247
LN +
Sbjct: 202 LLNSL 206
>UniRef100_Q6Z6B1 Hypothetical protein P0622F08.29 [Oryza sativa]
Length = 122
Score = 76.3 bits (186), Expect = 8e-13
Identities = 36/81 (44%), Positives = 49/81 (60%), Gaps = 8/81 (9%)
Query: 1 METDTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYE---EFNKSRDERGERQQQR--- 54
M DTTA YWLNWRF +CA+W+ M L FLIWKYE + + D G+ + R
Sbjct: 1 MAVDTTAPEYWLNWRFMLCAVWVYSCMVLACFLIWKYEGPSSQDGNGDGGGDSEDARLPW 60
Query: 55 --AGLLYEDELWNTCLKRIHP 73
+G+LY ++ W CL++IHP
Sbjct: 61 SASGVLYLEDCWKPCLEQIHP 81
>UniRef100_Q9C6E3 Hypothetical protein T32G9.33 [Arabidopsis thaliana]
Length = 1311
Score = 38.1 bits (87), Expect = 0.23
Identities = 32/119 (26%), Positives = 53/119 (43%), Gaps = 22/119 (18%)
Query: 75 WLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQ------------WTFTLVTIYFGLAS 122
WL +++ ++L L+ +VA GIL F W V + FG+ S
Sbjct: 39 WLWTLGVIAVVILSARLLVVVVATRGGILAFVLPDRRGLVTVEVRGWWLLAVRVRFGVLS 98
Query: 123 FFSIYGCFFKHDKFEGNTVGSACLNTERGTYVA-------PTIDGDTDIPHMYKSTDAY 174
FS+ + K G T GS L+ +RG +V+ P+ D+++P+ +TD Y
Sbjct: 99 SFSV--LIYSERKLLGLT-GSRFLSCKRGVFVSAGSTSQEPSSQWDSNLPNRLFATDHY 154
>UniRef100_Q7Q1E2 ENSANGP00000015709 [Anopheles gambiae str. PEST]
Length = 299
Score = 35.0 bits (79), Expect = 2.0
Identities = 20/53 (37%), Positives = 27/53 (50%), Gaps = 6/53 (11%)
Query: 59 YEDELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTF 111
Y L+N C KR+ VW+L +FIVL+GLL G+ +L W F
Sbjct: 71 YRFTLYNICYKRVLVVWIL----FAFIVLIGLLFSGFY--GITLLSLGVAWLF 117
>UniRef100_Q7Q1E1 ENSANGP00000015887 [Anopheles gambiae str. PEST]
Length = 303
Score = 35.0 bits (79), Expect = 2.0
Identities = 20/53 (37%), Positives = 27/53 (50%), Gaps = 6/53 (11%)
Query: 59 YEDELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTF 111
Y L+N C KR+ VW+L +FIVL+GLL G+ +L W F
Sbjct: 71 YRFTLYNICYKRVLVVWIL----FAFIVLIGLLFSGFY--GITLLSLGVAWLF 117
>UniRef100_UPI00003AA3B7 UPI00003AA3B7 UniRef100 entry
Length = 484
Score = 34.7 bits (78), Expect = 2.6
Identities = 27/107 (25%), Positives = 49/107 (45%), Gaps = 23/107 (21%)
Query: 24 IFTMGLGSFLIWKYEEFN-KSRDERGERQQQRAGLLYEDELWNTCLKRIHPVWLLAYRIV 82
+FT+ LG++ E N K+ G+R+ +R K+ V L +V
Sbjct: 147 VFTVALGAYWSGVAELENLKATASPGDRETRR--------------KKEENVTLTTLTVV 192
Query: 83 SFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLASFFSIYGC 129
F+V+ +++ +LYF+ +W ++ + F LAS S+Y C
Sbjct: 193 LFVVICCVMLI--------LLYFFYKWLVYVIILVFCLASAMSLYNC 231
>UniRef100_Q9A454 Hypothetical protein CC2989 [Caulobacter crescentus]
Length = 425
Score = 34.3 bits (77), Expect = 3.4
Identities = 24/75 (32%), Positives = 32/75 (42%), Gaps = 12/75 (16%)
Query: 5 TTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQQQRAGLLYEDELW 64
TT L + W +CA T G+G+FL W RG + Q +G L LW
Sbjct: 79 TTPLLFATRWITHVCAPAFALTAGVGAFLWW----------SRGRSRAQLSGFLVSRGLW 128
Query: 65 NTCLKRIHPVWLLAY 79
L+ + V LAY
Sbjct: 129 LMLLELV--VMRLAY 141
>UniRef100_UPI000042D911 UPI000042D911 UniRef100 entry
Length = 474
Score = 33.9 bits (76), Expect = 4.4
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 6/63 (9%)
Query: 71 IHPVWLLAYRIVS----FIVLLGLLIGNMVADGVGILY--FYTQWTFTLVTIYFGLASFF 124
I P W L Y +S +++LGL + N G + + + T WT+ + IY+ A+
Sbjct: 50 IIPQWFLIYLRLSVFLYLLIMLGLDVSNYYKLGYAMYWGVYVTNWTYAITIIYYFFATTS 109
Query: 125 SIY 127
+IY
Sbjct: 110 TIY 112
>UniRef100_UPI00002CCA07 UPI00002CCA07 UniRef100 entry
Length = 270
Score = 33.9 bits (76), Expect = 4.4
Identities = 22/71 (30%), Positives = 37/71 (51%), Gaps = 9/71 (12%)
Query: 73 PVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLASFFSIYGCFFK 132
P+++ ++S +VL LLIG+M G+ + + W L+ +YF LA F
Sbjct: 139 PLYISILSVISNVVLSLLLIGSMREMGIALATAISAWVNALL-LYFFLA--------FRN 189
Query: 133 HDKFEGNTVGS 143
H KF+G +G+
Sbjct: 190 HMKFDGLLIGN 200
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.328 0.143 0.473
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 476,609,044
Number of Sequences: 2790947
Number of extensions: 20013303
Number of successful extensions: 61877
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 61830
Number of HSP's gapped (non-prelim): 33
length of query: 265
length of database: 848,049,833
effective HSP length: 125
effective length of query: 140
effective length of database: 499,181,458
effective search space: 69885404120
effective search space used: 69885404120
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0008.11


