

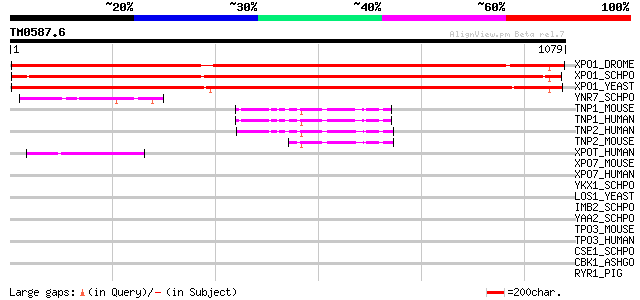
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0587.6
(1079 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
XPO1_DROME (Q9TVM2) Exportin 1 (Chromosome region maintenance 1 ... 983 0.0
XPO1_SCHPO (P14068) Exportin 1 (Chromosome region maintenance pr... 978 0.0
XPO1_YEAST (P30822) Exportin 1 (Chromosome region maintenance pr... 856 0.0
YNR7_SCHPO (Q9USZ2) Hypothetical protein C11G11.07 in chromosome II 60 3e-08
TNP1_MOUSE (Q8BFY9) Transportin 1 (Importin beta-2) (Karyopherin... 60 3e-08
TNP1_HUMAN (Q92973) Transportin 1 (Importin beta-)2 (Karyopherin... 60 3e-08
TNP2_HUMAN (O14787) Transportin 2 (Karyopherin beta-2b) 57 3e-07
TNP2_MOUSE (Q99LG2) Transportin 2 (Karyopherin beta-2b) 57 3e-07
XPOT_HUMAN (O43592) Exportin T (tRNA exportin) (Exportin(tRNA)) 52 1e-05
XPO7_MOUSE (Q9EPK7) Exportin 7 (Ran-binding protein 16) 38 0.13
XPO7_HUMAN (Q9UIA9) Exportin 7 (Ran-binding protein 16) 38 0.13
YKX1_SCHPO (O14116) Hypothetical protein C328.01c in chromosome I 38 0.17
LOS1_YEAST (P33418) LOS1 protein 36 0.49
IMB2_SCHPO (O14089) Putative importin beta-2 subunit (Karyopheri... 35 0.83
YAA2_SCHPO (Q09796) Hypothetical protein C22G7.02 in chromosome I 34 1.9
TPO3_MOUSE (Q6P2B1) Transportin 3 34 1.9
TPO3_HUMAN (Q9Y5L0) Transportin 3 (Transportin-SR) (TRN-SR) (Imp... 34 1.9
CSE1_SCHPO (O13671) Importin-alpha re-exporter (Cellular apoptos... 34 2.4
CBK1_ASHGO (Q754N7) Serine/threonine-protein kinase CBK1 (EC 2.7... 34 2.4
RYR1_PIG (P16960) Ryanodine receptor 1 (Skeletal muscle-type rya... 33 4.1
>XPO1_DROME (Q9TVM2) Exportin 1 (Chromosome region maintenance 1
protein) (Embargoed protein)
Length = 1063
Score = 983 bits (2541), Expect = 0.0
Identities = 508/1083 (46%), Positives = 704/1083 (64%), Gaps = 37/1083 (3%)
Query: 3 AEKLRDLSQPIDVPLLDATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQNTQ 62
A KL D SQ +D+ LLD V Y T EQ A IL L+ +P+ W +V IL+ +Q
Sbjct: 10 AGKLLDFSQKLDINLLDKIVEVVY-TAQGEQLRLAQSILTTLKEHPEAWTRVDSILEYSQ 68
Query: 63 NLNTKFFALQVLEGVIKYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNKLN 122
N TKF+ALQ+LE VIK RW LP Q +G+K ++ +I++ S + ++Y+NKLN
Sbjct: 69 NQRTKFYALQILEEVIKTRWKVLPRNQCEGIKKYVVSLIIKTSSDPIVMEQNKVYLNKLN 128
Query: 123 IILVQILKHEWPLRWRSFIPDLVSAAKTSETICENCMAILKLLSEEVFDFSRGEMTQQKI 182
+ILV ILK EWP W +FI D+V A+KT+E++C N M ILK LSEEVFDFS+G++TQ K
Sbjct: 129 MILVHILKREWPRNWETFISDIVGASKTNESLCMNNMVILKNLSEEVFDFSQGQITQTKA 188
Query: 183 KELKQSLNSEFQLIHELCLYVLSASQRAELIRATLSTLHAFLSWIPLGYIFESPLLETLL 242
K LK ++ SEF I LC +VL S A LI TL TL FL+WIPLGYIFE+ +ETL+
Sbjct: 189 KHLKDTMCSEFSQIFTLCSFVLENSMNAALIHVTLETLLRFLNWIPLGYIFETQQIETLI 248
Query: 243 -KFFPLPVYRNLTLKCLIEVAALQFGDYYDTQYTKMYNIFMVQLQTILLPTTNIPEAYAQ 301
KF +P++RN+TLKCL E+A L +Y D + ++ MVQL+ I+ N+ +
Sbjct: 249 FKFLSVPMFRNVTLKCLSEIAGLTAANY-DENFATLFKDTMVQLEQIVGQNMNMNHVFKH 307
Query: 302 GSSEEQTFIQNLALFFTSFYKVHIRILESTQENIAALLSGLEYLINISYVDDTEVFKVCL 361
GS EQ + NLA+F +F K H +++E + + L L YL+ IS V+D EVFK+CL
Sbjct: 308 GSDTEQELVLNLAMFLCTFLKEHGKLVEDAKY-VDYLNQALMYLVMISEVEDVEVFKICL 366
Query: 362 DYWNILVSELFEPHRSLENPAAAATNMLGNQQAPLMPPGMVDGLGSQLLQRRQLYAGPMS 421
+YWN LV +L+ + P + Q+ RR+ YA +S
Sbjct: 367 EYWNSLVEDLYN--------------------SEFFHPTLESTKRQQVYPRRRFYAPILS 406
Query: 422 KLRTLMICRMAKPEEVLIIEDENGNIVRETLKDNDVLVQYKIMRETLIYLSHLDHDDTER 481
K+R +MI RMAKPEEVL++E+ENG +VRE +KD + + YK MRETL++L+HLD DT+R
Sbjct: 407 KVRFIMISRMAKPEEVLVVENENGEVVREFMKDTNSINLYKNMRETLVFLTHLDSVDTDR 466
Query: 482 QMLRKLSKQLGGEDWTWNNLNTLCWAIGSISGSMMEDQENRFLVMVIRDLLNLCEFTKGK 541
M KL Q+ G +++W NLNTLCWAIGSISG+ E+ E RFLV VI+DLL LCE KGK
Sbjct: 467 IMTLKLLNQVNGSEFSWKNLNTLCWAIGSISGAFCEEDEKRFLVTVIKDLLGLCEQKKGK 526
Query: 542 DNKAVIASNIMYVVGQYPRFLRAHWKFLKTVVNKLFEFMHESHPGVQDMACDTFLKIVQK 601
DNKA+IASNIMYVVGQYPRFLRAHWKFLKTVVNKLFEFMHE+H GVQDMACDTF+KI K
Sbjct: 527 DNKAIIASNIMYVVGQYPRFLRAHWKFLKTVVNKLFEFMHETHDGVQDMACDTFIKIAIK 586
Query: 602 CKRKFVITQLGENEPFVSELLSGLSTTIVDLEPHQIHTFYESVGSMVQAESDSQKRDEYL 661
C+R FV Q E F+ E+L+ +S+ I DL+P Q+HTFYE+VG M+ A+ D ++D +
Sbjct: 587 CRRYFVTIQPNEACTFIDEILTTMSSIICDLQPQQVHTFYEAVGYMISAQVDQVQQDVLI 646
Query: 662 QRLMHLPNQKWLEIIGQARQNVDFLKDHDVIRTVLNILQTNTSVASSLGTYFLPQISLIF 721
+R M LPNQ W +II +A +NVDFLK+ ++ + +IL+TN + +LG ++ Q+ I+
Sbjct: 647 ERYMQLPNQVWDDIISRASKNVDFLKNMTAVKQLGSILKTNVAACKALGHAYVIQLGRIY 706
Query: 722 LDMLNVYRMYSELISKSIAEGGPYASRTSFVKLLRSVKRETLKLIETFLDKAEDQPQIGK 781
LDMLNVY++ SE I ++I G + +K + VK+ETL LI ++ ++ D +
Sbjct: 707 LDMLNVYKITSENIIQAIEVNGVNVNNQPLIKTMHVVKKETLNLISEWVSRSNDNQLVMD 766
Query: 782 QFVPPMMDPVLGDYAR-NVPDARESEVLSLFATIVNKYKAAMVEDVPRIFEAVFQCTLEM 840
F+PP++D +L DY R VP ARE +VLS A IV+K + + +VP+IF+AVF+CTL+M
Sbjct: 767 NFIPPLLDAILLDYQRCKVPSAREPKVLSAMAIIVHKLRQHITNEVPKIFDAVFECTLDM 826
Query: 841 ITKNFEDYPEHRLKFFSLLCAIATHCFHALICLSSQQLKFVMDSIIWAFRHTERNIAETG 900
I KNFED+P+HRL F+ LL A+ HCF A + + Q K V DS++WAF+HT RN+A+ G
Sbjct: 827 INKNFEDFPQHRLSFYELLQAVNAHCFKAFLNIPPAQFKLVFDSVVWAFKHTMRNVADMG 886
Query: 901 LNLLLEMLKKF-QATEFCNQFYRTYFLTIEQEIFAVLTDTFHKPGFKLHVLVLQHLFCLA 959
LN+L +ML+ Q FY+TYF I +IF+V+TDT H G H ++L ++F L
Sbjct: 887 LNILFKMLQNLDQHPGAAQSFYQTYFTDILMQIFSVVTDTSHTAGLPNHAIILAYMFSLV 946
Query: 960 ETGALTEPLWDAATNPYPYPSNAAFVLEYTIKLLSTSFPNMTAAEVTQFVNGLFESTNDL 1019
E +T N P P N F+ EY LL ++F +++ +V FV GLF ++
Sbjct: 947 ENRKIT-------VNLGPIPDNMIFIQEYVASLLKSAFTHLSDNQVKVFVTGLFNLDENV 999
Query: 1020 STFKNHIRDFLVQSKEFSAQDNKDLYAEE----AAAQRERERQRMLSVPGLIAPSELQDE 1075
FK H+RDFL+Q +E + +D+ DLY EE A ++ + Q ++PG++ P EL ++
Sbjct: 1000 QAFKEHLRDFLIQIREATGEDDSDLYLEEREAALAEEQSNKHQMQRNIPGMLNPHELPED 1059
Query: 1076 MLD 1078
M D
Sbjct: 1060 MQD 1062
>XPO1_SCHPO (P14068) Exportin 1 (Chromosome region maintenance protein
1) (Caffeine resistance protein 2)
Length = 1078
Score = 978 bits (2528), Expect = 0.0
Identities = 495/1077 (45%), Positives = 721/1077 (65%), Gaps = 16/1077 (1%)
Query: 4 EKLRDLSQPIDVPLLDATVAAFY-GTGSKEQRTAADQILRDLQNNPDMWLQVMHILQNTQ 62
E + + +DV LLD V FY G G+++Q+ A Q+L Q +PD W Q IL+ ++
Sbjct: 2 EGILAFDRELDVALLDRVVQTFYQGVGAEQQQ--AQQVLTQFQAHPDAWSQAYSILEKSE 59
Query: 63 NLNTKFFALQVLEGVIKYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNKLN 122
TK+ AL VL+ +I RW LP EQR G++N+I ++++ S +E + ++ ++NKL+
Sbjct: 60 YPQTKYIALSVLDKLITTRWKMLPKEQRLGIRNYIVAVMIKNSSDETVLQQQKTFLNKLD 119
Query: 123 IILVQILKHEWPLRWRSFIPDLVSAAKTSETICENCMAILKLLSEEVFDFSRGEMTQQKI 182
+ LVQILK EWP W +FIP++V A+KT+ ++CEN M +L+LLSEE+FD+S +MTQ K
Sbjct: 120 LTLVQILKQEWPHNWPNFIPEIVQASKTNLSLCENNMIVLRLLSEEIFDYSAEQMTQLKT 179
Query: 183 KELKQSLNSEFQLIHELCLYVLSASQRAELIRATLSTLHAFLSWIPLGYIFESPLLETLL 242
K LK + EF I +LC +L +Q+ LI+ATL TL FL+WIPLGYIFE+ ++E +
Sbjct: 180 KNLKNQMCGEFAEIFQLCSQILERAQKPSLIKATLGTLLRFLNWIPLGYIFETNIVELIT 239
Query: 243 -KFFPLPVYRNLTLKCLIEVAALQFGDYYDTQYTKMYNIFMVQLQTILLPTTNIPEAYAQ 301
+F +P +RN+T++CL E+A+L Y+ ++ M+N+ M + ++L T+ EAY +
Sbjct: 240 NRFLNVPDFRNVTIECLTEIASLTSQPQYNDKFVTMFNLVMTSVNSMLPLQTDFREAYEE 299
Query: 302 GSSEEQTFIQNLALFFTSFYKVHIRILESTQENIAALLSGLEYLINISYVDDTEVFKVCL 361
S+ EQ FIQNLALF +F+ H+R LE+ EN LL+ YL+NIS +++ E+FK+CL
Sbjct: 300 SSTNEQDFIQNLALFLCAFFSSHLRPLENP-ENQEVLLNAHSYLLNISRINEREIFKICL 358
Query: 362 DYWNILVSELFEPHRSLENPAAAATNMLGNQQAPLMPPGMVDGLGSQLLQRRQLYAGPMS 421
+YW+ LV++L+E ++ + N L N +P + + L R+ +Y +S
Sbjct: 359 EYWSKLVAQLYE---EMQQIPMSEMNPLLNLSSPTSLISSNPNMLANLPLRKHIYKDILS 415
Query: 422 KLRTLMICRMAKPEEVLIIEDENGNIVRETLKDNDVLVQYKIMRETLIYLSHLDHDDTER 481
LR +MI M KPEEVLI+E++ G IVRE +K+ D + YK MRE L+YL+HLD DTE
Sbjct: 416 TLRLVMIENMVKPEEVLIVENDEGEIVREFVKETDTITLYKSMREVLVYLTHLDVVDTEI 475
Query: 482 QMLRKLSKQLGGEDWTWNNLNTLCWAIGSISGSMMEDQENRFLVMVIRDLLNLCEFTKGK 541
M KL++ + G +W+W NLNTLCWAIGSISG+M E+ E RFLV VI+DLL LCE +GK
Sbjct: 476 VMTEKLARIVVGTEWSWQNLNTLCWAIGSISGAMNEEMEKRFLVNVIKDLLGLCEMKRGK 535
Query: 542 DNKAVIASNIMYVVGQYPRFLRAHWKFLKTVVNKLFEFMHESHPGVQDMACDTFLKIVQK 601
DNKAV+ASNIMYVVGQYPRFL+AHWKFLKTVVNKLFEFMHE H GVQDMACDTF+KI QK
Sbjct: 536 DNKAVVASNIMYVVGQYPRFLKAHWKFLKTVVNKLFEFMHEYHEGVQDMACDTFIKIAQK 595
Query: 602 CKRKFVITQLGENEPFVSELLSGLSTTIVDLEPHQIHTFYESVGSMVQAESDSQKRDEYL 661
C+R FV QLGE EPF++E++ L+ T DL P Q HTFYE+ G M+ A+ ++ +
Sbjct: 596 CRRHFVAQQLGETEPFINEIIRNLAKTTEDLTPQQTHTFYEACGYMISAQPQKHLQERLI 655
Query: 662 QRLMHLPNQKWLEIIGQARQNVDFLKDHDVIRTVLNILQTNTSVASSLGTYFLPQISLIF 721
LM LPNQ W I+ QA QN L D ++ + N+L+TN + +S+G+ F PQI+ +
Sbjct: 656 FDLMALPNQAWENIVAQAAQNAQVLGDPQTVKILANVLKTNVAACTSIGSGFYPQIAKNY 715
Query: 722 LDMLNVYRMYSELISKSIAEGGPYASRTSFVKLLRSVKRETLKLIETFLDKAEDQPQIGK 781
+DML +Y+ S LIS+ +A G A++T V+ LR++K+E LKL++ ++ +AED +G
Sbjct: 716 VDMLGLYKAVSGLISEVVAAQGNIATKTPHVRGLRTIKKEILKLVDAYISRAEDLELVGN 775
Query: 782 QFVPPMMDPVLGDYARNVPDARESEVLSLFATIVNKYKAAMVEDVPRIFEAVFQCTLEMI 841
+P + + VL DY +NVPDAR++EVL+L TIVN+ + + +P + +AVF CTLEMI
Sbjct: 776 TLIPALFEAVLLDYLQNVPDARDAEVLNLITTIVNQLSELLTDKIPLVLDAVFGCTLEMI 835
Query: 842 TKNFEDYPEHRLKFFSLLCAIATHCFHALICLSSQQLKFVMDSIIWAFRHTERNIAETGL 901
+K+F +YPEHR FF LL AI +CF AL+ + + Q K V++SI+W+F+H R+I ETGL
Sbjct: 836 SKDFSEYPEHRAAFFQLLRAINLNCFPALLNIPAPQFKLVINSIVWSFKHVSRDIQETGL 895
Query: 902 NLLLEMLKKF--QATEFCNQFYRTYFLTIEQEIFAVLTDTFHKPGFKLHVLVLQHLFCLA 959
N+LLE++ + N F++TY++++ Q+I VLTD+ HK GFKL L+L LF L
Sbjct: 896 NILLELINNMASMGPDVSNAFFQTYYISLLQDILYVLTDSDHKSGFKLQSLILARLFYLV 955
Query: 960 ETGALTEPLWDAATNPYPYPSNAAFVLEYTIKLLSTSFPNMTAAEVTQFVNGLFESTNDL 1019
E+ +T PL+D + P +N F+ +Y + LL T+FP++ ++ +FV + D
Sbjct: 956 ESNQITVPLYDPSQFPQEM-NNQLFLRQYIMNLLVTAFPHLQPIQIQEFVQTVLALNQDS 1014
Query: 1020 STFKNHIRDFLVQSKEFSAQDNKDLYAE----EAAAQRERERQRMLSVPGLIAPSEL 1072
FK +RDFL+Q KEF DN +LY E E AAQ++ + ++ ++VPG+I P ++
Sbjct: 1015 IKFKLALRDFLIQLKEFGG-DNAELYLEEKEQELAAQQKAQLEKAMTVPGMIKPVDM 1070
>XPO1_YEAST (P30822) Exportin 1 (Chromosome region maintenance protein
1)
Length = 1084
Score = 856 bits (2211), Expect = 0.0
Identities = 451/1087 (41%), Positives = 671/1087 (61%), Gaps = 20/1087 (1%)
Query: 4 EKLRDLSQPIDVPLLDATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQNTQN 63
E + D S +D+ LLD V+ FY GS Q+ A +IL Q+NPD W + ILQ + N
Sbjct: 2 EGILDFSNDLDIALLDQVVSTFY-QGSGVQQKQAQEILTKFQDNPDAWQKADQILQFSTN 60
Query: 64 LNTKFFALQVLEGVIKYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNKLNI 123
+KF AL +L+ +I +W LP + R G++NF+ +I+ + ++ F+ ++ +NK ++
Sbjct: 61 PQSKFIALSILDKLITRKWKLLPNDHRIGIRNFVVGMIISMCQDDEVFKTQKNLINKSDL 120
Query: 124 ILVQILKHEWPLRWRSFIPDLVSAAKTSETICENCMAILKLLSEEVFDFSRGEMTQQKIK 183
LVQILK EWP W FIP+L+ ++ +S +CEN M +LKLLSEEVFDFS +MTQ K
Sbjct: 121 TLVQILKQEWPQNWPEFIPELIGSSSSSVNVCENNMIVLKLLSEEVFDFSAEQMTQAKAL 180
Query: 184 ELKQSLNSEFQLIHELCLYVLSASQRAELIRATLSTLHAFLSWIPLGYIFESPLLETL-L 242
LK S++ EF+ I +LC VL + LI ATL +L +L WIP YI+E+ +LE L
Sbjct: 181 HLKNSMSKEFEQIFKLCFQVLEQGSSSSLIVATLESLLRYLHWIPYRYIYETNILELLST 240
Query: 243 KFFPLPVYRNLTLKCLIEVAAL---QFGDYYDTQYTKMYNIFMVQLQTILLPTT-NIPEA 298
KF P R +TLKCL EV+ L Q D Q + + Q+ T ++P T ++
Sbjct: 241 KFMTSPDTRAITLKCLTEVSNLKIPQDNDLIKRQTVLFFQNTLQQIATSVMPVTADLKAT 300
Query: 299 YAQGSSEEQTFIQNLALFFTSFYKVHIRILESTQENIAALLSGLEYLINISYVDDTEVFK 358
YA + +Q+F+Q+LA+F T++ + +LES + LL+ +YLI +S +++ E+FK
Sbjct: 301 YANANGNDQSFLQDLAMFLTTYLARNRALLESDESLRELLLNAHQYLIQLSKIEERELFK 360
Query: 359 VCLDYWNILVSELFEPHRSLENPAAAATNML----GNQQAPLMPPGMVDGLGSQLLQRRQ 414
LDYW+ LV++LF + L PA + ++ G+Q + + ++
Sbjct: 361 TTLDYWHNLVADLFYEVQRL--PATEMSPLIQLSVGSQAISTGSGALNPEYMKRFPLKKH 418
Query: 415 LYAGPMSKLRTLMICRMAKPEEVLIIEDENGNIVRETLKDNDVLVQYKIMRETLIYLSHL 474
+Y S+LR ++I M +PEEVL++E++ G IVRE +K++D + YK RE L+YL+HL
Sbjct: 419 IYEEICSQLRLVIIENMVRPEEVLVVENDEGEIVREFVKESDTIQLYKSEREVLVYLTHL 478
Query: 475 DHDDTERQMLRKLSKQLGGEDWTWNNLNTLCWAIGSISGSMMEDQENRFLVMVIRDLLNL 534
+ DTE M+ KL++Q+ G +W+W+N+NTL WAIGSISG+M ED E RF+V VI+DLL+L
Sbjct: 479 NVIDTEEIMISKLARQIDGSEWSWHNINTLSWAIGSISGTMSEDTEKRFVVTVIKDLLDL 538
Query: 535 CEFTKGKDNKAVIASNIMYVVGQYPRFLRAHWKFLKTVVNKLFEFMHESHPGVQDMACDT 594
+GKDNKAV+AS+IMYVVGQYPRFL+AHW FL+TV+ KLFEFMHE+H GVQDMACDT
Sbjct: 539 TVKKRGKDNKAVVASDIMYVVGQYPRFLKAHWNFLRTVILKLFEFMHETHEGVQDMACDT 598
Query: 595 FLKIVQKCKRKFVITQLGENEPFVSELLSGLSTTIVDLEPHQIHTFYESVGSMVQAESDS 654
F+KIVQKCK FVI Q E+EPF+ ++ + T DL+P Q+HTFY++ G ++ E
Sbjct: 599 FIKIVQKCKYHFVIQQPRESEPFIQTIIRDIQKTTADLQPQQVHTFYKACGIIISEERSV 658
Query: 655 QKRDEYLQRLMHLPNQKWLEIIGQARQNVDFLKDHDVIRTVLNILQTNTSVASSLGTYFL 714
+R+ L LM LPN W I+ Q+ N L D + ++ + NI++TN +V +S+G F
Sbjct: 659 AERNRLLSDLMQLPNMAWDTIVEQSTANPTLLLDSETVKIIANIIKTNVAVCTSMGADFY 718
Query: 715 PQISLIFLDMLNVYRMYSELISKSIAEGGPYASRTSFVKLLRSVKRETLKLIETFLDKAE 774
PQ+ I+ +ML +YR S +IS +A G A++T V+ LR++K+E LKL+ET++ KA
Sbjct: 719 PQLGHIYYNMLQLYRAVSSMISAQVAAEGLIATKTPKVRGLRTIKKEILKLVETYISKAR 778
Query: 775 DQPQIGKQFVPPMMDPVLGDYARNVPDARESEVLSLFATIVNKYKAAMVEDVPRIFEAVF 834
+ + K V P+++ VL DY NVPDAR++EVL+ T+V K + + V I ++VF
Sbjct: 779 NLDDVVKVLVEPLLNAVLEDYMNNVPDARDAEVLNCMTTVVEKVGHMIPQGVILILQSVF 838
Query: 835 QCTLEMITKNFEDYPEHRLKFFSLLCAIATHCFHALICLSSQQLKFVMDSIIWAFRHTER 894
+CTL+MI K+F +YPEHR++F+ LL I F A + L K +D+I WAF+H R
Sbjct: 839 ECTLDMINKDFTEYPEHRVEFYKLLKVINEKSFAAFLELPPAAFKLFVDAICWAFKHNNR 898
Query: 895 NIAETGLNLLLEMLKKFQA---TEFCNQFYRTYFLTIEQEIFAVLTDTFHKPGFKLHVLV 951
++ GL + L+++K + F N+F++ YF E F VLTD+ HK GF L+
Sbjct: 899 DVEVNGLQIALDLVKNIERMGNVPFANEFHKNYFFIFVSETFFVLTDSDHKSGFSKQALL 958
Query: 952 LQHLFCLAETGALTEPLWDAATNPYPYPSNAAFVLEYTIKLLSTSFPNMTAAEVTQFVNG 1011
L L L ++ PL+ A P SN ++ +Y +LS +FP++T+ ++ F++
Sbjct: 959 LMKLISLVYDNKISVPLYQEAEVPQG-TSNQVYLSQYLANMLSNAFPHLTSEQIASFLSA 1017
Query: 1012 LFESTNDLSTFKNHIRDFLVQSKEFSAQDNKDLYAEE----AAAQRERERQRMLSVPGLI 1067
L + DL FK +RDFLVQ KE L+AE+ Q ER++ + GL+
Sbjct: 1018 LTKQYKDLVVFKGTLRDFLVQIKEVGGDPTDYLFAEDKENALMEQNRLEREKAAKIGGLL 1077
Query: 1068 APSELQD 1074
PSEL D
Sbjct: 1078 KPSELDD 1084
>YNR7_SCHPO (Q9USZ2) Hypothetical protein C11G11.07 in chromosome II
Length = 955
Score = 60.1 bits (144), Expect = 3e-08
Identities = 64/291 (21%), Positives = 121/291 (40%), Gaps = 23/291 (7%)
Query: 20 ATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHIL-QNTQNLNTKFFALQVLEGVI 78
+ +A Y +EQ+ A+ L + Q +P W IL Q+ ++ K FA Q L I
Sbjct: 6 SALATLYANTDREQKLQANNYLEEFQKSPAAWQICFSILNQDDSSIEAKLFAAQTLRQKI 65
Query: 79 KYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNKLNIILVQILKHEWPLRWR 138
Y ++ LP E +N + + + A+ R + L + + I H W
Sbjct: 66 VYDFHQLPKETHIEFRNSLLQLFL------AAKDSPRPLLVSLAVCMAAIALHM--TEWH 117
Query: 139 SFIPDLVSAAKTSETICENCMAILKLLSEEVFDFSRGEMTQQKIK-ELKQSLNSEFQLIH 197
+ I D+ A + + + L +L EE D + ++ +++ + + L +
Sbjct: 118 NVIADVFQACSSKDPSGRCVLQFLSVLPEEASDPRKTSLSWEELCIRVDELLRDNGPAVL 177
Query: 198 ELCLYVLSA-----SQRAELIRATLSTLHAFLSWIPLGYIFESPLLETLLKFFPLPVYRN 252
EL + + A S + + L++L ++L IPL + SPL+E + + +
Sbjct: 178 ELLVQYVDAVRASGSPSSADLGLVLTSLISWLREIPLDKVMASPLIELAFR----SLDDD 233
Query: 253 LTLKCLIEVAALQFGDYYDTQYTK----MYNIFMVQLQTILLPTTNIPEAY 299
L L+ +E F + D T M +++LQ L+ + PE +
Sbjct: 234 LLLEDAVEFLCALFNETKDVDETTDAILMLYPRLLELQPKLIAACDDPETF 284
Score = 32.3 bits (72), Expect = 7.1
Identities = 40/179 (22%), Positives = 81/179 (44%), Gaps = 24/179 (13%)
Query: 482 QMLRKLSKQLGGEDWTWNNLNTLCWAIGSISGSMMEDQENRFLVMVIRDLLNLCEFTKGK 541
Q+++ L + G + W ++ +A+ +I G M+ E++ + + + L L E
Sbjct: 430 QLIKVLKIKESGLPYYWQDVEAPLFALRAI-GRMVPANEDQVIGSLFQILPQLPE----- 483
Query: 542 DNKAVIASNIMYVVGQYPRFLRAHWKFLKTVVNKLFEFMHESHPGVQDMACDTFLKIVQK 601
+NK A+ + +G+Y + H +FL+ +N + ++ VQ A
Sbjct: 484 NNKVRYAATLF--LGRYTEWTAQHSEFLELQLNYISAGFEVANKEVQSAAAQALKHFCYD 541
Query: 602 CKRKFV--ITQLG----------ENEPFVSELLSGLSTTIVDLEPHQIHTFYESVGSMV 648
C+ + V ++QL +P + E+ GL+ IVD++P + Y+SV S +
Sbjct: 542 CREQLVGHLSQLHMFYLNAKTYLAPDPLM-EVAQGLA-HIVDIQP--VANVYQSVHSFL 596
>TNP1_MOUSE (Q8BFY9) Transportin 1 (Importin beta-2) (Karyopherin
beta-2)
Length = 890
Score = 60.1 bits (144), Expect = 3e-08
Identities = 67/312 (21%), Positives = 137/312 (43%), Gaps = 53/312 (16%)
Query: 440 IEDENGNIVRETLKDNDVLVQYKIMR---ETLIYLSHLDHDDTERQMLRKLSKQLGGEDW 496
IE+E+ + + + D+D + + + + L L+++ D+ +L L + L +W
Sbjct: 354 IEEEDDD--DDEIDDDDTISDWNLRKCSAAALDVLANVYRDELLPHILPLLKELLFHHEW 411
Query: 497 TWNNLNTLCWAIGSISGSMMEDQENRFLVMVIRDLLNLCEFTKGKDNKAVIASNIMYVVG 556
L +G+I+ M+ +L +I L+ D KA++ S + +
Sbjct: 412 VVKESGIL--VLGAIAEGCMQGMIP-YLPELIPHLIQCLS-----DKKALVRSITCWTLS 463
Query: 557 QYPRFLRAHW-------KFLKTVVNKLFEFMHESHPGVQDMACDTFLKIVQKCKRKFVIT 609
+Y AHW +LK ++ +L + + +S+ VQ+ AC F + ++ + V
Sbjct: 464 RY-----AHWVVSQPPDTYLKPLMTELLKRILDSNKRVQEAACSAFATLEEEACTELV-- 516
Query: 610 QLGENEPFVSELLSGLSTTIVDLEPHQIHTFYESVGSMVQAESDSQKRDEYLQRLMHLPN 669
P+++ +L L + + Y+++G++ + + EY+Q LM
Sbjct: 517 ------PYLAYILDTLVFAFSKYQHKNLLILYDAIGTLADSVGHHLNKPEYIQMLMPPLI 570
Query: 670 QKWLEIIGQARQNVDFLKDHDVIRTVLNILQTNTSVASSLGTYFLPQISLIFLDMLNVYR 729
QKW + LKD D + + +L+ +SVA++L + FLP VY+
Sbjct: 571 QKW-----------NMLKDED--KDLFPLLECLSSVATALQSGFLPYCE-------PVYQ 610
Query: 730 MYSELISKSIAE 741
L+ K++A+
Sbjct: 611 RCVNLVQKTLAQ 622
>TNP1_HUMAN (Q92973) Transportin 1 (Importin beta-)2 (Karyopherin
beta-2) (M9 region interaction protein) (MIP)
Length = 890
Score = 60.1 bits (144), Expect = 3e-08
Identities = 67/312 (21%), Positives = 137/312 (43%), Gaps = 53/312 (16%)
Query: 440 IEDENGNIVRETLKDNDVLVQYKIMR---ETLIYLSHLDHDDTERQMLRKLSKQLGGEDW 496
IE+E+ + + + D+D + + + + L L+++ D+ +L L + L +W
Sbjct: 354 IEEEDDD--DDEIDDDDTISDWNLRKCSAAALDVLANVYRDELLPHILPLLKELLFHHEW 411
Query: 497 TWNNLNTLCWAIGSISGSMMEDQENRFLVMVIRDLLNLCEFTKGKDNKAVIASNIMYVVG 556
L +G+I+ M+ +L +I L+ D KA++ S + +
Sbjct: 412 VVKESGIL--VLGAIAEGCMQGMIP-YLPELIPHLIQCLS-----DKKALVRSITCWTLS 463
Query: 557 QYPRFLRAHW-------KFLKTVVNKLFEFMHESHPGVQDMACDTFLKIVQKCKRKFVIT 609
+Y AHW +LK ++ +L + + +S+ VQ+ AC F + ++ + V
Sbjct: 464 RY-----AHWVVSQPPDTYLKPLMTELLKRILDSNKRVQEAACSAFATLEEEACTELV-- 516
Query: 610 QLGENEPFVSELLSGLSTTIVDLEPHQIHTFYESVGSMVQAESDSQKRDEYLQRLMHLPN 669
P+++ +L L + + Y+++G++ + + EY+Q LM
Sbjct: 517 ------PYLAYILDTLVFAFSKYQHKNLLILYDAIGTLADSVGHHLNKPEYIQMLMPPLI 570
Query: 670 QKWLEIIGQARQNVDFLKDHDVIRTVLNILQTNTSVASSLGTYFLPQISLIFLDMLNVYR 729
QKW + LKD D + + +L+ +SVA++L + FLP VY+
Sbjct: 571 QKW-----------NMLKDED--KDLFPLLECLSSVATALQSGFLPYCE-------PVYQ 610
Query: 730 MYSELISKSIAE 741
L+ K++A+
Sbjct: 611 RCVNLVQKTLAQ 622
>TNP2_HUMAN (O14787) Transportin 2 (Karyopherin beta-2b)
Length = 897
Score = 57.0 bits (136), Expect = 3e-07
Identities = 69/315 (21%), Positives = 133/315 (41%), Gaps = 51/315 (16%)
Query: 441 EDENGNIVRETLKDNDVLVQYKIMR---ETLIYLSHLDHDDTERQMLRKLSKQLGGEDWT 497
E +G+ E D+D L + + + L L+++ ++ +L L L +W
Sbjct: 350 ERPDGSEDAEDDDDDDALSDWNLRKCSAAALDVLANVFREELLPHLLPLLKGLLFHPEWV 409
Query: 498 WNNLNTLCWAIGSISGSMMEDQENRFLVMVIRDLLNLCEFTKGKDNKAVIASNIMYVVGQ 557
L +G+I+ M+ +L +I L+ D KA++ S + + +
Sbjct: 410 VKESGIL--VLGAIAEGCMQGMVP-YLPELIPHLIQCLS-----DKKALVRSIACWTLSR 461
Query: 558 YPRFLRAHW-------KFLKTVVNKLFEFMHESHPGVQDMACDTFLKIVQKCKRKFVITQ 610
Y AHW LK ++ +L + + + + VQ+ AC F + ++ + V
Sbjct: 462 Y-----AHWVVSQPPDMHLKPLMTELLKRILDGNKRVQEAACSAFATLEEEACTELV--- 513
Query: 611 LGENEPFVSELLSGLSTTIVDLEPHQIHTFYESVGSMVQAESDSQKRDEYLQRLMHLPNQ 670
P++S +L L + + Y+++G++ + + EY+Q+LM Q
Sbjct: 514 -----PYLSYILDTLVFAFGKYQHKNLLILYDAIGTLADSVGHHLNQPEYIQKLMPPLIQ 568
Query: 671 KWLEIIGQARQNVDFLKDHDVIRTVLNILQTNTSVASSLGTYFLPQISLIFLDMLNVYRM 730
KW E LKD D + + +L+ +SVA++L + FLP VY+
Sbjct: 569 KWNE-----------LKDED--KDLFPLLECLSSVATALQSGFLPYCE-------PVYQR 608
Query: 731 YSELISKSIAEGGPY 745
L+ K++A+ Y
Sbjct: 609 CVTLVQKTLAQAMMY 623
>TNP2_MOUSE (Q99LG2) Transportin 2 (Karyopherin beta-2b)
Length = 887
Score = 56.6 bits (135), Expect = 3e-07
Identities = 50/211 (23%), Positives = 94/211 (43%), Gaps = 40/211 (18%)
Query: 542 DNKAVIASNIMYVVGQYPRFLRAHW-------KFLKTVVNKLFEFMHESHPGVQDMACDT 594
D KA++ S + + +Y AHW LK ++ +L + + + + VQ+ AC
Sbjct: 446 DKKALVRSIACWTLSRY-----AHWVVSQPPDMHLKPLMTELLKRILDGNKRVQEAACSA 500
Query: 595 FLKIVQKCKRKFVITQLGENEPFVSELLSGLSTTIVDLEPHQIHTFYESVGSMVQAESDS 654
F + ++ + V P++S +L L + + Y+++G++ +
Sbjct: 501 FATLEEEACTELV--------PYLSYILDTLVFAFGKYQHKNLLILYDAIGTLADSVGHH 552
Query: 655 QKRDEYLQRLMHLPNQKWLEIIGQARQNVDFLKDHDVIRTVLNILQTNTSVASSLGTYFL 714
+ EY+Q+LM QKW E LKD D + + +L+ +SVA++L + FL
Sbjct: 553 LNQPEYIQKLMPPLIQKWNE-----------LKDED--KDLFPLLECLSSVATALQSGFL 599
Query: 715 PQISLIFLDMLNVYRMYSELISKSIAEGGPY 745
P VY+ L+ K++A+ Y
Sbjct: 600 PYCE-------PVYQRCVTLVQKTLAQAMMY 623
>XPOT_HUMAN (O43592) Exportin T (tRNA exportin) (Exportin(tRNA))
Length = 962
Score = 51.6 bits (122), Expect = 1e-05
Identities = 52/236 (22%), Positives = 98/236 (41%), Gaps = 11/236 (4%)
Query: 34 RTAADQILRDLQNNPDMWLQVMHIL--QNTQNLNTKFFALQVLEGVIKYRWNALPLEQRD 91
R A L+ +PD W L + + + KFF QVLE +KY+++ L Q+
Sbjct: 18 RQRALAYFEQLKISPDAWQVCAEALAQRTYSDDHVKFFCFQVLEHQVKYKYSELTTVQQQ 77
Query: 92 GMKNFISDIIVQLSGNEASFRMERLYV-NKLNIILVQILKHEWPLRWRSFIPDLVSAAKT 150
++ +I L + + E+ ++ NK + + E+ +W F D++S
Sbjct: 78 LIRE---TLISWLQAQMLNPQPEKTFIRNKAAQVFALLFVTEYLTKWPKFFFDILSVVDL 134
Query: 151 SETICENCMAILKLLSEEVFDFSRGEMTQQKIKE--LKQSLNSE-FQLIHELCLYVLSAS 207
+ + + IL + E+ D +++ + +K ++ + + E +L
Sbjct: 135 NPRGVDLYLRILMAIDSELVDRDVVHTSEEARRNTLIKDTMREQCIPNLVESWYQILQNY 194
Query: 208 Q--RAELIRATLSTLHAFLSWIPLGYIFESPLLETLLKFFPLPVYRNLTLKCLIEV 261
Q +E+ L + A++SWI L I + LL + V R CL EV
Sbjct: 195 QFTNSEVTCQCLEVVGAYVSWIDLSLIANDRFINMLLGHMSIEVLREEACDCLFEV 250
>XPO7_MOUSE (Q9EPK7) Exportin 7 (Ran-binding protein 16)
Length = 1087
Score = 38.1 bits (87), Expect = 0.13
Identities = 62/307 (20%), Positives = 116/307 (37%), Gaps = 53/307 (17%)
Query: 26 YGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQNTQNLNTKFFALQVLEGVIKYRWNAL 85
Y T R A++ L + N+PD + +L+ + ++ A L ++ N L
Sbjct: 19 YETTDTTTRLQAEKALVEFTNSPDCLSKCQLLLERGSSSYSQLLAATCLTKLVSRTNNPL 78
Query: 86 PLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNKLNIILVQILKHEW------PLRWRS 139
PLEQR ++N++ + + ++ L + +I K W +R+
Sbjct: 79 PLEQRIDIRNYVLNYLATRP------KLATFVTQALIQLYARITKLGWFDCQKDDYVFRN 132
Query: 140 FIPDLVSAAKTSETICENCMAILKLLSEE---------------------------VFDF 172
I D+ + S C + IL L+ E +F
Sbjct: 133 AITDVTRFLQDSVEYCIIGVTILSQLTNEINQADTTHPLTKHRKIASSFRDSSLFDIFTL 192
Query: 173 SRGEMTQQKIKELKQSLNSEFQLIHELCLYVLSASQRA---ELIRATLSTLHAFLSWIPL 229
S + Q K L +LN E Q H L + +L + + I + L + +
Sbjct: 193 SCNLLKQASGKNL--NLNDESQ--HGLLMQLLKLTHNCLNFDFIGTSTDESSDDLCTVQI 248
Query: 230 GYIFESPLLE--TLLKFFPL-----PVYRNLTLKCLIEVAALQFGDYYDTQYTKMYNIFM 282
+ S L+ TL FF L P + L L CL+++A+++ + + + K + +
Sbjct: 249 PTSWRSAFLDSSTLQLFFDLYHSIPPSFSPLVLSCLVQIASVRRSLFNNAERAKFLSHLV 308
Query: 283 VQLQTIL 289
++ IL
Sbjct: 309 DGVKRIL 315
>XPO7_HUMAN (Q9UIA9) Exportin 7 (Ran-binding protein 16)
Length = 1087
Score = 38.1 bits (87), Expect = 0.13
Identities = 62/307 (20%), Positives = 116/307 (37%), Gaps = 53/307 (17%)
Query: 26 YGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQNTQNLNTKFFALQVLEGVIKYRWNAL 85
Y T R A++ L + N+PD + +L+ + ++ A L ++ N L
Sbjct: 19 YETTDTTTRLQAEKALVEFTNSPDCLSKCQLLLERGSSSYSQLLAATCLTKLVSRTNNPL 78
Query: 86 PLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNKLNIILVQILKHEW------PLRWRS 139
PLEQR ++N++ + + ++ L + +I K W +R+
Sbjct: 79 PLEQRIDIRNYVLNYLATRP------KLATFVTQALIQLYARITKLGWFDCQKDDYVFRN 132
Query: 140 FIPDLVSAAKTSETICENCMAILKLLSEE---------------------------VFDF 172
I D+ + S C + IL L+ E +F
Sbjct: 133 AITDVTRFLQDSVEYCIIGVTILSQLTNEINQADTTHPLTKHRKIASSFRDSSLFDIFTL 192
Query: 173 SRGEMTQQKIKELKQSLNSEFQLIHELCLYVLSASQRA---ELIRATLSTLHAFLSWIPL 229
S + Q K L +LN E Q H L + +L + + I + L + +
Sbjct: 193 SCNLLKQASGKNL--NLNDESQ--HGLLMQLLKLTHNCLNFDFIGTSTDESSDDLCTVQI 248
Query: 230 GYIFESPLLE--TLLKFFPL-----PVYRNLTLKCLIEVAALQFGDYYDTQYTKMYNIFM 282
+ S L+ TL FF L P + L L CL+++A+++ + + + K + +
Sbjct: 249 PTSWRSAFLDSSTLQLFFDLYHSIPPSFSPLVLSCLVQIASVRRSLFNNAERAKFLSHLV 308
Query: 283 VQLQTIL 289
++ IL
Sbjct: 309 DGVKRIL 315
>YKX1_SCHPO (O14116) Hypothetical protein C328.01c in chromosome I
Length = 1234
Score = 37.7 bits (86), Expect = 0.17
Identities = 34/158 (21%), Positives = 75/158 (46%), Gaps = 22/158 (13%)
Query: 30 SKEQRTAADQILRDLQNN---PDMWLQVMHILQNT-QNLNTKF-------FALQVLEGVI 78
S+E R +A Q+L +L+++ P + +Q++ + + +L K F+L + E +
Sbjct: 22 SRETRFSAQQLLDELKDSYSSPSVAIQLLELNEQAFSSLGCKLDIHIVQHFSLSLFETSV 81
Query: 79 KYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNKLNIILVQILKHEWPLRWR 138
W + ++++ + +F+ I ++ N S R +KL + ++I K +W WR
Sbjct: 82 GMNWKSFSNKEKESVTSFLCKISLE-DNNLLSVHFVR---SKLASVFIEIAKRDWYNTWR 137
Query: 139 ----SFIPDLVSAAKTSETICENCMAILKLLSEEVFDF 172
SF+ L S + + IL+ + E+++ +
Sbjct: 138 EEFDSFLQSLWSLSLQHRQLSS---LILRGIMEDLYQY 172
>LOS1_YEAST (P33418) LOS1 protein
Length = 1100
Score = 36.2 bits (82), Expect = 0.49
Identities = 59/305 (19%), Positives = 128/305 (41%), Gaps = 39/305 (12%)
Query: 4 EKLRDLSQPIDVPLLDATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQNTQN 63
E+++ L ++ P D +K Q ++ +N ++++ ++ I +N+ +
Sbjct: 3 ERIQQLVNAVNDPRSDVA--------TKRQAIELLNGIKSSENALEIFISLV-INENSND 53
Query: 64 LNTKFFALQVLEGVIKYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNKLNI 123
L KF+ L L ++ NA P + Q+ GN+ + ++ +NK++
Sbjct: 54 L-LKFYGLSTLIELMTEGVNANPNGLNLVKFEITKWLKFQVLGNKQT-KLPDFLMNKISE 111
Query: 124 ILVQILKHEWP----LRWRSFIPDLVSA-------AKTSETICENCMAILK-------LL 165
+L + + +W SF DL+S + TS + N + L+ ++
Sbjct: 112 VLTTLFMLMYSDCNGNQWNSFFDDLMSLFQVDSAISNTSPSTDGNILLGLEFFNKLCLMI 171
Query: 166 SEEVFD--FSRGEMTQQKIKELKQSL-NSEFQLIHELCLYVLS-----ASQRAELIRATL 217
+ E+ D F R + +Q K +K + +++ + + L SQ LI +TL
Sbjct: 172 NSEIADQSFIRSKESQLKNNNIKDWMRDNDIMKLSNVWFQCLKLDEQIVSQCPGLINSTL 231
Query: 218 STLHAFLSWIPLGYIFESP--LLETLLKFFPLPVYRNLTLKCLIEVAALQFGDYYDTQYT 275
+ +F+SWI + I ++ L+ + KF L + C++ + + + +
Sbjct: 232 DCIGSFISWIDINLIIDANNYYLQLIYKFLNLKETKISCYNCILAIISKKMKPMDKLAFL 291
Query: 276 KMYNI 280
M N+
Sbjct: 292 NMINL 296
>IMB2_SCHPO (O14089) Putative importin beta-2 subunit (Karyopherin
beta-2 subunit) (Importin 104) (Transportin) (TRN)
Length = 910
Score = 35.4 bits (80), Expect = 0.83
Identities = 43/241 (17%), Positives = 98/241 (39%), Gaps = 28/241 (11%)
Query: 441 EDENGNIVRETLKDNDVLVQYKIMRETLIYLSHLDHDDTERQM---LRKLSKQLGGEDWT 497
EDE+ + E +D+D + + + + + L L +R + L L + L EDW
Sbjct: 373 EDEDDD---EFDEDDDAFMDWNLRKCSAAALDVLSSFWKQRLLEIILPHLKQSLTSEDWK 429
Query: 498 WNNLNTLCWAIGSISGSMMEDQENRFLVMVIRDLLNLCEFTKGKDNKAVIASNIMYVVGQ 557
L A+G+I+ M+ ++L + L+L + K ++ + + +G+
Sbjct: 430 VQEAGVL--AVGAIAEGCMDGMV-QYLPELYPYFLSLLD-----SKKPLVRTITCWTLGR 481
Query: 558 YPRFLRA------HWKFLKTVVNKLFEFMHESHPGVQDMACDTFLKIVQKCKRKFVITQL 611
Y ++ K+ ++ L + +++ VQ+ C F + ++ V
Sbjct: 482 YSKWASCLESEEDRQKYFVPLLQGLLRMVVDNNKKVQEAGCSAFAILEEQAGPSLV---- 537
Query: 612 GENEPFVSELLSGLSTTIVDLEPHQIHTFYESVGSMVQAESDSQKRDEYLQRLMHLPNQK 671
P++ +L+ L+ + + Y++V ++ + Y++ L+ QK
Sbjct: 538 ----PYLEPILTNLAFAFQKYQRKNVLILYDAVQTLADYVGSALNDKRYIELLITPLLQK 593
Query: 672 W 672
W
Sbjct: 594 W 594
>YAA2_SCHPO (Q09796) Hypothetical protein C22G7.02 in chromosome I
Length = 990
Score = 34.3 bits (77), Expect = 1.9
Identities = 27/106 (25%), Positives = 45/106 (41%), Gaps = 6/106 (5%)
Query: 561 FLRAHWKFLKTVVNKLFEFMHESHPGVQDMACDTFLKIVQKCKRKFVITQLGENEPFVSE 620
FL+ H ++L + LF+ +H S +Q + + C ++T++ V E
Sbjct: 541 FLQNHPQYLNISLPVLFDALHISETSIQMTVSRSIHTLCTTCA-SHLLTEIDGFMAVVEE 599
Query: 621 LLSGLSTTIVDLEPHQIHTFYESVGSMVQAESDSQKRDEYLQRLMH 666
L L P + Y SVG + Q D + R YL RL++
Sbjct: 600 LTPKLVYV-----PSVLEKIYSSVGYVTQRIEDIELRISYLMRLLN 640
>TPO3_MOUSE (Q6P2B1) Transportin 3
Length = 923
Score = 34.3 bits (77), Expect = 1.9
Identities = 75/372 (20%), Positives = 138/372 (36%), Gaps = 41/372 (11%)
Query: 17 LLDATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQNTQNLNTKFFALQVLEG 76
L+ V A Y + A L +LQ + W +LQ Q++ + +FA Q ++
Sbjct: 10 LVYQAVQALYHDPDPSGKERASFWLGELQRSVHAWEISDQLLQIRQDVESCYFAAQTMKM 69
Query: 77 VIKYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNKLNIILVQILKHEWPLR 136
I+ + LP + +++ + I L + + V +L + + L + P
Sbjct: 70 KIQTSFYELPTDSHASLRDSLLTHIQNLKD------LSPVIVTQLALAIAD-LALQMP-S 121
Query: 137 WRSFIPDLVSAAKTSETICENCMAILKLLSEEVFDFSRGEMTQQKIKELKQSLNSEFQLI 196
W+ + LV T + IL +L EEV + L+ N ++I
Sbjct: 122 WKGCVQTLVEKYSNDVTSLPFLLEILTVLPEEVHS-----------RSLRIGANRRTEII 170
Query: 197 HELCLY-------VLSASQRAELIRATLSTLHAFL-SWIPLGYIFESPLLET-LLKFFPL 247
+L Y +++ ++A L + L SW LG + + + LL
Sbjct: 171 EDLAFYSSTVVSLLMTCVEKAGTDEKMLMKVFRCLGSWFNLGVLDSNFMANNKLLALLFE 230
Query: 248 PVYRNLTLKCLIEVA------ALQFGDYYDTQYTKMYNIFMVQLQTILLPTTNIPEAYAQ 301
+ ++ T L E A AL + +T +F Q +L T A A+
Sbjct: 231 VLQQDKTSSNLHEAASDCVCSALYAIENVETNLPLAMQLF----QGVLTLETAYHMAVAR 286
Query: 302 GSSEEQTFIQNLALFFTSFYKVHIRILESTQENIAALLSGLEYLINISYVDDTEVFKVCL 361
E+ + N FT + + + T L LE L+ + EV ++
Sbjct: 287 ---EDLDKVLNYCRIFTELCETFLEKIVCTPGQGLGDLRTLELLLICAGHPQYEVVEISF 343
Query: 362 DYWNILVSELFE 373
++W L L++
Sbjct: 344 NFWYRLGEHLYK 355
>TPO3_HUMAN (Q9Y5L0) Transportin 3 (Transportin-SR) (TRN-SR)
(Importin 12)
Length = 975
Score = 34.3 bits (77), Expect = 1.9
Identities = 75/372 (20%), Positives = 138/372 (36%), Gaps = 41/372 (11%)
Query: 17 LLDATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQNTQNLNTKFFALQVLEG 76
L+ V A Y + A L +LQ + W +LQ Q++ + +FA Q ++
Sbjct: 10 LVYQAVQALYHDPDPSGKERASFWLGELQRSVHAWEISDQLLQIRQDVESCYFAAQTMKM 69
Query: 77 VIKYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNKLNIILVQILKHEWPLR 136
I+ + LP + +++ + I L + + V +L + + L + P
Sbjct: 70 KIQTSFYELPTDSHASLRDSLLTHIQNLKD------LSPVIVTQLALAIAD-LALQMP-S 121
Query: 137 WRSFIPDLVSAAKTSETICENCMAILKLLSEEVFDFSRGEMTQQKIKELKQSLNSEFQLI 196
W+ + LV T + IL +L EEV + L+ N ++I
Sbjct: 122 WKGCVQTLVEKYSNDVTSLPFLLEILTVLPEEVHS-----------RSLRIGANRRTEII 170
Query: 197 HELCLY-------VLSASQRAELIRATLSTLHAFL-SWIPLGYIFESPLLET-LLKFFPL 247
+L Y +++ ++A L + L SW LG + + + LL
Sbjct: 171 EDLAFYSSTVVSLLMTCVEKAGTDEKMLMKVFRCLGSWFNLGVLDSNFMANNKLLALLFE 230
Query: 248 PVYRNLTLKCLIEVA------ALQFGDYYDTQYTKMYNIFMVQLQTILLPTTNIPEAYAQ 301
+ ++ T L E A AL + +T +F Q +L T A A+
Sbjct: 231 VLQQDKTSSNLHEAASDCVCSALYAIENVETNLPLAMQLF----QGVLTLETAYHMAVAR 286
Query: 302 GSSEEQTFIQNLALFFTSFYKVHIRILESTQENIAALLSGLEYLINISYVDDTEVFKVCL 361
E+ + N FT + + + T L LE L+ + EV ++
Sbjct: 287 ---EDLDKVLNYCRIFTELCETFLEKIVCTPGQGLGDLRTLELLLICAGHPQYEVVEISF 343
Query: 362 DYWNILVSELFE 373
++W L L++
Sbjct: 344 NFWYRLGEHLYK 355
>CSE1_SCHPO (O13671) Importin-alpha re-exporter (Cellular apoptosis
susceptibility protein homolog)
Length = 967
Score = 33.9 bits (76), Expect = 2.4
Identities = 26/97 (26%), Positives = 44/97 (44%), Gaps = 16/97 (16%)
Query: 57 ILQNTQNLNTKFFALQVLEGVIKYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERL 116
+ ++T ++N K A + IK W++ +G ISD + +L E + L
Sbjct: 42 VAEDTVDINIKLAASLYFKNYIKKHWDS-----EEGASIRISDEVAELIKRE----IINL 92
Query: 117 YVNKLNIILVQ-------ILKHEWPLRWRSFIPDLVS 146
+ II VQ I ++P RW + +PDL+S
Sbjct: 93 MLKSTTIIQVQLGEVIGYIANFDFPDRWDTLLPDLIS 129
>CBK1_ASHGO (Q754N7) Serine/threonine-protein kinase CBK1 (EC
2.7.1.37)
Length = 719
Score = 33.9 bits (76), Expect = 2.4
Identities = 33/158 (20%), Positives = 60/158 (37%), Gaps = 17/158 (10%)
Query: 379 ENPAAAATNMLGNQQAPLMP---PGMVDG-LGSQLLQRRQLYAGPMSKLRTLMIC----- 429
+ PA++A G ++PL P PG++DG LG Q L Q G M ++L
Sbjct: 150 QQPASSAYKQFGGSESPLQPAALPGLLDGSLGDQTLVGNQSSQGAMLSRQSLQCSSVPQS 209
Query: 430 -----RMAKPEEVLIIEDENGNIVRETLKDNDVLVQYKIMRETLIYLSHLDHDDTERQML 484
R + + ++ ++ ++ V+ K+ Y S ++H Q
Sbjct: 210 PNGGQRQTSGVGNYMYFERRPELLSKSTQEKAAAVKLKVEN---FYQSSVNHAIERNQRR 266
Query: 485 RKLSKQLGGEDWTWNNLNTLCWAIGSISGSMMEDQENR 522
+L QL W+ N ++G + + R
Sbjct: 267 VELESQLLSHGWSEERKNRQLSSLGKKESQFLRLRRTR 304
>RYR1_PIG (P16960) Ryanodine receptor 1 (Skeletal muscle-type
ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle
calcium release channel)
Length = 5035
Score = 33.1 bits (74), Expect = 4.1
Identities = 19/68 (27%), Positives = 35/68 (50%), Gaps = 4/68 (5%)
Query: 998 PNMTAAEVTQFVNGLFESTNDLSTFKNHIRDFLVQSK----EFSAQDNKDLYAEEAAAQR 1053
PN +A E+ + V +F + FK ++F+VQ++ F DNK A+ + +
Sbjct: 3428 PNPSAEELFRMVGEIFIYWSKSHNFKREEQNFVVQNEINNMSFLTADNKSKMAKSGGSDQ 3487
Query: 1054 ERERQRML 1061
ER +++ L
Sbjct: 3488 ERTKKKRL 3495
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 118,647,798
Number of Sequences: 164201
Number of extensions: 4880093
Number of successful extensions: 12720
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 12669
Number of HSP's gapped (non-prelim): 44
length of query: 1079
length of database: 59,974,054
effective HSP length: 121
effective length of query: 958
effective length of database: 40,105,733
effective search space: 38421292214
effective search space used: 38421292214
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0587.6


